Abstract
The use of reference genes is required for relative quantification in gene expression analysis and the stability of these genes can be variable depending on the experimental design. Therefore, it is indispensable to test the reliability of endogenous genes previously to their use. This study evaluated nine candidate reference genes to select the most stable genes to be used as reference in gene expression studies with the femoral cartilage of normal and epiphysiolysis-affected broilers. The femur articular cartilage of 29 male broilers with 35 days of age was collected, frozen and further submitted to RNA extraction and quantitative PCR (qPCR) analysis. The candidate reference genes evaluated were GAPDH, HMBS, HPRT1, MRPS27, MRPS30, RPL30, RPL4, RPL5, and RPLP1. For the gene stability evaluation, three software were used: GeNorm, BestKeeper and NormFinder, and a global ranking was generated using the function RankAggreg. In this study, the RPLP1 and RPL5 were the most reliable endogenous genes being recommended for expression studies with femur cartilage in broilers with epiphysiolysis and possible other femur anomalies.
Introduction
The use of gene expression analysis intends to clarify biological processes involved with several conditions in living organisms enabling the identification of diagnostic markers as therapeutic targets in the treatment of diseases [1]. The quantitative PCR (qPCR) is a fast, easy-to-use technique that provides simultaneous measurement of gene expression in many different samples for a limited number of genes [2]. In qPCR, fluorescent dyes are used to combine the amplification and detection steps of the PCR reaction in a single tube [3,4]. In addition, qPCR has been widely used for validating RNA-seq results due to its high sensitivity and precision [5]. When comparing to other techniques, it has advantages such as sensitivity, real-time detection of reaction progress, rapid results and accuracy in the measurement of the analyzed material [5,6]. Although the qPCR is a highly sensitive technique [7], its use must be standardized, especially considering the correct choice of reference genes to avoid mistaken results. The use of stable reference genes ensures the normalization in input RNA levels between samples, avoiding errors in the quantification [8]. Therefore, knowing the expression profile of these genes in each experimental design is crucial to obtain reliable results [7,9].
A valid reference gene should have its expression invariable between different experimental conditions, tissues or physiological state of the tissue or organism [8]. In relative quantification analyses, the use of reference genes is required to normalize the gene expression and to obtain the fold-change through mathematical algorithms [10–12]. Some of the most well-known reference genes are GAPDH (glyceraldehyde 3-phosphate dehydrogenase), PGK (phosphoglycerate kinase), UBQ (ubiquitin), RPL19 (ribosomal protein L19), 18S rRNA (ribosomal RNA 18S), β-actin and β-tubulin [7], which are used in several studies with many species. For instance, GAPDH and 18S rRNA were used as individual reference genes for normalization of qPCR data in multiple studies in chickens [13–17]. However, the stability of reference genes can be altered depending on the tissue, age, treatment and other conditions, which makes it indispensable to test the stability of several genes before using those as reference [18–20]. The selection of stable reference genes is critical for the reliable performance of qPCR experiments [21].
The epiphysiolysis or femur head separation (FHS) consists of the separation of the growth plate from the articular cartilage [22]. This condition is a risk factor for infection and may cause bacterial chondronecrosis with osteomyelitis (BCO) in broilers [22,23]. BCO is also named as femur head necrosis (FHN) and occurs in the proximal femoral head, beginning with the degeneration of the articular cartilage and the growth plate. The bone degeneration begins due to bone deprotection, which may promote a high incidence of other locomotor pathologies, becoming a gateway for viruses and bacteria. Animals have serious problems with movement, affecting them unilaterally or bilaterally [24]. Epiphysiolysis may be an early stage of BCO, due to the fact that its pathogenesis is possibly initiated by damage to poorly mineralized chondrocyte columns (cartilage cells) in the epiphyseal and physeal growth plates of the leg bones, followed by colonization of osteochondritic clefts by opportunistic bacteria [25,26].
Hence, expression studies are required to clarify the genetic mechanism involved with femur pathologies. Although some reference genes were described for bones, it is important to elucidate those for the cartilage as well, since both tissues are involved in the development of those disorders. However, studies on candidate reference genes for chicken cartilage are not available to date. Therefore, to obtain stable genes to be used as reference in expression studies related to bone/cartilage disorders in broilers, the present study evaluated nine endogenous candidate genes in the articular cartilage of chickens with 35 days of age.
Material and methods
Animals and sample collection
The Embrapa Swine and Poultry National Research Center Ethical Committee of Animal Use (CEUA/CNPSA) approved this study under the protocol number 012/2012. The samples used were previously collected from chickens at 35 days of age, as described in detail by Peixoto et al. [27]. Briefly, 29 male broilers from a commercial line (14 normal and 15 lameness) were selected and sent to the Embrapa Swine and Poultry National Research Center, in Concórdia, SC, Brazil for sampling. The femur proximal head was classified based on the clinical examination of the separation of the growth plate (GP) from the articular cartilage (AC), according to Wideman et al. [25]. The normal (control) group (CG) was characterized by good adhesion between the GP and the AC, and the epiphysiolysis-affected group (AG) presented epiphysiolysis and, consequently, separation between the GP and AC. From the 29 broilers, the AC of eight normal (with average body weight of 2,336.75 ± 233.43 g) and 8 affected broilers (2,189.62 ± 444.23 g) were collected, stored in liquid nitrogen and transferred to a freezer at -80°C until the samples were processed. The sample collection for the CG was performed by removing the AC from the bone carefully using a scalpel at the moment of necropsy.
RNA extraction
Eight samples of the AC from each group were ground in a mortar with liquid nitrogen. Then, Trizol reagent (Invitrogen, Carlsbad, CA, USA) was added for total RNA extraction, following the manufacturer's recommendations. To each 100mg of tissue, 1mL of Trizol was added and samples were vortexed and incubated for five minutes at room temperature. Then, 200μL of chloroform were added, the tubes were vigorously homogenized for 15 seconds and incubated at room temperature for five minutes. Centrifugation was performed at 16,000 xg at 4°C for fifteen minutes. The aqueous phase (containing the RNA) was separated into a new microtube, and 500μL of isopropanol were added, gently homogenized by inversion and incubated for 10 minutes at room temperature. Samples were centrifuged for 13,000 xg for 10 minutes at 4°C for RNA pellet formation. Subsequently, the supernatant was discarded and 1mL 75% Alcohol was added, homogenized and centrifuged at 10,500 xg for five minutes at 4°C. After discarding the supernatant, the remained pellet was dried for 15 to 20 minutes at room temperature and resuspended in 40μL of ultrapure water. To assure the samples quality and purity, the RNA clean up protocol was performed using the Qiagen Rneasy kit (Qiagen, Hilden, NRW, Germany), following the manufacturer´s instructions. The Biodrop (Biodrop, Cambridge, UK) spectrophotometer equipment was used to quantify the extracted samples, and samples with 260/280nm ratio above 1.8 were considered pure. To evaluate the integrity of each extracted sample, a 1.0% agarose gel was run and electrophoresed for 90 min.
Complementary DNA (cDNA) synthesis
The cDNA synthesis was performed using the SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen, Carlsbad, CA, USA), following the manufacturer's recommendations. Approximately 3 μg of total RNA were mixed with 1μL of 50μM OligoDT primer, and added 1 μL of Annealing buffer, completing the reaction up to 10μL. The samples were incubated in a thermocycler at 65°C for five minutes and chilled on ice for one minute. In sequence, 10 μL of the 2X First-Strand Reaction Enzyme Mix and 2μL of the SuperScript III/RNAseOUT Enzyme Mix (Invitrogen, Carlsbad, CA, USA) were added in the initial reaction for each sample. Subsequently, samples were incubated at 50°C for 50 minutes followed by 85°C for five minutes in the thermocycler. After, the samples were stored in a freezer -20°C for subsequent quantitative Polymerase Chain Reaction (qPCR).
Real-time qPCR
In order to perform the qPCR analyses, primers for nine genes: GAPDH, HMBS, HPRT1, MRPS27, MRPS30, RPL30, RPL4, RPL5 and RPLP1 were designed using NCBI Primer-BLAST (Table 1). These genes were selected based on previous studies in chicken. For the quantification analyses, the reactions were prepared using 7.5 μL of Master Mix SYBR Green 2x, 0,166 μM of forward primer, 0,166 μM of reverse primer, 2 μl of cDNA at the 1:10 dilution and ultrapure water to complete 15 μL. The reactions were distributed in 96-well plates of Microamp Fast 96-Well Reaction Plate (Applied Biosystems, Foster City, CA, USA) and then submitted to the QuantStudio 6 Real-Time PCR equipment (Applied Biosystems, Foster City, CA, USA) with a temperature cycling of 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 60°C for one minute. The melting curve was performed with the cycling of 95°C for 15 seconds, 60°C for 1 minute and 95°C for 15 seconds to evaluate the amplification specificities. Reactions were performed in duplicates to calculate the Ct (threshold cycle) mean, standard deviation and coefficient of variation between the two replicates.
Table 1. Primers for the candidate reference genes used for qPCR analysis in the femur articular cartilage of broilers.
| Gene/ Ensembl ID | Functions | Primer Sequences (5'-3') |
|---|---|---|
| HMBS1 Hydroxymethylbilane synthase | Heme synthesis, porphyrin metabolism, Third enzyme of the biosynthetic pathway of the Hemegroup | F: ACTAGTTCACTTCGGCGAGC |
| ENSGALG00000042939 | R: CTCAGGAGCTGACCTATGCG | |
| RPL52 Ribosomal Protein L5 | Responsible for the synthesis of proteins in the cell, structural constituent of ribosome, 5S rRNA binding | F: AATATAACGCCTGATGGGATGG |
| ENSGALG00000005922 | R: CTTGACTTCTCTCTTGGGTTTCT | |
| MRPS272 Mitochondrial Ribosomal Protein S27 | Mitochondrial ribosome binding, rRNA binding, tRNA binding | F: GCTCCCAGCTCTATGGTTATG |
| ENSGALG00000015002 | R: ATCACCTGCAAGGCTCTATTT | |
| MRPS302 Mitochondrial ribosomal protein S30 | Structural constituent of ribosome, RNA binding. | F: CCTGAATCCCGAGGTTAACTATT |
| ENSGALG00000014874 | R: GAGGTGCGGCTTATCATCTATC | |
| RPL43 Ribosomal Protein L4 | poly(U) RNA binding, rRNA binding, structural constituent of ribosome | F: TGTTTGCCCCAACCAAGACT |
| ENSGALG00000007711 | R: CTCCTCAATGCGGTGACCTT | |
| HPRT14 Hypoxanthine-guanine phosphoribosyltransferase | Purine synthesis in the salvage pathway | F: TGGGGATGACCTCTCAACCT |
| ENSGALG00000006098 | R: TCCAACAAAGTCTGGCCGAT | |
| GAPDH4 Glyceraldehyde-3-Phosphate Dehydrogenase | Transcription, RNA transport, DNA replication, and apoptosis. | F: TGGGAAGCTTACTGGAATGG |
| ENSGALG00000014442 | R: ATCAGCAGCAGCCTTCACTAC | |
| RPLP14 Ribosomal protein lateral stalk subunit P1 | Protein kinase activator activity, ribonucleoprotein complex binding, structural constituent of ribosome. | F: CCCTCATTCTCCACGACGACZ |
| ENSGALG00000030878 | R: CCAGAGCCTTAGCAAAGAGAC | |
| RPL303 Ribosomal Protein L30 | Antimicrobial humoral immune response mediated by an antimicrobial peptide, cytoplasmic translation, defense response to Gram-negative bacterium, the killing of cells of another organism. | F: ATGATTCGGCAAGGCAAAGC |
| ENSGALG00000029897 | R: GTCAGAGTCACCTGGGTCAA |
Stability evaluation of the candidate reference genes
To evaluate the candidate reference genes from this study, three popular algorithms were used to identify the most stable expressed genes: geNorm [28], NormFinder [29] and BestKeeper [30]. The BestKeeper is an Excel-based software that classifies genes through an index (gene power) composed of Ct, fold-change (FC), standard deviation (SD) and coefficient of variation (CV) values [30]. The most consistent genes present SD values of Cts less than 1 and SD of FC less than 2. The authors do not recommend using genes with SD of Cts above 1.5 [30]. The GeNorm is a program that calculates a measurement of stability of the internal control gene (M) for each combination of two control genes tested, obtaining a transformed expression rate, and then calculates a standard deviation of those combinations of paired genes. From the lowest values of M, the two most stable genes are determined and values smaller than 1.5 indicate stable genes [28]. The NormFinder is a program that automatically calculates the stability value based on intra- and inter-group variation of the tested genes, considering their co-regulation, classifying genes according to their expression stability and similarity. Lower stability values indicate better or more stable genes to be used as normalizers. The authors suggested using data transformed into log2 [29]. When comparing with GeNorm, Bestkeeper has the advantage of using both constitutive and target genes in the analysis, while GeNorm analyzes only reference genes [30].
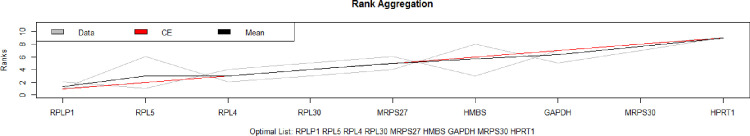
After performing each analysis, the stability values obtained in each tool were used to generate a ranking with the most stable genes using RankAggreg [31]. This is a package from the R environment that calculates the Spearman distance between two genes based on the Monte Carlo algorithm [32,33]. All of these analyses were performed using the endoGenes automatized pipeline available at https://github.com/hanielcedraz/endoGenes.
Results
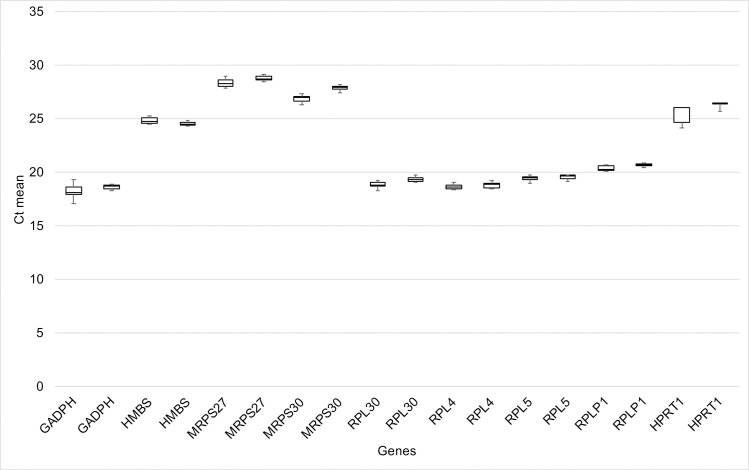
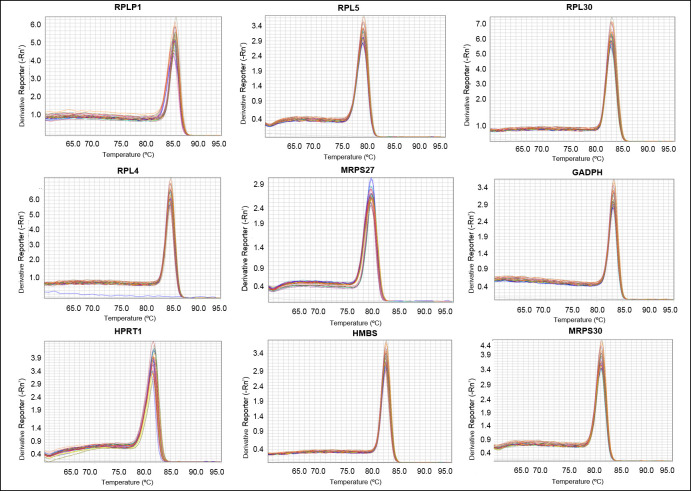
The total RNA average concentration was 195.92 ng/μL for the normal and 172.01 ng/μL for the epiphysiolysis-affected broilers. Regarding RNA quality, the mean A260/280 ratio was 2.07 for normal and 2.08 for the affected chickens. These values demonstrate good quality of samples. The Ct mean (± SD) values of the candidate reference genes ranged from approximately 18.51 ± 0.65 to 28.68 ± 0.59 (Fig 1 and S1 Table). According to the melting curve analysis, all genes showed specific amplification (Fig 2).
Fig 1. Cycle threshold (Ct) variation of the candidate reference gene in normal and proximal femoral epiphysiolysis-affected broilers.
CG: control group and AG: affected group.
Fig 2. Melting curve for the 9 candidate reference genes in the femur articular cartilage of broilers for primers specificity evaluation.
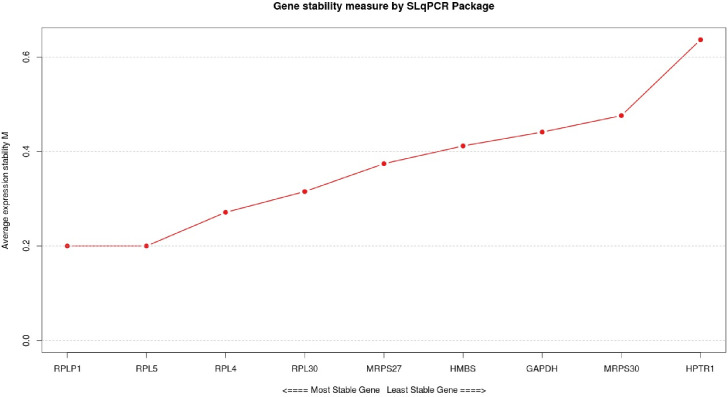
A similar expression profile among the RPLP1, MRPS30 and HPRT1 genes was obtained with the evaluated software (Fig 4 and Table 2). The genes RPL5 and RPLP1 were classified as the two most stable with the software BestKeeper (Table 2) and GeNorm (Fig 3 and Table 2), and RPLP1 and RPL4 with NormFinder (Table 2). The gene RPL5 was classified in the sixth position with the software NormFinder (Table 2) differing from the Bestkeeper and GeNorm results. The least stable gene was HPRT1 according to all the software evaluated (Figs 3 and 4 and Table 2). The gene HMBS had a divergent classification among BestKeeper, GeNorm and NormFinder, ranking this gene, respectively, in the third, sixth and eighth positions (Table 2).
Fig 4. Candidate reference genes ranked with the Rankaggreg function based on the geNorm, BestKeeper and NormFinder stability values.
Table 2. Gene classification values and ranking (in parenthesis) according to the three algorithms analyzed and the general rank generated with the RankAggreg.
| Gene | Bestkeeper (SD of [±Ct]) | NormFinder (S-value) | GeNorm (M-value) | RankAggreg (Ranking) |
|---|---|---|---|---|
| RPLP1 | 0.266 (2) | 0.15 (1) | 0.200 (1) | 1 |
| RPL5 | 0.239 (1) | 0.22 (6) | 0.200 (2) | 2 |
| RPL4 | 0.325 (4) | 0.15 (2) | 0.271 (3) | 3 |
| RPL30 | 0.367 (5) | 0.16 (3) | 0.315 (4) | 4 |
| MRPS27 | 0.433 (6) | 0.17 (4) | 0.374 (5) | 5 |
| HMBS | 0.311 (3) | 0.42 (8) | 0.412 (6) | 6 |
| GAPDH | 0.472 (7) | 0.22 (5) | 0.441 (7) | 7 |
| MRPS30 | 0.559 (8) | 0.39 (7) | 0.476 (8) | 8 |
| HPRT1 | 0.929 (9) | 0.48 (9) | 0.637 (9) | 9 |
Fig 3. Ranking of candidate reference genes based on the average expression M stability value using the geNorm software.
Since there were some variations in the classification of the reference genes in the results of each software separately, the RankAggreg was used to generate an overall classification, where RPLP1 and RPL5 genes were identified as the most stable ones (Fig 4 and Table 2). RankAggreg results were similar to those obtained with the evaluation of geNorm, BestKeeper and NormFinder regarding the MRPS30 and HPRT1 genes, which were classified as the least stable genes from those evaluated in our study (Figs 3 and 4 and Table 2).
Discussion
The evaluation of endogenous genes stability is an essential step in relative quantification analyses of gene expression and needs to be performed previously to its use to avoid biased results. Due to the widespread use of qPCR technique, several protocols, reagents, and methods of analysis have emerged, which can lead to contradictory results [8,38]. In order to obtain high quality results, the samples must have sufficient quantity and integrity of the RNA, generating results with high precision, sensitivity, and reproducibility for posterior analysis [29,30,39]. Another important factor is the evaluation of a set of candidate reference genes, which is fundamental for obtaining reliable results from qPCR studies [30,39]. It should be noted that different conditions and experiments require different genes to be used as normalizers, needing a specific search for genes with stable expression patterns [8,39]. Here, total RNA with good quality was used for the further analysis, according to the MIQE recommendations [40]. Nine candidate reference genes were evaluated, considering stability parameters and consistency of expression with four different specific software (Table 2) widely used in similar studies [30,37,41]. Although the search for the best reference genes is needed, there is no standard methodology established for this purpose, and a wide variety of approaches is available. Another issue is that there is no standardization of the values used by the algorithms, with some having a clear stability threshold value, while others do not, and this value is relevant for checking the stability of the gene. Thus, the use of several software could lead to a better selection of the most reliable reference genes.
In chickens, studies evaluating gene stability in several tissues, such as muscle, embryo, ovary, uterus, lung and heart fibroblasts have been reported [35,37,42–45]. However, there are no studies regarding the stability of endogenous genes in poultry articular cartilage tissue. Therefore, our study is important to characterize this type of sample, especially because this tissue is involved in several bone disorders or locomotor problems in commercial broilers.
Variations in the gene stability classification, such as the HMBS that was ranked as 3rd, 8th or 6th (Fig 3 and Table 2), was observed depending on the software used. Nascimento et al. [35] also found variation in the results obtained by the software used to evaluate reference genes in pectoralis major muscle of broilers. These differences can be explained by the fact that the software have different algorithms, and have been developed to address different types of experiments. The BestKeeper allows comparing the expression level of up to ten endogenous genes, generating an index combining the evaluated genes, with a high sensitivity to genes with very different expression levels [30]. The NormFinder automatically calculates the stability value (S) of all endogenous genes tested on a set of samples, regardless the number of samples and groups, showing a high sensitivity to co-regulating genes [29]. On the other hand, geNorm does not consider co-expression of candidate reference genes and classifies the two most stable reference genes among those tested [28].
Although there is a great variation in the classification of genes by different software, there is no recommendation for the best method of gene selection, nor a pattern that indicates good or poor stability [46]. After the analysis with BestKeeper, geNorm and NormFinder, the geNorm seemed to be the most suitable software to choose the most reliable genes, because its results were more similar to those found with RankAggreg. Through a general classification obtained with the RankAggreg function, RPLP1 and RPL5 were the most stable genes found in our study (Fig 4 and Table 2). Regarding the genes stability, those best classified in the general ranking also had a good stability in each software separately, since they had values within the parameters suggested by each software: S <0.5, M <1.5 and [± Ct] SD <1, for the NormFinder, GeNorm and Bestkeeper, respectively (Fig 3 and Table 2). The use of various tools to choose the reference genes allowed a broad check of the variation of the expression of these genes. Although RankAggreg provides a general classification of genes, it does not mean that all genes are stable or vice-versa [39]. The evaluation of the output from each software is needed to confirm whether the genes are indeed stable or not [8,39], as previously shown.
Several genes have been reported as reference genes. Ribosome proteins are suggested as good endogenous genes, because of their role in the production of ribosomes [47–49], since they are important components of the basic physiological processes in all cells [49]. Here, several known endogenous genes, such as GAPDH, HMBS and HPRT1, as well as many candidate ribosomal proteins (RPLs and MRPLs) were evaluated for their stability. The HPRT1 was the least stable gene, while RPLP1 was one of the most stable. These two genes were unstable at different stages of cardiac development in rats and were not indicated to be used as reference genes in cardiac tissue [50]. However, Nakayama et al. [51] and Nascimento et al. [35] found similar results from our study, where the RPLP1 was suitable for normalization of gene expression in nasal tissue in humans and muscle tissue in broilers, respectively. Furthermore, some studies that evaluated the stability of the HMBS gene found this gene to be one of the most stable when considering several tissues [34,35,49,52–54]. Zhang et al. [52], for example, evaluating the stability of eight reference genes in 10 types of Boer goat tissues found that the HMBS gene was the third most stable and therefore recommended this gene for calibrating gene expression analyses of goat tissues from this breed by real-time qPCR. In broiler chickens pectoralis major muscle, the HMBS gene was also found to be the most stable by Nascimento et al. [35]. According to these authors, the HMBS and HPRT1 genes were the most stable and could be used to normalize expression data in the pectoralis major muscle of chickens submitted to heat stress [51]. Zhang et al. [53] showed that HMBS was the most suitable gene for chickens gut, while Paludo et al. [34] used HMBS as an endogenous gene to study bones from broilers with 45 days of age affected with femoral head necrosis. This gene has been used as endogenous in many species, in different tissues and ages [34,49,54]. Here, the HMBS gene had a good classification in the Bestkeeper tool, but not in NormFinder and GeNorm (Table 2). These results reinforce that gene stability could be influenced by different factors, such as age, tissue and conditions [35,49]. Furthermore, the HPRT1 gene was classified as the least stable gene by the three software, although this gene has already been considered a good reference gene for different swine tissues [54] and mice [55], and is widely used in rats as a reference gene in qPCR studies.
The nuclear genes MRPS27 and MRPS30 encoding mitochondrial ribosomal proteins have high activity in muscle tissues and in the synthesis of proteins within the mitochondria [56]. In quail, the most stable genes evaluated in several tissues were MRPS30, EEF1 and HMBS in the thigh muscle, B2M, UBC and GAPDH in the brain, MRPS30, TFRC and HMBS in the heart, and EEF1, LDHA and HMBS in the spleen [57]. Furthermore, these authors also recommended testing the expression of endogenous genes that could vary between male and female quails [57]. When evaluating the muscle tissue of chickens under heat stress, the genes MRPS27, RPL5 and MRPS30 were considered stable according to the general classification of RankAggreg and can be used as normalizers in qPCR analysis of target genes in this condition [49].
Ribosomal proteins are crucial to the development and tissue homeostasis [58]. Wang et al. [59] suggest ribosomal proteins as good candidates to substitute the traditional reference genes as internal controls in real-time PCR assays. Previously, ribosomal proteins were recommended only for less sensitive detection methods like Northern blot [60]. However, recent studies evaluating the stability of reference genes have reported outstanding stabilities of ribosomal proteins in different cell lines and tissues of mammals [61–63], fish [64], shellfish [65] and plants [66]. The gene RPLP1 was studied as a reference gene in several animal species, such as Homo sapiens [58], Mus musculus [67], Rattus norvegicus [68,69], Gallus gallus [43,70], Bos Taurus [71] and Anopheles gambiae [72], having a role in the elongation step of protein synthesis [73]. This gene had a divergent stability ranking between neonatal and cardiosphere-derived adult cells, indicating that its gene expression was age-dependent [50]. RPLP1 expression seems to be tissue-dependent, because it was the least stable gene in breast muscle of chickens [37] while it was the most stable gene in the current study. Benak et al. (2019) [74], when selecting optimal reference genes for gene expression studies in chronically hypoxic rat heart, found that the gene RPLP1 was one of the most stable genes in the left and right ventricle, similarly to our study. This gene has a variable expression according to the tissue, age and species used in the experiment, reinforcing the importance of checking reference genes even when similar issues have already been addressed.
The RPL5, a gene that encodes a small protein that is a component of the 60S subunit and is responsible for transporting the 5S rRNA to the nucleolus, was the 2nd most stable gene in the general ranking in the current study. Our results corroborate with Oliveira et al. [49], which evaluated the RPL5 gene in chicken muscle tissue of males and females, and was indicated as suitable for normalization of gene expression. The RPL5 is a constitutive protein in the large ribosomal subunit that catalyzes mRNA-directed protein synthesis [60]. When evaluating assays of the cornea in various murine disease models, Ren et al. [55] recommended RPL5 as a reference gene, showing stability in that study. RPL5 was also a stable gene when studying heart failure of the right ventricles in humans [75] and tissues of red abalone Haliotis rufescens (Mollusca, Vetigastropoda) [59,76]. Kim et al. [77] used RPL5 as a reference gene to calibrate the reverse-transcribed cDNA templates for samples of cerebral ganglion, pleuro-pedal ganglion, ovary, gills, intestine and adductor muscle of Pacific abalone (Haliotis discus hannai). Marciano et al. [37], when studying breast muscle of chicken also found RPL5 as one of the most stable genes. These results corroborate with those found in our study, showing that this gene is suitable as normalizer for several species, including chickens.
In the current study, we showed that RPLP1 and RPL5 were ranked as the most stable genes when femur head’s articular cartilage of broilers were evaluated, contributing to the understanding of gene expression profiles of candidate endogenous genes in chickens. These results can also help to clarify the etiology of bone-related problems in avian and other species. Despite the indication of using two or more reference genes in gene expression studies, it is common to find the use of only one gene, and based merely in the literature, not following the MIQE recommendations [39,40]. Therefore, in order to obtain the best reference genes, it is necessary to evaluate a broad panel of genes and software, considering the complexity of the experimental designs and tissues [39]. Thus, the choise of the most reliable reference genes for relative quantification analysis reduces the selection of false endogenous genes, improving the accuracy of the results [78].
Conclusions
The RPL5 and RPLP1 were the most reliable endogenous genes for qPCR analyses in the femur articular cartilage tissue of normal and epiphysiolysis-affected broilers. This is the first study evaluating reference genes in the chicken articular cartilage. Our results can be useful for investigating articular disorders in chickens and other species by the analysis of articular cartilage gene expression.
Supporting information
(DOCX)
Acknowledgments
The authors are grateful to A. L. Tessmann for technical assistance. We thank CAPES (Finance Code 001) for the free access to the journals used in the literature review.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
IRS was supported by a Graduate Teaching Assistant Scholarship (PROMOP)/Udesc #18001/2020, Brazil. MRS received a scholarship from the Coordination for the Improvement of Higher Education Personnel/The Santa Catarina State Research Foundation (CAPES/FAPESC) #0337/2018, Brazil. DEPM is a recipient of a PIBIC scholarship #135359/2018-0 from the National Council of Scientific and Technological Development (CNPq) at Embrapa Swine and Poultry National Research Center, Brazil. MCL is recipient of a productivity fellowship #307026/2017-5 from CNPq. This study was supported by project # 01.11.07.002.04.03 from the Brazilian Agricultural Research Corporation (EMBRAPA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hu M, Polyak K. Serial analysis of gene expression. Nat Protoc. 2006;1: 1743–1760. 10.1038/nprot.2006.269 [DOI] [PubMed] [Google Scholar]
- 2.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1: 1559–1582. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- 3.Higuchi R, Dollinger G, Sean Walsh P, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology. 1992;10: 413–417. 10.1038/nbt0492-413 [DOI] [PubMed] [Google Scholar]
- 4.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11: 1026–1030. 10.1038/nbt0993-1026 [DOI] [PubMed] [Google Scholar]
- 5.Schnerr H, Niessen L, Vogel RF. Real time detection of the tri5 gene in fusarium species by lightcyclerTM-PCR using SYBR® green I for continuous fluorescence monitoring. Int J Food Microbiol. 2001;71: 53–61. 10.1016/s0168-1605(01)00579-7 [DOI] [PubMed] [Google Scholar]
- 6.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing sytem: From gene expression to physiology. Chromosoma. 2004. pp. 103–112. 10.1007/s00412-004-0296-2 [DOI] [PubMed] [Google Scholar]
- 7.Rebouças E de L, Costa JJ do N, Passos MJ, Passos JR de S, Hurk R van den, Silva JRV. Real time PCR and importance of housekeepings genes for normalization and quantification of mRNA expression in different tissues. Brazilian Arch Biol Technol. 2013;56: 143–154. 10.1590/S1516-89132013000100019 [DOI] [Google Scholar]
- 8.Kozera B, Rapacz M. Reference genes in real-time PCR. Journal of Applied Genetics. 2013. pp. 391–406. 10.1007/s13353-013-0173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37: 112–119. 10.2144/04371RR03 [DOI] [PubMed] [Google Scholar]
- 10.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29: 45e – 45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 13.Adams SC, Xing Z, Li J, Cardona CJ. Immune-related gene expression in response to H11N9 low pathogenic avian influenza virus infection in chicken and Pekin duck peripheral blood mononuclear cells. Mol Immunol. 2009;46: 1744–1749. 10.1016/j.molimm.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 14.Hong YH, Lillehoj HS, Lee SH, Dalloul RA, Lillehoj EP. Analysis of chicken cytokine and chemokine gene expression following Eimeria acervulina and Eimeria tenella infections. Vet Immunol Immunopathol. 2006;114: 209–223. 10.1016/j.vetimm.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 15.Hrabia A, Leśniak A, Sechman A. In vitro Effects of TCDD, PCB126 and PCB153 on Estrogen Receptors, Caspases and Metalloproteinase-2 mRNA Expression in the Chicken Shell Gland. Folia Biol. 2013;61: 277–282. 10.3409/fb61_3-4.277 [DOI] [PubMed] [Google Scholar]
- 16.Richards MP, Poch SM, McMurtry JP. Characterization of turkey and chicken ghrelin genes, and regulation of ghrelin and ghrelin receptor mRNA levels in broiler chickens. Gen Comp Endocrinol. 2006;145: 298–310. 10.1016/j.ygcen.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Sargis RM, Volden PA, Carmean CM, Sun XJ, Brady MJ. PCB 126 and Other Dioxin-Like PCBs Specifically Suppress Hepatic PEPCK Expression via the Aryl Hydrocarbon Receptor. Andrabi SA, editor. PLoS One. 2012;7: e37103 10.1371/journal.pone.0037103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uddin MJ, Cinar MU, Tesfaye D, Looft C, Tholen E, Schellander K. Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res Notes. 2011;4: 441 10.1186/1756-0500-4-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: Comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med. 2012;126: 943–952. 10.1007/s00414-012-0774-7 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Zhao Y, Li J, Liu H, Ernst CW, Liu X, et al. Evaluation of housekeeping genes for normalizing real-time quantitative PCR assays in pig skeletal muscle at multiple developmental stages. Gene. 2015;565: 235–241. 10.1016/j.gene.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 21.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5: 516–535. 10.1038/nprot.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddell C, King MW, Gunasekera KR. Pathology of the Skeleton and Tendons of Broiler Chickens Reared to Roaster Weights. II. Normal Chickens. Avian Dis. 1983;27: 980–991. 10.2307/1590199 [DOI] [PubMed] [Google Scholar]
- 23.Packialakshmi B, Rath NC, Huff WE, Huff GR. Poultry Femoral Head Separation and Necrosis: A Review. Avian Dis. 2015;59: 349–354. 10.1637/11082-040715-Review.1 [DOI] [PubMed] [Google Scholar]
- 24.Kealy JK. Diagnostic radiology of the dog and cat Diagnostic radiology of the dog and cat. Philadelphia: W.B. Saunders.; 1979. [Google Scholar]
- 25.Wideman RF, Hamal KR, Stark JM, Blankenship J, Lester H, Mitchell KN, et al. A wire-flooring model 1 for inducing lameness in broilers: Evaluation of probiotics as a prophylactic treatment. Poult Sci. 2012;91: 870–883. 10.3382/ps.2011-01907 [DOI] [PubMed] [Google Scholar]
- 26.Wideman R. F.; Prisby RD. Bone circulatory disturbances in the development of spontaneous bacterial chondronecrosis with osteomyelitis: a translational model for the pathogenesis of femoral head necrosis. Front Endocrinol (Lausanne). 2013;183 10.3389/fendo.2012.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Oliveira Peixoto J, Savoldi IR, Ibelli AMG, Cantão ME, Jaenisch FRF, Giachetto PF, et al. Proximal femoral head transcriptome reveals novel candidate genes related to epiphysiolysis in broiler chickens. BMC Genomics. 2019;20 10.1186/s12864-019-6411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64: 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26: 509–515. 10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 31.Pihur V, Datta S, Datta S. RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics. 2009;10 10.1186/1471-2105-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pihur V, Datta S, Datta S. Weighted rank aggregation of cluster validation measures: a Monte Carlo cross-entropy approach. Bioinformatics. 2007;23: 1607–1615. 10.1093/bioinformatics/btm158 [DOI] [PubMed] [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 34.Paludo E, Ibelli AMG, Peixoto JO, Tavernari FC, Lima-Rosa CAV, Pandolfi JRC, et al. The involvement of RUNX2 and SPARC genes in the bacterial chondronecrosis with osteomyelitis in broilers. Animal. 2017;11: 1063–1070. 10.1017/S1751731116002433 [DOI] [PubMed] [Google Scholar]
- 35.Nascimento CS, Barbosa LT, Brito C, Fernandes RPM, Mann RS, Pinto APG, et al. Identification of suitable reference genes for real time quantitative polymerase chain reaction assays on Pectoralis major muscle in chicken (Gallus gallus). PLoS One. 2015;10 10.1371/journal.pone.0127935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petry B, Savoldi IR, Ibelli AMG, Paludo E, de Oliveira Peixoto J, Jaenisch FRF, et al. New genes involved in the Bacterial Chondronecrosis with Osteomyelitis in commercial broilers. Livest Sci. 2018;208: 33–39. 10.1016/j.livsci.2017.12.003 [DOI] [Google Scholar]
- 37.Marciano CMM, Ibelli AMG, Peixoto J de O, Savoldi IR, do Carmo KB, Fernandes LT, et al. Stable reference genes for expression studies in breast muscle of normal and white striping-affected chickens. Mol Biol Rep. 2020;47: 45–53. 10.1007/s11033-019-05103-z [DOI] [PubMed] [Google Scholar]
- 38.Bustin S, Huggett J. qPCR primer design revisited Biomolecular Detection and Quantification. Elsevier GmbH; 2017. pp. 19–28. 10.1016/j.bdq.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman JR, Waldenström J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS One. 2015;10 10.1371/journal.pone.0141853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55: 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 41.Lorenzetti WR, Mercia A, Ibelli G, De Oliveira Peixoto J, Antonio M, Mores Z, et al. Identification of endogenous normalizing genes for expression studies in inguinal ring tissue for scrotal hernias in pigs. 2018. 10.1371/journal.pone.0204348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X-S. Bat Algorithm: Literature Review and Applications. J Bio-Inspired Comput. 2013;5: 141–149. 10.1504/IJBIC.2013.055093 [DOI] [Google Scholar]
- 43.Katarzyńska-Banasik D, Grzesiak M, Sechman A. Selection of reference genes for quantitative real-time PCR analysis in chicken ovary following silver nanoparticle treatment. Environ Toxicol Pharmacol. 2017;56: 186–190. 10.1016/j.etap.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 44.Hassanpour H.; Bahadoran S.; Farhadfar F.; Chamali Z. F.; Nazari H.; Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in lung and heart of pulmonary hypertensive chickens. Poult Sci. 2018;97: 4048–4056. 10.3382/ps/pey258 [DOI] [PubMed] [Google Scholar]
- 45.Hassanpour H, Aghajani Z, Bahadoran S, Farhadi N, Nazari H, Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in ovary and uterus of laying hens under heat stress. Stress. 2019;22: 387–394. 10.1080/10253890.2019.1574294 [DOI] [PubMed] [Google Scholar]
- 46.Mosley Y-YC, HogenEsch H. Selection of a suitable reference gene for quantitative gene expression in mouse lymph nodes after vaccination. BMC Res Notes. 2017;10: 689 10.1186/s13104-017-3005-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Jonge HJM, Fehrmann RSN, de Bont ESJM, Hofstra RMW, Gerbens F, Kamps WA, et al. Evidence Based Selection of Housekeeping Genes. Lichten M, editor. PLoS One. 2007;2: e898 10.1371/journal.pone.0000898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14: 1918–1929. 10.1261/rna.1132008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Oliveira HC, Garcia AAP, Gromboni JGG, Filho RVF, Do Nascimento CS, Wenceslau AA. Influence of heat stress, sex and genetic groups on reference genes stability in muscle tissue of chicken. PLoS One. 2017;12 10.1371/journal.pone.0176402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan SC, Carr CA, Yeoh KK, Schofield CJ, Davies KE, Clarke K. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Mol Biol Rep. 2012;39: 4857–4867. 10.1007/s11033-011-1281-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakayama T, Okada N, Yoshikawa M, Asaka D, Kuboki A, Kojima H, et al. Assessment of suitable reference genes for RT-qPCR studies in chronic rhinosinusitis. Sci Rep. 2018;8 10.1038/s41598-018-19834-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Zhang XD, Liu X, Li YS, Ding JP, Zhang XR, et al. Reference gene screening for analyzing gene expression across goat tissue. Asian-Australasian J Anim Sci. 2013;26: 1665–1671. 10.5713/ajas.2013.13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Gao Y-Y, Huang Y-Q, Fan Q, Lu X-T, Wang C-K. Selection of housekeeping genes for quantitative gene expression analysis in yellow-feathered broilers. Ital J Anim Sci. 2018;17: 540–546. 10.1080/1828051X.2017.1365633 [DOI] [Google Scholar]
- 54.Nygard AB, Jørgensen CB, Cirera S, Fredholm M. Selection of rNygard A. B., Jørgensen C. B., Cirera S., & Fredholm M. (2007). Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Molecular Biology, 8. 10.1186/1471-2199-8-67eference genes. BMC Mol Biol. 2007;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren S, Zhang F, Li C, Jia C, Li S, Xi H, et al. Selection of housekeeping genes for use in quantitative reverse transcription PCR assays on the murine cornea. Mol Vis. 2010;16: 1076–1086. [PMC free article] [PubMed] [Google Scholar]
- 56.Davies SMK, Lopez Sanchez MIG, Narsai R, Shearwood AMJ, Razif MFM, Small ID, et al. MRPS27 is a pentatricopeptide repeat domain protein required for the translation of mitochondrially encoded proteins. FEBS Lett. 2012;586: 3555–3561. 10.1016/j.febslet.2012.07.043 [DOI] [PubMed] [Google Scholar]
- 57.Macario M. Genes de referência para expressão gênica em codornas de corte. UNIVERSIDADE FEDERAL DO SERGIPE; 2016. [Google Scholar]
- 58.Perucho L, Artero-Castro A, Guerrero S, Ramón Y Cajal S, LLeonart ME, Wang ZQ. RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS One. 2014;9 10.1371/journal.pone.0099956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan Q, Whang I, Choi CY, Lee JS, Lee J. Validation of housekeeping genes as internal controls for studying biomarkers of endocrine-disrupting chemicals in disk abalone by real-time PCR. Comp Biochem Physiol—C Toxicol Pharmacol. 2011;153: 259–268. 10.1016/j.cbpc.2010.11.009 [DOI] [PubMed] [Google Scholar]
- 60.Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, et al. Housekeeping genes as internal standards: Use and limits. J Biotechnol. 1999;75: 291–295. 10.1016/s0168-1656(99)00163-7 [DOI] [PubMed] [Google Scholar]
- 61.Brinkhof B, Spee B, Rothuizen J, Penning LC. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem. 2006;356: 36–43. 10.1016/j.ab.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 62.Janovick-Guretzky NA, Dann HM, Carlson DB, Murphy MR, Loor JJ, Drackley JK. Housekeeping gene expression in bovine liver is affected by physiological state, feed intake, and dietary treatment. J Dairy Sci. 2007;90: 2246–2252. 10.3168/jds.2006-640 [DOI] [PubMed] [Google Scholar]
- 63.Spinsanti G, Panti C, Lazzeri E, Marsili L, Casini S, Frati F, et al. Selection of reference genes for quantitative RT-PCR studies in striped dolphin (Stenella coeruleoalba) skin biopsies. BMC Mol Biol. 2006;7 10.1186/1471-2199-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Infante C, Matsuoka MP, Asensio E, Cañavate JP, Reith M, Manchado M. Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Mol Biol. 2008;9 10.1186/1471-2199-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siah A, Dohoo C, McKenna P, Delaporte M, Berthe FCJ. Selecting a set of housekeeping genes for quantitative real-time PCR in normal and tetraploid haemocytes of soft-shell clams, Mya arenaria. Fish Shellfish Immunol. 2008;25: 202–207. 10.1016/j.fsi.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 66.Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG. Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol Biol. 2008;10 10.1186/1471-2199-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ishii K, Washio T, Uechi T, Yoshihama M, Kenmochi N, Tomita M. Characteristics and clustering of human ribosomal protein genes. BMC Genomics. 2006;7 10.1186/1471-2164-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taulan M, Paquet F, Maubert C, Delissen O, Demaille J, Romey MC. Renal toxicogenomic response to chronic uranyl nitrate insult in mice. Environ Health Perspect. 2004;112: 1628–1635. 10.1289/txg.7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wool IG, Chan YL, Glück A, Suzuki K. The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie. 1991;73: 861–870. 10.1016/0300-9084(91)90127-m [DOI] [PubMed] [Google Scholar]
- 70.Ferro JA, Reinach FC. The complete sequence of a chicken‐muscle cDNA encoding the acidic ribosomal protein P1. Eur J Biochem. 1988;177: 513–516. 10.1111/j.1432-1033.1988.tb14402.x [DOI] [PubMed] [Google Scholar]
- 71.Muramatsu Y, Lejukole HY, Taniguchi Y, Konfortov BA, Yamada T, Yasue H, et al. Chromosomal assignments of expressed sequence tags for ACTG1, AHSG, COL1a1, GNAS1, and RPLP1 expressed abundantly in the bovine foetus. Anim Genet. 2002;33: 230–231. 10.1046/j.1365-2052.2002.t01-1-00876.x [DOI] [PubMed] [Google Scholar]
- 72.Holt RA, Mani Subramanian G, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science (80-). 2002;298: 129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- 73.Du YJ, Luo XY, Hao YZ, Zhang T, Hou WR. cDNA cloning and overexpression of acidic ribosomal phosphoprotein P1 gene (RPLP1) from the Giant Panda. Int J Biol Sci. 2007;3: 428–433. 10.7150/ijbs.3.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benak D, Sotakova-Kasparova D, Neckar J, Kolar F, Hlavackova M. Selection of optimal reference genes for gene expression studies in chronically hypoxic rat heart. Mol Cell Biochem. 2019;461: 15–22. 10.1007/s11010-019-03584-x [DOI] [PubMed] [Google Scholar]
- 75.Li M, Rao M, Chen K, Zhou J, Song J. Selection of reference genes for gene expression studies in heart failure for left and right ventricles. Gene. 2017;620: 30–35. 10.1016/j.gene.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 76.López-Landavery EA, Portillo-López A, Gallardo-Escárate C, Del Río-Portilla MA. Selection of reference genes as internal controls for gene expression in tissues of red abalone Haliotis rufescens (Mollusca, Vetigastropoda; Swainson, 1822). Gene. 2014;549: 258–265. 10.1016/j.gene.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 77.Kim KS, Kim MA, Sohn YC. Molecular characterization, expression analysis, and functional properties of multiple 5-hydroxytryptamine receptors in Pacific abalone (Haliotis discus hannai). Gen Comp Endocrinol. 2019;276: 52–59. 10.1016/j.ygcen.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 78.Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, et al. Careful Selection of Reference Genes Is Required for Reliable Performance of RT-qPCR in Human Normal and Cancer Cell Lines. PLoS One. 2013;8 10.1371/journal.pone.0059180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.