Supplemental Digital Content is Available in the Text.
There is a similar genetic association among different phenotypes of central serous chorioretinopathy and variants in the complement system genes. However, this study was unable to demonstrate an association with central serous chorioretinopathy severity. The relevance of these findings is that different central serous chorioretinopathy phenotypes may share a similar genetic predisposition and possibly also pathophysiology, whereas other genetic or nongenetic risk factors may play a larger role in determining the clinical course of central serous chorioretinopathy.
Key words: acute central serous chorioretinopathy, ARMS2, chronic CSC, complement factor H, complement component 4, genetic association, NR3C2, severe CSC
Abstract
Purpose:
To study genetic predispositions and differences between severe chronic central serous chorioretinopathy (cCSC), nonsevere cCSC, and acute central serous chorioretinopathy (aCSC).
Methods:
One hundred seventy-three severe cCSC patients, 272 nonsevere cCSC patients, 135 aCSC patients, and 1,385 control individuals were included. Eight single-nucleotide polymorphisms were genotyped in the ARMS2 (rs10490924), CFH (rs800292, rs1061170, rs1065489, rs1329428, rs2284664, rs3753394), and NR3C2 (rs2070951). Additionally, C4B gene copy numbers were analyzed.
Results:
A significant association in 5 single-nucleotide polymorphisms in the CFH gene could be reproduced among severe cCSC patients, including rs800292 (P = 0.0014; odds ratio [OR] = 1.93; 95% confidence interval [CI] = 1.51–2.47), rs1065489 (P = 2.22 × 10−4; OR = 0.49; 95% CI = 0.34–0.72), rs1329428 (P = 0.001; OR = 1.89; 95% CI = 1.49–2.40), rs2284664 (P = 1.21× 10−4; OR = 1.65; 95% CI = 1.28–2.13), and rs3753394 (P = 6.10× 10−4; OR = 0.61; 95% CI = 0.46–0.81). Carrying three C4B copies was protective for severe cCSC (P = 0.001; OR = 0.29; 95% CI = 0.14–0.61). No significant differences in allele frequencies could be found among the CSC phenotypes.
Conclusion:
Acute CSC, nonsevere cCSC, and severe cCSC all showed a similar association with the CFH and C4B genes, and the three phenotypes could not be distinguished based on the genetics. This shows that despite the differences in clinical presentation and severity, there is an overlap in the genetic predisposition of different CSC phenotypes. Nongenetic factors may play a more important role in determining the clinical course of CSC.
Central serous chorioretinopathy (CSC) is a chorioretinal disease, characterized by serous fluid accumulation in the subretinal space, often affecting the macula with subsequent visual impairment.1 The underlying pathophysiology of CSC is not fully understood. However, a congested, hyperpermeable, and leaking choroid, together with a damaged and dysfunctional retinal pigment epithelium (RPE) are thought to underlie the subretinal fluid (SRF) accumulation in CSC.2
At least two different CSC phenotypes can be distinguished: acute and chronic CSC. Acute CSC (aCSC) is generally considered self-limiting with a near-complete visual recovery, thus not requiring treatment in most cases. In contrast, chronic CSC (cCSC) often has persistent SRF with more extensive atrophic RPE changes, in which treatment can be beneficial.1 There is no consensus on the duration threshold that distinguishes acute and chronic CSC, but an arbitrary period of 4 to 6 months of duration of active disease (SRF leakage) is often considered for the definition of chronicity.1 Apart from chronic SRF leakage, patients with cCSC may present with a wide spectrum of retinal abnormalities. In mild cCSC cases, there are limited areas of RPE atrophy, few RPE detachments, and a circumscribed area of leakage.3 More severe cCSC cases show widespread or multifocal (or both) areas of RPE atrophy, more numerous RPE detachments, diffuse areas of leakage, and intraretinal cystoid degeneration.4–6 Moreover, this spectrum of severe cCSC was previously shown to have the worst visual prognosis among all cCSC cases, even after treatment and complete resolution of SRF.7 Therefore, severe cCSC may be considered a distinct clinical subgroup within the spectrum of CSC.
Recently, specific single-nucleotide polymorphisms (SNPs) in the age-related maculopathy susceptibility 2 (ARMS2), the complement factor H (CFH), and the nuclear receptor subfamily three Group C member 2 (NR3C2) genes were found to be associated with the risk of cCSC.8–10 Genomic copy number variations in the complement component 4 (C4B) gene were also shown to be associated with cCSC.11 As aCSC, “nonsevere” cCSC, and “severe” cCSC appear substantially distinct CSC subgroups with regard to clinical manifestation and prognosis (Figure 1), these different CSC forms may also have different genetic risk profiles.
Fig. 1.
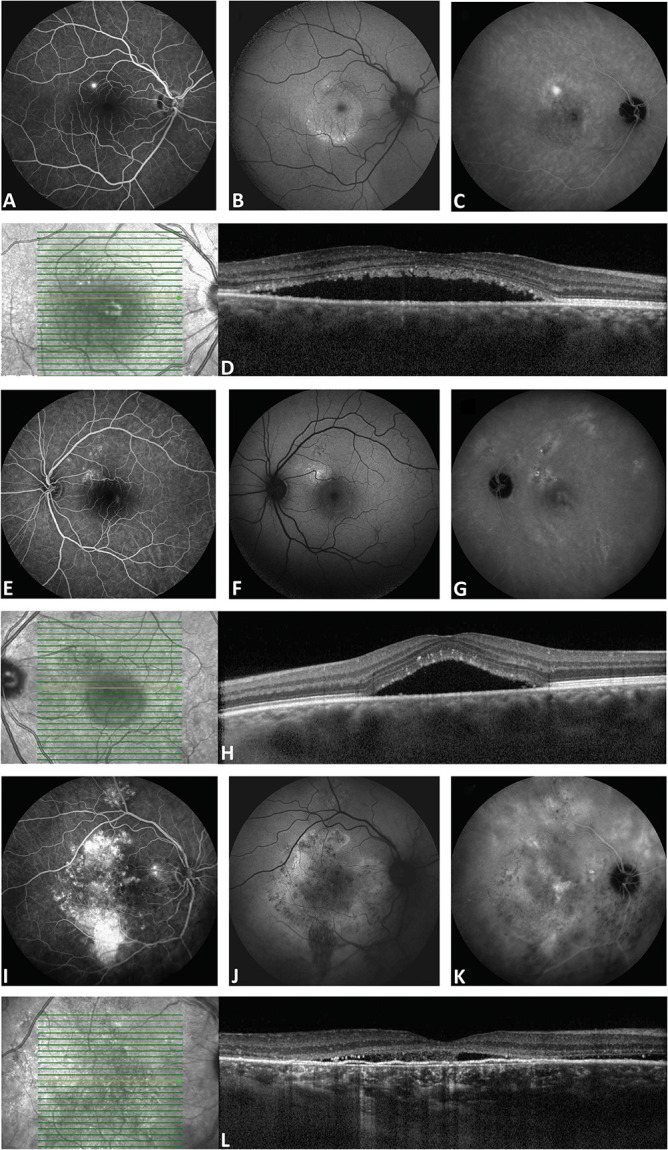
Clinical features on multimodal imaging in different CSC phenotypes. The right eye of a 34-year-old man with aCSC is shown in A–D. In E–H, the left eye of a 43-year-old male patient with nonsevere cCSC is shown. In I–L, the right eye of a 61-year-old male patient with severe cCSC is shown. Fluorescein angiography imaging revealed a single “hot spot” of leakage and no atrophic RPE changes in the aCSC patient (A). Fluorescein angiography in the nonsevere cCSC showed a leakage spot and multifocal small areas of RPE changes (E), whereas in the severe cCSC case, large and widespread RPE atrophy and diffuse leaking areas were seen (I). On midphase indocyanine green angiography, in the aCSC case, a small hyperfluorescent lesion was observed at the site of the “hot spot” on fluorescein angiography (C). In contrast, indocyanine green angiography in the severe and nonsevere cCSC patients showed more extensive multifocal hyperfluorescent changes (G and K). Fundus autofluorescence imaging showed a mix of granular hyperautofluorescent and hypoautofluorescent changes, which were most prominent in the severe cCSC patient (B, F, and J). Optical coherence tomography scan at first presentation revealed a subretinal serous fluid accumulation and subretinal debris in all patients (D, H, and L). Furthermore, a typical irregular shallow RPE detachment was present in the severe cCSC case (L), which is often observed in combination with chronic SRF leakage.
In the present study, we analyzed the association of SNPs in the ARMS2, CFH, and NR3C2 genes, and copy numbers of C4B gene, in a cohort of cCSC patients who showed a severe disease presentation based on previously published disease characteristics.7 In addition, we analyzed and compared the association of the aforementioned risk SNPs between three Caucasian CSC subgroups, including aCSC, cCSC without characteristics of severity, and cCSC patients with severity characteristics.
Materials and Methods
In total, 173 white subjects with a severe cCSC phenotype were included, originating from four tertiary referral centers: 65 patients from the Department of Ophthalmology of Leiden University Medical Center (Leiden, the Netherlands), 67 patients from the Radboud University Medical Center (Nijmegen, the Netherlands), 24 patients from the Rotterdam Eye Hospital (Rotterdam, the Netherlands), and 17 patients from the University Eye Hospital of Cologne (Cologne, Germany).
Patients were phenotyped by two experienced retina specialists (S.Y. and C.J.F.B.). For phenotyping, a complete ophthalmological examination was used, including fundoscopy, optical coherence tomography, fluorescein angiography (FA), and, when available, indocyanine green angiography. White patients were included in the severe group of cCSC when they had a history of active disease for more than 6 months, in combination with at least one of the following abnormalities: 1) cumulative areas of larger than five optic disk diameters of diffuse atrophic RPE alterations visible on midphase FA, 2) at least 2 “hot spots” of leakage on midphase FA, 3) an area of diffuse fluorescein leakage larger than one disk diameter on midphase FA, without an evident leaking focus, or 4) presence of posterior cystoid retinal degeneration assessed on optical coherence tomography.7,12 Subjects were excluded when there was a suspicion of a (secondary) choroidal neovascularization, aneurysmal choroidal vasculopathy, age-related macular degeneration, multifocal choroiditis, retinal vascular occlusions, or high myopia. The presumably steroid-induced CSC cases (steroid use within 3 months before CSC diagnosis) were not excluded from analysis.
The cohort of severe cCSC was genetically compared with a cohort of 272 white patients with nonsevere cCSC, who had a history of persistent disease but did not have any of the 4 previously mentioned characteristics of severity. Additionally, severe cCSC was compared with 135 white patients with aCSC, defined as a combination of 1) documented serous SRF accumulation on optical coherence tomography, 2) a single focal leakage point on FA, and 3) atrophic RPE alterations limited to less than one disk diameter in size. The control group included white individuals enrolled in the European Genetic Database (EUGENDA; www.eugenda.org), in whom no signs of macular disease were found on multimodal imaging, and 176 subjects included in the blood bank of the Radboud University Medical Center. Approval for this study was obtained at the local institutional review boards in all participating centers, and the study adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained from all subjects before blood collection for genetic analysis.
Single-Nucleotide Polymorphism Genotyping
DNA was isolated from peripheral blood using standard procedures. The most relevant genetic variants to be analyzed were chosen based on findings in earlier genetic studies in CSC and included the following variants: ARMS2 (rs10490924), CFH (rs800292, rs1061170, rs1065489, rs1329428, rs2284664, rs3753394), and NR3C2 (rs2070951), and copy number variations in the C4B gene.8–11 KASP assays (LGC Genomics, Berlin, Germany) were used for SNP genotyping, as described previously and according to manufacturer's instructions. A 7900HT Fast Real-Time PCR system (Applied Biosystems by Life Technologies, Austin, TX) was used to read out the genotyping data. Data analysis was performed with SDS (version 2.4, Applied Biosystems). A TaqMan genotyping assay (Hs07226350_cn; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA) with RNaseP as a reference assay was used to measure C4B gene copy numbers, as described previously.
Statistical Analysis
The allele frequency of the SNPs in severe cCSC patients was compared with unaffected controls, nonsevere cCSC, or aCSC subjects using a two-sided Pearson's chi-square test (IBM SPSS Statistics, version 22; SPSS, Inc, Chicago, IL). The C4B copy numbers distribution was compared with a two-sided Fisher's exact test. Additionally, a logistic model correcting for gender was designed, and two copies of C4B were set as a reference.11 P values of <0.0056 were considered statistically significant after a Bonferroni correction for multiple testing for 9 variants. Haplotype analysis correcting for gender was performed to assess the combined effect of the selected six variants in CFH using R (R Core Team, v3.0.2) with the haplo.stats package (v1.7.7). As a reference, the two most frequent haplotypes were used in the haplo.glm command to determine odds ratios (ORs) for the haplotypes with a frequency >5% and the aggregate of the haplotypes with a frequency of <5%.
Results
In the present study, we included 173 patients with severe cCSC (mean age: 54 ± 10 years; 151 [87%] males), 272 patients with nonsevere cCSC (mean age: 51 ± 10 years; 216 [79%] males), and 135 patients with aCSC (mean age: 47 ± 10 years; 92 [68%] males). The demographic characteristics are summarized in Table 1.
Table 1.
Demographic Characteristics of the Study Population and Control Subjects per Tested Gene
| Severe cCSC | Non-severe cCSC | aCSC | Controls ARMS2 and CFH | Controls C4B | Controls NR3C2 | |
| No. of subjects | 173 | 272 | 135 | 826 | 250 | 1,385 |
| No. of male subjects | 151 (87%) | 216 (79%) | 92 (68%) | 424 (51%) | 198 (79%) | 635 (46%) |
| Mean age ± SD (years) | 54 ± 10 | 51 ± 10 | 47 ± 10 | 64 ± 12 | 51 ± 10 | 51 ± 10 |
ARMS2, age-related maculopathy susceptibility 2; CFH, complement factor H; C4B, complement component 4; NR3C2, nuclear receptor subfamily 3 Group C member 2.
Association With Single-Nucleotide Polymorphisms in the ARMS2, NR3C2, and CFH Genes
No significant association was found with the rs10490924 variant in ARMS2 gene or with the rs2070951 variant in the NR3C2 gene in the severe cCSC group after correction for multiple testing (Table 2). Also, no difference was observed in allele frequencies of these tested variants when comparing severe cCSC with nonsevere cCSC or aCSC (Table 3). An association could be found in six tested variants in the CFH gene in the severe cCSC group (Table 2). Associations of five CFH variants remained significant after correction for multiple testing: rs800292 (P = 0.0014; OR = 1.93; 95% confidence interval [CI] = 1.51 to 2.47), rs1065489 (P = 2.22 × 10−4; OR = 0.49; 95% CI = 0.34–0.72), rs1329428 (P = 0.001; OR = 1.89; 95% CI = 1.49–2.40), rs2284664 (P = 1.21× 10−4; OR = 1.65; 95% CI = 1.28–2.13), and rs3753394 (P = 6.10× 10−4; OR = 0.61; 95% CI = 0.46–0.81). No difference was observed when comparing allele frequencies of the six tested variants in the CFH gene between severe cCSC and either nonsevere cCSC or aCSC (Table 3).
Table 2.
Analysis of Eight SNPs in Severe cCSC
| SNP (Locus) | Alleles in Controls (Major/Minor) | Severe cCSC (n) | MAF in Severe cCSC | Controls (n) | MAF Among Controls | Unadjusted Allelic P | Allelic OR (95% CI) |
| rs10490924 (ARMS2) | G/T | 171 | 0.187 | 812 | 0.217 | 0.214 | 0.83 (0.62–1.11) |
| rs2070951 (NR3C2) | C/G | 172 | 0.494 | 1,385 | 0.468 | 0.350 | 1.11 (0.89–1.39) |
| rs800292 (CFH) | G/A | 172 | 0.372 | 798 | 0.235 | 0.0014* | 1.93 (1.51–2.47) |
| rs1061170 (CFH) | T/C | 172 | 0.282 | 803 | 0.353 | 0.012 | 0.72 (0.56–0.93) |
| rs1065489 (CFH) | G/T | 172 | 0.096 | 794 | 0.177 | 2.22 × 10−4* | 0.49 (0.34–0.72) |
| rs1329428 (CFH) | C/T | 171 | 0.588 | 787 | 0.429 | 0.0010* | 1.89 (1.49–2.40) |
| rs2284664 (CFH) | C/T | 171 | 0.316 | 805 | 0.219 | 1.21 × 10−4* | 1.65 (1.28–2.13) |
| rs3753394 (CFH) | C/T | 171 | 0.202 | 800 | 0.293 | 6.10 × 10−4* | 0.61 (0.46–0.81) |
Two-sided P values < 0.00556 were considered significant after correction for multiple testing.
ARMS2, age-related maculopathy susceptibility 2; CFH, complement factor H; MAF, minor allele frequency; NR3C2, nuclear receptor subfamily 3 Group C member 2.
Table 3.
Comparison of Allele Frequencies in Severe cCSC Versus Nonsevere cCSC and aCSC
| SNPs (Locus) | Group 1* | Group 2† | Group 3‡ | Group 2 Versus 1 | Group 3 Versus 1 | |||||
| Severe cCSC (n) | MAF | Non-severe cCSC (n) | MAF | aCSC (n) | MAF | Unadjusted Allelic P | Allelic OR (95% CI) | Unadjusted Allelic P | Allelic OR (95% CI) | |
| rs10490924 (ARMS2) | 171 | 0.187 | 243 | 0.193 | 132 | 0.174 | 0.821 | 0.96 (0.67–1.37) | 0.683 | 1.09 (0.72–1.66) |
| rs2070951 (NR3C2) | 172 | 0.494 | 269 | 0.520 | 132 | 0.538 | 0.447 | 0.90 (0.69–1.18) | 0.216 | 0.82 (0.59–1.13) |
| rs800292 (CFH) | 172 | 0.372 | 245 | 0.296 | 133 | 0.320 | 0.021 | 1.41 (1.05–1.89) | 0.177 | 1.26 (0.90–1.77) |
| rs1061170 (CFH) | 172 | 0.282 | 245 | 0.320 | 133 | 0.259 | 0.235 | 0.83 (0.62–1.13) | 0.534 | 1.12 (0.78–1.61) |
| rs1065489 (CFH) | 172 | 0.096 | 244 | 0.133 | 134 | 0.119 | 0.101 | 0.69 (0.44–1.08) | 0.350 | 0.78 (0.47–1.31) |
| rs1329428 (CFH) | 171 | 0.588 | 244 | 0.510 | 133 | 0.579 | 0.0275 | 1.37 (1.04–1.81) | 0.828 | 1.04 (0.75–1.43) |
| rs2284664 (CFH) | 171 | 0.316 | 244 | 0.275 | 134 | 0.287 | 0.199 | 1.22 (0.90–1.65) | 0.448 | 1.14 (0.81–1.62) |
| rs3753394 (CFH) | 171 | 0.202 | 242 | 0.273 | 131 | 0.263 | 0.0192 | 0.67 (0.48–0.94) | 0.073 | 0.71 (0.48–1.0) |
Two-sided P values < 0.00556 were considered significant after correction for multiple testing.
Group 1: severe cCSC.
Group 2: nonsevere cCSC.
Group 3: aCSC.
ARMS2, age-related maculopathy susceptibility 2; CFH, complement factor H; MAF, minor allele frequency; NR3C2, nuclear receptor subfamily 3 Group C member 2.
Association With CFH Haplotypes
Five haplotypes in the CFH gene with a frequency higher than 5% and an aggregate of the haplotypes with a frequency lower than 5% were identified. When using the most common haplotype (H1) as a reference and correcting for gender, severe cCSC showed an association with H2, H3, H4, H5, and the low frequency aggregated haplotypes (Table 4). However, only H2 remained significant after correction for multiple testing, which was risk carrying for severe cCSC (P = 0.001; OR = 1.73; 95% CI = 1.24–2.41; Table 4). Using the H2 haplotype as a reference, H1 and H3 were found to be associated with severe cCSC after correction for multiple testing, carrying a protective effect (P = 0.0013; OR = 0.58; 95% CI = 0.41–0.81 and P = 4.14 × 10−6; OR = 0.30; 95% CI = 0.18–0.50, respectively; Table 4). When comparing the haplotype frequencies of severe cCSC to this frequencies in nonsevere cCSC and aCSC, no significant differences were found after correction for multiple testing (see Tables 1 and 2, Supplemental Digital Content 1, http://links.lww.com/IAE/B132 and http://links.lww.com/IAE/B133, respectively).
Table 4.
Complement Factor H Haplotypes in Severe cCSC
| Haplotypes | Variants | HF Among Controls | HF Among Severe cCSC | Unadjusted Allelic, P | Allelic OR (95% CI) | Unadjusted Allelic, P | Allelic OR (95% CI) | |||||
| rs3753394 | rs800292 | rs1061170 | rs2284664 | rs1329428 | rs1065489 | |||||||
| H1 | C | G | C | C | C | G | 0.329 | 0.255 | Base | Base | 1.25 × 10−3* | 0.58 (0.41–0.81) |
| H2 | C | A | T | T | T | G | 0.209 | 0.300 | 0.0013* | 1.73 (1.24–2.41) | Base | Base |
| H3 | T | G | T | C | C | T | 0.158 | 0.065 | 0.012 | 0.52 (0.31–0.87) | 4.14 × 10−6* | 0.30 (0.18–0.50) |
| H4 | C | G | T | C | T | G | 0.133 | 0.164 | 0.030 | 1.57 (1.05–2.35) | 0.615 | 0.90 (0.61–1.34) |
| H5 | T | G | T | C | T | G | 0.072 | 0.094 | 0.011 | 1.91 (1.16–3.15) | 0.697 | 1.10 (0.67–1.82) |
| Rare | * | * | * | * | * | * | 0.098 | 0.122 | 0.028 | 1.65 (1.06–2.57) | 0.829 | 0.95 (0.61–1.49) |
P-values < 0.0083 were considered significant after correction for multiple testing.
HF, haplotype frequency; MAF, minor allele frequency.
C4B Copy Number Determination
The distribution of C4B copy numbers was significantly different in severe cCSC compared with controls after correction for multiple testing (P = 0.0020) (see Figure 1, Supplemental Digital Content 3, http://links.lww.com/IAE/B134). A logistic regression model showed that carrying three C4B copies was protective for severe cCSC (P = 0.001; OR = 0.29; 95% CI = 0.14–0.61) (Table 5). The distribution of C4B copy numbers was not significantly different between severe cCSC, nonsevere cCSC, and aCSC groups (see Figure 1, Supplemental Digital Content 3, http://links.lww.com/IAE/B134). In addition, the overall logistic regression model for effect size was not significant when comparing severe cCSC with nonsevere cCSC (P = 0.665) or when comparing severe cCSC with aCSC (P = 0.551) (see Tables 4 and 5, Supplemental Digital Content 4, http://links.lww.com/IAE/B135 and http://links.lww.com/IAE/B136, respectively).
Table 5.
Logistic Regression Model for C4B Load in Severe cCSC Patients
| Overall Significance Model P = 0.007 | ||||
| Controls (n = 250) | Severe cCSC (n = 164) | P | OR (95% CI) | |
| Male sex | 198 (79%) | 143 (87%) | 0.010 | 0.48 (0.27–0.84) |
| C4B copy number | ||||
| 0 | 6 (2.4%) | 4 (2%) | 0.781 | 0.83 (0.23–3.04) |
| 1 | 55 (22%) | 51 (31%) | 0.225 | 1.33 (0.84–2.12) |
| 2 | 142 (57%) | 99 (60%) | Base | Base |
| 3 | 44 (18%) | 10 (6%) | 0.001* | 0.29 (0.14–0.61) |
| 4 | 3 (1.2%) | 0 | 0.999 | NA |
P-values < 0.0055 were considered significant after correction for multiple testing.
C4B, complement component 4; NA, not annotated.
Discussion
There is a wide variety in the clinical presentation of CSC, ranging from aCSC to severe chronic CSC,1,7,12–14 and it is unclear whether these subgroups are different with regard to pathogenesis and genetic background. In the present study, we analyzed specific genetic risk factors in severe cCSC patients and compared them with nonsevere cCSC and aCSC patients. Our data showed that in patients with severe cCSC, three variants (rs800292, rs1329428, and rs2284664) in the CFH gene were significantly associated with an increased risk of the disease, whereas two variants (rs1065489 and rs3753394) were protective. Also, having three copies of the C4B gene was protective against severe cCSC. However, no differences were identified between severe CSC, nonsevere CSC, and aCSC phenotypes.
A comparison of the genetic associations in the three phenotypic subgroups indicated similar risk and protective profiles in the CFH gene variants, CFH haplotypes, and C4B gene copy numbers. Interestingly, although the groups were not significantly different, the genetic effect size, in terms of protective or risk-conferring ORs, was systematically larger in the severe cCSC subgroup compared with nonsevere cCSC and aCSC subgroups. This was also true when comparing the genetic effect size of CFH variants rs800292, rs1329428, and rs1065489 in severe cCSC with cCSC patients in the literature.9 Severe cCSC may therefore have a stronger genetic predisposition than milder CSC subtypes. Our findings indicate that there is a significant overlap in the known genetic risk factors and therefore likely also pathophysiological overlap between CSC subtypes, despite clinical differences.
A role for the complement system, and the CFH gene in particular, in the pathogenesis of CSC was suggested previously based on genetic association studies.8,9,11 Our present study confirms this association in all three CSC phenotypic subtypes. The choroid and choriocapillaris play a central role in the pathogenesis of CSC; although complement activity is abundant in choroidal tissue,15 complement system dysregulation may be a key factor in CSC disease mechanism. A range of variants in genes involved in the complement system have also been identified in age-related macular degeneration.16,17 In contrast to age-related macular degeneration, no systemic complement abnormalities were found in a relatively small group of cCSC patients.18,19 Local complement system effects may be more important in CSC, rather than systemic complement system abnormalities. However, larger studies on systemic complement differences in cCSC patients are necessary.
Patients with CSC share certain clinical characteristics with age-related macular degeneration, such as macular fluid leakage and RPE abnormalities, as well as possible complication of choroidal neovascularization,20 but there are also clear differences such as an earlier age at onset, an absence of drusen, the presence of pachychoroid, and association with steroid use. The CFH variants reported in this study seem to have opposite effects in CSC compared with age-related macular degeneration, which may point to a different role of the complement system in the pathophysiology of these diseases as suggested before.8 In our current cohorts, we could not replicate the associations with the ARMS2 gene and NR3C2 gene variants as demonstrated previously.8,10 This lack of a significant association may be explained by the smaller sample size of the subgroups.
In the present study, a possible role of other, currently unknown, genetic variants cannot be excluded. Other factors may have a more prominent role than genetic factors in determining the course and severity of the disease. Daruich et al21 suggested that older age (>40 years), presence of high (>50 µm) RPE detachments, and a thickened (>500 µm) choroid are significantly correlated with a prolonged episode of aCSC. Long-term steroid use has been suggested not only to increase the risk of CSC but also to cause a more severe bilateral chronic disease with multiple RPE leaking sites, more extended areas of RPE atrophy, and even bullous retinal detachments.22–24 Piccolino et al12 have shown that presence of posterior cystoid retinal degeneration, which was considered a sign of severity in our study, is specifically associated with steroid use, longer duration of symptoms, and subretinal fibrin accumulation. Furthermore, severe cCSC presentations were previously described in pregnant women25 and among certain ethnic groups.26 Our findings suggest that the profile of known genetic risk SNPs between phenotypically different CSC patients is similar, and therefore, it is likely that other factors, such as described above, determine disease course and outcome.
In conclusion, associations between CFH genetic variants and C4B copy numbers and severe CSC were demonstrated, but no marked genetic differences were found between acute, nonsevere, and severe chronic phenotypes of CSC in the tested variants. This study indicates that different phenotypes of CSC may not develop as a result of genetic predisposition, at least among the currently known CSC-associated CFH variants. Presumably, other nongenetic risk factors such as environmental factors or currently unknown genetic variants may play a role in the clinical course of CSC. Future genetic and clinical studies in larger cohorts may provide important clues about the different risk factors associated with CSC disease severity.
Supplementary Material
Acknowledgments
Dr. Magda A. Meester-Smoor, PhD, from the Department of Ophthalmology of the Erasmus University Medical Center, and coordinator of the CORRBI biobank in Rotterdam Eye Hospital, is thanked for her assistance in the storage and preparation of DNA materials. Also, Prof. Dr. Hein W. Verspaget, PhD, from the Department of Gastroenterology and Hepatology, and chairman of the biobank facilities of the Leiden University Medical Center, is thanked for his assistance and support.
Footnotes
Supported by the following funding sources: Stichting Leids Oogheelkundig Ondersteuningsfonds, Rotterdamse Stichting Blindenbelangen, Stichting Wetenschappelijk Onderzoek Het Oogziekenhuis, Macula Fonds, Landelijke Stichting voor Blinden en Slechtzienden, Retina Netherlands, and BlindenPenning. C. J. F. Boon. was supported by a Gisela Thier Fellowship from the Leiden University and a ZonMw VENI grant from the Netherlands Organization for Scientific Research. These sponsors and funding organizations played no role in the design or conduct of this research.
None of the authors has any conflicting interests to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.retinajournal.com).
S. Yzer and C. J. F. Boon are shared last authors.
References
- 1.Daruich A, Matet A, Dirani A, et al. Central serous chorioretinopathy: recent findings and new physiopathology hypothesis. Prog Retin Eye Res 2015;48:82–118. [DOI] [PubMed] [Google Scholar]
- 2.Yannuzzi LA, Slakter JS, Sorenson JA, et al. Digital indocyanine green videoangiography and choroidal neovascularization. Retina 1992;12:191–223. [PubMed] [Google Scholar]
- 3.Breukink MB, Dingemans AJ, den Hollander AI, et al. Chronic central serous chorioretinopathy: long-term follow-up and vision-related quality of life. Clin Ophthalmol 2017;11:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iida T, Yannuzzi LA, Spaide RF, et al. Cystoid macular degeneration in chronic central serous chorioretinopathy. Retina 2003;23:1–7; quiz 137-8. [DOI] [PubMed] [Google Scholar]
- 5.Castro-Correia J, Coutinho MF, Rosas V, et al. Long-term follow-up of central serous retinopathy in 150 patients. Doc Ophthalmol 1992;81:379–386. [DOI] [PubMed] [Google Scholar]
- 6.Yannuzzi LA, Shakin JL, Fisher YL, et al. Peripheral retinal detachments and retinal pigment epithelial atrophic tracts secondary to central serous pigment epitheliopathy. Retina 1984;91:1554–1572. [DOI] [PubMed] [Google Scholar]
- 7.Mohabati D, van Rijssen TJ, van Dijk EHC, et al. Clinical characteristics and long-term visual outcome of severe phenotypes of chronic central serous chorioretinopathy. Clin Ophthalmol 2018;12:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong EK, Breukink MB, Schellevis RL, et al. Chronic central serous chorioretinopathy is associated with genetic variants implicated in age-related macular degeneration. Ophthalmology 2015;122:562–570. [DOI] [PubMed] [Google Scholar]
- 9.Miki A, Kondo N, Yanagisawa S, et al. Common variants in the complement factor H gene confer genetic susceptibility to central serous chorioretinopathy. Ophthalmology 2014;121:1067–1072. [DOI] [PubMed] [Google Scholar]
- 10.van Dijk EHC, Schellevis RL, van Bergen M, et al. Association of a haplotype in the NR3C2 gene, encoding the mineralocorticoid receptor, with chronic central serous chorioretinopathy. JAMA Ophthalmol 2017;135:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breukink MB, Schellevis RL, Boon CJ, et al. Genomic copy number variations of the complement component C4B gene are associated with chronic central serous chorioretinopathy. Invest Ophthalmol Vis Sci 2015;56:5608–5613. [DOI] [PubMed] [Google Scholar]
- 12.Piccolino FC, De La Longrais RR, Manea M, et al. Posterior cystoid retinal degeneration in central serous chorioretinopathy. Retina 2008;28:1008–1012. [DOI] [PubMed] [Google Scholar]
- 13.Otsuka S, Ohba N, Nakao K. A long-term follow-up study of severe variant of central serous chorioretinopathy. Retina 2002;22:25–32. [DOI] [PubMed] [Google Scholar]
- 14.von Winning CH, Oosterhuis JA, Renger-van Dijk AH, et al. Diffuse retinal pigment epitheliopathy. Ophthalmologica 1982;185:7–14. [DOI] [PubMed] [Google Scholar]
- 15.Whitmore SS, Sohn EH, Chirco KR, et al. Complement activation and choriocapillaris loss in early AMD: implications for pathophysiology and therapy. Prog Retin Eye Res 2015;45:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boon CJ, van de Kar NC, Klevering BJ, et al. The spectrum of phenotypes caused by variants in the CFH gene. Mol Immunol 2009;46:1573–1594. [DOI] [PubMed] [Google Scholar]
- 17.Geerlings MJ, de Jong EK, den Hollander AI. The complement system in age-related macular degeneration: a review of rare genetic variants and implications for personalized treatment. Mol Immunol 2017;84:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk EHC, Tsonaka R, Klar-Mohamad N, et al. Systemic complement activation in central serous chorioretinopathy. PLoS One 2017;12:e0180312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersten E, Paun CC, Schellevis RL, et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv Ophthalmol 2018;63:9–39. [DOI] [PubMed] [Google Scholar]
- 20.Peiretti E, Ferrara DC, Caminiti G, et al. Choroidal neovascularization in Caucasian patients with longstanding central serous chorioretinopathy. Retina 2015;35:1360–1367. [DOI] [PubMed] [Google Scholar]
- 21.Daruich A, Matet A, Marchionno L, et al. Acute central serous chorioretinopathy: factors influencing episode duration. Retina 2017;37:1905–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gass JD, Little H. Bilateral bullous exudative retinal detachment complicating idiopathic central serous chorioretinopathy during systemic corticosteroid therapy. Ophthalmology 1995;102:737–747. [DOI] [PubMed] [Google Scholar]
- 23.Quillen DA, Gass DM, Brod RD, et al. Central serous chorioretinopathy in women. Ophthalmology 1996;103:72–79. [DOI] [PubMed] [Google Scholar]
- 24.Stefaniotou M, Vourda E, Katsanos A, et al. Multifocal central serous chorioretinopathy associated with steroids in a patient with myasthenia gravis. Case Rep Ophthalmol 2013;4:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggio E, Polito A, Freno MC, et al. Multimodal imaging findings in a case of severe Central Serous Chorioretinopathy in an uncomplicated pregnancy. BMC Ophthalmol 2015;15:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai UR, Alhalel AA, Campen TJ, et al. Central serous chorioretinopathy in African Americans. J Natl Med Assoc 2003;95:553–559. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


