Abstract
Osteosarcoma (OS) is one of the most common malignant bone tumours and generally occurs in children and adolescents. Increasing evidence has demonstrated that dysregulated long non-coding RNAs (lncRNAs) play crucial roles in the progression of various human neoplasms. Among these, tumour suppressor candidate 8 (TUSC8) is a novel lncRNA and has been reported to function as a tumour suppressor in cervical cancer. However, the exact role of TUSC8 in OS remains largely unknown. In the present study, it was observed that TUSC8 was markedly downregulated in OS tissues and cell lines. Functional experiments demonstrated that the overexpression of TUSC8 significantly suppressed the proliferation, migration, invasion and epithelial-mesenchymal transition (EMT), whereas it accelerated the apoptosis of OS cells. Mechanistically, TUSC8 served as a sponge for miR-197-3p, and EH-domain containing 2 (EHD2) was identified as a downstream target molecule of miR-197-3p. Further investigations indicated that EHD2 knockdown significantly reversed the effects on OS cellular processes induced by TUSC8 overexpression. On the whole, these findings indicate that TUSC8 functions as a competing endogenous RNA (ceRNA) to suppress OS cell growth and EMT via the miR-197-3p/EHD2 axis. TUSC8 may thus function as a potential therapeutic target in OS treatment.
Keywords: tumour suppressor candidate 8, miR-197-3p, EH-domain containing 2, osteosarcoma
Introduction
Osteosarcoma (OS) is a common primary malignant tumour that severely affects the growth of bones and occurs mostly in patients under the age of 25 years (1,2). Although advanced therapeutic methods (particularly limb salvage combined with neoadjuvant and adjuvant chemotherapy) have markedly improved the limb salvage rates and long-term survival of patients with OS, the risks of relapse and metastasis in patients with OS remain high (3-5). Therefore, it is necessary to explore novel prognostic molecular markers and the underlying mechanisms of OS.
Long non-coding RNAs (lncRNAs) are endogenous RNA molecules of >200 nucleotides in length and lack the ability to encode proteins (6,7). It has been demonstrated that lncRNAs are involved in the development of multiple tumours. For example, lncRNA RP4 inhibits colorectal cancer cell proliferation and tumour growth (8). LUCAT1 plays an oncogenic role in ovarian cancer by promoting cell proliferation, migration and invasion (9). lncRNA-LOWEG inhibits the progression of gastric cancer by suppressing cell invasion (10). Moreover, increasing it has been demonstrated that lncRNAs exert important functions in OS development. For instance, lncRNA miR210HG has been shown to enhance cell invasion and metastasis by sponging miR-503 in OS (11). CAT104 has also been shown to promote OS development by sponging miR-381 (12). TUG1 knockdown also suppresses OS cell proliferation and invasion via miR-153 (13). Indeed, recent evidence has revealed that TUSC8 serves as a tumour suppressor in the development of cervical cancer by regulating the miR-641/PTEN pathway (14). However, as a novel lncRNA, the specific role of TUSC8 in OS has not yet been clarified.
MicroRNAs (miRNAs or miRs) are a category of small non-coding RNAs approximately 20 nucleotides in length (15). Previous studies have indicated that miRNAs, such as miR-214, miR-153 and miR-146a-5p, play important roles during the progression of various types of cancer (16-18). As a type of miRNA, miR-197-3p has been reported to facilitate the proliferation of breast cancer cells (19). miR-197-3p functions as a tumour promoter in bladder cancer by accelerating cell proliferation, migration and invasion (20). It is well known that lncRNAs function as competitive endogenous RNAs (ceRNAs) and regulate protein expression by sponging miRNAs (21). For example, EPB41L4A-AS2 suppresses the progression of hepatocellular carcinoma by regulating the miR-301a-5p/FOXL1 axis (22). MAGI2-AS3 inhibits bladder cancer development by sponging miR-15b-5p and modulating CCDC1 (23). SNHG16 facilitates the proliferation of OS cells via the miR-205/ZEB1 pathway (24). Nevertheless, little is known about the exact regulatory mechanisms of TUSC8 in OS.
Therefore, the present study aimed to elucidate the biological role and the underlying molecular mechanisms of TUSC8 in OS. The results revealed that TUSC8 inhibits the development of OS by modulating the miR-197-3p/EH-domain containing 2 (EHD2) pathway, suggesting that TUSC8 may function as a novel target for the treatment of OS.
Materials and methods
Tissue samples
A total of 52 OS tissues and normal bone tissues were collected from patients at The Second Hospital of Jilin University (Changchun, China) via surgery, and none of the patients had been treated with chemotherapy before surgery. The cancerous and normal tissues were identified according to the Tumor Node Metastasis (TNM) Classification of Malignant Tumours (6th edition) from the Union for International Cancer Control (UICC) by 2 experienced pathologists. All collected tissues were rapidly frozen in liquid nitrogen at −80°C. The present study was approved by the Human Research Ethics Committee of The Second Hospital of Jilin University (Changchun, China). Written informed consent was signed by each patient.
Cell culture and transfection
OS cell lines (MG-63, U2OS, Saos-2 and HOS) and the osteoblastic cell line, OB3 (hFOB 1.19), were provided by the Cell Bank of the Chinese Academy of Sciences (http://www.cellbank.org.cn/). These cells were incubated in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% foetal bovine serum (FBS; Gibco) and maintained in a humid atmosphere with 5% CO2 at 37°C.
For TUSC8 overexpression experiments, the full-length sequence of TUSC8 was subcloned into the pcDNA3.1 plasmid (Sangon Biotech Co., Ltd.) to generate the pcDNA3.1/TUSC8 construct. Short hairpin RNAs (shRNAs) targeting EHD2 were purchased from Shanghai GenePharma Co., Ltd. to knock-down EHD2. miR-197-3p mimics and controls (NC mimics) were purchased from Shanghai GenePharma Co., Ltd. Lipofectamine 2,000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell transfection following the provided instructions. The mass of the miR-197-3p mimics or miR-197-3p inhibitor was 20 µl. The co-transfection of plasmid DNA and shRNA was performed using Lipofectamine 2,000 reagent by the addition if 30 pmol of shRNA per 1 µg of DNA. Following 48 h of incubation at 37°C, cells were harvested and used in the subsequent experiments.
Reverse transcription-quantitative PCR (RT-qPCR) assay
Total RNA was extracted from tissues and cells (OB3, MG-63, U2OS, Saos2 and HOS) using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into complementary DNA (cDNA) using PrimeScript RT reagent kits (Takara Biotechnology, Ltd.). A SYBR® Premix Ex Taq™ II reagent kit (Takara Biotechnology, Ltd.) was utilized to perform qPCR. The mirVana™ qRT-PCR microRNA Detection kit (Ambion Inc.) was used for miRNA detection. U6 or GAPDH was regarded as the internal reference. Relative quantification was evaluated by the 2−∆∆Cq method (25). The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 3 min, followed by 40 cycles at 95°C for 5 sec and at 60°C for 30 sec. The sequences of the primers are presented in Table SI.
Bioinformatics analysis
The starBase website (http://starbase.sysu.edu.cn/) was used to predict potential miRNAs which interacted with TUSC8, and two candidate miRNAs were predicted. Moreovoer, 6 candidate genes that containing binding sites with miR-197-3p were screened out by overlap-ping the bioinformatics prediction results of PITA and RNA22 under the condition of Pan-Cancer (10 cancer types).
Luciferase reporter assay
The pmirGLO-TUSC8-Wt, pmirGLO-TUSC8-Mut, pmirGLO-EHD2-Wt and pmirGLO-EHD2-Mut vectors were co-transfected with miR-197-3p mimic or NC mimic into the MG-63 and U2OS cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h of incubation at room temperature, the luciferase activity of the reporter plasmids was detected using the Dual Luciferase Reporter Assay System (Promega Corp.). The relative Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
RIP assay was performed with the EZ-Magna RIP kit (EMD Millipore). Cells were cultivated with RIP buffer containing magnetic beads conjugated with anti-Ago2 antibodies (1:20 dilution, ab32381, Abcam). Anti-IgG (1:20 dilution, ab190475 Abcam) acted as a negative control. The coprecipitated RNAs were then eluted from the beads and measured by RT-qPCR.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was analysed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). Briefly, the cells were seeded into 96-well plates (1×103 cells/well) and maintained in RPMI-1640 medium supplemented with 10% FBS. CCK-8 solution (10 µl) was added at 24, 48, 72 and 96 h. The absorption was recorded at 450 nm using a microplate reader (EL340; BioTek Instruments, Inc.).
Colony formation assay
Transfected cells were seeded into 6-well plates at a density of approximately 1×103 cells per well. The cells were then cultured at 37°C for 2 weeks. Subsequently, the cells were fixed with 5% paraformaldehyde for 15 min and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology, Inc.) for 15 min at room temperature. Finally, the colonies were visible and were counted under a light microscope (Olympus Corp.).
Cell apoptotic analysis
Transfected cells were harvested and resuspended in phosphate-buffered saline (PBS). The cells were then double-stained with propidium iodide (PI) and Annexin V-fluorescein isothiocyanate (Beyotime Institute of Biotechnolgoy, Inc.) according to the manufacturer's instructions. Cell apoptosis was assessed using a flow cytometer (BD Biosciences).
Transwell assay
Cells at a density of 1×105 cells per well were added into the upper chamber, which had already been coated with Matrigel and contained serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). DMEM containing 10% FBS was added to the lower chamber. Following 24 h of incubation, the non-invaded cells were removed, and the invaded cells were fixed with methanol and stained with crystal violet (Beyotime Institute of Biotechnology, Inc.) for 15 min at room temperature. The number of invaded cells was counted using an inverted microscope (Olympus Corp.) to measure the invasive ability. Cell migration was assessed in a similar manner, with the exception that the upper chambers were not coated with Matrigel.
Western blot analysis
Cells were lysed in RIPA buffer, and the protein concentration was measured using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Inc.). The proteins (30 µg) were then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore). The membranes were cultured at 4°C overnight with primary antibodies against E-cadherin (1:50 dilution, ab1416; Abcam), N-cadherin (1:500 dilution, ab18203; Abcam), EHD2 (1:50,000 dilution, ab23935; Abcam) and GAPDH (1:10,000 dilution, ab181602; Abcam). GAPDH antibody served as a control. The following day, the bands were incubated with HRP-conjugated goat anti-rabbit secondary antibodies (1:10,000 dilution, ab205718; Abcam) or HRP-conjugated goat anti-mouse secondary antibodies (1:10,000 dilution, ab205719; Abcam) for 1 h at room temperature and rinsed with TBST solution 3 times. Finally, proteins were visualized using an ECL chemiluminescent detection system (Thermo Fisher Scientific, Inc.).
Fluorescence in situ hybridization (FISH)
FISH was conducted using the Ribo™ Fluorescent In Situ Hybridization kit (Guangzhou RiboBio Co., Ltd.) as previously described (26). The TUSC8 probe was designed and synthesized by Guangzhou RiboBio Co., Ltd. DAPI (Guangzhou RiboBio Co., Ltd.) was used to stain the nuclei. Images were obtained using a fluorescence microscope (Zeiss AG).
Statistical analysis
Data are presented as the means ± standard deviation (SD) and were analysed using SPSS v17.0 software (SPSS, Inc.). The statistical significance of the data was determined by one-way analysis of variance (ANOVA) or a Student's t-test (P<0.05). The post hoc test used following one-way ANOVA was Tukey's test. Each experiment was repeated in triplicate.
Results
TUSC8 is prominently downregulated in OS tissues and cells
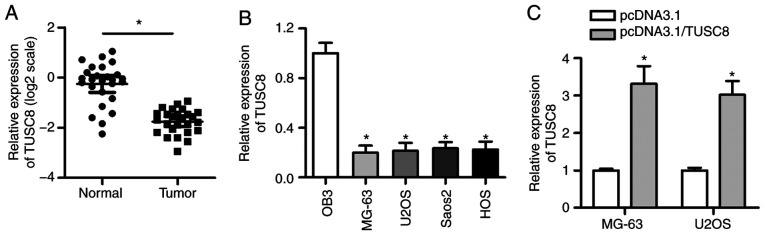
To investigate the clinical significance of TUSC8 in the development of OS, an RT-qPCR assay was conducted to detect the expression status of TUSC8 in OS tissues and corresponding non-cancerous tissues. As shown in Fig. 1A, the expression of TUSC8 was significantly decreased in OS tissues compared with normal tissues. Moreover, the TUSC8 level was consistently downregulated in the OS cell lines (MG-63, U2OS, Saos-2 and HOS) compared with the osteoblastic cell line OB3 (Fig. 1B). To verify the overexpression efficiency of pcDNA3.1/TUSC8 in the MG-63 and U2OS cells, RT-qPCR assays were carried out. As shown in Fig. 1C, the transfection of pcDNA3.1/TUSC8 triggered an obvious increase in TUSC8 expression in the OS cells, suggesting that pcDNA3.1/TUSC8 could be used for follow-up experiments. Thus, these data indicate that TUSC8 is prominently downregulated in OS tissues and cells.
Figure 1.
TUSC8 is prominently downregulated in OS tissues and cells. (A) RT-qPCR was carried out to determine TUSC8 expression in OS tissues and adja-cent normal tissues. (B) The level of TUSC8 in OS cell lines (MG-63, U2OS, Saos-2 and HOS) and the osteoblastic cell line OB3 was analysed by RT-qPCR. (C) The overexpression efficiency of pcDNA3.1/TUSC8 was detected by RT-qPCR. *P<0.05 vs. OB3 cells or control vector-transfected cells.
TUSC8 suppresses the progression of OS
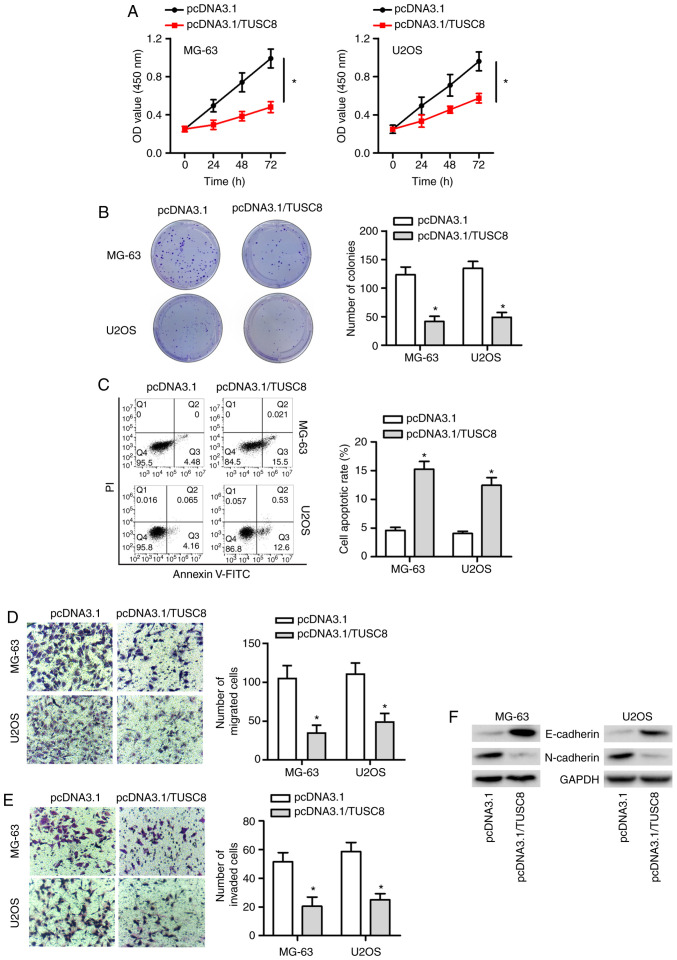
To confirm whether endogenous TUSC8 plays a role in OS development, a series of functional assays were conducted. As presented in Fig. 2A and B, CCK-8 and colony formation assays revealed that TUSC8 overexpression markedly suppressed OS cell proliferation. Subsequently, flow cytometric analysis demonstrated that the overexpression of TUSC8 increased the apoptosis of the MG-63 and U2OS cells (Fig. 2C). Furthermore, it was found that the enhancement of TUSC8 expression markedly suppressed the migratory and invasive capacity of the OS cells (Fig. 2D and E). It is well known that EMT plays a crucial role in cancer development. Therefore, western blot analysis was also conducted to validate whether TUSC8 affected the EMT process in OS. As depicted in Fig. 2F, the protein expression of E-cadherin (the epithelial marker) was markedly increased and the protein level of N-cadherin (the mesenchymal marker) was notably decreased by TUSC8 overexpression in the OS cells. Taken together, TUSC8 suppressed the progression of OS by inhibiting cell proliferation, migration, invasion and EMT, as well as by promoting cell apoptosis in OS.
Figure 2.
TUSC8 suppresses the progression of OS. (A and B) The effect of TUSC8 overexpression on OS cell proliferation was assessed by CCK-8 and colony formation assays. (C) Cell apoptosis was measured by flow cytometry. (D and E) Transwell assays were conducted in MG-63 and U2OS cells. (F) Western blot analysis was utilized to examine the expression of epithelial-mesenchymal transition-related proteins. *P<0.05 vs. control vector-transfected cells.
TUSC8 functions as a sponge of miR-197-3p
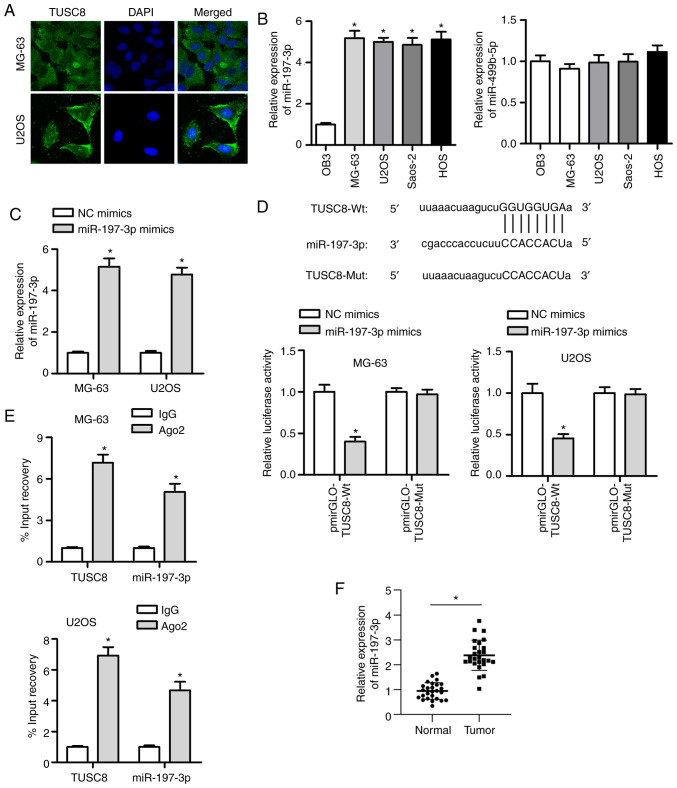
Based on the above-mentoined functional experiments, the antioncogenic role of TUSC8 was elucidated. To further explore the under-lying mechanisms, the distribution of TUSC8 was measured in the MG-63 and U2OS cells by FISH assay. The results indicated that TUSC8 was predominantly distributed in the cytoplasm (Fig. 3A). Bioinformatics software (http://starbase.sysu.edu.cn/) was used to identify 2 candidate miRNAs (miR-197-3p and miR-499b-5p) that exhibited binding sites for TUSC8. Subsequently, the expression of these 2 miRNAs was analysed in OS cell lines and osteoblastic cell lines. As shown by RT-qPCR, miR-197-3p was prominently upregulated in OS cell lines compared with the osteoblastic cell line, while no significant difference was found in the level of miR-499b-5p (Fig. 3B). It has been reported that miR-197-3p serves as a tumour promoter in various types of cancer (19,27); thus, miR-197-3p was analysed in subsequent explorations. It was observed that miR-197-3p expression was substantially increased by transfection with miR-197-3p mimic in the MG-63 and U2OS cells (Fig. 3C). A luciferase reporter assay was employed to verify the interaction between TUSC8 and miR-197-3p. It was evident that the luciferase activity of the pmirGLO-TUSC8-Wt vector was markedly suppressed by transfection with miR-197-3p mimic (Fig. 3D). However, no significant changes were observed in cells transfected with the pmirGLO-TUSC8-Mut vector. Moreover, RIP assay revealed that both TUSC8 and miR-197-3p were more enriched in Ago2-containing miRNA ribonucleoprotein complexes than in IgG immunoprecipitates (Fig. 3E). In addition, it was found that miR-197-3p expression was evidently higher in the OS tissues than in matched normal tissues (Fig. 3F). Furthermore, the transfection efficiency of miR-197-3p was confirmed by RT-qPCR (Fig. S1A). The inhibition of miR-197-3p mark-edly suppressed cell proliferation, migration and invasion in OS (Fig. S1B-D). Overall, the obtained findings suggest that TUSC8 can directly bind with miR-197-3p.
Figure 3.
TUSC8 functions as a sponge of miR-197-3p. (A) FISH staining revealed that TUSC8 was mainly distributed in the cytoplasm. (B) RT-qPCR was employed to detect the level of candidate miRNAs in the osteoblastic cell line, OB3, and in OS cell lines (MG-63, U2OS, Saos-2 and HOS). (C) miR-197-3p expression in OS cells was enhanced by transfection of miR-197-3p mimic. (D and E) Luciferase reporter and RNA immunoprecipitation (RIP) assays were carried out to confirm the interaction between miR-197-3p and TUSC8. (F) RT-qPCR was performed to measure miR-197-3p expression in OS tissues and adjacent normal tissues. *P<0.05 vs. NC mimics or IgG.
EHD2 is a downstream target of miR-197-3p
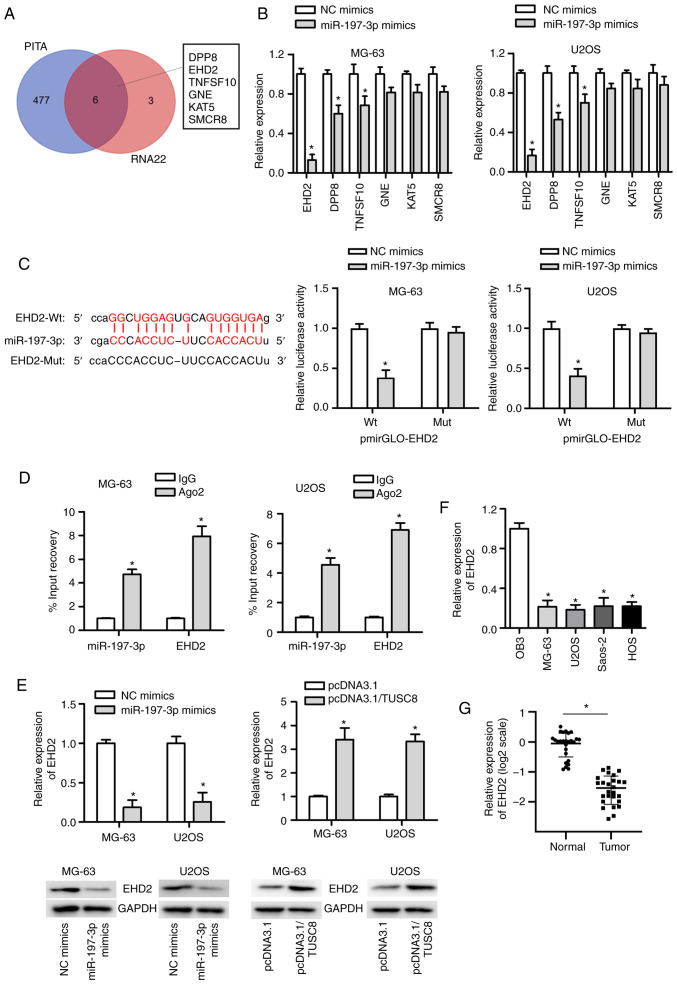
It is well known that miRNAs exert their effects on cancer development by targeting specific genes. To identify the potential target genes of miR-197-3p, the starBase website was used, and the most likely mRNAs (DPP8, EHD2, TNFSF10, GNE, KAT5 and SMCR8) were identified (Fig. 4A). Subsequently, the expression of these 6 mRNAs was analysed in OS cells transfected with miR-197-3p mimic, and it was found that the expression of EHD2 exhibited the most significant reduction (Fig. 4B). As displayed in Fig. 4C, miR-197-3p had a binding site for EHD2, and the luciferase reporter assay suggested that the luciferase activity of the pmirGLO-EHD2-Wt vector was markedly decreased by co-transfection with miR-197-3p mimic, whereas no significant change was observed in cells transfected with the pmirGLO-EHD2-Mut vector. In addition, RIP assay further confirmed the interactions between miR-197-3p and EHD2 (Fig. 4D). Furthermore, it was observed that transfection with miR-197-3p mimics markedly diminished EHD2 mRNA expression, and the protein level of EHD2 was decreased in the U2OS and MG63 cells. However, the opposite tendency was observed in the pcDNA3.1/TUSC8-transfected U2OS and MG63 cells (Fig. 4E). In addition, the expression of EHD2 in OS cell lines was much lower than that in the normal osteoblastic cell line OB3 (Fig. 4F). Compared with the non-cancerous tissues, OS tissues exhibited a lower expression of EHD2 (Fig. 4G). In summary, miR-197-3p targets EHD2 in OS.
Figure 4.
EHD2 is a downstream target of miR-197-3p. (A) The Venn diagram shows the potential downstream target genes of miR-197-3p predicted by starBase. (B) The expression of candidate target genes following transfection of miR-197-3p mimic was measured by RT-qPCR. (C and D) The potential interaction between miR-197-3p and EHD2 is shown, and the association between them was further confirmed by luciferase reporter and RIP assays. (E) The mRNA and protein expression of EHD2 were detected in cells transfected with miR-197-3p mimic or pcDNA3.1/TUSC8. (F) The level of EHD2 was detected in OS cell lines and osteoblastic cell lines. (G) The expression of EHD2 in OS tissues and matched non-cancer tissues was estimated by RT-qPCR. *P<0.05 vs. NC mimics, IgG or OB3 cells.
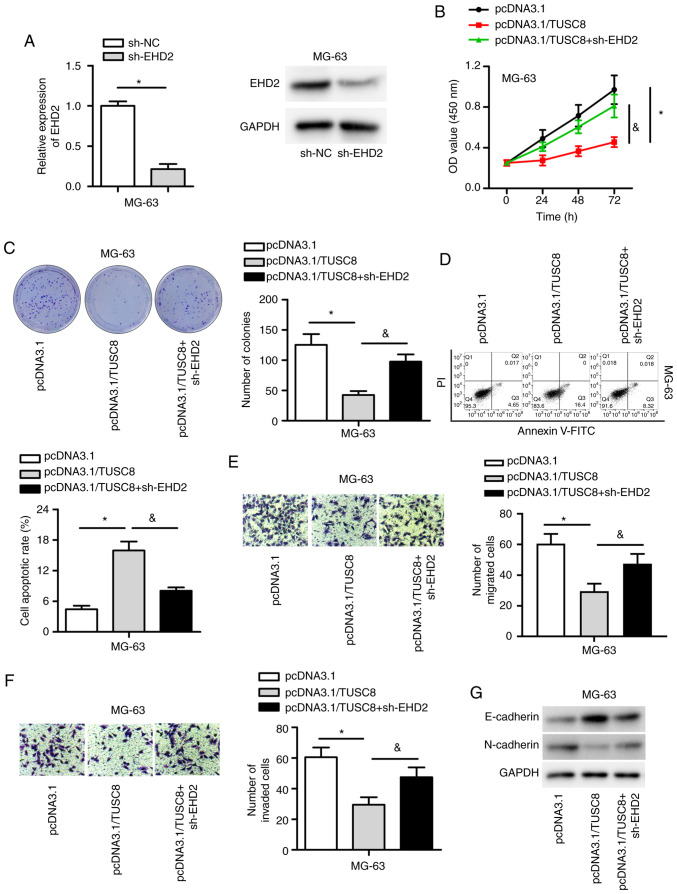
TUSC8 inhibits OS development by regulating EHD2
To determine whether TUSC8 induces its suppressive effects on OS by regulating EHD2, rescue assays were conducted. First, EHD2 was knocked down by transfection with sh-EHD2 into the MG-63 cells, which resulted in a notable decrease in both the mRNA and protein expression of EHD2 (Fig. 5A). As depicted in Fig. 5B and C, EHD2 depletion markedly reversed the decreased proliferative capability of MG-63 cells induced by TUSC8 overexpression. Flow cytometric analysis also indicated that EHD2 knockdown significantly reversed the elevated percentage of apoptotic cells triggered by TUSC8 overexpression (Fig. 5D). In addition, the data demonstrated that EHD2 silencing reversed the inhibitory effects of TUSC8 overexpression on the migration and invasion of OS cells (Fig. 5E and F). Moreover, the pcDNA3.1/TUSC8-mediated decline in N-cadherin expression and the increase in E-cadherin expression were partly reversed by EHD2 knock-down (Fig. 5G). Thus, TUSC8 inhibits OS development by regulating EHD2.
Figure 5.
TUSC8 inhibits OS development by regulating EHD2. (A) The mRNA and protein levels of EHD2 were evaluated in sh-EHD2-transfected cells. (B and C) CCK-8 and colony formation assays were used to assess cell proliferation in transfected cells. (D) Flow cytometry was used to measure cell apoptosis. (E and F) Transwell assays were carried out to evaluate cell migration and invasion. (G) Western blot analysis was used to evaluate epithelial-mesenchymal transition. *P<0.05 and &P<0.05.
Discussion
Osteosarcoma (OS) is a common bone malignancy that accounts for approximately 20% of all bone tumours and occurs predominantly in the femur (28,29). The incidence of OS is increasing rapidly at a rate of approximately 1.4% per year, and the prognosis of patients with OS is poor due to the high risks of relapse and distant metastasis (30-32). Hence, there is an urgent need for the identification of novel therapeutic targets for OS treatment.
Long non-coding RNAs (lncRNAs) are expressed in specific differentiated tissues or cancers (33-35). TUSC8 is a novel lncRNA, and its role in cancer progression is largely unknown. Increasing evidence has revealed that the dysregulated expression of TUSC8 may be considered as a potential biomarker in several types of cancer. A recent study demon-strated that TUSC8 was downregulated in cervical cancer and suppressed cell invasion and migration (14). The upregulation of TUSC8 has been shown to significantly suppress tumour growth and the metastasis of breast cancer (36). Likewise, the results of the present study indicated that the expression of TUSC8 was markedly decreased in OS tissues and cell lines. Furthermore, it was discovered that the enhanced expression of TUSC8 suppressed the proliferation, migration, invasion and EMT, whereas it promoted the apoptosis of OS cells.
miRNAs are small non-coding RNA molecules of 20-24 nucleotides in length that play significant roles in the progression of tumours, including OS (15). For example, miR-214 overexpression suppresses cell migration and invasion in gastric cancer (37). miR-148a suppresses the metastasis of non-small cell lung cancer via Wnt1 (38). miR-708 regulates cell proliferation and apoptosis by targeting CUL4B in OS (39). It has been demonstrated that lncRNAs regulate the development of multiple types of cancer by sponging specific miRNAs. For example, HOXA11-AS contributes to the tumorigenesis of glioma by sponging miR-140-5p (40). PVT1 functions as a sponge for miR-152 in gastric cancer (41). lncRNA RP4 suppresses the development of colorectal cancer by acting as a sponge for miR-7-5p (8). It is worth noting that TUSC8 plays an anti-oncogenic role in cervical cancer by sponging miR-641 (14). Moreover, a recent study indicated that TUSC8 functions as a ceRNA of MYLIP by competitively binding with miR-190b-5p to inhibit breast cancer growth (36). In the present study, TUSC8 was found to be predominantly distributed in the cytoplasm, which provided the possibility of ceRNA mechanism research. Subsequently, a TUSC8 binding site was predicted in two candidate miRNAs (miR-197-3p or miR-499b-5p) by the starBase website. Owing to the differential expression of miR-197-3p in OS cell lines, the underlying regulatory mechanisms were further analysed in OS. As expected, the interaction between miR-197-3p and TUSC8 was confirmed by luciferase reporter and RIP assays. According to previous studies, miR-197-3p plays an oncogenic role in breast cancer, bladder cancer and thyroid cancer (19,20,42,43). However, the anti-oncogenic role of miR-197-3p has been found in prostate cancer and gastric cancer (44,45). In the present study, the findings were consisted with those of previous studies, which suggests the promoting effects of miR-197-3p on the progression of OS.
EHD2 is a plasma membrane-associated member of the EHD family and is related to the actin cytoskeleton (46). It has been reported that the dysregulated expression of EHD2 is closely associated with the metastasis of cancer. For example, EHD2 inhibits the metastasis of hepatocellular carcinoma (47). EHD2 knockdown promotes cell migration in oesophageal squamous cell carcinoma (48). Extensive studies have shown that lncRNAs containing miRNA binding sites can function as ceRNAs to regulate mRNAs in cancers, including OS (49-51). The ceRNA pattern mediated by TUSC8 also functions in other types of cancer. For example, TUSC8 sponges miR-190b-5p and targets MTLIP to suppresses breast cancer growth and metastasis (36). TUSC8 inhibits cell migration and invasion by regulating the miR-641/PTEN axis in cervical cancer (14). The present study, to the best of our knowledge, is the first to certify that TUSC8 possesses the capacity to modulate EHD2 expression in OS. Through mechanistic analysis, EHD2 was identified by starBase and further proved to bind with miR-197-3p. Furthermore, the present study elucidated that the decreased EHD2 expression notably reversed the TUSC8 overexpression-mediated effects on OS cellular processes.
Overall, the present study demonstrates that the upregulation of TUSC8 is negatively associated with OS cell proliferation, migration and invasion, whereas it is positively associated with cell apoptosis. Mechanistically, TUSC8 serves as competitive ceRNA to sponge miR-197-3p and target EHD2. These findings indicate that TUSC8 may be a promising prognostic biomarker and therapeutic target for OS therapy. However, further experiments are required to investigate the mechanisms of TUSC8 in OS in the future, including rescue assays of miR-197-3p on TUSC8 and in vivo experiments.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported and funded by the National Nature Science Foundation of China (grant no. 81601908) and the Outstanding Youth Foundation from the Science and Technology Department of Jilin Province (grant no. 20180520108JH).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
HF and SZ conceived and designed the experiments. HF, TL and HT performed the experiments. HF and SZ analyzed the data. HF, TL and SZ drafted the manuscript and reviewed the final manuscript. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
The present study was approved by the Human Research Ethics Committee of The Second Hospital of Jilin University (Changchun, China). Written informed consent was signed by each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82:690–698. [PubMed] [Google Scholar]
- 3.Wu PK, Chen WM, Chen CF, Lee OK, Haung CK, Chen TH. Primary osteogenic sarcoma with pulmonary metastasis: Clinical results and prognostic factors in 91 patients. Jpn J Clin Oncol. 2009;39:514–522. doi: 10.1093/jjco/hyp057. [DOI] [PubMed] [Google Scholar]
- 4.Fan XL, Cai GP, Zhu LL, Ding GM. Efficacy and safety of ifosfamide-based chemotherapy for osteosarcoma: A meta-analysis. Drug Des Devel Ther. 2015;9:5925–5932. doi: 10.2147/DDDT.S91217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong XD, Ren X, Cai MY, Yang JW, Liu X, Yang JM. Long non-coding RNAs: An emerging powerhouse in the battle between life and death of tumor cells. Drug Resist Updat. 2016;26:28–42. doi: 10.1016/j.drup.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Liu ML, Zhang Q, Yuan X, Jin L, Wang LL, Fang TT, Wang WB. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World J Gastroenterol. 2018;24:1004–1012. doi: 10.3748/wjg.v24.i9.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503:2095–2100. doi: 10.1016/j.bbrc.2018.07.165. [DOI] [PubMed] [Google Scholar]
- 10.Zhao JH, Sun JX, Song YX, Chen XW, Yang YC, Ma B, Wang J, Gao P, Wang ZN. A novel long noncoding RNA-LOWEG is low expressed in gastric cancer and acts as a tumor suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol. 2016;142:601–609. doi: 10.1007/s00432-015-2071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Wu QM, Wang XQ, Zhang CQ. Long Noncoding RNA miR210HG Sponges miR-503 to facilitate osteosarcoma cell invasion and metastasis. DNA Cell Biol. 2017;36:1117–1125. doi: 10.1089/dna.2017.3888. [DOI] [PubMed] [Google Scholar]
- 12.Xia B, Wang L, Feng L, Tian B, Tan Y, Du B. Knockdown of long noncoding RNA CAT104 inhibits the proliferation, migration, and invasion of human osteosarcoma cells by regulating MicroRNA-381. Oncol Res. 2018;27:89–98. doi: 10.3727/096504018X15199511344806. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang H, Yu Y, Fan S, Luo L. Knockdown of long noncoding RNA TUG1 inhibits the proliferation and cellular invasion of osteosarcoma cells by sponging miR-153. Oncol Res. 2018;26:665–673. doi: 10.3727/096504017X14908298412505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Liu B, Zhang P, Zhang J, Wang L. LncRNA TUSC8 inhibits the invasion and migration of cervical cancer cells via miR-641/PTEN axis. Cell Biol Int. 2019;43:781–788. doi: 10.1002/cbin.11152. [DOI] [PubMed] [Google Scholar]
- 15.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Wang R, Sun Y, Yu W, Yan Y, Qiao M, Jiang R, Guan W, Wang L. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res. 2019;38:20. doi: 10.1186/s13046-018-0995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C. miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis. 2018;21:849–860. doi: 10.1007/s10456-018-9630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iacona JR, Monteleone NJ, Lemenze AD, Cornett AL, Lutz CS. Transcriptomic studies provide insights into the tumor suppressive role of miR-146a-5p in non-small cell lung cancer (NSCLC) cells. RNA Biol. 2019;16:1721–1732. doi: 10.1080/15476286.2019.1657351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu F, Li H, Hu C. LIFR-AS1 modulates Sufu to inhibit cell proliferation and migration by miR-197-3p in breast cancer. Biosci Rep. 2019;39:BSR20180551. doi: 10.1042/BSR20180551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Li Z, Hong S, Liu Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun. 2018;503:1825–1829. doi: 10.1016/j.bbrc.2018.07.120. [DOI] [PubMed] [Google Scholar]
- 21.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sc. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YG, Wang T, Shi M, Zhai B. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin Cancer Res. 2019;38:153. doi: 10.1186/s13046-019-1128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Zu Y, Zhu S, Yang Y, Huang W, Xie H, Li G. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem Biophys Res Commun. 2018;507:231–235. doi: 10.1016/j.bbrc.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z. Long Noncoding RNA SNHG16 promotes cell proliferation by sponging MicroRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem. 2018;51:429–440. doi: 10.1159/000495239. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Dai C, Wu Q, Liu H, Li F. Expression profiling of long noncoding RNA identifies lnc-MMP31 as a prognostic biomarker in external auditory canal squamous cell carcinoma. Cancer Med. 2017;6:2541–2551. doi: 10.1002/cam4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Wei T, Li W, Zhang R, Chen M. Circular RNA hsa_circ_0002024 suppresses cell proliferation, migration, and invasion in bladder cancer by sponging miR-197-3p. Am J Transl Res. 2019;11:1644–1652. [PMC free article] [PubMed] [Google Scholar]
- 28.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 29.Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O'Leary M, Smith FO, Reaman GH. Outcomes for children and adolescents with cancer: Challenges for the twenty-first century. J Clin Onco. 2010;28:2625–2634. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mialou V, Philip T, Kalifa C, Perol D, Gentet JC, Marec-Berard P, Pacquement H, Chastagner P, Defaschelles AS, Hartmann O. Metastatic osteosarcoma at diagnosis: prognostic factors and long-term outcome-the French pediatric experience. Cancer. 2005;104:1100–1109. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 31.Xiao X, Wang W, Wang Z. The role of chemotherapy for metastatic, relapsed and refractory osteosarcoma. Paediatr Drugs. 2014;16:503–512. doi: 10.1007/s40272-014-0095-z. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Zhang Y, Wan S, Li H, Li D, Xia J, Yuan Z, Ren M, Yu S, Li S, et al. A comparative study between limb-salvage and amputation for treating osteosarcoma. J Bone Oncol. 2016;5:15–21. doi: 10.1016/j.jbo.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin PC, Huang HD, Chang CC, Chang YS, Yen JC, Lee CC, Chang WH, Liu TC, Chang JG. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:583. doi: 10.1186/s12885-016-2569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Zhang B, Jia Y, Shi H, Wang H, Guo Q, Li H. LncRNA LOXL1-AS1 regulates the tumorigenesis and development of lung adenocarcinoma through sponging miR-423-5p and targeting MYBL2. Cancer Med. 2019;9:689–699. doi: 10.1002/cam4.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Liu G, Ye W, Xie J, Shao C, Wang X, Li X. ZEB2-AS1 Accelerates epithelial/mesenchymal transition through miR-1205/CRKL pathway in colorectal cancer. Cancer Biother Radiopharm. 2019;35:153–162. doi: 10.1089/cbr.2019.3000. [DOI] [PubMed] [Google Scholar]
- 36.Zhao L, Zhou Y, Zhao Y, Li Q, Zhou J, Mao Y. Long non-coding RNA TUSC8 inhibits breast cancer growth and metastasis via miR-190b-5p/MYLIP axis. Aging (Albany NY) 2020;12:2974–2991. doi: 10.18632/aging.102791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wang X, Tang J, Su X, Miao Y. The study of mechanism of miR-34c-5p targeting FLOT2 to regulate proliferation, migration and invasion of osteosarcoma cells. Artif Cells Nanomed Biotechno. 2019;47:3559–3568. doi: 10.1080/21691401.2019.1640714. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Min L, Ren C, Xu X, Yang J, Sun X, Wang T, Wang F, Sun C, Zhang X. miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 2017;12:e0171751. doi: 10.1371/journal.pone.0171751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Zhou H. MiRNA-708/CUL4B axis contributes into cell proliferation and apoptosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:5452–5459. doi: 10.26355/eurrev_201809_15805. [DOI] [PubMed] [Google Scholar]
- 40.Cui Y, Yi L, Zhao JZ, Jiang YG. Long noncoding RNA HOXA11-AS Functions as miRNA sponge to promote the glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol. 2017;36:822–828. doi: 10.1089/dna.2017.3805. [DOI] [PubMed] [Google Scholar]
- 41.Li T, Meng XL, Yang WQ. Long noncoding RNA PVT1 acts as a 'sponge' to inhibit microRNA-152 in gastric cancer cells. Dig Dis Sci. 2017;62:3021–3028. doi: 10.1007/s10620-017-4508-z. [DOI] [PubMed] [Google Scholar]
- 42.Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, Xie X, Tang H. circFBXW7 inhibits malignant progression by sponging miR-197-3p and Encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu K, Huang W, Yan DQ, Luo Q, Min X. Overexpression of long intergenic noncoding RNA LINC00312 inhibits the invasion and migration of thyroid cancer cells by down-regulating microRNA-197-3p. Biosci Rep. 2017;37:BSR20170109. doi: 10.1042/BSR20170109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Huang Q, Ma B, Su Y, Chan K, Qu H, Huang J, Wang D, Qiu J, Liu H, Yang X, Wang Z. miR-197-3 prepresses the proliferation of prostate cancer by regulating the VDAC1/AKT/β-catenin signaling axis. Int J Biol Sci. 2020;16:1417–1426. doi: 10.7150/ijbs.42019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Ju H, Zhao T, Yu S, Li P, Jia J, Li N, Jing X, Tan B, Li Y. hsa_circ_0092306 Targeting miR-197-3p promotes gastric cancer development by regulating PRKCB in MKN-45 cells. Mol Ther Nucleic Acids. 2019;18:617–626. doi: 10.1016/j.omtn.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guilherme A, Soriano NA, Bose S, Holik J, Bose A, Pomerleau DP, Furcinitti P, Leszyk J, Corvera S, Czech MP. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J Biol Chem. 2004;279:10593–10605. doi: 10.1074/jbc.M307702200. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Ni W, Qu L, Cui X, Lin Z, Liu Q, Zhou H, Ni R. Decreased expression of EHD2 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Dig Dis Sci. 2016;61:2554–2567. doi: 10.1007/s10620-016-4202-6. [DOI] [PubMed] [Google Scholar]
- 48.Li M, Yang X, Zhang J, Shi H, Hang Q, Huang X, Liu G, Zhu J, He S, Wang H. Effects of EHD2 interference on migration of esophageal squamous cell carcinoma. Med Oncol. 2013;30:396. doi: 10.1007/s12032-012-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C, Liu S, Wang L, Wang Y, Li Y, Cui Y. Effect of EH domain containing protein 2 on the biological behavior of clear cell renal cell carcinoma. Hum Exp Toxicol. 2019;38:927–937. doi: 10.1177/0960327119842241. [DOI] [PubMed] [Google Scholar]
- 50.Qu F, Cao P. Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp Cell Res. 2018;369:187–196. doi: 10.1016/j.yexcr.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Gu M, Liu C, Wan X, Shi Q, Chen Q, Wang Z. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37–42. doi: 10.1016/j.gene.2018.10.085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.