Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the progressive loss of upper and lower motor neurons. Although its precise aetiopathogenesis remains unclear, cellular hallmarks of the disease include the deregulation of RNA metabolism and the mislocalization of RNA binding proteins (RBPs) from the nucleus to the cytoplasm (Butti and Patten, 2018). We recently showed that FUS exhibits nuclear loss in sporadic ALS (Tyzack et al., 2019). This finding builds on our earlier discovery of nuclear loss of SFPQ in ALS (Luisier et al., 2018) and the established hallmark of TAR DNA-binding protein 43 (TDP-43, encoded by TARDBP) nuclear-to-cytoplasmic mislocalization (Neumann et al., 2006). These three proteins are components of paraspeckles, nuclear ribonucleoprotein membraneless compartments (or granules) formed in the interchromatin space together with the long non-coding RNA (lncRNA) NEAT1_2 (Clemson et al., 2009; Sasaki et al., 2009; Mao et al., 2011).
Paraspeckles are involved in the regulation of normal gene expression via sequestration of transcription factors and RBPs, in the nuclear retention of some specific classes of transcripts, and in pri-miRNA processing (Nakagawa et al., 2018). While in healthy motor neurons paraspeckles are absent due to the lack of expression of NEAT1_2, enhanced paraspeckle formation has been observed in both sporadic (Nishimoto et al., 2013; Shelkovnikova et al., 2018) and familial ALS cases (Shelkovnikova et al., 2018; An et al., 2019), and is proposed to represent a protective neuronal response to stress.
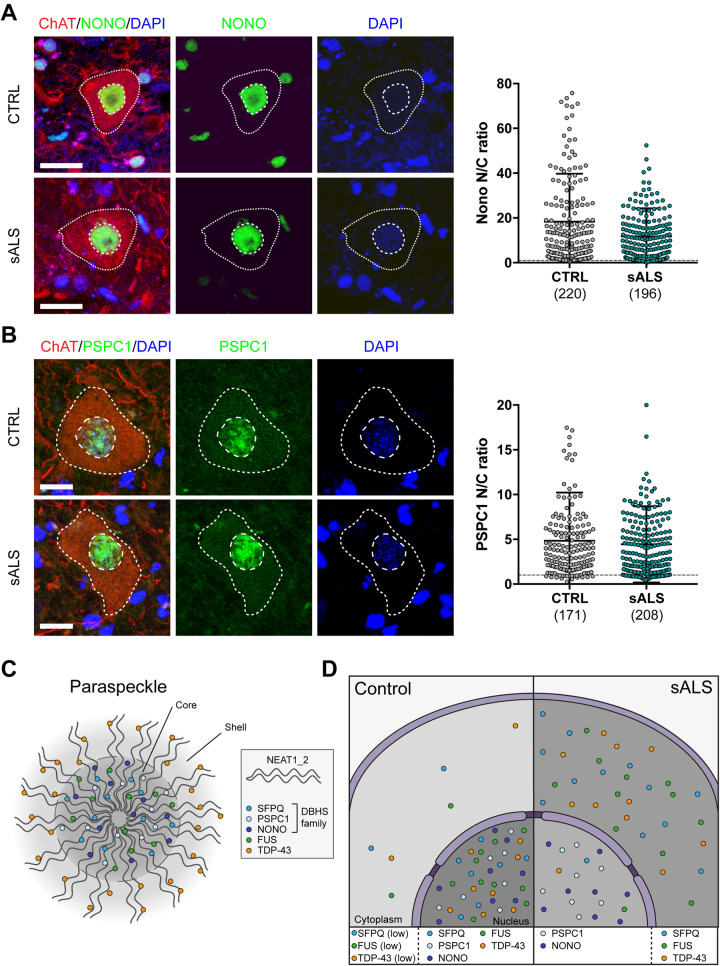
In paraspeckles, RBPs are arranged with a core and shell structure (Fig. 1C). RBPs in the core are fundamental for the structural integrity of paraspeckles and include FUS and the DBHS family members SFPQ, NONO and PSPC1, while TDP-43 is found in the shell (West et al., 2016). Proteins in this family share a similar protein domain architecture and evolutionary origin, and they are known to form homodimers or heterodimers with other DBHS proteins (Knott et al., 2016). Noting that at least three paraspeckle components are lost from the nuclei of ALS motor neurons (Neumann et al., 2006; Luisier et al., 2018; Tyzack et al., 2019), we sought to address the hypothesis that nuclear dislocation is a more widespread phenomenon affecting other paraspeckle proteins, specifically NONO and PSPC1.
Figure 1.
NONO and PSPC1 are not lost from motor neuron nuclei in sporadic ALS. (A and B) Analysis of the subcellular localization of NONO (A) and PSPC1 (B) in motor neurons in the ventral spinal cord of healthy control subjects (CTRL, n = 8) and patients with sporadic ALS (sALS, n = 12) from the same cohort analysed in our previous study (Tyzack et al., 2019). Motor neuron cytoplasm was identified by ChAT immunohistochemistry (red), nuclei were counterstained with DAPI (blue). NONO and PSPC1 are shown in green in A and B, respectively. Images were acquired as confocal z-stacks using a Zeiss 710 confocal microscope with a z step of 1 μm, processed to obtain a maximum intensity projection and analysed using Fiji. The nuclear and cytoplasmic areas were manually drawn based on DAPI and ChAT staining, respectively. For each region of interest, the average immunoreactivity intensity for each RBP was measured, the background was subtracted, and the ratio between nuclear and cytoplasmic average intensity was calculated. Data shown in the dot plots are nuclear/cytoplasmic (N/C) ratio (mean ± SD) per cell, with the dashed line set at a N/C ratio of 1. The total number of cells analysed is shown for each group. Scale bar = 20 μm. Linear mixed effect analysis was used to test the relationship between NONO or PSPC1 localization and sporadic ALS, thus accounting for individual case-based idiosyncratic variation. P-values were obtained by likelihood ratio tests of the full model with the effect in question against the model without the effect in question: P-value (NONO) = 0.37 and P-value (PSPC1) = 0.78. (C) Paraspeckle structure showing the localization of different RBPs in the core or shell. (D) Schema summarizing the findings of N/C distribution of different paraspeckle components in human sporadic ALS.
Here, we examined the nuclear/cytosolic ratio of NONO and PSPC1 in lumbar spinal cord motor neurons of eight sporadic ALS cases and 12 age- and gender-matched controls using immunohistochemistry, as previously described (Tyzack et al., 2019). In contrast to the reported nuclear displacement of FUS and SFPQ, we observed comparable nuclear/cytosolic distributions of both NONO and PSPC1 in all samples (NONO: control = 18.35 ± 21.38, sporadic ALS = 11.79 ± 12.5; PSPC1: control = 4.84 ± 5.39, sporadic ALS = 4.44 ± 4.25) (Fig. 1A and B). Taken together with aforementioned studies, these data suggest that some paraspeckle components (TDP-43, SFPQ and FUS), but not others (NONO and PSPC1) exhibit nuclear loss in sporadic ALS (Fig. 1C and D).
To our knowledge this is the first study investigating the subcellular localization of PSPC1 in ALS-affected motor neurons. Previous work by Shelkovnikova et al. (2014) described NONO co-localization with cytoplasmic FUS aggregates in mouse models and ALS patients carrying FUS mutations. However, in line with our findings, this was not detected in the cytosol of sporadic cases, or SOD1-ALS patients, suggesting that sequestration of NONO to cytoplasmic aggregates represents a pathological feature that is specific to FUS mutations (Shelkovnikova et al., 2014). The fact that, while sharing structure and function with the other DBHS proteins, SFPQ is the only RBP of this family that is lost from the nucleus of ALS motor neurons is somewhat surprising. Despite substantial overlap, there are also some remarkable differences in the role of DBHS proteins in regulating gene expression, splicing, transcript localization and stability (Knott et al., 2016). In specific circumstances, and in a cell type-specific manner, this is also reflected in a differential localization of these proteins. For example, PSPC1, but not SFPQ nor NONO, is localized to the cytoplasm during adipocyte differentiation (Wang et al., 2017). In neurons, in addition to its nuclear localization, SFPQ is also found in axons, where it is required for correct axon development and maintenance of axon viability through the local transport of specific classes of transcripts (Cosker et al., 2016; Thomas-Jinu et al., 2017). The ability of SFPQ to shuttle between the nucleus and the cytoplasm, and its physiological cytoplasmic roles in mature neurons, might, at least in part, explain its susceptibility to nuclear loss under pathological conditions.
Lastly, our observation that NONO and PSPC1 retain a predominantly nuclear localization in ALS does not necessarily exclude a loss of their nuclear physiological functions. Their sequestration within subnuclear bodies is a possibility that remains open, and yet to be fully addressed. For example, paraspeckle components, including SFPQ, FUS, NONO, and PSPC1 are sequestered by G4C2 nuclear foci in C9ORF72 patient-derived fibroblasts (Bajc Česnik et al., 2019). Similarly, in light of the obligatory dimerization of DBHS proteins, the nuclear loss of SFPQ in ALS might alter the balance and composition of these dimers, potentially affecting the function of nuclear NONO and PSPC1.
Data availability
Data supporting the findings of this study are available from the corresponding authors, upon reasonable request.
Funding
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC010110), the UK Medical Research Council (FC010110), and the Wellcome Trust (FC010110). R.P. holds an MRC/MND Association Lady Edith Wolfson Senior Clinical Fellowship [MR/S006591/1] and is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. N.M.L. is supported by a Wellcome Trust Senior Investigator Award [103760/Z/14/Z], an MRC eMedLab Medical Bioinformatics Infrastructure Award to N.M.L. (MR/L016311/1). N.M.L. is a Winton Group Leader in recognition of the Winton Charitable Foundation’s support towards the establishment of the Francis Crick Institute. R.L. is supported by the Idiap Research Institute.
Competing interests
The authors report no competing interests.
References
- An H, Skelt L, Notaro A, Highley JR, Fox AH, La Bella V, et al. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol Commun 2019; 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajc Česnik A, Darovic S, Prpar Mihevc S, Štalekar M, Malnar M, Motaln H, et al. Nuclear RNA foci from expansion mutation form paraspeckle-like bodies. J Cell Sci 2019; 132: jcs224303. [DOI] [PubMed] [Google Scholar]
- Butti Z, Patten SA.. RNA dysregulation in amyotrophic lateral sclerosis. Front Genet 2018; 9: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33: 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA.. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci 2016; 19: 690–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GJ, Bond CS, Fox AH.. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res 2016; 44: 3989–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisier R, Tyzack GE, Hall CE, Mitchell JS, Devine H, Taha DM, et al. Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat Commun 2018; 9: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL.. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Yamazaki T, Hirose T.. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol 2018; 8: 180150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Nishimoto Y, Nakagawa S, Hirose T, Okano H, Takao M, Shibata S, et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain 2013; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YTF, Ideue T, Sano M, Mituyama T, Hirose T.. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 2009; 106: 2525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, Kukharsky MS, An H, Dimasi P, Alexeeva S, Shabir O, et al. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol Neurodegener 2018; 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL.. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum Mol Genet 2014; 23: 2298–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Jinu S, Gordon PM, Fielding T, Taylor R, Smith BN, Snowden V, et al. Non-nuclear pool of splicing factor SFPQ regulates axonal transcripts required for normal motor development. Neuron 2017; 94: 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzack GE, Luisier R, Taha DM, Neeves J, Modic M, Mitchell JS, et al. Widespread FUS mislocalization is a molecular hallmark of amyotrophic lateral sclerosis. Brain 2019; 142: 2572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rajbhandari P, Damianov A, Han A, Sallam T, Waki H, et al. RNA-binding protein PSPC1 promotes the differentiation-dependent nuclear export of adipocyte RNAs. J Clin Invest 2017; 127: 987–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol 2016; 214: 817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding authors, upon reasonable request.