We thank Tábuas-Pereira and colleagues for their interest in our study demonstrating a causative role for CYLD mutations in frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (Dobson-Stone et al., 2020). Tábuas-Pereira et al. (2020) report the identification of rare CYLD missense variants in 2 of 65 Portuguese clinically diagnosed FTD patients. Neither variant was observed in the patient cohorts we examined previously. However, it is of interest that the p.P229S variant affects the amino acid immediately adjacent to the p.P228L variant identified in the ALSdb database (http://alsdb.org/) (Dobson-Stone et al., 2020). CYLD missense mutations have also been reported in CYLD cutaneous syndrome (Bignell et al., 2000; Rajan and Ashworth, 2015). CYLD cutaneous syndrome-causative mutations have the opposite effect on CYLD deubiquitinase activity to the FTD-ALS-causative p.M719V mutation and are not associated with dementia (Dobson-Stone et al., 2020). We reviewed the scientific literature and the Leiden Open Variation Database (https://www.lovd.nl/) and confirmed that neither p.P229S nor p.S615F have been reported in CYLD cutaneous syndrome patients.
Both patients described by Tábuas-Pereira et al. exhibited severe episodic memory impairments, and early memory impairment was also a feature for several patients in the FTD-ALS family with the CYLD p.M719V mutation in our study. As the authors note, this is an Alzheimer’s disease-like phenotype and it is possible that other patients with an Alzheimer’s disease clinical diagnosis harbour pathogenic CYLD variants. Subsequent to the acceptance of our manuscript, we examined the Alzheimer’s Disease Sequencing Project dataset [accessed through the Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/], comprising whole exome sequencing from 10 913 individuals. We used the peddy ancestry prediction program (Pedersen and Quinlan, 2017) to filter for individuals with European ancestry, leaving 5544 cases and 4531 controls. We performed gene burden testing of CYLD using the TRAPD software package (Guo et al., 2018) using previously described parameters (Dobson-Stone et al., 2020) and removing variants within the ReFiNE full 0.01 blacklist (Maffucci et al., 2019). No enrichment of qualifying variants (missense variants with maximum population minor allele frequency < 0.0001 in gnomAD non-neurological cohort) was observed in Alzheimer’s disease cases (16/5544, 0.29%) relative to controls (21/4531, 0.46%; Fisher’s exact test two-sided P = 0.185). Interestingly, one of the variants observed in an Alzheimer’s disease case was p.P229S, the variant observed in Patient 1 of Tábuas-Pereira et al. (2020). TRAPD analysis restricted to the deubiquitinase domain of CYLD (amino acids 593–948) in the Alzheimer’s Disease Sequencing Project dataset revealed a significant enrichment of rare missense variants, but in controls rather than cases (cases 1/5544, 0.02%; controls 8/4531, 0.18%; P = 0.014). The reason for this is unclear, but it does emphasize the necessity of functional assays to aid in the interpretation of pathogenicity of any CYLD variant, given that they may have opposing effects on gene function and disease phenotypes.
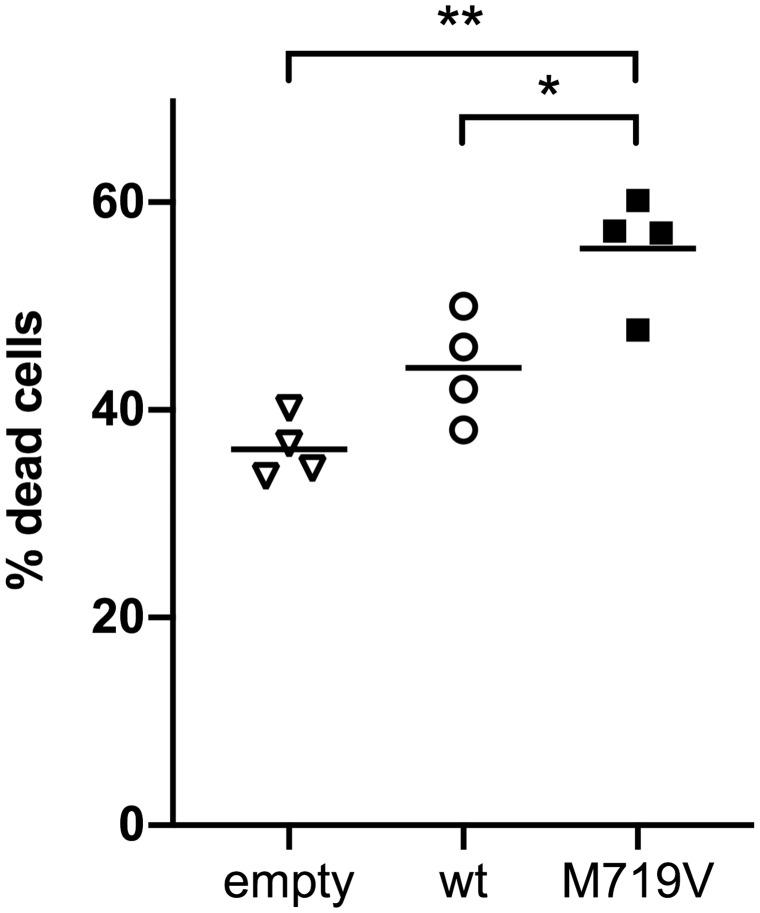
Further, in silico analysis may have limited predictive value for FTD-ALS-causative CYLD mutations: the CYLD p.M719V mutation was not predicted to be deleterious by common pathogenicity prediction programs PolyPhen-2 and SIFT (Dobson-Stone et al., 2013), in contrast to most CYLD cutaneous syndrome-causative mutations. Also, no family members were available for either patient reported by Tábuas-Pereira et al. to determine segregation of the variants with disease. Since our manuscript was accepted, we have examined the effect of CYLD p.M719V on cell death in L929 cells by detection of dead cells with SYTOXTM Green (Invitrogen) after transfection with wild-type or mutant CYLD cDNA constructs (Fig. 1). Transfection of CYLDM719V led to a significantly higher percentage of dead cells [mean ± standard error of the mean (SEM) 55.5 ± 2.7%] relative to wild-type CYLD (44.0 ± 2.6%, t = 3.653, P = 0.035) and empty vector (36.2 ± 1.5%, t = 7.636, P = 0.0047). We examined L929 cells as they have been extensively studied with respect to necroptosis (Degterev et al., 2014), a cell death pathway that CYLD has been implicated in Hitomi et al. (2008) and O’Donnell et al. (2011); however, we note that they are subcutaneous fibroblastoid cells and thus are not closely related to the cell types affected in FTD and ALS. Nevertheless, we share this assay in the hope that it could be used to help prioritize CYLD variants to be screened in other functional assays to determine pathogenicity.
Figure 1.
Expression of CYLDM719V increases cell death. L929 cells were seeded into 96-well plates at 104 cells/well and transfected with 100 ng CYLD wild-type (wt) or mutant cDNA construct per well 16–24 h after seeding using X-tremeGENETM HP (Merck). Dead cells were detected 48 h after transfection by incubation with 125 nM SYTOXTM Green nucleic acid reagent (Invitrogen) and fluorescence detection on the CLARIOStar microplate reader (BMG Labtech). Dead cells were calculated as a percentage of the total cell count, with the latter determined by SYTOXTM fluorescence after 1 h treatment with 1% TritonTM X-100. Horizontal bars indicate mean values from four independent experiments, with transfections performed in quadruplicate per experiment. *P < 0.05; **P < 0.01.
Data availability
The authors are willing to provide raw data related to this study upon request.
Acknowledgements
We thank the contributing teams of the Alzheimer’s Disease Sequencing Project (ADSP) for access to whole-exome sequencing data (dbGaP accession number phs000572.v7.p4). The ADSP is comprised of two Alzheimer’s disease genetics consortia and three National Human Genome Research Institute-funded large-scale sequencing and analysis centres. The two Alzheimer’s disease genetics consortia are the Alzheimer’s Disease Genetics Consortium funded by the National Institute on Aging (NIA, U01AG032984), and the Cohorts for Heart and Aging Research in Genomic Epidemiology funded by NIA (R01AG033193), the National Heart, Lung, and Blood Institute, other National Institute of Health institutes and other foreign governmental and non-governmental organizations. The discovery phase analysis of sequence data is supported through UF1AG047133 (to Drs Schellenberg, Farrer, Pericak-Vance, Mayeux, and Haines); U01AG049505 to Dr Seshadri; U01AG049506 to Dr. Boerwinkle; U01AG049507 to Dr Wijsman; and U01AG049508 to Dr Goate and the discovery extension phase analysis is supported through U01AG052411 to Dr Goate, U01AG052410 to Dr Pericak-Vance and U01AG052409 to Dr. Seshadri and Fornage. Data generation and harmonisation in the follow-up phases is supported by U54AG052427 (to Drs Schellenberg and Wang). A full acknowledgement statement for the ADSP can be found here: https://www.niagads.org/adsp/content/acknowledgement-statement
Funding
This research was funded by the National Health and Medical Research Council of Australia (NHMRC) Project Grant 1140708 (to C.D.S. and J.B.K.). C.D.S. is funded by an NHMRC Boosting Dementia Research Leadership Fellowship (1138223). N.R. is funded by a Wellcome Trust Intermediate Clinical Fellowship (WT097163MA), the Wellcome Trust and UK Department of Health under the Health Innovation Challenge Fund (100935/Z/13/Z), the Newcastle NIHR Biomedical Research Centre (BRC) and the Newcastle MRC/EPSRC Molecular Pathology Node. Z.C. is supported by a University of Sydney Postdoctoral Fellowship.
Competing interests
The authors report no competing interests.
References
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 2000; 25: 160–5. [DOI] [PubMed] [Google Scholar]
- Degterev A, Zhou W, Maki JL, Yuan J.. Assays for necroptosis and activity of RIP kinases. Methods Enzymol 2014; 545: 1–33. [DOI] [PubMed] [Google Scholar]
- Dobson-Stone C, Hallupp M, Shahheydari H, Ragagnin AMG, Chatterton Z, Carew-Jones F, et al. CYLD is a causative gene for frontotemporal dementia – amyotrophic lateral sclerosis. Brain 2020; 143: 783–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson-Stone C, Luty AA, Thompson EM, Blumbergs P, Brooks WS, Short CL, et al. Frontotemporal dementia-amyotrophic lateral sclerosis syndrome locus on chromosome 16p12.1-q12.2: genetic, clinical and neuropathological analysis. Acta Neuropathol 2013; 125: 523–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF.. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet 2018; 103: 522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135: 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci P, Bigio B, Rapaport F, Cobat A, Borghesi A, Lopez M, et al. Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc Natl Acad Sci USA 2019; 116: 950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 2011; 13: 1437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. Who’s who? Detecting and resolving sample anomalies in human DNA sequencing studies with peddy. Am J Hum Genet 2017; 100: 406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan N, Ashworth A.. Inherited cylindromas: lessons from a rare tumour. Lancet Oncol 2015; 16: e460–e9. [DOI] [PubMed] [Google Scholar]
- Tábuas-Pereira M, Santana I, Kun-Rodrigues C, Bras J, Guerreiro R.. CYLD variants in frontotemporal dementia associated with severe memory impairment in a Portuguese cohort. Brain 2020; 143: e67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors are willing to provide raw data related to this study upon request.