Abstract
Trastuzumab has led to a marked improvement in the outcomes of patients with human epidermal growth factor receptor 2 (HER-2)-positive breast cancer. However, the effects of trastuzumab on HER-2-positive breast cancer are limited by the emergence of its cardiotoxicside effects. MicroRNA (miR)-135b-5p has been shown to inhibit tumor metastasis in breast cancer. The present study aimed to explore the effects of miR-135b-5p overexpression on the efficacy of trastuzumab in HER-2-positive breast cancer. Reverse transcription-quantitative PCR was performed to detect the levels of miR-135b-5p. Cell viability was evaluated with a Cell Counting Kit-8 assay. Annexin V/propidium iodide staining was employed to detect the number of apoptotic cells. Flow cytometry assay was performed to investigate the cell cycle. Western blotting was used to detect the expression levels of Bax, cleaved caspase-3, Bcl-2, cyclin D2, p27Kip1 and cyclin E1. Cell migration and invasion were detected by Transwell assay. Luciferase assays were conducted to identify the target gene of miR-135b-5p. In addition, an in vivo tumor xenograft model was established. miR-135b-5p agomir significantly enhanced the anti-proliferative effect of trastuzumab on HER-2-positive breast cancer cells via the induction of apoptosis, whereas the anti-metastatic effect of trastuzumab was enhanced by miR-135b-5p agomir treatment. Subsequently, luciferase assays indicated that cyclin D2 was the direct target of miR-135b-5p, whereas overexpression of the latter arrested cell cycleduring the G0/G1 phase. Moreover, miR-135b-5p agomir notably increased the antitumor effect of trastuzumab in vivo. The data demonstrated that miR-135b-5p sensitized HER-2-positive breast cancer cells to trastuzumab in vitro and in vivo by directly binding to cyclin D2. These results suggested that the combination of miR-135b-5p with trastuzumab may be a therapeutic strategy for patients with HER-2-positive breast cancer.
Keywords: trastuzumab, HER-2, BT-474 cells, cyclin D2, microRNA-135b-5p
Introduction
Breast cancer is a complex and heterogeneous disease that is considered the leading cause of cancer-related death among females worldwide (1). In the past decades, advances in molecular biology have contributed to significant improvements in the diagnosis and classification of breast cancer (2,3). Surgery, radiotherapy and chemotherapy (hormone and targeted) have shown remarkable survival benefits in patients with breast cancer (4). Approximately 30% of breast tumors overexpress human epidermal growth factor receptor 2 (HER-2) (5). These cells are characterized by high rates of cell proliferation, metastasis and low overall survival (5). The humanized HER-2 monoclonal antibody trastuzumab, also known as Herceptin, targets HER-2 and has been approved by the United States Food and Drug Administration (6). Trastuzumab drug has become the standard treatment for patients with HER-2-positive cancer in early or advanced breast cancer (7). Trastuzumab treatment has led to a marked improvement in the outcomes of patients with HER-2-positive breast cancer (8). It was reported that trastuzumab markedly inhibits tumor proliferation, angiogenesis and metastasis via downregulation of HER-2 (9-12). Although this antibody has led to optimal stratification of breast cancer, while the prognosis of patients with HER-2 breast cancer remains poor (13,14).
MicroRNAs (miRs/miRNAs) are small regulatory RNAs that modulate the expression of their target genes (15). Several miRNAs have shown preferentially conserved interactions with the majority of human mRNAs; they can influence specific developmental processes and the progression of several diseases (16). Notably, miRNAs are well known for their roles in regulating cell growth, which is critical to cancer development (17). Several studies have investigated the combination of miRNAs with trastuzumab for the treatment of patients with HER-2-positive breast cancer. It was reported that miR-16 (18), miR-26a, miR-30b (1), miR-141-3p (19), miR-770-5p (20) and miR-205-5p (21) are involved in mediating trastuzumab response in HER-2-positive breast cancer. Moreover, repression of miR-135b-5p was reported to promote metastasis of early-stage breast cancer (22), whereas it was also shown to enhance the antitumor effect of doxorubicin in breast cancer cells by targeting anterior gradient 2 (23). The present study hypothesized that miR-135b-5p may be associated with regulating the sensitivity of breast cancer cells to trastuzumab. Therefore, the focus of the current study was to investigate of the expression of miR-135b-5p, which was markedly down-regulated in HER-2-positive breast cancer cells. In addition, the mechanism by which miR-135b-5p improved trastuzumab sensitivity was investigated.
Materials and methods
Cell culture and transfection
HER-2-positive breast cancer cells BT-474 and SK-BR-3 and the normal breast epithelial cell line MCF-10A were obtained from the American Type Culture Collection. Both cell lines were grown in Roswell Park Memorial Institute medium (Thermo Fisher Scientific, Inc.), which was supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin. The cells were maintained at 37°C in a humidified atmosphere at 5% CO2.
miR-135b-5p agomir (5′-UAU GGC UUU UUA UUC CUG UGU GA-3′) and its negative control (NC; 5′-UUU GUA CUA CAC AAA AGU ACU G-3′) were synthesized and purchased from Guangzhou RiboBio Co., Ltd. The cells were transfected with 50 nM miR-135b-5p agomir and its NC (50 nM) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h according to the manufacturer's protocol. Cells in the blank group were not given any treatment.
pcDNA3.1-cyclin D2 was purchased from Shanghai GenePharma Co., Ltd. BT-474 cells were transiently transfected with 2 µg/ml pcDNA3.1-NC or 2 µg/ml pcDNA3.1-cyclin D2 using Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h according to the manufacturer's instructions.
Cell viability
The cell viability of BT-474 and SK-BR-3 cells was evaluated using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. The cells were seeded into 96-well plates at a density of 5×104 cells/ml overnight. Subsequently, the cells were treated with 10 nM miR-135b-5p agomir or agomir NC for 48 h. Finally, 10 µl CCK-8 (5 mg/ml) solution was added to the culture medium in each well and the cells were incubated for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm using a multifunctional microplate reader (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative PCR(RT-qPCR)
miR-135b-5p levels were quantified using RT-qPCR. Total RNA was extracted from BT-474, SK-BR-4 or MCF-10A cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA synthesis was performed using a High Capacity cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) with miRNA-specific primers. The following specific primers were used for RT: U6, 5′-AAC GCT TCA CGA ATT TGC GT′-3′ and miR-135b-5p, 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG TCA CAT AG-3′. qPCR was performed on an Applied Biosystems 7500 Real-Time PCR machine (Thermo Fisher Scientific, Inc.) with the EnTurbo™ SYBR Green PCR SuperMix (ELK Biotechnology Co., Ltd.). The following primer pairs were used for the qPCR: miR-135b-5p forward, 5′-AGC TAT GGC TTT TCA TTC CTA TG-3′ and reverse, 5′-CTC AAC TGG TGT CGT GGA GTC -3′; U6 forward, 5′-CTC GCT TCG GCA GCA CAT -3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3; GAPDH forward, 5′-CGG ACC AAT ACG ACC AAA TCC G-3′ and reverse, 5′-AGC CAC ATC GCT CAG ACA CC-3′; and cyclin D2 forward, 5′-GAA CTC GAG GAG AGC CAT CT-3′ and reverse, 5′-AGT TCG AAT CTG CAC CGT AG-3′. The relative level of miR-135b-5p was normalized to U6, while the relative level of cyclin D2 normalized to GAPDH according to the 2−ΔΔCq method (24). miRNA RT-qPCR primer sets (one RT primer and one pair of qPCR primers) specific for miR-135b-5p, and mRNA RT-qPCR primers specific for cyclin D2 were obtained from Genecreate. Cyclin D2 levels were analyzed by RT-qPCR in BT-474 breast cancer cells transfected with miR-135b-5p agomir for 24 and 48 h.
Detection of apoptosis and cell cycle analysis
Apoptosis induction was analyzed using the Annexin V-FITC apoptosis detection kit (BD Biosciences) according to the manufacturer's protocol. The cells were harvested and washed with PBS twice. Subsequently, they were stained with propidium iodide (PI) and Annexin V. Following 15 min of incubation in the dark at room temperature, cell apoptosis was detected by flow cytometry (FACSCalibur; BD Biosciences) using the CellQuest Pro software (version 3.3; BD Biosciences).
For cell cycle analysis, the cells were harvested and resuspended at 1×106 cells/ml in modified Krishan's buffer (0.1% sodium citrate, 0.3% NP-40, 0.02 mg/ml RNase and 0.05 mg/ml PI). The stained cells were detected by flow cytometry (FACSCalibur; BD Biosciences) using CellQuest Pro software (version 3.3; BD Biosciences).
Transwell assay
Migration and invasion assays were performed using a 24-well Transwell chamber (Sigma-Aldrich; Merck KGaA) with or without a Matrigel-coated membrane. The cells (5×104/well) were plated in serum-free medium in the upper chamber with a non-coated membrane (24-well insert, 8-µm pore size) in order to assess migration, whereas a matrigel-coated membrane was used for the invasion assay. The lower chamber was filled with medium containing 10% fetal bovine serum (Thermo Fisher Scientific, Inc.). After 24 h of incubation, the cells on the upper surface were removed by a cotton swab and the cells on the lower surface were stained with 10% w/v aqueous Giemsa's solution (VWR International, LLC) for 30 min at room temperature. The stained cells were imaged and counted under a fluorescence microscope (Nikon Eclipse Ti-E; Nikon Corporation) in five randomly selected fields at ×200 magnification.
Dual-luciferase reporter assay
TargetScan (http://www.targetscan.org/vert_71/) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) online tools were used to predict target genes of miR-135b-5p. The predicted miR-135b-5p binding sequence (wild-type; WT) or a mismatch sequence (mutant type) in the 3′-untranslated region (3′-UTR) of cyclin D2 mRNA were synthesized and cloned separately into the multiple cloning site of the luciferase miRNA expression reporter vector (Promega Corporation). The sequences of these synthesized oligonucleotides are as follows: Forward WT: 5′-GGC UCA GGU UUU GAG AAG CCA UC-3′ and mutant 5′-GGC UCA GGU UUU GAG GUU AAG GC-3′. Subsequently, cells were co-transfected with pRL-TK-cyclin D2-WT or pRL-TK-cyclin D2-MT plasmid and miR-135b-5p agomir using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Additionally, cells were co-transfected with pRL-TK-cyclin D2-WT or pRL-TK-cyclin D2-MT plasmid and NC using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), which was considered as the vector-control group. Meanwhile, cells transfected with pRL-TK-cyclin D2-WT or pRL-TK-cyclin D2-MT plasmid alone was considered as the control group. Following transfection, the cells were cultured for 6 h at 37°C and the transfection medium was removed and replenished with DMSO. At 48 h post-transfection, luciferase activity was measured using the dual-luciferase reporter assay system (Promega Corporation) and normalized to Renilla luciferase activity.
Western blotting
Total protein was extracted from the cells with lysis buffer (Cell Signaling Technology, Inc.). The lysates were centrifuged at 10,000 × g for 10 min at 4°C. The protein concentration was determined using a bicinchoninic acid protein quantification kit (Promega Corporation). Equal amounts of cell extracts (30 µg/well) were subjected to 10% SDS-PAGE and transferred to PVDF membranes (EMD Millipore) for anti-body blotting. The membranes were blocked with 5% bovine serum albumin (Thermo Fisher Scientific, Inc.) for 1 h at room temperature and subsequently incubated with β-actin (1:1,000; dilution, cat. no. ab179467), Bax (1:1,000; cat. no. ab32503), cleaved caspase 3 (1:1,000; cat. no. ab2303), Bcl-2 (1:1,000; cat. no. ab59348), cyclin D2 (1:1,000; cat. no. ab207604), p27kip1(1:1,000; cat. no. ab32034), cyclin E1 (1:1,000; cat. no. ab33911), phosphorylated (p)-Akt (1:1,000; cat. no. 38449) and Akt (1:1,000; cat. no. ab8850) primary antibodies (all from Abcam) overnight at 4°C. Subsequently, the membranes were incubated with a horseradish peroxidase-conjugated anti-mouse (1:5,000; cat. no. ab97040; Abcam) or an anti-rabbit secondary antibody (1:5,000; cat. no. ab7090; Abcam) at room temperature for 1 h. Protein bands were visualized using an enhanced chemiluminescence reagent kit (GE Healthcare) on a Tanon 5200 Chemiluminescent imaging system (Tanon Science and Technology Co., Ltd.) according to the manufacturer's protocol. ImageJ software (v1.8.0.112; National Institutes of Health) was used for quantification of protein expression with β-actin as the reference protein.
Tumor xenograft model
A total of 24 female BALB/c nude mice (18-20 g, 4-5 weeks) were obtained from Vital River. The animals were housed at a temperature of 23±1°C, and a humidity of 50-60% for 4 days in a 12-h light/dark cycle prior to the initiation of the experiment. For induction of anesthesia, the mice were anesthetized by isoflurane (3% v/v) inhalation. Nude mice were inoculated with 17β-estradiol pellets 48 h before implantation of BT-474 cells. Subsequently, BT-474 cells (5×106 cells/ml in 100 µl PBS mixed with Matrigel matrix) were subcutaneously injected into the mice. Animal health and behavior were monitored twice a day. All animal procedures were approved by the Seventh People's Hospital of Shanghai University of Traditional Chinese Medicine Committee in June 2019. The National Institute of Health Guide for the Care and Use of Laboratory Animals was followed (25). The mice were randomly divided into four groups (n=6) when the tumor volume reached 100 mm3: Control, miR-135b-5p agomir, trastuzumab and trastuzumab + miR-135b-5p agomir. The tumor volume was estimated according to the following equation: Volume= Length x Width2/2. Trastuzumab (10 mg/kg; ChemBest), miR-135b-5p agomir (50 nM) or the combination treatment of trastuzumab with miR-135b-5p agomir were administered as follows: Trastuzumab (10 mg/kg) was administered intraperitoneally once per week whilemiR-135b-5p agomir was directly injected into the implanted tumor at a dose of 50 nM (in 20 µl PBS) per mouse twice a week. After 3 weeks of treatment, the mice were euthanized using CO2 at a displacement rate of 20% of the chamber volume/min (CO2 flow rate, 2.5 l/min). The tumors were removed and weighted. The duration of the experiment is 6 weeks, and no mice died during the experimental process. Humane endpoints were determined as previously described (26). Criteria for judging the death of animals is continuous no spontaneous breathing for 2-3 min and no blink reflex.
Immunohistochemistry (IHC) assay
The tumor tissues were fixed in 4% paraformaldehyde for 48 h at room temperature, embedded in paraffin and cut into 4-µm sections. Deparaffinized tissue sections were incubated with Ki67 monoclonal antibody (1:500; cat. no. ab15580; Abcam) over-night at 4°C. Subsequently, the sections were incubated with biotinylated goat anti-rabbit immunoglobulin G (1:1,000; cat. no. ab6721; Abcam) at room temperature for 30 min. IHC reactions were visualized by using the IHC detection system (EnVision kit; Dako; Agilent Technologies, Inc.). The images were captured under a fluorescence microscope (Nikon Eclipse Ti-E; Nikon Corporation) at ×200 magnification. ImageJ software (v1.8.0.112; National Institutes of Health) was used for quantification of protein expression.
Statistical analysis
All experiments were repeated three times and the results are expressed as the mean ± SD. One-way ANOVA followed by Tukey's post hoc test was performed to analyze differences among multiple groups (>2 groups). Statistical analysis was performed using GraphPad Prism (version 7; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-135b-5p agomir enhances the anti-proliferative effect of trastuzumab in HER-2-positive breast cancer cells
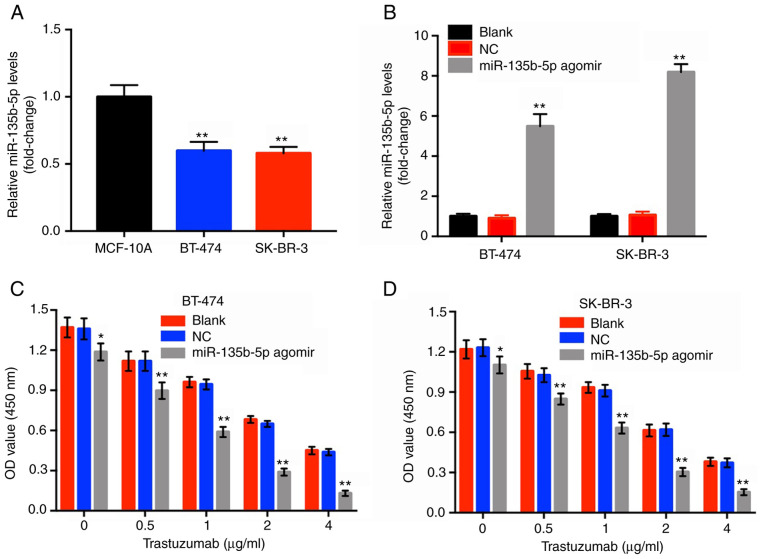
To investigate the effects of miR-135b-5p agomir in BT-474 and SK-BR-3 cells, RT-qPCR assay was performed. The results indicated that the levels of miR-135b-5p in BT-474 and SK-BR-3 cells were significantly lower compared within the normal breast epithelial cell line MCF-10A (Fig. 1A). Moreover, miR-135b-5p agomir caused a significant upregulation in miR-135b-5p expression in these two breast cancer cell lines compared with the blank group (Fig. 1B). Subsequently, a CCK-8 assay was performed to investigate the cytotoxic effects of trastuzumab or of the combination treatment (trastuzumab plus miR-135b-5p agomir) in HER-2-positive breast cancer cells. Trastuzumab alone inhibited the proliferation of HER-2-positive breast cancer cells in a dose-dependent manner, which was significantly enhanced in the presence of miR-135b-5p agomir compared with the blank groups (Fig. 1C and D). Since trastuzumab (1 µg/ml) combined with miR-135b-5p agomir induced ~50% inhibition of cellular growth, this dose was applied for subsequent cell assays. In addition, BT-474 cells were more sensitive to the combination treatment compared with SK-BR-3 cells and therefore this cell line was employed in the following experiments. The results suggested that miR-135b-5p significantly increased the anti-proliferative effect of trastuzumab in HER-2-positive breast cancer cells.
Figure 1.
miR-135b-5pagomirenhancesthe anti-proliferation effect oftrastuzumab in HER-2 positive breast cancer cells. (A) The levels of miR-135b-5p in BT-474, SK-BR-3 and MCF-10A cells were evaluated using RT-qPCR. **P<0.01 vs. MCF-10A. (B) The cells were transfected with either agomir or NC for 48 h. The levels of miR-135b-5p in BT-474 and SK-BR-3 cells after transfection was detected using RT-qPCR. **P<0.01 vs. blank. Breast cancer cells (C) BT-474 and (D) SK-BR-3 were treated with trastuzumab (0, 0.5, 1, 2 or 4 µg/ml) and miR-135b-5p agomirfor 48 h. Cell viability was detected using a Cell-Counting Kit 8 assay. *P<0.05 and **P<0.01 vs. blank. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; OD, optical density.
Trastuzumab-induced apoptosis is enhanced by miR-135b-5p agomir in BT-474 cells
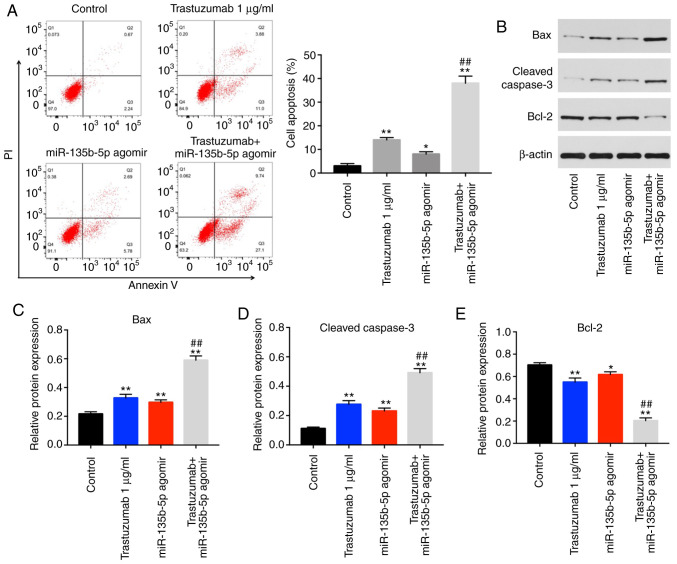
Annexin V/PI staining was performed to detect the mechanism by which miR-135b-5p enhanced the anti-proliferative effect of trastuzumab. The results indicated that trastuzumab or miR-135b-5p agomir treatment significantly induced cell apoptosis compared with the control group (Fig. 2A). In addition, trastuzumab-induced apoptosis was significantly enhanced by miR-135b-5p agomir in BT-474 cells (Fig. 2A). Moreover, the expression levels of apoptotic-associated proteins were examined in BT-474 cells by western blotting. The combination of miR-135b-5p agomir with trastuzumab significantly increased the expression levels of cleaved caspase 3 and Bax in BT-474 cells compared cells treated miR-135b-5p agomir or trastuzumab alone (Fig. 2B-D). Furthermore, trastuzumab-induced Bcl-2 downregulation was significantly enhanced in the presence of miR-135b-5p agomir (Fig. 2E). Taken together, the data demonstrated that miR-135b-5p agomir could enhance trastuzumab-induced apoptosis via activation of the intrinsic apoptotic pathway.
Figure 2.
miR-135b-5p agomir enhances the anti-proliferation effects of trastuzumab via inducing apoptosis. (A) Annexin V/PI staining in BT-474 cells was detected by flow cytometry. (B) Expression levels of Bax, cleaved caspase 3 and Bcl-2 in BT-474 cells were detected using western blotting. The relative expression levels of (C) Bax, (D) cleaved caspase-3 and (E) Bcl-2 were quantifiedby normalizing to β-actin levels. *P<0.05 and **P<0.01 vs. control; ##P<0.01 vs. trastuzumab. miR, microRNA; PI, propidium iodide.
Anti-migration and anti-invasion effects of trastuzumab is enhanced by miR-135b-5p agomir
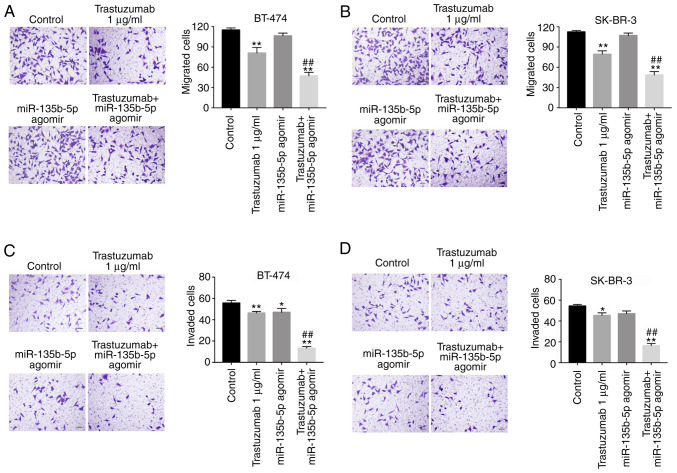
Following the initial investigation of the effects of miR-135b-5p on trastuzumab-induced apoptosis, its effectson the inhibition breast cancer cell migration and invasion were also investigated. Transwell assays indicated that trastuzumab significantly inhibited migration and invasion abilities in BT-474 and SK-BR-3 cells compared with the control group (Fig. 3A-D). Furthermore, the inhibitory effects of trastuzumab on the migration and invasion of BT-474 and SK-BR-3 cells were significantly enhanced in the presence of miR-135b-5p agomir (Fig. 3A-D). These results suggested that miR-135b-5p agomir could increase the anti-metastatic effects of trastuzumab in HER-2-positive breast cancer cells.
Figure 3.
Anti-migration and anti-invasion effects of trastuzumab on breast cancer cells is enhanced by miR-135b-5p agomir. BT-474 and SK-BR-3 cells were transfected with 1 µg/ml trastuzumab, 50 nM miR-135b-5p agomir or a combination of trastuzumab and miR-135b-5p agomir for 24 h. Cell migration assay was performed to evaluate the migration ability of (A) BT-474 and (B) SK-BR-3 cells. Magnification ×200. Cell invasion assay was performed to evaluate the invasion ability of (C) BT-474 and (D) SK-BR-3 cells. Magnification, ×200. *P<0.05 and **P<0.01 vs. control; ##P<0.01 vs. trastuzumab. miR, microRNA.
Cyclin D2 is a direct target of miR-135b-5p in BT-474 breast cancer cells
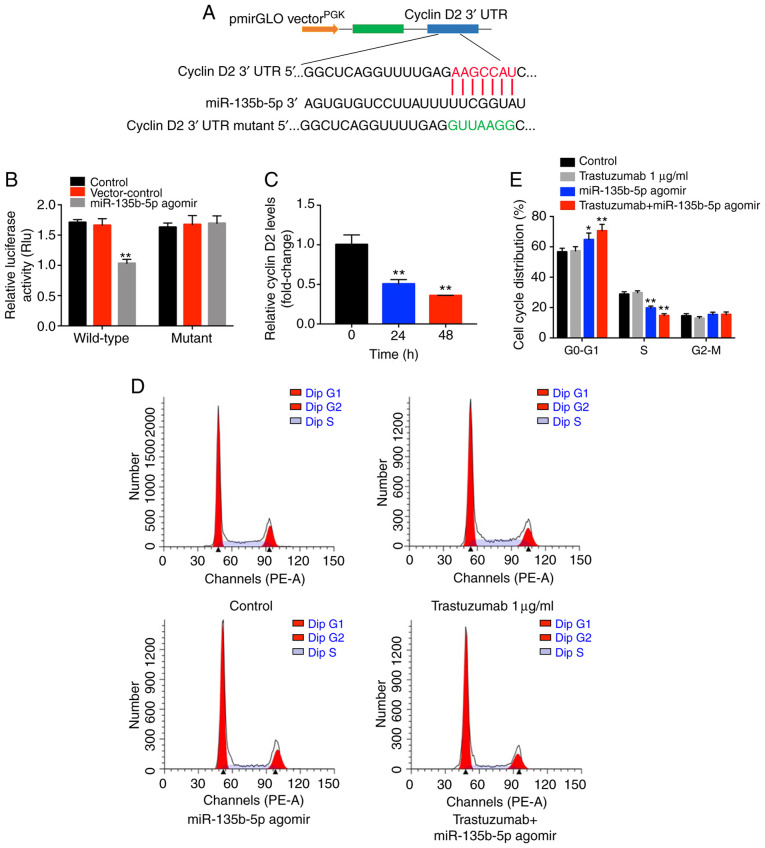
To investigate the underlying mechanisms by which miR-135b-5p agomir increased the antitumor effects of trastuzumab in breast cancer cells, TargetScan and miRWalk online tools were used. A putative miR-135b-5p-binding site was identified at a specific position in the 3′-UTR of cyclin D2 mRNA. To further confirm the direct interaction between miR-135b-5p and cyclin D2 mRNA, the predicted binding site of miR-135b-5p to cyclin D2 mRNA was cloned into a luciferase reporter vector. miR-135b-5p agomir significantly inhibited luciferase activity compared with the control group (Fig. 4B), suggesting that it could interact directly with the 3′-UTR of cyclin D2 mRNA. In contrast to these observations, miR-135b-5p agomir did not affect the luciferase activity of the mutated cyclin D2. In addition, the qPCR data indicated that cyclin D2 mRNA synthesis was significantly inhibited when the cells were treated with miR-135b-5p agomir, and cyclin D2 levels decreased over time following agomir transfection (Fig. 4C). Taken together, the data indicated that cyclin D2 was a direct target of miR-135b-5p in BT-474 breast cancer cells.
Figure 4.
miR-135b-5pinduces G0/G1 cell cycle arrest in BT-474 cells via directly targeting cyclin D2. (A) The target sequences for miR-135b-5p within the 3′-UTR of cyclin D2. (B) Comparison of luciferase activity in cells of different groups that were co-transfected with wild-type or mutant 3′-UTR cyclin D2. **P<0.01 vs. control. (C) The levels of cyclin D2 mRNA was evaluated using reverse transcription-quantitative PCR. **P<0.01 vs. 0 h. (D) Cell cycle analysis was performed to determine the effect of miR-135b-5p agomir on cell cycle distribution. (E) Quantification of cell cycle distribution ratios. *P<0.05 and **P<0.01 vs. control. 3′UTR, 3′-untranslated region; miR, microRNA.
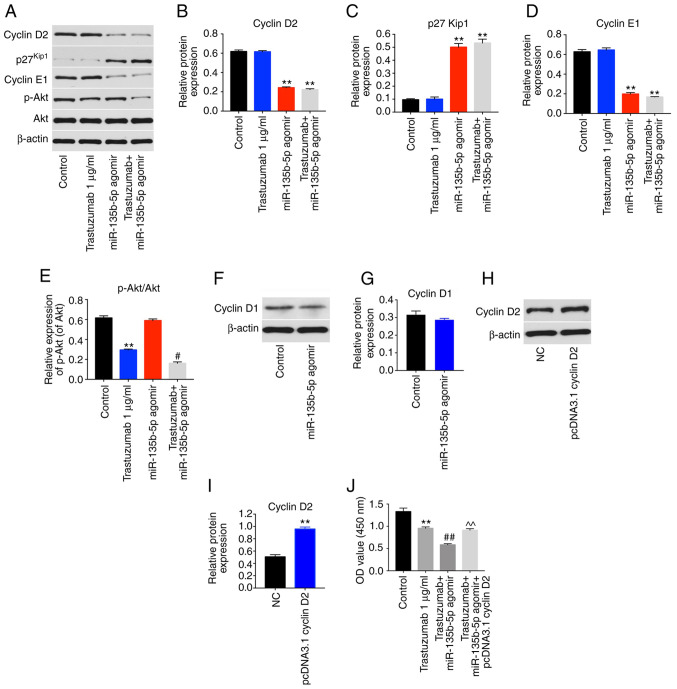
Since cyclin D2 mRNA was demonstrated to directly bind to miR-135b-5p, the cell cycle ofmiR-135b-5p agomir-treated cells was examined. miR-135b-5p-overexpressing BT-474 cells displayed a significant increase in the percentage of cells in the G0/G1-phase but significantly decreased proportions in S-phase cells compared with controls, while trastuzumab exhibited no effects (Fig. 4D and E). In addition, western blot analysis illustrated that trastuzumab treatment had no effect on the expression levels of cyclin D2, p27kip1 and cyclin E1 in BT-474 cells. However, miR-135b-5p agomir or combination treatment significantly inhibited the expression levels of cyclin D2 and cyclin E1 and significantly increased p27kip1 levels in BT-474 cells compared with the control group (Fig. 5A-D). Moreover, trastuzumab treatment significantly downregulated the expression of phosphorylated (p)-Akt in BT-474 cells, while overexpression of miR-135b-5p did not affect p-Akt activation in BT-474 cells (Fig. 5A and E). Interestingly, trastuzumab-induced p-Akt downregulation was enhanced in the presence of miR-135b-5p agomir (Fig. 5A and E). Meanwhile, overexpression of miR-135b-5p did not show any significant effect on the expression of cyclin D1 in BT-474 cells (Fig. 5F and G). Taken together, the data demon-strated that miR-135b-5p agomir could induce G0/G1 arrest in BT-474 cells by directly binding to cyclin D2. Furthermore, the overexpression efficiency of cyclin D2 was confirmed by western blotting (Fig. 5H and I). Overexpression of cyclin D2 significantly reversed the growth-inhibitory activity of miR-135b-5p in combination with trastuzumab in BT-474 cells (Fig. 5J). These data indicated that miR-135b-5p agomir enhanced the anti-proliferative effects of trastuzumab in BT-474 cells via downregulation of cyclin D2.
Figure 5.
miR-135b-5p agomir regulates cyclin D2, p27Kip1 and cyclin E1 signaling pathways. BT-474 cells were transfected with 1 µg/ml trastuzumab, 50 nM miR-135b-5p agomir or a combination of trastuzumab and miR-135b-5p agomir for 48 h. (A) Expression levels of cyclin D2, p27Kip1, cyclin E1 and p-Akt in BT-474 cells were detected with western blotting. The relative levels of (B) cyclin D2, (C) p27Kip1, (D) cyclin E1 and (E) p-Akt were quantified by normalizing to β-actin levels. (F) BT-474 cells were transfected with miR-135b-5p agomir or NC for 48 h. The levels of cyclin D1 in BT-474 cells was detected using western blotting. (G) Quantification of cyclin D1 expression. (H) BT-474 cells were transfected with NC or pcDNA 3.1-cyclin D2 for 48 h. (I) The level of cyclin D2 in BT-474 cells was detected with western blotting. (J) BT-474 cells were transfected with pcDNA 3.1-cyclin D2 and miR-135b-5p agomir for 48 h in the presence of trastuzumab. The cell viability was detected using a Cell Counting Kit-8 assay. **P<0.01 vs. control; #P<0.05 and ##P<0.01 vs. trastuzumab; ^^P<0.01 vs. trastuzumab + miR-125b-3p agomir. p-Akt, phosphorylated Akt; miR, microRNA; OD, optical density; NC, negative control.
miR-135b-5p agomir enhances the antitumor effect of trastuzumab in vivo
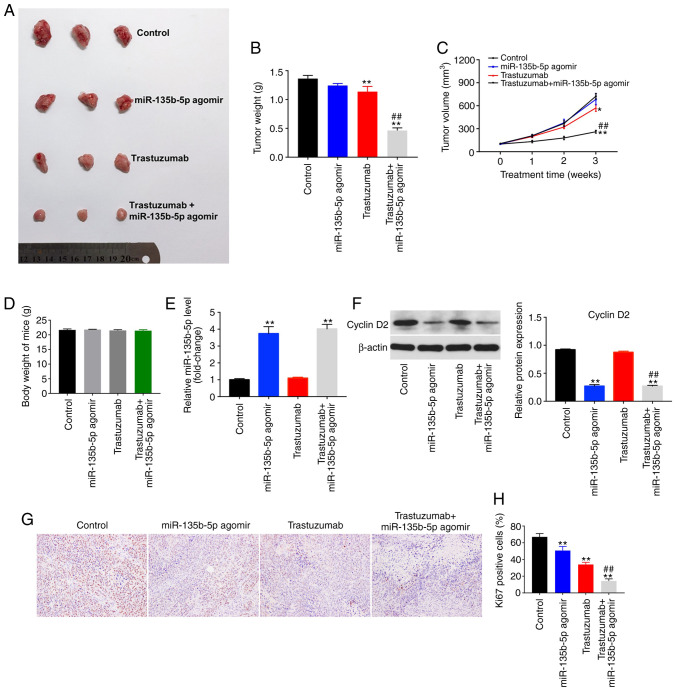
To explore whether miR-135b-5p could increase the antitumor effects of trastuzumab in vivo, a breast cancer xenograft model was established. The results indicated that trastuzumab treatment alone was able to decrease tumor size compared with controls (Fig. 6A). Tumor weight and tumor volume were significantly reduced in the combination treatment group compared with mice treated with trastuzumab or miR-135b-5p agomir alone (Fig. 6A-C). However, miR-135b-5p agomir alone showed no significant difference on tumor volume. In addition, the body weight changes indicated the safety and tolerability of the combination treatment (Fig. 6D). Furthermore, the expression levels of miR-135b-5p was significantly upregulated in the miR-135b-5p agomir or combination treatment group compared with the control group, indicating that miR-135b-5p is stably expressed in tumor tissues (Fig. 6E). Additionally, trastuzumab had no effect on cyclin D2 expression in tumor tissues, whereas the level of cyclin D2 significantly decreased in the presence of miR-135b-5p agomir compared with controls (Fig. 6F). IHC assay indicated that miR-135b-5p agomir or trastuzumab treatment significantly inhibited proliferation in tumor tissues, compared with the control group. As expected, combination treatment significantly inhibited proliferation in tumor tissues compared with the trastuzumab treatment group (Fig. 6G and H). These data were consistent with the in vitro results. Taken together, these results confirmed that miR-135b-5p agomir could enhance the antitumor effects of trastuzumab in vivo.
Figure 6.
miR-135b-5p agomir enhances the antitumor effects of trastuzumab in vivo. (A) The mice were treated with saline solution (control), trastuzumab, miR-135b-5p agomir or a combination of trastuzumab and miR-135b-5p agomir for 3 weeks. (B) Tumor weight in each group was detected after 3 weeks. (C) Tumor volumes of nude mice were monitored weekly. (D) Body weight of each group of nude mice was evaluated at day 27. (E) The levels of miR-135b-5p in tumor tissues was detected using reverse transcription-quantitative PCR. (F) The expression levels of cyclin D2 in tumor tissues was detected using western blotting. Relative protein expression was quantified by normalizing to β-actin levels. (G) Immunohistochemistry images and (H) quantification of proliferation in tumor tissues. Magnification ×200. *P<0.05 and **P<0.01 vs. control; ##P<0.01 vs. trastuzumab. miR, microRNA.
Discussion
In the present study, the combined antitumor effects of trastuzumab with miR-135b-5p agomir were investigated in vitro and in vivo. miR-135b-5p agomir enhanced the antitumor effect of trastuzumab in breast cancer cells in vitro and in vivo. A previous study reported that overexpression of miR-135b-5p suppressed migration and invasion in breast cancer (22). The present results indicated that miR-135b-5p agomir suppressed proliferation, migration and invasion in BT-474 and SK-BR-3 HER-2-positive breast cancer cells, consistent with previous reports (27,28). Agomir is a type of specially labeled and chemically modified double-stranded microRNA that regulates the biological function of target genes by mimicking endogenous microRNA (29). Compared with common miRNA mimics, miRNA agomir has a higher affinity for the cell membrane (29). In addition, agomir is especially suitable for animal in vivo experiments and have higher stability (30). These data showed that miR-135b-5p exerted anti-proliferation, anti-migration and anti-invasion effects in HER-2-positive breast cancer cells. Moreover, miR-135b-5p agomir potentiated the antitumor effect of trastuzumab by inducing apoptosis. In addition, the inhibition of breast cell migration and invasion by trastuzumab was enhanced by miR-135b-5p agomir transfection. Tang et al (31) indicated that overexpression of miR-200c increased drug sensitivity to trastuzumab in HER-2-positive breast cancer cells, consistent with the data reported here. miR-135b-5p agomir enhanced the antitumor effect of trastuzumab in vivo and in vitro.
The target predicting databases TargetScan and miRWalk were used to investigate the mechanism by which miR-135b-5p increases the antitumor effect of trastuzumab. Cyclin D2 was identified as a potential target of miR-135b-5p. Cyclin D2 protein is involved in the regulation of the cell cycle at the point of transition from G1 to DNA synthesis (32). Zhou et al (33) indicated that overexpression of miR-206 blocked G1/S transition and inhibited the proliferation of breast cancer cells via downregulation of cyclin D2 expression. In the present study, miR-135b-5p agomir suppressed the proliferation of breast cancer cells by inhibiting cyclin D2 expression, which was consistent with the previous study. Zhang et al (23) reported that miR-135b-5p enhanced doxorubicin-sensitivity of ER-positive breast cancer cells by targeting anterior gradient 2. The differences noted on the targeting of proteins may be associated with the breast cancer subtypes, namely estrogen (ER) or HER-2, which are upstream regulators of miR-135b-5p. Therefore, in addition to ER positive breast cancer, miR-135b also plays a role as a regulator of HER-2-positive breast cancer. These findings suggested that miR-135b-5p may act as regulator in different types of breast cancer.
Overexpression of ErbB2 (HER-2) leads to the potentiation of cyclin E-Cdk2 activity by sequestration of the cyclin-dependent kinase inhibitor p27kip1 (10). A previous study reported that trastuzumab inhibited mitogen-activated protein kinase and PI3K/AKT pathways, leading to cell cycle arrest (34). However, in the present study, the HER-2 antibody trastuzumab exhibited limited effects on the expression of cyclin D2 or p27Kip1, while miR-135b-5p acted as a regulator of cyclin D2 and p27Kip1 expression levels. The correlation between p27Kip1 expression and the PI3K/Akt pathway in the combination treatment of miR-135b-5p with trastuzumab can be further explored in future studies. In addition, a number of miRNAs have been shown to suppress cyclin D2 expression in various tumor types (35-38), including miR-133, miR-203, miR-204 and miR-206. To the best of our knowledge, the present study was the first to demonstrate that miR-135b-5p directly suppressed cyclin D2 expression in HER-2-positive breast cancer.
However, the present study had several limitations. The present study indicated that miR-770-5p agomir in combination with trastuzumab treatment blocked the migration and invasion capacities of HER-2-positive breast cancer cells in vitro. However, the correlation between miR-770-5p agomir in combination with trastuzumab and breast cancer metastasis in vivo is still unclear and need to be investigated. In addition, the present study provided evidence to support the anti-cancer properties of miR-135b-5p, but little is known regarding the clinical significance of miR-135b-5p in human breast cancer. Therefore, the clinical application value of miR-135b-5p in breast cancer should be investigated in the future. Moreover, the present study only detected the expression of p-Akt in HER-2-positive breast cancer cells. However, expression of other proteins associated with the PI3K/Akt pathway, such as PI3K and mTOR should be investigate using western blotting to demonstrate whether miR-135b-5p agomir enhances the antitumor effect of trastuzumab in HER-2-positive breast cancer cells via the PI3K/Akt pathway. Meanwhile, rescue experiments are needed to further prove whether miR-135b-5p agomir could enhance the antitumor effect of trastuzumab in HER-2-positive breast cancer cells via targeting cyclin D2.
In conclusion, miR-135b-5p agomir enhanced the anti-tumor effect of trastuzumab in BT-474 cells in vitro and in vivo. miR-135b-5p induced cell cycle arrest by binding to cyclin D2. Therefore, the combination of miR-135b-5p agomir with trastuzumab may be a potential strategy for the treatment of patients with HER-2-positive breast cancer.
Acknowledgments
Not applicable.
Funding
The study was supported by the Pudong New Area Science and Technology Development Fund (grant no. PKJ2017-Y12), Important Weak Subject of the Pudong New Area Health and Family Planning Commission (grant no. PWZbr2017-01) and the National Natural Science Foundation Youth Project (grant no. 81702422).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZL and YQ made major contributions to the conception, design and manuscript drafting of this study. PC, QL and HS were responsible for data acquisition, data analysis, data interpretation and manuscript revision. XJ made substantial contributions to conception and design of the study and revised the manuscript critically for important intellectual content. All authors agreed to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal procedures were approved by the Seventh People's Hospital of Shanghai University of Traditional Chinese Medicine Committee. National Institutes of Health Guide for the Care and Use of Laboratory Animals was followed strictly.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tormo E, Adam-Artigues A, Ballester S, Pineda B, Zazo S, González-Alonso P, Albanell J, Rovira A, Rojo F, Lluch A, Eroles P. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: From early detection to new therapies. Radiologia. 2017;59:368–379. doi: 10.1016/j.rx.2017.06.003. In English, Spanish. [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Jung SY, Kim K. Survival benefit of radiotherapy after surgery in de novo stage IV breast cancer: A population-based propensity-score matched analysis. Sci Rep. 2019;9:8527. doi: 10.1038/s41598-019-45016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen PL, Taghian AG, Katz MS, Niemierko A, Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL, Harris JR. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26:2373–2378. doi: 10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 6.Chavez-Blanco A, Perez-Sanchez V, Gonzalez-Fierro A, Vela-Chavez T, Candelaria M, Cetina L, Vidal S, Dueñas-Gonzalez A. HER2 expression in cervical cancer as a potential therapeutic target. BMC Cancer. 2004;4:59. doi: 10.1186/1471-2407-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baselga J, Swain SM. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 8.Horton J. Trastuzumab use in breast cancer: Clinical issues. Cancer Control. 2002;9:499–507. doi: 10.1177/107327480200900607. [DOI] [PubMed] [Google Scholar]
- 9.Sarup JC, Johnson RM, King KL, Fendly BM, Lipari MT, Napier MA, Ullrich A, Shepard HM. Characterization of an anti-p185HER2 monoclonal antibody that stimulates receptor function and inhibits tumor cell growth. Growth Regul. 1991;1:72–82. [PubMed] [Google Scholar]
- 10.Lane HA, Beuvink I, Motoyama AB, Daly JM, Neve RM, Hynes NE. ErbB2 potentiates breast tumor proliferation through modulation of p27(Kip1)-Cdk2 complex formation: Receptor overexpression does not determine growth dependency. Mol Cell Biol. 2000;20:3210–3223. doi: 10.1128/MCB.20.9.3210-3223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit AM, Rak J, Hung MC, Rockwell P, Goldstein N, Fendly B, Kerbel RS. Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: Angiogenic implications for signal transduction therapy of solid tumors. Am J Pathol. 1997;151:1523–1530. [PMC free article] [PubMed] [Google Scholar]
- 12.Sliwkowski MX, Lofgren JA, Lewis GD, Hotaling TE, Fendly BM, Fox JA. Nonclinical studies addressing the mechanism of action of trastuzumab (Herceptin) Semin Oncol. 1999;26:60–70. [PubMed] [Google Scholar]
- 13.Van Swearingen AED, Siegel MB, Deal AM, Sambade MJ, Hoyle A, Hayes DN, Jo H, Little P, Dees EC, Muss H, et al. LCCC 1025: A phase II study of everolimus, trastuzumab, and vinorelbine to treat progressive HER2-positive breast cancer brain metastases. Breast Cancer Res Treat. 2018;171:637–648. doi: 10.1007/s10549-018-4852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding K, Wu Z, Li X, Sheng Y, Wang X, Tan S. LMO4 mediates trastuzumab resistance in HER2 positive breast cancer cells. Am J Cancer Res. 2018;8:594–609. [PMC free article] [PubMed] [Google Scholar]
- 15.Tan H, Huang S, Zhang Z, Qian X, Sun P, Zhou X. Pan-Cancer analysis on microRNA-associated gene activation. EBioMedicine. 2019;43:82–97. doi: 10.1016/j.ebiom.2019.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider A, Victoria B, Lopez YN, Suchorska W, Barczak W, Sobecka A, Golusinski W, Masternak MM, Golusinski P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci Rep. 2018;8:675. doi: 10.1038/s41598-017-18945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venturutti L, Cordo Russo RI, Rivas MA, Mercogliano MF, Izzo F, Oakley RH, Pereyra MD, Proietti CJ, Yankilevich P, Roa JC, et al. miR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene. 2016;35:6189–6202. doi: 10.1038/onc.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song W, Wu S, Wu Q, Zhou L, Yu L, Zhu B, Gong X. The microRNA-141-3p/CDK8 pathway regulates the chemosensitivity of breast cancer cells to trastuzumab. J Cell Biochem. 2019;120:14095–14106. doi: 10.1002/jcb.28685. [DOI] [PubMed] [Google Scholar]
- 20.Noyan S, Gurdal H, Gur Dedeoglu B. Involvement of miR-770-5p in trastuzumab response in HER2 positive breast cancer cells. PLoS One. 2019;14:e0215894. doi: 10.1371/journal.pone.0215894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Cola A, Volpe S, Budani MC, Ferracin M, Lattanzio R, Turdo A, D'Agostino D, Capone E, Stassi G, Todaro M, et al. miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 2015;6:e1823. doi: 10.1038/cddis.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pu T, Shen M, Li S, Yang L, Gao H, Xiao L, Zhong X, Zheng H, Liu Y, Ye F, Bu H. Repression of miR-135b-5p promotes metastasis of early-stage breast cancer by regulating downstream target SDCBP. Lab Invest. 2019;99:1296–1308. doi: 10.1038/s41374-019-0258-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Xia F, Zhang F, Cui Y, Wang Q, Liu H, Wu Y. miR-135b-5p enhances doxorubicin-sensitivity of breast cancer cells through targeting anterior gradient 2. J Exp Clin Cancer Res. 2019;38:26. doi: 10.1186/s13046-019-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.National Institutes of Health . Guide for the Care and Use of Laboratory Animals. The National Academies Press; Washington, DC: 2011. p. p246. [Google Scholar]
- 26.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DAH, Glennie MJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, Zhang S, Wu Y, Calin GA, Bast RC., Jr Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One. 2012;7:e41170. doi: 10.1371/journal.pone.0041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lulli V, Buccarelli M, Martini M, Signore M, Biffoni M, Giannetti S, Morgante L, Marziali G, Ilari R, Pagliuca A, et al. miR-135b suppresses tumorigenesis in glioblastoma stem-like cells impairing proliferation, migration and self-renewal. Oncotarget. 2015;6:37241–37256. doi: 10.18632/oncotarget.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao M, Cai J, Cai L, Jia J, Xie L, Zhu Y, Huang B, Jin D, Wang Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J Ovarian Res. 2017;10:24. doi: 10.1186/s13048-017-0321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 31.Tang H, Song C, Ye F, Gao G, Ou X, Zhang L, Xie X, Xie X. miR-200c suppresses stemness and increases cellular sensitivity to trastuzumab in HER2+ breast cancer. J Cell Mol Med. 2019;23:8114–8127. doi: 10.1111/jcmm.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K, Ji G. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207–212. doi: 10.1016/j.bbrc.2013.02.084. [DOI] [PubMed] [Google Scholar]
- 34.Vu T, Claret FX. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front Oncol. 2012;2:62. doi: 10.3389/fonc.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu WF, Wang HM, Lu BC, Zhang GZ, Ma HM, Wu ZY. miR-206 inhibits human laryngeal squamous cell carcinoma cell growth by regulation of cyclin D2. Eur Rev Med Pharmacol Sci. 2015;19:2697–2702. [PubMed] [Google Scholar]
- 36.Wu X, Zeng Y, Wu S, Zhong J, Wang Y, Xu J. miR-204, down-regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting cyclin D2 and MMP-9. FEBS Lett. 2015;589:645–650. doi: 10.1016/j.febslet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 37.Pan JL, Yuan DZ, Zhao YB, Nie L, Lei Y, Liu M, Long Y, Zhang JH, Blok LJ, Burger CW, Yue LM. Progesterone-induced miR-133a inhibits the proliferation of endometrial epithelial cells. Acta Physiol (Oxf) 2017;219:683–692. doi: 10.1111/apha.12762. [DOI] [PubMed] [Google Scholar]
- 38.Zhao S, Han J, Zheng L, Yang Z, Zhao L, Lv Y. MicroRNA-203 regulates growth and metastasis of breast cancer. Cell Physiol Biochem. 2015;37:35–42. doi: 10.1159/000430331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.