Abstract
Antimicrobial peptides (AMPs) have been regarded as a potential weapon to fight against drug-resistant bacteria, which is threating the globe. Thus, more and more AMPs had been designed or identified. There is a need to integrate them into a platform for researchers to facilitate investigation and analyze existing AMPs. The AMP database has become an important tool for the discovery and transformation of AMPs as agents. A database linking antimicrobial peptides (LAMPs), launched in 2013, serves as a comprehensive tool to supply exhaustive information of AMP on a single platform. LAMP2, an updated version of LAMP, holds 23 253 unique AMP sequences and expands to link 16 public AMP databases. In the current version, there are more than 50% (12 236) sequences only linking a single database and more than 45% of AMPs linking two or more database links. Additionally, updated categories based on primary structure, collection, composition, source and function have been integrated into LAMP2. Peptides in LAMP2 have been integrated in 8 major functional classes and 38 functional activities. More than 89% (20 909) of the peptides are experimentally validated peptides. A total of 1924 references were extracted and regarded as the evidence for supporting AMP activity and cytotoxicity. The updated version will be helpful to the scientific community.
Keywords: antimicrobial peptides, database, LAMP
Introduction
During the past 30 years, antimicrobial peptides (AMPs) have been considered as a potential source for the development of new antibacterial drugs against drug-resistant bacteria (1, 2). Currently, a lot of potential AMPs have been designed or identified. But most of them have failed in clinical trials. A thorough understanding of the role of sequence of AMPs on their specificity and activity is essential to exploit them as antimicrobial agents. The AMP database has become an important tool for the discovery and transformation of AMPs (3). Now, there are more and more AMP databases that had been developed. Some of them are general databases, such as APD (4), DBAASP (5), CAMP (6) and dbAMP (7), whereas others are specialized databases, such as CancerPPD, Hemolytik, THPdb, InverPep and AntiTbPdb. CancerPPD (8) is a database of anticancer peptides and proteins, Hemolytik (9) is a database of experimentally determined hemolytic and non-hemolytic peptides, THPdb (10) is a database of FDA-approved peptide and protein therapeutics, InverPep (11) is a database of invertebrate AMPs and AntiTbPdb is a database of experimentally verified anti-tubercular or anti-mycobacterial peptides.
The increase in the number of databases is on the one hand, the increase in the amount of AMP sequences is on the other. For example, the number of records in CAMP was about 3782 in the 2009 version (12). In 2013, the number of records exceeds 6756 (13). And in 2015, the number of records exceeds 10 000 (6). It is too difficult for researchers to obtain all the properties of AMPs from dozens of various databases and tens of thousands of sequences. As of date, the most entries databases, such as dbAMP (12 389 AMP sequences), CAMPR3 (10 247 AMP sequences) and SATPdb (10 585 AMP sequences) (14), have their own characteristics or limitations. It creates a need to establish a comprehensive data platform integrating the latest AMP database information. LAMP, a database linking AMP, serves as a tool to provide a full collection of AMPs with cross-linking between existing databases. Compared with other available AMP databases, significant improvements available in LAMP include not only significantly more AMPs but also the unique cross-linking and top similar AMP (Topview) functions. LAMP is a comprehensive platform integrating majority of the peptides from various databases. Since the original version of LAMP was released online in 2013 (15), LAMP has been widely accepted and utilized. In the past, a significant number of AMPs have been discovered. It is proper to include the newly identified AMPs into the updating LAMP. Here, we present LAMP2, an update to the existing LAMP database. LAMP2 would help researchers work on AMPs more efficiently and conveniently.
Materials and Methods
Data collection and compilation
All the AMP sequences were collected from the scientific literatures or authoritative public AMP-related databases. In this updated LAMP2, the public AMP databases contain 16 public databases (4–9, 14, 16–24). We obtained AMP information using export option provided by databases or using the ‘wget’ program. Then, we selected identical AMP sequences of less than 100 amino acid residues long exclude the existing AMPs in previous LAMP database and integrated them into the LAMP2 platform after annotating the physicochemical properties, activities, functions, references and other basic information for each AMP. Structural information of AMPs was retrieved from protein databases of PDB. A total of 1924 references were extracted and regarded as the evidence for supporting AMP activity and cytotoxicity. All sequences of AMPs are presented in FASTA format.
Results and Discussion
Data update
Sequences and links to databases update
After manual checking and removal of redundancy, the current updating LAMP2 holds 23 253 unique AMP sequences. The cross-linking databases expand to 16 different published AMP databases including the latest or the most commonly used AMP databases, such as APD, CAMP, DBAASP, dbAMP (2019), SATPdb, DRAMP (2019) (Figure 1). Compared with the previous version, LAMP2 has added almost 20 000 new entries (Table 1). Most entries in LAMP2 are cross-linked with other individual databases. The cross-linking databases comprised 16 public AMP databases, which are nearly three times of the previous version.
Figure 1. A total of 23 253 entries are distributed in 16 different databases in LAMP2.
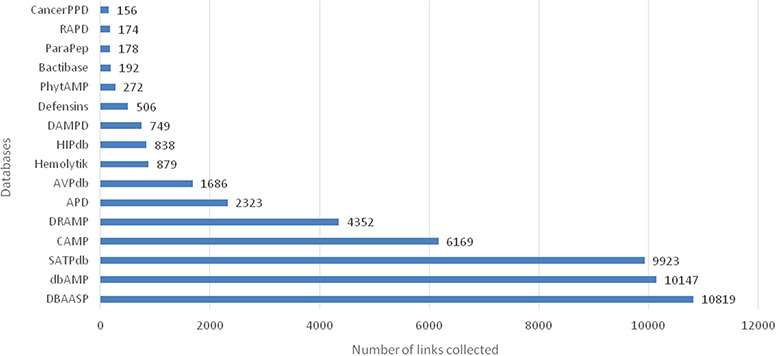
Table 1. Comparison of data in two versions of LAMP database.
| Group | LAMP | LAMP2 | |
|---|---|---|---|
| By primary structure | Linear | / | 22 934 |
| Cyclic | 319 | ||
| By collection | Experimental validated peptides | 3203 | 20 909 |
| Patent peptides | 1491 | 1491 | |
| Predicted peptides | 853 | 853 | |
| By composition | Peptides having D-amino acids | / | 839 |
| Peptides having natural amino acids | 21 691 | ||
| Peptides having modified residues | 723 | ||
| By source | Synthetic peptides | 1643 | 15 429 |
| Natural peptides | 3904 | 7824 | |
Categories of entries update
To meet the needs of different researchers for different options, the categories of updated database are more abundant. We added two types of new classification and expand the activity group. The new categories were based on primary structures and the composition of special amino acid. LAMP2 include linear (22934) and cyclic (319) based on primary structures, peptides having D-amino acids (839), peptides having natural amino acids (21691) and peptides having modified residues (723) based on the composition (Table 1). The section of functional update list expanded groups.
Functional update
Based on curated information of functional activity in 16 databases, peptides in LAMP2 have been integrated in 8 major functional classes and 38 functional activities (Table 2). The maximum numbers of peptides are in the class of antibacterial peptides (67.9%), followed by new mechanism-associated peptides (17.3%), antiviral peptides (15.1%), antifungal peptides (14.0%), toxic peptides (7.9%), disease-associated peptides (6.7%), antiparasitic peptides (3.7%) and new functional peptides (0.5%) (Table 2).
Table 2. New categories based on functional activity in LAMP2.
| Function classes | Functional activity | Numbers of AMPs |
|---|---|---|
| Antibacterial | Antibacterial | 15 844 |
| Anti-Gram positive | 7026 | |
| Anti-Gram negative | 7101 | |
| Antifungal | Antifungal | 2147 |
| Antiviral | Antiviral | 4241 |
| Antiparasitic | Antiparasitic | 374 |
| New functional peptides | Wound healing | 19 |
| Antioxidant | 22 | |
| Spermicidal | 13 | |
| Enzyme inhibitor | 25 | |
| Mollicute | 16 | |
| Antihypertensive | 15 | |
| Disease-associated peptides | Anti-MRSA | 114 |
| Anti-HIV | 105 | |
| Anti-inflammatory | 2 | |
| Anticancer | 1314 | |
| Anti-tumor | 22 | |
| New mechanism-associated peptides | Chemotactic | 50 |
| Sodium channel blocker | 2 | |
| Surface immobilized | 10 | |
| Anti-biofilm | 34 | |
| Cancer cells | 223 | |
| Mammalian cells | 3682 | |
| Blood-brain barrier | 10 | |
| Cell penetrating | 12 | |
| Tumor homing | 2 | |
| Hormones | 3 | |
| Quorum sensing | 5 | |
| Toxic | Hemolytic | 1601 |
| Cytotoxic | 244 |
Cross-linking update and new statistics
Many peptides in LAMP2 belong to two or more databases. More than 45% of AMPs have two or more database links, and at most, one AMP sequence link to nine different databases (Figure 2).
Figure 2. Cross-links to other databases in LAMP2.
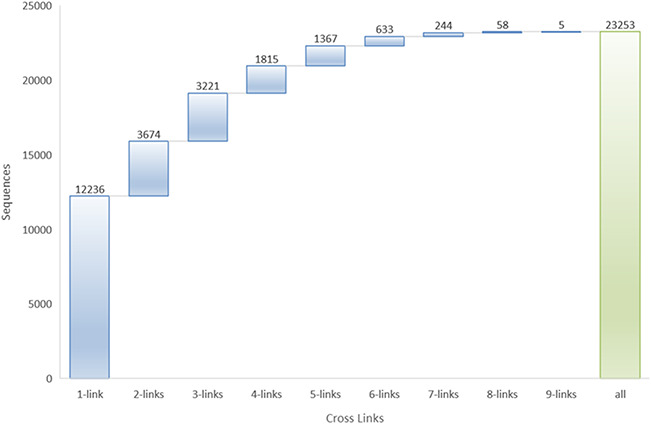
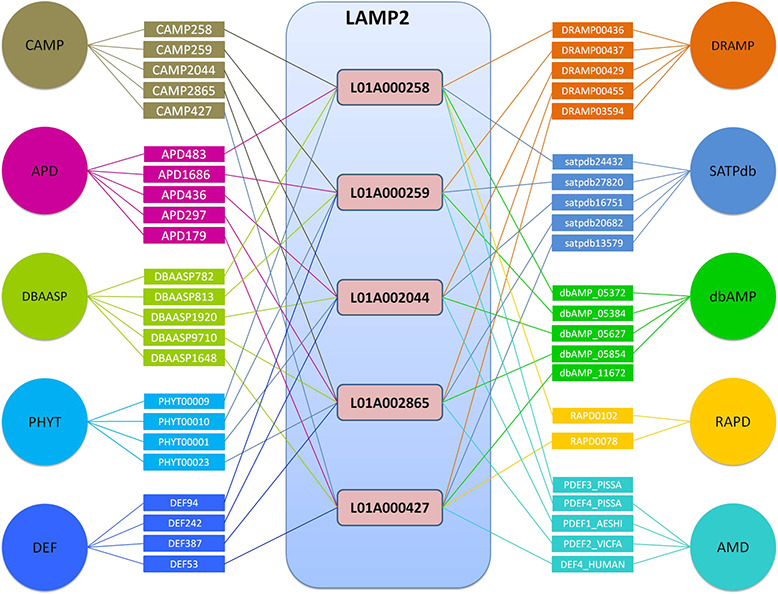
The top 5 links to databases were 10 819 links to the DBAASP, 10 147 links to dbAMP, 9923 links to SATPdb, 6169 links to the CAMP and 4352 links to DRAMP (Figure 1). The cross-links among these top 5 databases have been demonstrated by Venn diagram (Figure 3). Although we have included almost all of the important databases so far, there are still more than 50% (12 236) sequences only link to a single database, indicating that these AMP sequences are somewhat unique or not representative and require further research (Figure 2). At the same time, it also shows that the databases holding these sequences are created on their own and are not universal. The top 5 AMPs in the number of cross-links are listed in Figure 4. More links often demonstrate the more comprehensive, complete information about the sequence.
Figure 3. Venn diagram demonstrating peptides cross links among the top 5 databases in LAMP2 (plotted using http://bioinformatics.psb.ugent.be/webtools/Venn/).
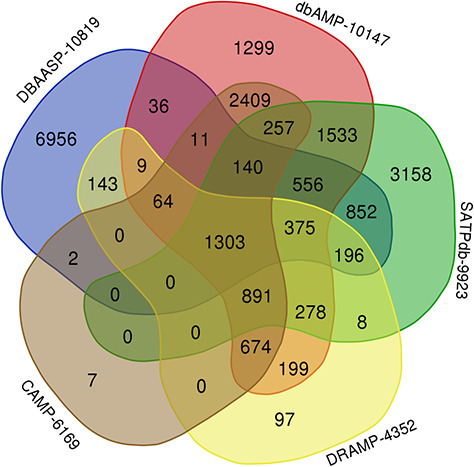
Figure 4. Top 5 AMPs in the number of cross-links in LAMP2.
Utility of database
LAMP2 can be used to get exhaustive information of AMP on a single platform. For example, input ‘defensing’ in the protein name inputbox given at database search page. By a simple click on the search button, you will be directed toward a list of 30 entries stored in LAMP2, which are differentiated with a unique ID. A click on each ID will direct to a detailed display page, providing all the information about general information (LAMP_ID, name, full name, source, mass, sequence, sequence length, isoelectric point, activity and function), structure, activity (MICs), toxicity and reference. LAMP2 contains almost all the data currently included in the commonly used published databases. Take DEF1_PEA as an example: cross-linking provides hyperlinks to other public databases, such as SWISS P8192, CAMP 258, AMD PDEF3_PISSA, PHY PHYT00009, RAP RAPD0102, APD 00483, DBAASP782, dbAMP_05372, DRAMP00436 and SATPdb24432, which allows additional information on the AMP to be easily obtained. For example, if you are interested in family signatures of DEF1_PEA, you can click on the hyperlink CAMP 258. Or you want to know other physical-chemical parameters, you can click on the hyperlink DBAASP 782. Databases that did not appear above basically indicate that they did not include the AMP sequence. Topview function provides the top similar AMPs produced by the BLASTP program. Equipped with the detailed antimicrobial activity and cytotoxicity data, the cross-linking and Topview functions will serve the study of sequence-activity better.
More functions, statistical description and findings were described in our original article (15). Here, we take a successful example of utilizing the database in a real drug discovery. We previously investigated the LAMP database and selected CP-P (lamp-id: L08AP00001) as a template and designed a series of its derivatives considering structure-activity relationship (α-helical structure and hydrophobicity for peptide activity) and AMP statistics (the amino acid composition of AMPs, average net charge, sequence length) after using the Topview function in LAMP. Among them, the derivative peptide K11 exhibited low MICs (less than 10 μg/mL) and broad spectrum antimicrobial activity, especially exhibited strong therapeutic effect on antibiotic resistant clinical isolates of both Gram-positive and Gram-negative bacteria (25).
Conclusion
LAMP2 currently holds the most entries (a total of 23 253 unique AMPs, until November 2019) and is the unique cross-linking AMP database. All entries in LAMP2 are cross-linked with individual databases to provide an option for easy switching to the individual databases for extensive insight to them. The updated database includes details about primary structure, collection, composition, source and function, especially highlighting 8 major functional classes and 38 functional activities. The updated version will be helpful to the scientific community.
Acknowledgements
We thank all of our colleagues at the State Key Laboratory of Genetic Engineering, Fudan University and at Shanghai High-Tech United BioTechnological R&D Co., Ltd, China, and at Kushan BioGreen Technology Co., Ltd, China, for their contributions to the literature search and discussions regarding this manuscript.
Funding
National Key Research and Development Program of China (2016YFD0501308); Jiangsu Province Key Research and Development Program (BE2017314); National Mega-project for Innovative Drugs (2019ZX09721001).
Conflict of interest
None declared.
Database URL: http://biotechlab.fudan.edu.cn/database/lamp/index.php
References
- 1. Pfalzgraff A., Brandenburg K. and Weindl G. (2018) Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol., 9, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vila J., Moreno-Morales J. and Balleste-Delpierre C. (2019) Current landscape in the discovery of novel antibacterial agents. Clin. Microbiol. Infect. 26, 596–603. [DOI] [PubMed] [Google Scholar]
- 3. Porto W.F., Pires A.S. and Franco O.L. (2017) Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol. Adv., 35, 337–349. [DOI] [PubMed] [Google Scholar]
- 4. Wang G.S., Li X. and Wang Z. (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res., 44, D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pirtskhalava M., Gabrielian A., Cruz P. et al. (2016) DBAASP v.2: an enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res., 44, D1104–D1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waghu F.H., Barai R.S., Gurung P. et al. (2016) CAMP(R3): a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res., 44, D1094–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jhong J.H., Chi Y.H., Li W.C. et al. (2019) dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res., 47, D285–D297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tyagi A., Tuknait A., Anand P. et al. (2015) CancerPPD: a database of anticancer peptides and proteins. Nucleic Acids Res., 43, D837–D843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gautam A., Chaudhary K., Singh S. et al. (2014) Hemolytik: a database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res., 42, D444–D449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Usmani S.S., Bedi G., Samuel J.S. et al. (2017) THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One, 12, e0181748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez E.A., Giraldo P. and Orduz S. (2017) InverPep: a database of invertebrate antimicrobial peptides. J. Glob. Antimicrob. Resist., 8, 13–17. [DOI] [PubMed] [Google Scholar]
- 12. Thomas S., Karnik S., Barai R.S. et al. (2010) CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res., 38, D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waghu F.H., Gopi L., Barai R.S. et al. (2014) CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res., 42, D1154–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh S., Chaudhary K., Dhanda S.K. et al. (2016) SATPdb: a database of structurally annotated therapeutic peptides. Nucleic Acids Res., 44, D1119–D1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao X., Wu H., Lu H. et al. (2013) LAMP: a database linking antimicrobial peptides. PLoS One, 8, e66557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang X., Dong F., Shi C. et al. (2019) DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data, 6, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qureshi A., Thakur N., Tandon H. et al. (2014) AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res., 42, D1147–D1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehta D., Anand P., Kumar V. et al. (2014) ParaPep: a web resource for experimentally validated antiparasitic peptide sequences and their structures. Database, 2014, bau051. 10.1093/database/bau051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qureshi A., Thakur N. and Kumar M. (2013) HIPdb: a database of experimentally validated HIV inhibiting peptides. PLoS One, 8, e54908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundararajan V.S., Gabere M.N., Pretorius A. et al. (2012) DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res., 40, D1108–D1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hammami R., Ben Hamida J., Vergoten G. et al. (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res., 37, D963–D968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y. and Chen Z. (2008) RAPD: a database of recombinantly-produced antimicrobial peptides. FEMS Microbiol. Lett., 289, 126–129. [DOI] [PubMed] [Google Scholar]
- 23. Gupta S., Kapoor P., Chaudhary K. et al. (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS One, 8, e73957.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammami R., Zouhir A., Le Lay C. et al. (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol., 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin-Jiang H., Jin-Chun L., Min L. et al. (2012) The design and construction of K11: a novel alpha-helical antimicrobial peptide. Int. J. Microbiol., 2012, 764834. [DOI] [PMC free article] [PubMed] [Google Scholar]


