Abstract
Angiogenesis and vascular maturation play important roles in tumorigenesis and tumor development. The expression of neuropilin 1 (NRP1) is closely associated with angiogenesis in tumors; however, the molecular mechanisms of action in angiogenesis and tumor maturation, as well as the potential clinical value of NRP1 remain unclear. The importance of NRP1 expression in tumor progression was determined using The Cancer Genome Atlas (TCGA) database analysis. Gain- and loss-of-function experiments of NRP1 were performed in vascular endothelial cells (ECs) to investigate the functions in angiogenesis. CCK-8, flow cytometry, Transwell experiments and a series of in vitro experiments were used to detect cell functions. A combination of angiogenesis antibody arrays and RNA-Seq analyses were performed to reveal the proangiogenic mechanisms of action. The function of semaphorin 4D (SEMA4D) was also investigated separately. NRP1 mRNA levels were significantly increased in primary tumors compared with normal tissues based on TCGA data (P<0.01) and were associated with tumor development in patients. Gain- and loss-of-function experiments highlighted the function of NRP1 in promoting EC proliferation, motility and capillary-like tube formation and in reducing apoptosis. NRP1 overexpression led to significantly decreased EC markers (PECAM-1, angiogenin, PIGF and MMP-9) expression levels and reduced the vascular maturity. MAPK7, TPM1, RRBP1, PTPRK, HSP90A, PRKD2, PFKFB3, RGS4 and SPARC were revealed to play important roles in this process. SEMA4D was revealed to be a key protein associated with NRP1 in ECs. These data indicated that NRP1-promoted angiogenesis may be induced at the cost of reducing maturity of the ECs. NRP1 may also be a therapeutic target for antiangiogenic strategies and a candidate prognostic marker for tumors.
Keywords: angiogenesis, vascular maturation, neuropilin 1, semaphorin 4D
Introduction
Angiogenesis is the process of forming new blood vessels from existing blood vessels, which can supply oxygen and nutrients to cells, remove metabolic waste and deliver necessary immune cells or molecules into tissues to provide immune surveillance to prevent disease (1,2). Vascular maturation is a complex process of vascular remodeling through the interactions between endothelial cells (ECs) and perivascular cells, in the late stage of angiogenesis. It includes partial degeneration of microvessels and the construction of the vascular basement membrane, as well the gradual maturation of ECs from their active proliferation stage to a relatively static condition (3). The mature vascular ECs will establish a relationship with the vascular basement membrane and various peripheral cells in order to improve the hemodynamic characteristics and diffusion function (4,5). Angiogenesis and maturation are essential for tumorigenesis and development, and they play an important role in the occurrence, development and metastasis of numerous malignant tumors, which can take advantage of abnormal angiogenesis for rapid growth, metastasis and death (6,7). Rapidly growing tumors are in continuous demand for oxygen and nutrients, which create an imbalance in pro- and anti-angiogenic signaling and reduce vascular maturation (8). As antiangiogenic drugs, which were designed to starve tumors by cutting off the vascular supply of cancer cells, only provide modest survival benefits in the order of weeks to months in most patients with cancer, tumor vascular normalization has developed as an alternative strategy for anti-angiogenic cancer treatment (9). Therefore, factors associated with reduced vascular maturity in tumors may be important targets for anti-angiogenic drugs.
Neuropilin 1 (NRP1) is a non-tyrosine kinase trans-membrane glycoprotein and is a co-receptor of semaphorin (SEMA)3A and vascular endothelial growth factor (VEGF) on the cell membrane. NRP1 plays an important role in angiogenesis, cell survival, migration and invasion, and could be a novel tumor marker (10). The expression of NRP1 is closely related to angiogenesis (11). NRP1 overexpression can induce excessive blood vessel formation, especially capillaries, and knockdown of NRP1 can cause severe vascular developmental defects, which can cause death in mice (12). The interaction between NRP1 and VEGF has demonstrated that the VEGF/NRP1 signaling pathway in ECs is essential for angiogenesis (13). Blocking SEMA3A/NRP1 signaling can inhibit angiogenesis and also can control the heterogeneity of tumor-associated macrophages (14). The NRP1:SEMA4A axis can regulate stability of T cells, which may be related to certain inflammatory sites (15). These studies suggested an important role of NRP1 in angiogenesis; however, the underlying molecular mechanisms remain unclear and require further study.
NRP1 has been revealed to be strongly expressed in tumor ECs of pancreatic ductal adenocarcinoma as well as liver cancer (10,16). Our previous study found that NRP1 was predominantly expressed in ECs from the paratumor of lung squamous cell carcinomas and could be identified as a potential biomarker for anti-angiogenic therapies (17). To further study the effects of NRP1 on angiogenesis, the present study was designed. An overexpression NRP1 model and siRNA NRP1 model were established in vascular ECs [microvascular EC (MVEC) and Ealy926], and the effects of NRP1 on cell angiogenesis-related functions, including proliferation, apoptosis, migration and tube-forming activity, were examined at the cell level. The key factors related to NRP1-induced promotion of angiogenesis were identified. Possible molecular mechanisms of action underlying the effects of NRP1 on angiogenesis and vascular maturity were analyzed using omics research.
Materials and methods
Database analysis
The association between NRP1 and tumor grades or the prognosis was analyzed using UALCAN (http://ualcan.path.uab.edu), which is an interactive web-portal that allows cancer researchers and clinicians to analyze the relative expression levels of a query gene(s) across tumor and normal samples, and to evaluate relative clinicopathological parameters in various individual cancer types within TCGA database (18). The variation in expression levels between normal and different tumor grades were analyzed using the in-built statistical methods of the UALCAN web-software and a P<0.05 was considered to indicate a statistically significant difference.
Survival analysis of differentially expressed NRP1 levels in cancer tissues were performed using the Kaplan-Meier Plotter database (http://kmplot.com/analysis/) (19). The Kaplan-Meier Plotter was established using gene expression data and survival information for cancer survival rates. The cancer patients were divided into high- and low-expression value groups, and the survival analysis was carried out using the Kaplan-Meier method. Hazard ratios with 95% confidence intervals and log-rank P-values were calculated.
Cell culture
An inferior mesenteric artery vascular EC line (Ealy926; cat. no. 20911549; Shanghai Aolu Biological Technology Co., Ltd.), and human MVECs (HMEC-1; plat-form no. bio-106081; Biobw.org) were purchased and cultured in DMEM-F12 medium supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). Primary human umbilical vein ECs (HUVECs) were prepared by the present laboratory (previously purchased and strictly preserved at our laboratory; cat. no. C-003-5C; Invitrogen; Thermo Fisher Scientific, Inc.) and cultured in Medium 131 (Thermo Fisher Scientific, Inc.) supplemented with 10% microvascular growth supplement (Invitrogen; Thermo Fisher Scientific, Inc.). The mediums were supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were maintained in a humidified chamber at 37°C in 5% CO2. The seeding cells of primary HUVECs were stored using liquid nitrogen, and after each resuscitation, and only ~3-4 generations were used.
To detect the general angiogenesis function of NRP1 in ECs, all of these three EC lines were used for identification. The primary HUVECs, which were not easy to culture and closer to the state of cells in vivo, were just used for gene expression profile analysis.
NRP1 silencing and overexpression
The siRNA targeting NRP1 was designed and synthesized by Sigma-Aldrich; Merck KGaA. Transfections of the siRNA were performed using Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) in 6-well plates, according to the manufacturer's protocol. Briefly, the NRP1 siRNA was diluted in Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated with Lipofectamine® RNAiMAX for 20 min before being added to the cultures for transfections. After a 6-h culture, the medium was replaced with new complete medium cultured for another 48 h. The efficiency of siRNA transfections was verified by reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis. The siRNA sequences that resulted in efficient NRP1 knockdown were as follows: NRP1 siRNA-1, 5′-CUG AAU GUU CCC AGA ACU AdT dT-3′; NRP1 siRNA-2, 5′-GAA GUA UAC GGU UGC AAG AdT dT-3′; NRP1 siRNA-3, 5′-UGU UGU GGU UGC AGU AUU CdT dT-3′; NRP1 forward, 5′-GCA GGA TTT TCC ATA CGT TAT-3′ and reverse, 5′-AAA TTC CAG GAT AAT CTC TGA C-3′; 18srRNA forward, 5′-CCT GGA TAC CGC AGC TAG GA-3′ and reverse, 5′-GCG GCG CAA TAC GAA TGC CCC-3′. siRNA sequences which resulted in efficient SEMA4D knockdown were also prepared: SEMA4D siRNA-1, 5′-GGA AGG TCT CAG AAG ACA A-3′; SEMA4D siRNA-2, 5′-CCT TGA ATT TGC CAG ACA A-3′; SEMA4D siRNA-3, 5′-GGA CAC CTT GTA CAT AGG T-3′; SEMA4D forward, 5′-GCT ACA CAT CCG TCA TGG TT-3′ and reverse, 5′-AGA CAC CTC CGT GAA GAA GA-3′.
A lentiviral expression vector (pLVX-IRES-Neo) was used for NRP1 gene delivery and stable overexpression. The human NRP1 gene (NM_001024628.2) was PCR-amplified from the human 293T cell cDNA library. PCR primers were designed and the XhoI and BamHI restriction endonuclease sites were introduced into the plasmid as follows: NRP1-XhoI forward, 5′-ccg ctc gag gcc acc ATG GAG AGG GGG CTG CCG CTC CTC TGC-3′; NRP1-BamHI reverse, 5′-cgc gga tcc TTA TTT GAT ACC TGA TTG TAT GGT GCT G-3′. The plasmid was doubly digested using XhoI and BamHI (New England Biolabs, Inc.). The PCR product recovery and enzymatic-digested plasmid were purified using a DNA Gel Extraction kit (Guangzhou Dongsheng Biotech Co., Ltd.). Then they were ligated (T4 DNA Ligase was purchased from Takara Biotechnology Co., Ltd.) and the ligation mixture was transformed into competent E.coli DH5α cells (Invitrogen; Thermo Fisher Scientific, Inc.). The clones were selected using double enzyme digestion and sequencing, and the recombinant plasmid was extracted for transfections. The plasmids were transfected into the MVEC and Ealy926 cells according to the instructions of Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
RT-qPCR and western blotting
Total RNA was extracted from ECs using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. DNase I (Promega Corporation) was used to remove DNA. Reverse transcription was performed using GoScript™ reverse transcriptase (Promega Corporation). RT-qPCR was carried out using SYBR Green qPCR SuperMix (Invitrogen; Thermo Fisher Scientific, Inc.) and an ABI PRISM® 7500 Sequence Detection System. The primers used were as follows: NRP1 forward, 5′-GCA GGA TTT TCC ATA CGT TAT-3′ and reverse, 5′-AAA TTC CAG GAT AAT CTC TGA C-3′; 18srRNA forward, 5′-CCT GGA TAC CGC AGC TAG GA-3′ and reverse, 5′-GCG GCG CAA TAC GAA TGC CCC-3′. The thermocycling conditions were as follows: 95°C for 3 min; followed by 95°C for 15 sec and 60°C for 32 sec, for 40 cycles. The 2−ΔΔCq method (20) was employed to perform the analysis of differential gene expression.
For western blotting, 1×106 cells of each group were collected for total protein extraction using RIPA buffer (product no. R0278) with protease inhibitor cocktail (1:100; product no. P8340; both from Sigma-Aldrich; Merck KGaA), and the concentration was detected using Bradford assay (cat. no. KGPBCA; KeyGen Biotech Co., Ltd.). Equal amounts of lysates (40 µg) were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes and blocked using 5% skim milk at room temperature for 1 h. Subsequently, the membranes were incubated with following primary antibodies: Anti-NRP1 (1:1,000; product code ab25998), anti-SEMA4D (1:2,000; product code ab134128) and anti-GAPDH-HRP (1:10,000; product code 9485; all from Abcam) at 4°C overnight. Then the secondary antibody, HRP-conjugated goat anti-rabbit IgG (H+L) (1:20,000; cat. no. 4050-05; SouthernBiotech) was added and the membranes were incubated at 37°C for 1 h. Immobilon western chemiluminescent HRP substrate (cat. no. WBKLS0500; Millipore; Merck KGaA) was used for protein band visualization, and ImageJ 1.4.4 software (National Institutes of Health) for densitometric analysis.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assays were employed to detect the effects of NRP1 overexpression/knockdown on the cell proliferative ability of MVEC and Ealy926 cells. Cells were seeded in 96-well plates at 4.0×103 cells/well and the cell viability was evaluated through the CCK-8 assay (10 µl of solution reagent for each well of a 96-well assay plate containing the samples in 100 µl of culture medium) (cat. no. G3582; Promega Corporation) after the cells were cultured for 0, 24, 48, 72 and 96 h, following the manufacturer's protocol.
Flow cytometric analysis
A total of 3×105 cells/well were cultured in 12-well cell culture plates and were harvested after 48 h of incubation. Annexin V-FITC and propidium iodide were used to stain the cells, following the protocol of Cellular Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.). Apoptosis was determined using flow cytometry (BD Biosciences) and the flow rate and cell concentration of the samples were adjusted to ensure an acquisition <500 cells/sec. At least 104 cells were acquired for analysis. Data were collected and further analyzed using FlowJo 7.0 (FlowJo LLC).
Transwell assay
Cell migration assays were performed using Transwell chambers (cat. no. 353097; Corning, Inc.). MVEC, Ealy926 and HUVEC cells (with overexpression/knockdown of NRP1), as well as Ealy926 cells (with knockdown of SEMA4D and SEMA4D knockdown + NRP1 overexpression) were cultured in serum-free DMEM-F12 medium (5×104 cells/ml). A total of 300 µl of the cell suspension was added into the upper chamber and 600 µl DMEM-F12 medium with 10% FBS was added into the lower chamber at 37°C for 24 h. A cotton-tipped swab was used to wipe out the cells that did not migrate through the membrane of the upper chamber. The filters were fixed using 4% paraformaldehyde for 20 min at room temperature, stained using 1% crystal violet for 20 min at room temperature (product no. 61135; Sigma-Aldrich; Merck KGaA) and observed using a light microscope (CKX41, U-CTR30-2; Olympus Corporation, ×200). Each sample was assayed in triplicate.
Tube-forming assay
MVEC, Ealy926 or HUVEC cells (2×104) were cultured in the 96-well plates, which were pre-coated with 50 µl of 10 mg/ml solution of Matrigel (BD Biosciences) and allowed to polymerize at 37°C for 2 h. After 6 h of culture on the Matrigel-coated wells at 37°C, cell angiogenesis, including junction, segment and node formation, were observed and analyzed using light microscopy (×40) and ImageJ V1.44 (National Institutes of Health).
Human angiogenesis antibody array
The human angiogenesis array kit was purchased from RayBiotech, Inc. (cat. no. QAH-ANG-1000). Control and overexpression NRP1 HUVEC cells were harvested from 6-well plates after 48 h. The angiogenesis antibody array analysis procedure was performed according to the manufacturer's instructions. The signals were visualized using a laser scanner equipped with a Cy3 wavelength (green channel), along with Axon GenePix (Axon Instruments). Data was analyzed using GenePix. Proteins densities exhibiting a >1.5-fold or a <0.67-fold change were defined as differentially expressed proteins.
RNA-seq analysis
HUVEC cells infected with the lentiviral expression vector for the increased expression of NRP1 or with mock vectors were prepared for RNA extraction and transcriptomic sequencing analysis. Total RNA was extracted using TRIzol® according to the manufacturer's protocol. The RNA-seq libraries were constructed using an Ultra RNA sample preparation kit (Illumina) and sequencing was performed using an Illumina HiSeq™ according to the manufacturer's protocol. The differentially expressed genes (DEGs) were identified using two criteria: i) A false discovery rate of <0.05 and ii) A log2 fold change of >1. Heatmap, volcano plot map, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed. A protein-protein interaction network was constructed using the STITCH website, version 5.0 (http://stitch.embl.de).
Statistical analysis
The results are presented as the mean ± SD/SEM and analyzed using SPSS 22 (IBM Corp.) and GraphPad Prism 7 (GraphPad Software, Inc.). Differences among groups were analyzed using one-way ANOVAs, two-tailed Student's t-tests, Mann-Whitney U tests or Fisher's exact test when applicable. P<0.05 was considered to indicate a statistically significant difference.
Results
Increased expression level of NRP1 is associated with increasing tumor grades
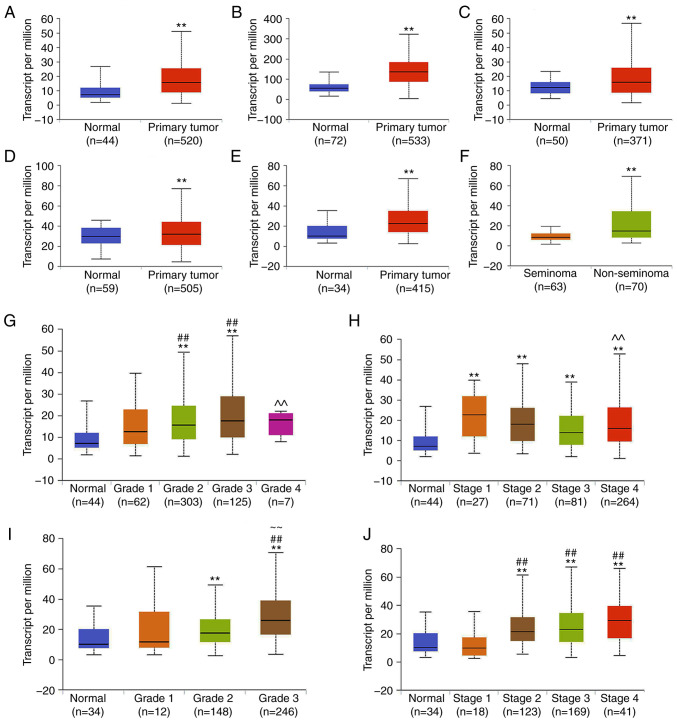
Data extracted from the UALCAN database demonstrated that NRP1 was significantly higher in the 6 tumor tissues compared with the matched TCGA normal tissue (P<0.01; Fig. 1A-F). NRP1 was expressed at higher levels in primary tumor tissue than in normal tissue in head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma, liver hepatocellular carcinoma, thyroid carcinoma and stomach adenocarcinoma (STAD), as well as being expressed at a higher level in non-seminoma tissue than seminoma tissue in testicular germ cell tumors.
Figure 1.
Expression of NRP1 in tumors using the UALCAN database analysis. Expression of NRP1 in (A) HNSC, (B) KIRC, (C) LIHC, (D) THCA and (E) STAD samples based on sample types of normal and primary tumor TCGA samples. (F) Expression of NRP1 in TGCTs based on tumor histology of seminoma and non-seminoma TCGA samples. Expression of NRP1 in HNSC based on the (G) tumor grade and (H) individual cancer stages in TCGA samples. Expression of NRP1 in STAD samples based on the (I) tumor grade and (J) individual cancer stages in TCGA samples. Grade 1, well differentiated (low grade); grade 2, moderately differentiated (intermediate grade); grade 3, poorly differentiated (high grade); grade 4, undifferentiated (high grade). **P<0.01 vs. Normal; ##P<0.01 vs. Grade 1 or Stage 1; ~~P<0.01 vs. Grade 2 or Stage 2; ^^P<0.01 vs. Grade 3 or Stage 3. NRP1, neuropilin 1; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; THCA, thyroid carcinoma; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas; TGCT, testicular germ cell tumor.
The expression of NRP1 on the basis of the pathological grades of patients and individual cancer stages in TCGA cancer types were also investigated. In HNSC, the NRP1 expression levels were significantly higher at grades 2 and 3 than the normal and grade 1 samples. NRP1 levels in grade 4 samples were significantly lower than in grade 3 samples (P<0.01; Fig. 1G). In regard to the stages, NRP1 expression in all stages (stages 1-4) was higher than in the normal samples. However, there was a reduced level of NRP1 in the stage 4 samples compared with the stage 3 samples (P<0.01; Fig. 1H). In STAD, NRP1 expression levels in the grade 2 and 3 samples were significantly higher than in the normal samples, and in the grade 3 samples, NRP1 expression was also higher than in the grade 1 and 2 samples (P<0.01; Fig. 1I). Additionally, for the stages in STAD, NRP1 expression levels in stage 2, 3 and 4 samples were significantly higher than in the normal and stage 1 samples (P<0.01; Fig. 1J). This indicated that the increased expression of NRP1 occurred mainly in the mid-grades (grades 2 and 3), and in stages that were higher than normal.
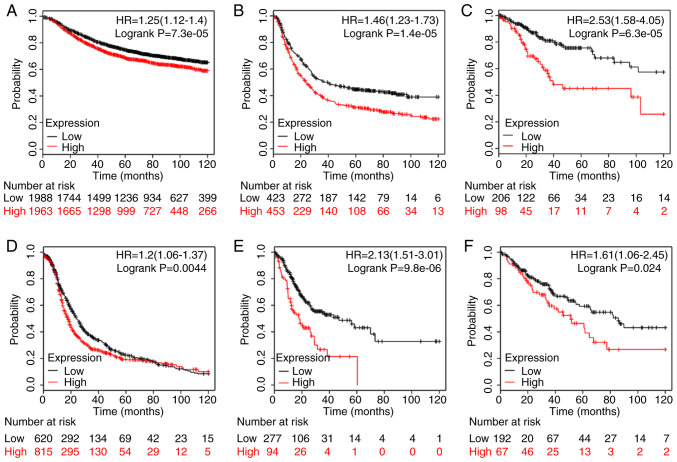
Survival analysis using the Kaplan-Meier Plotter database results revealed that an increased NRP1 expression level predicted a poorer prognosis in breast cancer (P<0.01; Fig. 2A), gastric cancer (P<0.01; Fig. 2B), cervical squamous cell carcinoma (P<0.01; Fig. 2C), ovarian cancer (P<0.01; Fig. 2D), STAD (P<0.01; Fig. 2E) and sarcoma (P<0.05; Fig. 2F) for 120 months. Overexpression of NRP1 was significantly related to a shorter survival time and there were significant differences between the high and low NRP1 groups. These data indicated the important roles of NRP1 in the development of various tumors and more studies are warranted to investigate the function of NRP1 in tumor angiogenesis.
Figure 2.
Prognostic significance of NRP1 expression in various tumor types of patients with (A) breast cancer (n=3951), (B) gastric cancer (n=876), (C) cervical squamous cell carcinoma (n=304), (D) ovarian cancer (n=1435), (E) stomach adenocarcinoma (n=371), and (F) sarcoma (n=259). The analysis was carried using the Kaplan-Meier plotter database (http://kmplot.com/analysis/). Using the Kaplan-Meier plots, hazard ratios with 95% confidence intervals and log-rank P-values were calculated. NRP1, neuropilin 1.
NRP1 promotes endothelial angiogenesis
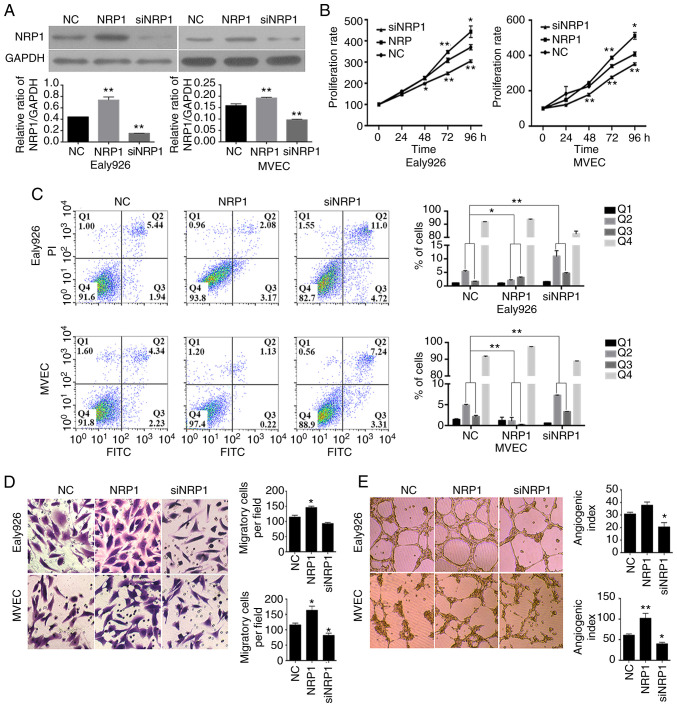
The overexpression and knockdown of NRP1 in MVEC and Ealy926 cells were carried out using pLVX-IRES-Neo-NRP1 vectors and siNRP1 transfections. The effects of regulating NRP1 levels were detected using western blotting and the results are presented in Fig. 3A. It was demonstrated that the overexpression and knockdown of NRP1 could effectively increase or decrease the protein levels, respectively, in MVEC and Ealy926 cells (P<0.01). These data demonstrated that the expression NRP1 was effectively regulated in MVEC and Ealy926 cells in the present study, for the subsequent experiments.
Figure 3.
In vitro assays of NRP1 functions in the Ealy926 and MVEC cell lines with overexpression or knockdown of NRP1. (A) Efficient NRP1 overexpression or knockdown was confirmed using western blotting. GAPDH was used as the loading control. (B) Proliferative rates of Ealy926 and MVEC cell lines with overexpression or knockdown of NRP1, detected using CCK-8 assays. (C) Apoptosis assays with Ealy926 and MVEC cell lines after NRP1 overexpres-sion and knockdown as determined using flow cytometry. (D) Migration (×200) and (E) in vitro tube-forming activity (×40) of Ealy926 and MVEC cells after overexpression or knockdown of NRP1. The number of cells or tubes were counted in five randomly selected fields. NC represents the control group and NRP1 represents the NRP1 overexpression group. *P<0.05 and **P<0.01 vs. NC. NRP1, neuropilin 1; siNRP1, siRNA knockdown of the NRP1 group; CCK-8, Cell Counting Kit-8.
A CCK-8 assay was employed to detect the proliferative rates of MVEC and Ealy926 cells in the control group, overexpression-NRP1 group and the knockdown-NRP1 group. The results demonstrated that NRP1 overexpression could significantly increase the cell proliferative rate after 72 h (P<0.01) and at 96 h (P<0.05) of culture in both the MVEC and Ealy926 cells (Fig. 3B). While the cells with low-NRP1 expression levels (knockdown) demonstrated a lower proliferative rate than the control after 48 h (P<0.05 for Ealy926 cells and P<0.01 for MVEC cells), 72 h (P<0.01 for both cell lines) and 96 h (P<0.01 for both cell lines) of culture.
The results of the flow cytometric apoptosis analysis of the MVEC and Ealy926 cells in the three groups are presented in Fig. 3C. The siNRP1 group had a greater proportion of apoptotic cells than the control in the MVEC and Ealy926 cells after 48 h of culture. The early and late apoptotic cells (Q2 + Q3; Fig. 3C) in the siNRP1 cells were significantly higher than in the control group (P<0.01). While the apoptotic cells in the overexpression-NRP1 groups both in the MVEC and Ealy926 cells were significantly lower than the control (P<0.05 for Ealy926 cells and P<0.01 for MVEC cells). According to the CCK-8 and flow cytometric results, the high NRP1 expression levels promoted the proliferation of vascular ECs, while low expression was not conducive to vascular EC growth.
To examine the effect of NRP1 on vascular EC motility, Transwell migration assays with MVEC, Ealy926 and HUVEC cell lines were performed. The experiments revealed that NRP1 knockdown resulted in fewer cells migrating to the bottom of the chamber compared with the control (P<0.05 for MVEC cell lines, Fig. 3D; P<0.01 for the HUVEC cell line, Fig. S1A). Following NRP1 overexpression, there was a greater number of cells which migrated to the bottom chamber (P<0.05 for Ealy926 cells and P<0.01 for MVEC cell lines, Fig. 3D; P<0.01 for the HUVEC cell line, Fig. S1A). Although overexpression and knockdown of NRP1 had no significant effect on MVEC and Ealy926 cell proliferation after 24 h of culture (Fig. 3B), migration assays revealed that knockdown of NRP1 inhibited EC migration and that overexpression of NRP1 promoted cell migration.
Angiogenesis and counting results are presented in Figs. 3E and S1B. The NRP1-knockdown reduced the cord forming ability of ECs (P<0.05 for both Ealy926 and MVEC cell lines, Fig. 3E; P<0.01 for the HUVEC cell line, Fig. S1B) while overexpression of NRP1 significantly increased the angiogenesis ability of MVEC and HUVEC cells, with the difference observed in the counting analysis being significant (P<0.01 for the MVEC cell line, Fig. 3E; P<0.01 for the HUVEC cell line, Fig. S1B). It was revealed that the expression levels of NRP1 were positively associated with vascular EC proliferation, migration and angiogenesis.
Potential proangiogenic mechanism of NRP1
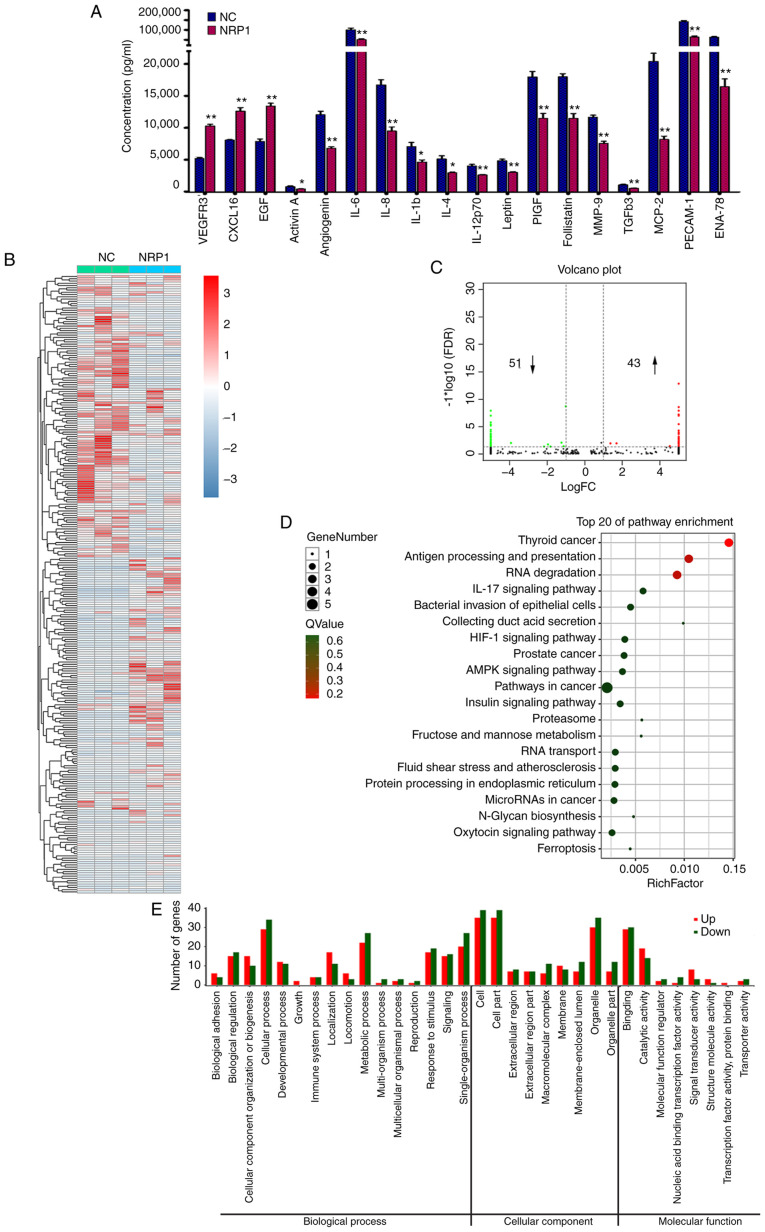
To further investigate the mechanisms of action involved in the proangiogenic activity of NRP1, a human angiogenesis antibody array (Fig. 4A) and RNA-Seq (Fig. 4B-E) analysis were performed in HUVECs with the control group and the overexpression NRP1 groups. The human angiogenesis antibody array could concurrently detect the expression of 60 angiogenesis-related proteins. Three proteins, VEGF receptor 3 (VEGFR3), CXCL16 and epidermal growth factor (EGF), were significantly upregulated after NRP1 overexpression (P<0.01; Fig. 4A). Fifteen proteins, including activin A, angiogenin (ANG), interleukin (IL)-6, IL-8, IL-1b, IL-4, IL-12p70, leptin, PIGF, follistatin, MMP-9, TGFb3, MCP-2, PECAM-1 and ENA-78, were downregulated by NRP1 overexpression in HUVECs (P<0.05 for activin A, IL-1b and IL-4; P<0.01 for the other proteins). Proteins with band densities exhibiting a fold increase >1.5 or a fold decrease <0.67 were defined as differentially expressed between the control and NRP1 overexpression groups in HUVECs.
Figure 4.
Analysis of potential mechanisms of action involved in the proangiogenic activity of NRP1 in HUVECs. (A) The various expression levels of angio-genesis-related proteins in the control (NC) and NRP1-overexpression (NRP1) HUVEC groups were detected using the RayBiotech Human Angiogenesis Antibody Array C Series 1000 kit. (B) The heatmap plot of RNA-Seq results of HUVEC cells in the control (NC) and NRP1-overexpression (NRP1) groups. (C) The Volcano plot of RNA-Seq results. (D) Top 20 KEGG pathways enriched in the DEGs identified by RNA-Seq. (E) Comparison of GO terms of DEGs identified by RNA-Seq in HUVECs. Proteins with band densities exhibiting a fold increase of >1.5 or a fold decrease of <0.67 were defined as differentially expressed. DEGs of RNA-Seq were identified with two criteria: i) An FDR of <0.05 and a |log2fold change| of >1. NC represents the control group and NRP1 represents the NRP1 overexpression group. *P<0.05 and **P<0.01. NRP1, neuropilin 1; HUVEC, human umbilical vein endothelial cells; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed gene; GO, Gene Ontology; FDR, false discovery rate.
To obtain a more comprehensive understanding of the NRP1-associated molecular mechanism of action, RNA-Seq analysis was used for transcriptomic profiling. The overall Q30 percentage was >90.43 and >98.87% of the reads were mapped to reference genes in all groups. The gene expression levels were presented as fragments per kilobase of transcript per million mapped read values and a correlation coefficient >0.99 was observed between samples of the same group. The heatmap of significantly expressed genes is presented in Fig. 4B. Overall, 94 DEGs, including 45 upregulated (Table I, not including NRP1) and 48 downregulated (Table II), were identified between the control and NRP1 overexpression groups in HUVECs (Fig. 4C). DEGs were further analyzed using the KEGG and GO databases for pathway and functional annotations. The KEGG enrichment pathway analysis matched to 88 pathways (Table SI). The top 20 KEGG pathways most significantly enriched in DEGs are displayed in Fig. 4D. The 10 most enriched pathways with the greatest distribution of DEGs were the following: Thyroid cancer; antigen processing and presentation; RNA degradation; IL-17 signaling pathway; bacterial invasion of epithelial cells; collecting duct acid secretion; HIF-1 signaling pathway; prostate cancer; AMPK signaling pathway; and pathways in cancer. Three of them were directly associated with some types of cancer. The DEGs were assigned into 33 sub-categories from the three main GO functional categories, namely: Biological process (sixteen), cellular component (nine) and molecular function (eight) (Fig. 4E).
Table I.
Upregulated genes by overexpression of NRP1 using RNA-Seq.
| No. | Symbol | Description | FPKM
|
P-value | |
|---|---|---|---|---|---|
| Control | NRP1 | ||||
| 1 | MAPK7 | Mitogen-activated protein kinase 7 | 0.001 | 0.543 | 2.718×10−06 |
| 2 | CDC42BPA | CDC42 binding protein kinase α | 0.001 | 0.267 | 5.426×10−05 |
| 3 | CAMKK2 | Calcium/calmodulin-dependent protein kinase kinase 2 | 0.001 | 0.687 | 1.347×10−05 |
| 4 | APLP2 | Amyloid β precursor like protein 2 | 0.230 | 11.753 | 1.687×10−08 |
| 5 | RHOT1 | Ras homolog family member T1 | 0.001 | 1.370 | 1.032×10−11 |
| 6 | NRDC | Nardilysin convertase | 0.001 | 1.950 | 3.621×10−07 |
| 7 | TPM1 | Tropomyosin 1 | 0.001 | 0.900 | 5.959×10−06 |
| 8 | CASP10 | Caspase-10 | 0.001 | 0.263 | 1.351×10−06 |
| 9 | PCMTD1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 | 0.001 | 1.280 | 2.477×10−06 |
| 10 | RRBP1 | Ribosome binding protein 1 | 0.001 | 0.727 | 5.203×10−05 |
| 11 | HIVEP2 | Human immunodeficiency virus type I enhancer-binding protein 2 | 1.813 | 0.837 | 6.189×10−05 |
| 12 | PTPRK | Protein tyrosine phosphatase, receptor type K | 0.001 | 0.383 | 5.873×10−07 |
| 13 | ZNF462 | Zinc finger protein 462 | 0.001 | 0.417 | 6.091×10−08 |
| 14 | NRP1 | Neuropilin 1 | 0.930 | 960.273 | 2.646×10−18 |
| 15 | ABCF1 | ATP binding cassette subfamily F member 1 | 0.003 | 1.083 | 1.727×10−11 |
| 16 | NSD2 | Nuclear receptor binding SET domain protein 2 | 0.001 | 0.253 | 8.008×10−07 |
| 17 | MGAT1 | Mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase | 0.001 | 1.430 | 1.994×10−11 |
| 18 | LRRC37A | Leucine rich repeat containing 37A | 0.001 | 0.253 | 1.017×10−05 |
| 19 | ELP5 | Elongator acetyltransferase complex subunit 5 | 0.001 | 1.007 | 2.366×10−05 |
| 20 | NEB | Nebulin | 0.001 | 0.053 | 4.932×10−06 |
| 21 | DIAPH3 | Diaphanous related formin 3 | 0.001 | 0.777 | 1.002×10−07 |
| 22 | SEMA4D | Semaphorin 4D | 0.001 | 1.027 | 1.650×10−13 |
| 23 | ZNF566 | Zinc finger protein 566 | 0.007 | 0.490 | 5.861×10−05 |
| 24 | VRK2 | Vaccinia related kinase 2 | 0.001 | 1.510 | 8.116×10−05 |
| 25 | LANCL1 | LanC like 1 | 0.001 | 0.570 | 7.683×10−05 |
| 26 | 8 Sep | Septin 8 | 0.001 | 1.700 | 1.013×10−06 |
| 27 | OSBPL9 | Oxysterol binding protein like 9 | 0.001 | 0.507 | 3.895×10−06 |
| 28 | TACC3 | Transforming acidic coiled-coil protein 3 | 0.001 | 2.230 | 4.007×10−07 |
| 29 | SLBP | Stem-loop binding protein | 1.940 | 0.040 | 2.892×10−05 |
| 30 | HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H1 | 0.001 | 6.140 | 1.272×10−12 |
| 31 | ASPH | Aspartate β-hydroxylase | 0.113 | 2.607 | 6.574×10−05 |
| 32 | DNPEP | Aspartyl aminopeptidase | 0.001 | 3.310 | 2.407×10−06 |
| 33 | ATP2C1 | ATPase secretory pathway Ca2+ transporting 1 | 0.843 | 2.737 | 1.329×10−05 |
| 34 | SLC38A2 | Solute carrier family 38 member 2 | 2.913 | 7.513 | 1.511×10−05 |
| 35 | HSP90AA1 | Heat shock protein 90 α family class A member 1 | 19.937 | 5.660 | 8.247×10−05 |
| 36 | C16orf58 | Chromosome 16 open reading frame 58 | 0.007 | 1.153 | 4.260×10−07 |
| 37 | NCOA4 | Nuclear receptor coactivator 4 | 0.001 | 1.097 | 3.774×10−08 |
| 38 | RPL17-C18orf32 | RPL17-C18orf32 readthrough | 0.001 | 2.440 | 7.856×10−12 |
| 39 | NCOA4 | Nuclear receptor coactivator 4 | 0.001 | 0.847 | 1.586×10−06 |
| 40 | FCHO1 | FCH domain only 1 | 0.001 | 0.913 | 8.297×10−10 |
| 41 | PRKD2 | Protein kinase D2 | 0.001 | 1.423 | 7.957×10−06 |
| 42 | GAS2L1 | Growth arrest specific 2 like 1 | 0.001 | 3.313 | 4.079×10−07 |
| 43 | RPH3AL | Rabphilin 3A like (without C2 domains) | 0.010 | 0.550 | 5.177×10−05 |
| 44 | MAPKAPK3 | Mitogen-activated protein kinase-activated protein kinase 3 | 0.003 | 0.960 | 8.374×10−06 |
| 45 | TCF7L2 | Transcription factor 7 like 2 | 0.001 | 0.363 | 1.569×10−05 |
| 46 | EAW79261 | hCG2022618 [Homo sapiens] | 0.001 | 0.293 | 5.914×10−05 |
Table II.
Downregulated genes by overexpression of NRP1 using RNA-Seq.
| No. | Symbol | Description | FPKM
|
P-value | |
|---|---|---|---|---|---|
| Control | NRP1 | ||||
| 1 | COQ8B | Coenzyme Q8B | 1.053 | 0.001 | 3.004×10−07 |
| 2 | TUBGCP2 | Tubulin γ complex associated protein 2 | 0.527 | 0.001 | 2.474×10−05 |
| 3 | SPINK5 | Serine peptidase inhibitor, Kazal type 5 | 0.320 | 0.001 | 4.827×10−05 |
| 4 | GLIS2 | GLIS family zinc finger 2 | 0.253 | 0.001 | 3.011×10−05 |
| 5 | ZNF217 | Zinc finger protein 217 | 0.833 | 0.003 | 2.186×10−05 |
| 6 | PTPRA | Protein tyrosine phosphatase, receptor type A | 0.537 | 0.001 | 3.686×10−07 |
| 7 | ATP2C1 | ATPase secretory pathway Ca2+ transporting 1 | 1.157 | 0.001 | 1.334×10−12 |
| 8 | MBD1 | Methyl-CpG binding domain protein 1 | 0.763 | 0.001 | 6.928×10−07 |
| 9 | GPSM1 | G protein signaling modulator 1 | 1.623 | 0.001 | 2.892×10−06 |
| 10 | ELP5 | Elongator acetyltransferase complex subunit 5 | 0.857 | 0.001 | 2.144×10−06 |
| 11 | PFKFB3 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1.353 | 0.001 | 5.488×10−05 |
| 12 | PJA1 | Praja ring finger ubiquitin ligase 1 | 0.530 | 0.001 | 7.216×10−06 |
| 13 | PISD | Phosphatidylserine decarboxylase | 3.573 | 0.001 | 2.896×10−07 |
| 14 | SFXN3 | Sideroflexin 3 | 3.280 | 0.001 | 1.663×10−07 |
| 15 | ARL6 | ADP ribosylation factor like GTPase 6 | 0.813 | 0.001 | 1.320×10−05 |
| 16 | GLT8D1 | Glycosyltransferase 8 domain containing 1 | 3.860 | 0.860 | 7.515×10−05 |
| 17 | UBFD1 | Ubiquitin family domain containing 1 | 3.867 | 1.633 | 8.530×10−06 |
| 18 | KIF16B | Kinesin family member 16B | 0.280 | 0.001 | 3.078×10−05 |
| 19 | TSNAX | Translin associated factor X | 1.720 | 0.001 | 2.502×10−05 |
| 20 | CDK19 | Cyclin dependent kinase 19 | 0.457 | 0.001 | 5.554×10−05 |
| 21 | RGS4 | Regulator of G protein signaling 4 | 0.807 | 0.001 | 1.101×10−09 |
| 22 | TMED2 | Transmembrane p24 trafficking protein 2 | 4.257 | 0.001 | 6.963×10−05 |
| 23 | CCM2 | CCM2 scaffolding protein | 0.627 | 0.001 | 8.447×10−05 |
| 24 | MARK2 | Microtubule affinity regulating kinase 2 | 0.420 | 0.001 | 1.108×10−05 |
| 25 | FBXL5 | F-box and leucine-rich repeat protein 5 | 1.970 | 0.001 | 1.773×10−11 |
| 26 | PAICS | Phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase | 9.247 | 0.010 | 4.015×10−10 |
| 27 | PABPC1 | Poly(A) binding protein cytoplasmic 1 | 6.967 | 0.001 | 2.650×10−05 |
| 28 | TNIP1 | TNFAIP3 interacting protein 1 | 1.540 | 0.001 | 1.133×10−07 |
| 29 | COPS7A | COP9 signalosome subunit 7A | 1.113 | 0.001 | 5.359×10−07 |
| 30 | ARL13B | ADP ribosylation factor like GTPase 13B | 0.287 | 0.001 | 4.100×10−05 |
| 31 | INPPL1 | Inositol polyphosphate phosphatase like 1 | 0.510 | 0.001 | 8.327×10−05 |
| 32 | ENO2 | Enolase 2 | 1.200 | 0.001 | 1.513×10−07 |
| 33 | PITPNA | Phosphatidylinositol transfer protein α | 1.487 | 0.001 | 1.251×10−05 |
| 34 | SPARC | Secreted protein acidic and cysteine rich | 11.103 | 0.733 | 1.092×10−05 |
| 35 | ZNF839 | Zinc finger protein 839 | 0.727 | 0.001 | 2.689×10−08 |
| 36 | PSME2 | Proteasome activator subunit 2 | 14.677 | 3.767 | 2.264×10−05 |
| 37 | B2M | β-2-Microglobulin | 248.433 | 123.977 | 7.467×10−14 |
| 38 | NBPF14 | Neuroblastoma breakpoint family member 14 | 0.650 | 0.001 | 1.319×10−06 |
| 39 | NFE2L1 | Nuclear factor, erythroid 2 like 1 | 6.493 | 0.001 | 7.856×10−08 |
| 40 | ZNF17 | Zinc finger protein 17 | 0.583 | 0.001 | 9.434×10−06 |
| 41 | TPR | Translocated promoter region, nuclear basket protein | 1.153 | 0.001 | 6.454×10−05 |
| 42 | NAGK | N-acetylglucosamine kinase | 1.793 | 0.001 | 2.326×10−05 |
| 43 | ATP6V0E2 | ATPase H+ transporting V0 subunit e2 | 1.050 | 0.001 | 1.849×10−09 |
| 44 | CNOT9 | CCR4-NOT transcription complex subunit 9 | 1.210 | 0.001 | 1.082×10−07 |
| 45 | CBL | Cbl proto-oncogene | 0.293 | 0.001 | 5.579×10−06 |
| 46 | PPT1 | Palmitoyl-protein thioesterase 1 | 1.003 | 0.001 | 1.167×10−08 |
| 47 | PLEKHM1 | hCG2002091, isoform CRA_c, partial | 0.193 | 0.001 | 3.445×10−05 |
| 48 | Uncharacterized protein | LOC102723360 | 0.573 | 0.001 | 2.123×10−06 |
Most critical molecule and signaling pathway related to the angiogenic function of NRP1
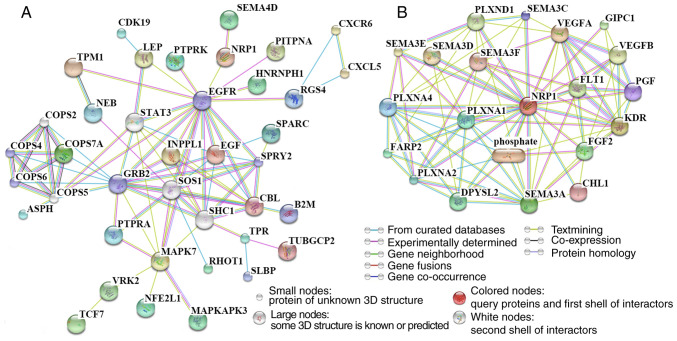
To identify the most critical molecule and signaling pathway related to the angiogenic function of NRP1, the relationship between 7 differentially regulated proteins from the angiogenesis antibody array and DEGs identified by RNA-Seq (including 36 upregulated and 35 downregulated DEGs) was analyzed using the STITCH website. A protein-protein interaction network was constructed, with isolated proteins not being shown (Fig. 5A). The maximum number of interactors for the 1st shell and 2nd shells were set as no more than 5, with the score of the predicted functional partners being >0.999. The minimum required interaction score was a medium confidence (0.400). The EGF receptor (EGFR) was an important node in the protein-protein network and NRP1 was found to have a 'textmining' interaction with EGFR and an 'experimentally determined' interaction with SEMA4D.
Figure 5.
Signaling pathways related to the angiogenic function of NRP1 as determined using the STITCH website. (A) Pathway analysis of differentially expressed proteins detected using the human angiogenesis antibody array and DEGs from RNA-Seq. The maximum number of interactors for the 1st shell and 2nd shells were set as no more than 5 and the score of predicted functional partners was >0.999. (B) The top 20 proteins which were most closely related to NRP1. The maximum number of interactors for the 1st shell was no more than 20 interactors and the score of predicted functional partners was >0.935. The minimum required interaction score was a medium confidence of 0.400. NRP1, neuropilin 1; DEG, differentially expressed gene; GRB2, growth factor receptor-bound protein 2; EGFR, epidermal growth factor receptor; COPS6, COP9 constitutive photomorphogenic homolog subunit 6; SPRY2, sprout homolog 2; COPS4, COP9 constitutive photomorphogenic homolog subunit 4; VEGFA, vascular endothelial growth factor A; KDR, kinase insert domain receptor; FLT1, fms-related tyrosine kinase 1; SEMA3A, semaphorin 3A; PLXNA1, plexin A1, coreceptor for SEMA3A, SEMA3C, SEMA3F and SEMA6D; PLXNA2, plexin A2, coreceptor for SEMA3A and SEMA6A; PLXNA4, plexin A4, coreceptor for SEMA3A; SEMA3C, semaphorin 3C; PGF, placental growth factor; CHL1, close homolog of L1; SEMA3F, semaphorin 3F; phosphate, phosphoric acid; SEMA3D, semaphorin 3D; SEMA3E, semaphorin 3E; VEGFB, vascular endothelial growth factor B; PLXND1, plexin D1, cell surface receptor for SEMA4A and for class 3 semaphorins; GIPC1, GIPC PDZ domain-containing family, member 1; FGF2, fibroblast growth factor 2; DPYSL2, dihydropyrimidinase-like 2; FARP2, FERM, RhoGEF and pleckstrin domain protein 2.
To observe NRP1-related proteins in human cells, the STITCH website also was employed to determine the top 20 proteins which were most closely related to NRP1 (Fig. 5B). The predicted functional partners score was >0.935. There were nine SEMA family related proteins: Five SEMA family members including SEMA3A, SEMA3C, SEMA3F, SEMA3D and SEMA3E; and four coreceptors for SEMA proteins, PLXNA1, PLXNA2, PLXNA4 and PLXND1. There was a potential close association between NRP1 and the SEMA family identified.
Key differentially expressed genes related to vascular maturation
PECAM-1 has often been used as one of the indicators of vascular maturation (21) and a previous study found that the ability of tumor-associated macrophages to produce SEMA4D was critical for tumor angiogenesis and vessel maturation (22). The effects of NRP1 on angiogenesis may be achieved by affecting vascular maturity; therefore, the present study reviewed in detail the functions of each DEG and protein. A total of 10 genes which had been reported to be closely associated to angiogenesis and maturation were found. The 10 genes are listed in Table III, including 7 upregulated (MAPK7, TPM1, RRBP1, PTPRK, SEMA4D, HSP90AA1 and PRKD2) and 3 downregulated (PFKFB3, RGS4 and SPARC) factors. The related functions, including fluid shear stress (MAPK7) (23), EC-cell junctions (TPM1) (24), maintenance of cell adherens junction (PTPRK) (25), support for tumor angiogenesis (PRKD2, HSP90A, PFKFB3 and RGS4) (26-29), critical and support for vessel maturation (SEMA4D, RRBP1) (22,30-31) and suppression of angiogenesis (SPARC) (32) were revealed. These DEGs provided supporting evidence for the hypothesis that NRP1 impacts angiogenesis by altering vascular maturity.
Table III.
Differentially expressed genes from RNA-seq related to angiogenesis and maturation.
| No. | Name | Function (related to angiogenesis and maturation) | Regulated by NRP1 | (Refs.) |
|---|---|---|---|---|
| 1 | MAPK7 | Response to fluid shear stress in endothelial cells | Up | (20) |
| 2 | TPM1 | Protects endothelial cell-cell junctions | Up | (21) |
| 3 | RRBP1 | Novel biomarker of intestinal epithelial cell maturation | Up | (37) |
| 4 | PTPRK | Dissolution of adherens junctions in a rat model of pancreatitis | Up | (32) |
| 5 | SEMA4D | Critical for tumor angiogenesis and vessel maturation; enhances angiogenesis | Up | (19,27) |
| 6 | HSP90AA1 | HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization | Up | (23) |
| 7 | PRKD2 | Up | ||
| 8 | PFKFB3 | Tumor angiogenesis | Down | (24,25) |
| 9 | RGS4 | Angiogenesis-related | Down | (26) |
| 10 | SPARC | Inhibition of angiogenesis | Down | (29) |
SEMA4D plays a key role in the angiogenic function of NRP1
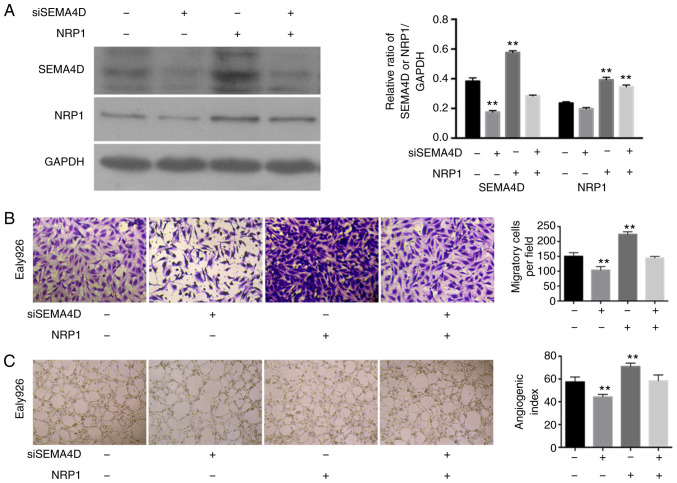
NRP1 overexpression significantly increased the levels of SEMA4D (P<0.01) (Table III and Fig. 6A), and siSEMA4D could not significantly regulate the expression levels of NRP1 (P>0.05) in the Ealy926 cell line. However, SEMA4D knockdown could not effectively downregulate the expression levels of SEMA4D when NRP1 was overexpressed concurrently (Fig. 6A). SEMA4D knockdown with normal NRP1 expression levels maintained significantly decreased the migratory ability of the Ealy926 cell line compared with the control (P<0.01). While NRP1 overexpression increased the cell migration, SEMA4D knockdown with NRP1-overexpression did not result in a significant change in the cell migration of Ealy926 cells compared with the control (P>0.05; Fig. 6B). SEMA4D knock-down significantly reduced the cord forming ability (P<0.01; Fig. 6C) compared with the control in the Ealy926 cell lines. However, SEMA4D-knockdown with NRP1-overexpression did not cause a change in the cord forming ability compared with the control (P>0.05; Fig. 6C). SEMA4D may have played an important role in the process whereby NRP1 affects the angiogenesis and maturation of ECs.
Figure 6.
SEMA4D mediates the angiogenic function of NRP1 in the Ealy926 cell line. (A) Western blotting for the protein levels of NRP1 and SEMA4D in Ealy926 cell line treated with control, SEMA4D knockdown, NRP1 overexpression and siSEMA4D + NRP1 overexpression. GAPDH was used as the loading control. (B) Migration activity of the Ealy926 cell line with control, SEMA4D knockdown, NRP1 overexpression and siSEMA4D + NRP1 overexpression (×200). (C) In vitro tube-forming activity of the Ealy926 cell line with control, SEMA4D knockdown, NRP1 overexpression and siSEMA4D + NRP1 overexpression (×40). The number of cells or tubes were counted in five randomly selected fields. **P<0.01 vs. NC. NRP1, neuropilin 1; SEMA4D, semaphoring 4D; si, siRNA.
Discussion
According to the TCGA database analysis, NRP1 mRNA expression levels were significantly higher in primary tumor tissues than in normal tissues for various tumor types. In addition, NRP1 was abundantly expressed in the mid-grade and persistently overexpressed throughout the process of tumor development. Moreover, high NRP1 expression levels were associated to the survival time of patients with various types of tumors. Therefore, the present data indicated a pro-carcinogenic effect of NRP1 in tumors, which was also supported by previous studies (10,33).
The proangiogenic function of NRP1 may be produced by promoting the endothelial tip cell function during angiogenesis, to promote neovascularization (34). NRP1 over-expression significantly promoted EC proliferation, migration and angiogenesis, but these processes were inhibited after NRP1 downregulation.
According to the antibody array analysis results, the levels of angiogenic cytokines and chemokines (activin A, IL-6, IL-8, IL-1b, IL-4, IL-12p70, leptin, follistatin, TGFβ3, MCP-2 and ENA78) and EC markers (PECAM-1, ANG, PIGF and MMP-9) were significantly downregulated by NRP1 overexpression. PECAM-1 (CD31) has been reported as one of the commonly used markers of vascular ECs during the process of angiogenesis in tumors (35), and is one of the main components of the EC intercellular junction (36). ANG was first isolated and identified solely by its ability to induce new blood vessel formation (37) and also was reported to have a potential control over vascular homeostasis through the maintenance of EC self-renewal (38). PIGF is a homodimeric glycoprotein, belonging to the vascular EGF sub-family and is a potent angiogenic factor (39). MMP-9, is a key regulator of the extracellular matrix, involved in the degradation of various extracellular matrix proteins (40). The EC-cell junction and matrix maintain the connections between ECs and control vascular permeability and leukocyte migration. Downregulation of ANG and PIGF may result in damage to the endothelial or vascular homeostasis. Downregulation of PECAM-1 and MMP-9 were revealed to play an important role in regulating the EC network, both in terms of the formation and migration (41), indicating a reduced connection and support among ECs, and a function of these molecules in reducing the vascular maturity.
High expression levels of NRP1 in ECs also significantly regulated other key factors which were related to the effects on angiogenesis and maturation. MAPK7 has been reported as essential for EC function, such as the response to fluid shear stress and angiogenesis (23,42). TPM1 protects EC-cell junctions through the stabilization of F-actin-dependent cell-cell junctions in the H1299 and EA.hy926 cell lines (24). RRBP1 is a novel marker for intestinal epithelial cell maturation, as revealed using proteomic detection (31). PTPRK is a negative regulator of adhesion, invasion and the proliferative capacity of cancer cells (43), and interacts, as determined by co-immunoprecipitation, with E-cadherin, α-catenin and β-catenin prior to the dissolution of adherens junctions in a rat model of pancreatitis (25). The HSP90AA1 gene encodes for the HSP90A protein, which is essential for malignant transformation and progression (44). Signals from hypoxia and the HSP90 pathways are interconnected and funneled by PRKD2 into the nuclear factor-κB/VEGF-A signaling axis to promote tumor angiogenesis (26,45). The enrichment of PFKFB3 may promote HUVEC angiogenesis (27), with its blockade inhibiting cancer cell proliferation, causing tumor vessel disintegration and suppressing EC growth (28). RGS4 levels are decreased when angiogenesis is induced using nitric oxide (29). SPARC expression may reduce the extent of angiogenesis, and SPARC silencing increased angiogenesis. These observations were found by regulating the expression levels of VEGF and MMP-7 (32). The angiogenesis and reduced vascular maturity of ECs generated by NRP1 overexpression were also found to be closely associated with MAPK7, TPM1, RRBP1, PTPRK, HSP90A, PRKD2, PFKFB3, RGS4 and SPARC expression levels.
Furthermore, the results of an integrated analysis of anti-body array and RNA-Seq data suggested that SEMA4D may be the critical molecule which mediates the angiogenic and maturation related to NRP1 regulation. SEMA4D is a protein of the semaphorin family, which plays an important role in the tumor microenvironment and neoplastic angiogenesis, and may markedly enhance angiogenic potential (30). When SEMA4D was lacking in the tumor microenvironment, the ability of cancer cells to generate tumor masses and metastasize was severely impaired, which was determined to be due to a defective vascularization inside the tumor, and the ability of tumor-associated macrophages to produce SEMA4D was found to be critical for tumor angiogenesis and vessel maturation (22). NRPs are also closely associated to the SEMA protein family members, especially NRP1 (46). In the present study, SEMA4D knockdown effectively reduced the migratory and cord forming ability of ECs, and this downregulation was prevented when NRP1 was overexpressed concurrently. NRP1 overexpression was also found to increase the expression levels of SEMA4D, while SEMA4D knockdown did not affect the expression levels and the angiogenic function of NRP1. The results of these assays further revealed the critical role of SEMA4D in the angiogenic and maturation activity of NRP1. These findings indicated the potential therapeutic value of NRP1 in combating angiogenesis in tumors.
In conclusion, the present study demonstrated that NRP1 was significantly upregulated in solid primary tumors compared with that of normal tissues, and was significantly associated with tumor development. The results of gain- and loss-of-function experiments emphasized the function of NRP1 in promoting EC proliferation, motility and capillary-like tube formation, as well as in reducing apoptosis. NRP1 overexpression led to significantly decreased expression levels of EC markers (PECAM-1, ANG, PIGF and MMP-9) to reduce the vascular maturity. MAPK7, TPM1, RRBP1, PTPRK, HSP90A, PRKD2, PFKFB3, RGS4 and SPARC were revealed to play important roles in this process. SEMA4D was the key molecule associated with the angiogenic function of NRP1 in ECs. NRP1 may therefore be both a therapeutic target in combination with current antiangiogenic strategies and a candidate prognostic marker for tumors.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Fujian Province (grant nos. 2016J01618 and 2017J01380), and the Projects for Technology Plan of Xiamen in China (grant nos. 3502Z20174076 and 3502Z20174077).
Availability of data and materials
The datasets generated and/or analyzed during this study are available from the corresponding author on reasonable request.
Authors' contributions
HZ and HP conceived and designed the study. ZL, ZY, HJ, HP, KH and HJ performed the experiments. HP, HJ and HJ processed and analyzed the data. ZL, HP and HZ wrote, reviewed, and/or revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moriya J, Minamino T. Angiogenesis, cancer, and vascular aging. Front Cardiovasc Med. 2017;4:65. doi: 10.3389/fcvm.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gacche RN, Meshram RJ. Targeting tumor micro-environment for design and development of novel anti-angiogenic agents arresting tumor growth. Prog Biophys Mol Biol. 2013;113:333–354. doi: 10.1016/j.pbiomolbio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Sun X, Evren S, Nunes SS. Blood vessel maturation in health and disease and its implications for vascularization of engineered tissues. Crit Rev Biomed Eng. 2015;43:433–454. doi: 10.1615/CritRevBiomedEng.2016016063. [DOI] [PubMed] [Google Scholar]
- 4.Jain RK. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 5.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, Jain RK. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 7.Ronca R, Benkheil M, Mitola S, Struyf S, Liekens S. Tumor angiogenesis revisited: Regulators and clinical implications. Med Res Rev. 2017;37:1231–1274. doi: 10.1002/med.21452. [DOI] [PubMed] [Google Scholar]
- 8.De Bock K, Cauwenberghs S, Carmeliet P. Vessel abnormalization: Another hallmark of cancer? Molecular mechanisms and therapeutic implications. Curr Opin Genet Dev. 2011;21:73–79. doi: 10.1016/j.gde.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Cantelmo AR, Pircher A, Kalucka J, Carmeliet P. Vessel pruning or healing: Endothelial metabolism as a novel target? Expert Opin Ther Targets. 2017;21:239–247. doi: 10.1080/14728222.2017.1282465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Zhang Y, Wu J, Li L, Chen N, Ni P, Song L, Liu X. Neuropilin 1 (NRP1) is a novel tumor marker in hepatocellular carcinoma. Clin Chim Acta. 2018;485:158–165. doi: 10.1016/j.cca.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 11.Lampropoulou A, Ruhrberg C. Neuropilin regulation of angiogenesis. Biochem Soc Trans. 2014;42:1623–1628. doi: 10.1042/BST20140244. [DOI] [PubMed] [Google Scholar]
- 12.Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji JI, Hirota S, Kitamura Y, et al. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc Natl Acad Sci USA. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu C, Rodriguez ER, Reimert DV, Shu T, Fritzsch B, Richards LJ, Kolodkin AL, Ginty DD. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/S1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, Deschoemaeker S, Van Ginderachter JA, Tamagnone L, Mazzone M. Impeding macrophage entry into hypoxic tumor areas by sema3a/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D, Bonnevier J, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501:252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin E, Sjöberg E, Tjomsland V, Testini C, Lindskog C, Franklin O, Sund M, Öhlund D, Kiflemariam S, Sjöblom T, Claesson-Welsh L. VEGF receptor-2/neuropilin 1 trans-complex formation between endothelial and tumor cells is an independent predictor of pancreatic cancer survival. J Pathol. 2018;246:311–322. doi: 10.1002/path.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuo H, Lyu Z, Su J, He J, Pei Y, Cheng X, Zhou N, Lu X, Zhou S, Zhao Y. Effect of lung squamous cell carcinoma tumor microenvironment on the CD105+ endothelial cell proteome. J Proteome Res. 2014;13:4717–4729. doi: 10.1021/pr5006229. [DOI] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Jin H, Cheng X, Pei Y, Fu J, Lyu Z, Peng H, Yao Q, Jiang Y, Luo L, Zhuo H. Identification and verification of transgelin-2 as a potential biomarker of tumor-derived lung-cancer endothelial cells by comparative proteomics. J Proteomics. 2016;136:77–88. doi: 10.1016/j.jprot.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Sierra JR, Corso S, Caione L, Cepero V, Conrotto P, Cignetti A, Piacibello W, Kumanogoh A, Kikutani H, Comoglio PM, et al. Tumor angiogenesis and progression are enhanced by sema4d produced by tumor-associated macrophages. J Exp Med. 2008;205:1673–1685. doi: 10.1084/jem.20072602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histonemethyl transferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis. 2016;19:9–24. doi: 10.1007/s10456-015-9485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagat M, Grzanka D, Izdebska M, Sroka WD, Hałas- Wiśniewska M, Grzanka A. Tropomyosin-1 protects transformed alveolar epithelial cells against cigarette smoke extract through the stabilization of F-actin-dependent cell-cell junctions. Acta Histochem. 2016;118:225–235. doi: 10.1016/j.acthis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Schnekenburger J, Mayerle J, Krüger B, Buchwalow I, Weiss FU, Albrecht E, Samoilova VE, Domschke W, Lerch MM. Protein tyrosine phosphatase kappa and SHP-1 are involved in the regulation of cell-cell contacts at adherens junctions in the exocrine pancreas. Gut. 2005;54:1445–1155. doi: 10.1136/gut.2004.063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azoitei N, Diepold K, Brunner C, Rouhi A, Genze F, Becher A, Kestler H, van Lint J, Chiosis G, Koren J, III, et al. HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization. Cancer Res. 2014;74:7125–7136. doi: 10.1158/0008-5472.CAN-14-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu M, Li L, Zhang Z, Chen J, Zhang W, Zhang J, Han L, Tang M, You B, Zhang Q, You Y. PFKFB3 promotes proliferation, migration and angiogenesis in nasopharyngeal carcinoma. J Cancer. 2017;8:3887–3896. doi: 10.7150/jca.19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conradi LC, Brajic A, Cantelmo AR, Bouché A, Kalucka J, Pircher A, Brüning U, Teuwen LA, Vinckier S, Ghesquière B, et al. Tumor vessel disintegration by maximum tolerable PFKFB3 blockade. Angiogenesis. 2017;20:599–613. doi: 10.1007/s10456-017-9573-6. [DOI] [PubMed] [Google Scholar]
- 29.Jaba IM, Zhuang ZW, Li N, Jiang Y, Martin KA, Sinusas AJ, Papademetris X, Simons M, Sessa WC, Young LH, Tirziu D. NO triggers RGS4 degradation to coordinate angiogenesis and cardiomyocyte growth. J Clin Invest. 2013;123:1718–1731. doi: 10.1172/JCI65112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou T, Dissanayaka WL, Jiang S, Wang S, Heng BC, Huang X, Zhang C. Semaphorin 4D enhances angiogenic potential and suppresses osteo-/odontogenic differentiation of human dental pulp stem cells. J Endod. 2017;43:297–305. doi: 10.1016/j.joen.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Chang J, Chance MR, Nicholas C, Ahmed N, Guilmeau S, Flandez M, Wang D, Byun DS, Nasser S, Albanese JM, et al. Proteomic changes during intestinal cell maturation in vivo. J Proteomics. 2008;71:530–546. doi: 10.1016/j.jprot.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JL, Chen GW, Liu YC, Wang PY, Wang X, Wan YL, Zhu J, Gao HQ, Yin J, Wang W, Tian ML. Secreted protein acidic and rich in cysteine (SPARC) suppresses angiogenesis by down-regulating the expression of VEGF and MMP-7 in gastric cancer. PLoS One. 2012;7:e44618. doi: 10.1371/journal.pone.0044618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leng Q, Woodle MC, Mixson AJ. NRP1 transport of cancer therapeutics mediated by tumor-penetrating peptides. Drugs Future. 2017;42:95–104. doi: 10.1358/dof.2017.042.02.2564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fantin A, Vieira JM, Plein A, Denti L, Fruttiger M, Pollard JW, Ruhrberg C. NRP1 acts cell autonomously in endothelium to promote tip cell function during sprouting angiogenesis. Blood. 2013;121:2352–2362. doi: 10.1182/blood-2012-05-424713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman PJ, Berndt MC, Gorski J, White GC, II, Lyman S, Paddock C, Muller WA. PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247:1219–1222. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 36.Lertkiatmongkol P, Liao D, Mei H, Hu Y, Newman PJ. Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31) Curr Opin Hematol. 2016;23:253–259. doi: 10.1097/MOH.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 38.Sheng J, Xu Z. Three decades of research on angiogenin: A review and perspective. Acta Biochim Biophys Sin (Shanghai) 2016;48:399–410. doi: 10.1093/abbs/gmv131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athanassiades A, Lala PK. Role of placenta growth factor (PIGF) in human extravillous trophoblast proliferation, migration and invasiveness. Placenta. 1998;19:465–473. doi: 10.1016/S0143-4004(98)91039-6. [DOI] [PubMed] [Google Scholar]
- 40.Appleby TC, Greenstein AE, Hung M, Liclican A, Velasquez M, Villaseñor AG, Wang R, Wong MH, Liu X, Papalia GA, et al. Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9. J Biol Chem. 2017;292:6810–6820. doi: 10.1074/jbc.M116.760579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chistiakov DA, Orekhov AN, Bobryshev YV. Endothelial PECAM-1 and its function in vascular physiology and atherogenic pathology. Exp Mol Pathol. 2016;100:409–415. doi: 10.1016/j.yexmp.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Roberts OL, Holmes K, Müller J, Cross DA, Cross MJ. ERK5 and the regulation of endothelial cell function. Biochem Soc Trans. 2019;37:1254–1259. doi: 10.1042/BST0371254. [DOI] [PubMed] [Google Scholar]
- 43.Sun PH, Ye L, Mason MD, Jiang WG. Protein tyrosine phos-phatase kappa (PTPRK) is a negative regulator of adhesion and invasion of breast cancer cells, and associates with poor prognosis of breast cancer. J Cancer Res Clin Oncol. 2013;139:1129–1139. doi: 10.1007/s00432-013-1421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuehlke AD, Beebe K, Neckers L, Prince T. Regulation and function of the humanHSP90AA1 gene. Gene. 2015;570:8–16. doi: 10.1016/j.gene.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azoitei N, Fröhling S, Scholl C, Seufferlein T. PRKD2: A two-pronged kinase crucial for the tumor-supporting activity of HSP90. Mol Cell Oncol. 2015;2:e981444. doi: 10.4161/23723556.2014.981444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alto LT, Terman JR. Semaphorins and their signaling mechanisms. Methods Mol Biol. 2017;1493:1–25. doi: 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during this study are available from the corresponding author on reasonable request.