Abstract
Background
Malaria is still a heavy public health concern in Madagascar. Few studies combining parasitology and entomology have been conducted despite the need for accurate information to design effective vector control measures. In a Malagasy region of moderate to intense transmission of both Plasmodium falciparum and P. vivax, parasitology and entomology have been combined to survey malaria transmission in two nearby villages.
Methods
Community-based surveys were conducted in the villages of Ambohitromby and Miarinarivo at three time points (T1, T2 and T3) during a single malaria transmission season. Human malaria prevalence was determined by rapid diagnostic tests (RDTs), microscopy and real-time PCR. Mosquitoes were collected by human landing catches and pyrethrum spray catches and the presence of Plasmodium sporozoites was assessed by TaqMan assay.
Results
Malaria prevalence was not significantly different between villages, with an average of 8.0% by RDT, 4.8% by microscopy and 11.9% by PCR. This was mainly due to P. falciparum and to a lesser extent to P. vivax. However, there was a significantly higher prevalence rate as determined by PCR at T2 ( = 7.46, P = 0.025). Likewise, mosquitoes were significantly more abundant at T2 ( = 64.8, P < 0.001), especially in Ambohitromby. At T1 and T3 mosquito abundance was higher in Miarinarivo than in Ambohitromby ( = 14.92, P < 0.001). Of 1550 Anopheles mosquitoes tested, 28 (1.8%) were found carrying Plasmodium sporozoites. The entomological inoculation rate revealed that Anopheles coustani played a major contribution in malaria transmission in Miarinarivo, being responsible of 61.2 infective bites per human (ib/h) during the whole six months of the survey, whereas, it was An. arabiensis, with 36 ib/h, that played that role in Ambohitromby.
Conclusions
Despite a similar malaria prevalence in two nearby villages, the entomological survey showed a different contribution of An. coustani and An. arabiensis to malaria transmission in each village. Importantly, the suspected secondary malaria vector An. coustani, was found playing the major role in malaria transmission in one village. This highlights the importance of combining parasitology and entomology surveys for better targeting local malaria vectors. Such study should contribute to the malaria pre-elimination goal established under the 2018–2022 National Malaria Strategic Plan. 
Keywords: Anopheles coustani, Anopheles arabiensis, Plasmodium falciparum, Plasmodium vivax, Vector biology dynamics, Andriba, Madagascar
Background
Malaria remains a major public health concern in Madagascar, with an increase in the number of cases and deaths in years 2017 and 2018, compared to the year 2016 [1]. Malaria epidemiology in Madagascar is highly heterogenous and varies according to the climatic and ecological environment that allows a stratification in bioclimatic zones and ecozones [2, 3]. All four human malaria species are circulating, with Plasmodium falciparum being the most prevalent. Across the ecozones, the average prevalence of P. falciparum varies from 2 to 12% [3] but can reach 30% in some areas [4]. Among the 26 Anopheles species described in the country, six have been reported as malaria vectors with different role according to geography and behaviour [5]. Three species belong to the Anopheles gambiae complex: Anopheles gambiae (sensu stricto), An. arabiensis and An. merus, the latter having a minor role in malaria transmission and being restricted to the most southern region of Madagascar [6]. Of the two other members of the An. gambiae complex, An. arabiensis is prevalent throughout Madagascar and plays a major role in malaria transmission along with An. funestus [7]. Anopheles mascarensis, endemic to Madagascar, and An. coustani, act as local or minor vectors [8–10].
Recent surveys of malaria incidence and prevalence between 2010–2016 confirm the heavy malaria burden for the population living in the western part of Madagascar [3, 4, 11]. In the Tsiroanomandidy district which constitutes a bridge area between the low transmission Central Highlands and the high endemic western region, both P. falciparum and P. vivax circulate. In this area, malaria prevalence and Anopheles species distribution have been described in detail [7, 12, 13]. Such combined parasitological and entomological information is critical for developing effective strategies for interrupting malaria transmission and moving towards malaria elimination which has been set up on the agenda of the 2018–2022 Malagasy Malaria Strategic Plan as geographically progressive elimination. Toward this goal, is reported here a combined parasitological and entomological survey in the Maevatanana district, located in the northwestern ecozone of Madagascar, and which faces a high malaria burden due to both P. falciparum and P. vivax. The study was conducted in two neighbouring villages in Andriba, a rural commune located at the transition between the western fringe of the Central Highlands (low malaria prevalence) and the northwestern ecozone (moderate to high prevalence) according to Howes et al. [3]. The main goal of this study was to determine which Anopheles species contribute to the local malaria transmission with the aim for providing targeted vector control recommendation to the local authorities. To our knowledge, no such combined parasitological and entomological survey has ever been performed in that region.
Methods
Study design and setting
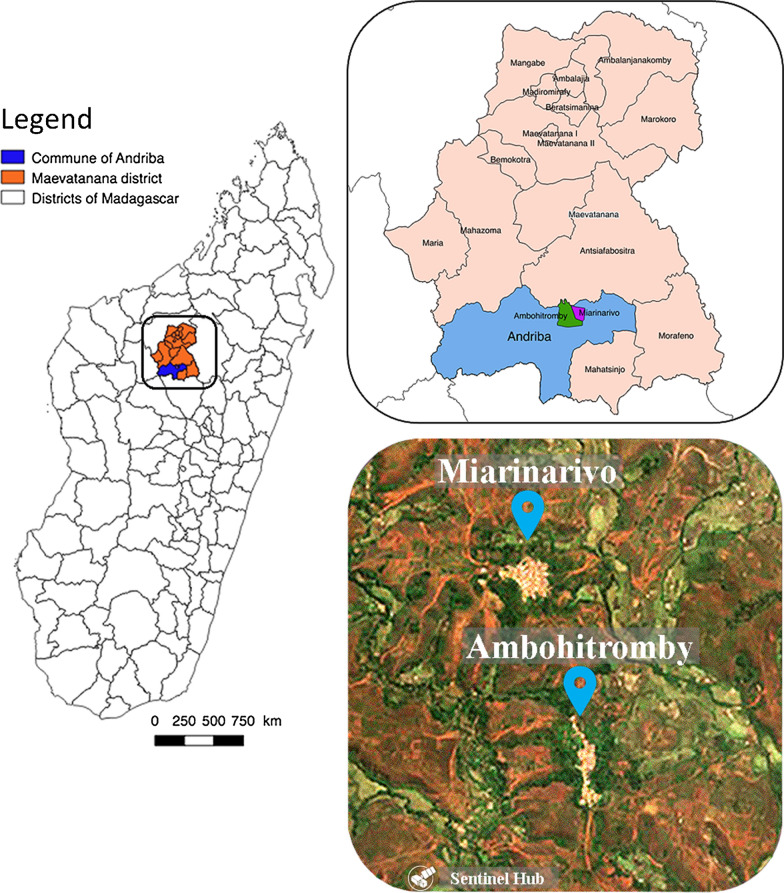
The study was conducted in two villages of the rural commune of Andriba (Maevatanana district, Madagascar) which is located in the tropical northwest region of Madagascar. Andriba is characterised by a dry season that generally lasts from April to October and a rainy season from November to April; the average annual temperature is 24 °C and the average annual rainfall is 1828 mm [14]. The two villages, Ambohitromby (17°34′23.7″S, 46°55′21.4″E) and Miarinarivo (17°33′56.7″S, 46°55′10.8″E) are located 1.5 km apart and 6 km from Andriba town hall (Fig. 1), with a population of 384 and 302 inhabitants, respectively. The houses in the two villages were of typical Malagasy construction common in the rural areas of the Maevatanana district: thatched roofs, adobe walls and composed of 1 to 2 rooms (Fig. 2). Parasitological and entomological data were collected at three time points during a single malaria transmission season: at the onset of the season (November and December 2016, labelled T1); mid-season (February 2017, labelled T2); and late-season (April and May 2017, labelled T3). The latter time point was selected to correspond with the cessation of malaria transmission, but this depends upon annual climatic variation. Additional parasitological data were recorded in March 2016 and in March 2018 as part of active malaria parasite surveillance in school-age children (Bourgouin et al. unpublished data).
Fig. 1.
Study site. The map of Madagascar is depicted in the left panel with a focus on the Andriba region presented in more details in the upper right panel. The bottom right panel is a satellite image of the study villages, Ambohitromby and Miarinarivo
(Modified Copernicus Sentinel data [2019]/Sentinel Hub)
Fig. 2.

Typical Malagasy houses in Andriba rural area. The houses are built with adobe walls and thatched roofs, and usually composed of one or two rooms. The picture was taken in the village of Ambohitromby located in the rural commune of Andriba, Madagascar
Malaria prevalence in the human population
Blood sample collection
Study participants included village residents among whom volunteers were involved in human landing catches (HLCs). The week before each survey, a community sensitisation was organized with the help of the staff of the Andriba health centre and the presence of the head of the village explaining the benefit of malaria parasite detection. On the day of the survey, people gathered at a central point proposed by the head of the village, usually the local school. Each participant was provided a short questionnaire to collect name, age, sex, and any health conditions that might exclude them from the survey (such as dizziness or heavy treatment; neither of them was encountered). Blood samples were collected from both children and adults without clinical signs of malaria at the time of the survey (asymptomatic individuals). According to the study protocol, any individual who would have come with malaria symptoms would have been referred to the Andriba health centre. We did not encounter such a situation. Temperature and weight where recorded for each participant showing a positive rapid diagnostic test (RDT) for malaria. Participant’s names were used for longitudinal follow-up of their participation at the next surveys, but names were encoded for data analysis.
Blood samples were obtained by finger prick to perform RDTs (SD Bioline Malaria Ag P.f/Pan; Standard Diagnostics Inc., Suwon City, South Korea), thick and thin blood smears, and blood spots on filter paper (standard Whatman 3MM filter paper). The Bioline RDT enabled specific detection of P. falciparum, and any of P. vivax, P. ovale and P. malariae, as all four species are present in Madagascar. Plasmodium species, parasite stages and parasite density were further determined by microscopic observation of the blood smears stained with 10% Giemsa, using a light microscope (100×). A thin blood smear slide was declared malaria-negative when Plasmodium parasites were not detected after examination of 100 high power microscopic fields. Slides were read for asexual parasites and gametocytes, enumerated against 500 leucocytes and expressed as density/μl assuming an average leucocyte count of 8,000/μl of blood (data not shown). Individuals with positive RDT were treated with artemisinin-based combination therapy (ACT), according to national guidelines.
Detection of Plasmodium parasites by PCR using dried blood spots
Dried blood spots (DBS) were lysed overnight at 4 °C in 150 μl per microtube of 1× HBS buffer (Hepes buffered saline) supplemented with 0.5% saponin, final concentration. Samples were then washed twice with 1× PBS and DNA extracted with Instagene® Matrix resin (Bio-Rad Laboratories Inc., Hercules, California, USA) according to manufacturer’s instructions. Molecular detection and species identification of Plasmodium parasites were performed in two steps as previously described by Canier et al. [15]. Plasmodium spp. were first detected by a real-time PCR using genus-specific primers targeting the Plasmodium cytochrome b gene. Then, Plasmodium species identification was performed on DNA samples identified as positive for Plasmodium using a nested real-time PCR assay [15].
Entomological data
Mosquito collection
Mosquitoes were collected by HLCs and indoor pyrethrum spray catches (PSCs), following WHO protocols [16]. For both HLCs and PSCs, houses were chosen randomly at the first time point, and then according to the availability of the houses. Therefore, some houses were sampled with repetition. For each of the three surveys throughout the malaria season, adult volunteers performed HLCs from 18:00 h to 06:00 h in 2 houses per night, for 3 consecutive nights. For each house, one volunteer sat inside and another outside at more than 15 m from the house; capture stations in the village were distributed ensuring a distance of more than 15 m between volunteers. The indoor and outdoor volunteers changed places every hour to minimise individual bias. The entire night was worked in two shifts, with two volunteers working between 18:00 h and 24:00 h, and a second set of volunteers from 24:00 h to 06:00 h. Thus, for each sampling period, 12 human-nights (HNs) of data were collected from each village, with a total of 72 HNs for the entire study.
PSCs were conducted in 5 houses/day/village, choosing houses that were not used for HLCs and in which no insecticide or repellent had been used during the previous week. Some of the houses were using insecticide-treated bed nets. The GPS (global positioning system) coordinates of each house was recorded as well as the date at which the PSC was performed. PSCs were done on 3 consecutive days from 06:00 h to 08:00 h each morning following the HLCs. No insecticide residual spraying had occurred in the villages from 2016 till 2018. PSCs were performed using a pyrethroid mixture of prallethrin, tetramethrin, and deltamethrin. Spraying was performed from outside of the houses, into openings, holes in walls and eaves, then in the rooms, following WHO procedures. Knockdown mosquitoes were then collected by hand picking.
Mosquito species identification
All mosquitoes were identified morphologically using the determination keys of Grjebine [17] and De Meillon [18]. To discriminate An. gambiae from An. arabiensis, a TaqMan assay was used targeting the intergenic spacer region of rDNA as described by Walker et al. [19], following the initial work of Scott et al. [20]. Primers and sequences used are listed in Additional file 1: Table S1. PCR reactions (20 μl) contained 5 μl of genomic DNA (see extraction procedure below), 4 μl of 5× HOT FIREPol® Probe qPCR Mix Plus/no ROX (Solis Biodyne, Tartu, Estonia), 300 nM of each primer and 200 nM of each probe. Reactions were run on a StepOnePlus (Applied Biosystems, Waltham, Massachusetts, USA) using the following temperatures: an initial step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 20 s and annealing/elongation at 60 °C for 1 min.
Plasmodium detection in Anopheles mosquitoes
DNA extraction and quality control
Genomic DNA from Anopheles head-thorax was extracted using the DNAzol® Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Briefly, the head-thorax from each mosquito was put individually in a tube; care was taken to rinse the dissecting equipment in 70% ethanol between each mosquito. A volume of 150 µl of DNAzol was added and mosquito tissues were crushed using an individual conical plastic pestle. DNA was then extracted following the manufacturer’s protocol. After precipitation, the DNA pellet was suspended in a final volume of 50 µl of nuclease-free water. DNA quality was controlled using a SYBR Green real-time PCR assay targeting the ribosomal S7 protein encoding gene. This gene is highly conserved among species belonging to the same genus. Primers previously designed against the An. gambiae S7 gene [21] were aligned against all available Anopheles S7 sequences to ensure that those primers will efficiently amplify the S7 gene fragment from any Anopheles captured in the field. Amplification conditions were validated on a subset of laboratory and field-collected mosquito samples including species of the subgenera Anopheles, Cellia and Nyssorhynchus (not shown). Amplification using the PowerSYBER® Green Master mix (Applied Biosystems) was performed as follows: an initial step at 95 °C for 15 min; followed by 40 cycles of denaturation at 95 °C for 45 s, annealing at 55 °C for 30 s and elongation at 60 °C for 45 s. Specificity of the amplification was assessed by viewing the melting curves.
Plasmodium detection
The detection of human Plasmodium gDNA in mosquitoes was performed in 2 steps. The first step used a TaqMan PCR assay targeting a region of the 18S rRNA gene conserved among the human infecting Plasmodium species. For this assay, primers and probe previously described [22] have been used, with a MGB probe as in Taylor et al. [23]; this combination was previously validated for Plasmodium detection in mosquitoes [24]. The Plasmodium TaqMan probe was labelled with 5’ NED. PCR reactions (20 μl) contained 5 μl of mosquito genomic DNA, 4 μl of 5× HOT FIREPol® Probe qPCR Mix Plus/no ROX (Solis Biodyne), 300 nM of each primer and 200 nM of probe. Reactions were run on a StepOnePlus (Applied Biosystems) using the following conditions: an initial step at 95 °C for 10 min; followed by 50 cycles of denaturation at 95 °C for 20 s and annealing/elongation at 60 °C for 1 min. Plasmodium falciparum genomic DNA extracted from NF54 parasite cultures was used as a positive control. Any sample with amplification signal before the 38th cycle was considered positive. For high throughput screening, a pool strategy was used [25]; equal volumes of genomic DNA (extracted as described above) from 6 mosquitoes of the same species were pooled. The Plasmodium TaqMan assay was run using 5 µl of each DNA pool in triplicate. Mosquitoes from positive pools were then analysed individually using the same protocol as for pools. All samples positive in the TaqMan assay were then analysed for the identification of P. falciparum and P. vivax species. For each positive sample, 2 distinct real-time SYBR Green PCR assays were done using species-specific primers targeting the cytochrome b gene. Purified gDNA from P. falciparum and P. vivax were used as positive controls. Each reaction was run in triplicate. The real-time PCR conditions were as previously described [15]: an initial step at 95 °C for 15 min; followed by 40 cycles of denaturation at 95 °C for 20 s and annealing/elongation at 60 °C for 1 min. Sequences of the primers and TaqMan probes used for the Plasmodium detection in Anopheles mosquitoes are listed in Additional file 1: Table S1.
Statistical analysis
Plasmodium spp. prevalence rates, as determined by microscopy, RDT and PCR were analysed by fitting a generalized linear mixed model (GLMM) with binomial error structure (i.e. a logistic regression) with individual as the random factor and village, time point (and their interaction), age and sex as explanatory variables. Total mosquito numbers from HLC were analysed by fitting a GLMM with Poisson error structure (i.e. a loglinear regression) with house identification number as the random factor and village, time point (and their interaction) as explanatory variables. For endophagy, mosquito species-specific analyses were similarly performed individually for all mosquito species for which more than a grand total of 50 individual mosquitoes were collected. In addition to village and time point (and their interaction), place of capture (indoors vs outdoors) was also included in the model. All GLMM analyses were performed in GenStat version 15 (GenStat for Windows 15th Edition, VSN International Ltd., Hemel Hempstead, UK.). To account for any underdispersion or overdispersion in the data, a dispersion heterogeneity was used in the analyses.
Results
Plasmodium carriage in the human population
The parasitological survey involved 380 individuals (218 in Ambohitromby and 162 in Miarinarivo, fairly representative of more than 50% of the population, ranging from 5 months to 68 years-old (Additional file 1: Table S2). No participant with clinical signs of malaria was encountered during the surveys. A total of 590 samples (351 in Ambohitromby and 239 in Miarinarivo) were analysed by RDT, microscopy and real-time PCR. Human malaria prevalence was 8.0% by RDT, 4.8% by microscopy and 11.9% by real-time PCR over the whole study (Table 1). There were no significant associations for any variables for Plasmodium spp. prevalence rates when determined by microscopy or RDT. However, there was a significantly higher prevalence rate as determined by PCR at T2 ( = 7.46, P = 0.025). There were no differences between villages, nor by age or sex. Except for T3 in Miarinarivo, the PCR technique, as expected, was able to detect a greater number of parasite carriers over RDT and microscopy, revealing a substantial proportion of sub-microscopic parasite carriers. The lower proportion of PCR-positive samples at T3 in Miarinarivo, compared to T1 and T2, might result from inadequate conservation of the blood spots (12 of them) before PCR processing.
Table 1.
Prevalence of Plasmodium infections in asymptomatic individuals assessed by RDT, microscopy and real-time PCR
| RDT | Microscopy | Real-time PCR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | Total | T1 | T2 | T3 | Total | T1 | T2 | T3 | Total | ||
| Ambohitromby | n | 173 | 79 | 99 | 351 | 173 | 79 | 99 | 351 | 173 | 79 | 99 | 351 |
| Positive | 13 | 10 | 5 | 28 | 9 | 3 | 3 | 15 | 20 | 12 | 10 | 42 | |
| Prevalence (%) | 7.5 | 12.7 | 5.1 | 8.0 | 5.2 | 3.8 | 3.0 | 4.3 | 11.6 | 15.2 | 10.1 | 12.0 | |
| Miarinarivo | n | 122 | 65 | 52 | 239 | 122 | 65 | 49 | 236 | 122 | 65 | 52a | 239 |
| Positive | 9 | 7 | 3 | 19 | 7 | 3 | 3 | 13 | 14 | 12 | 2 | 28 | |
| Prevalence (%) | 7.4 | 10.8 | 5.8 | 7.9 | 5.7 | 4.6 | 6.1 | 5.5 | 11.5 | 18.5 | 3.8 | 11.7 | |
| Prevalence of Plasmodium infections in the two villages | 7.5 | 11.8 | 5.3 | 8.0 | 5.4 | 4.2 | 4.1 | 4.8 | 11.5 | 16.7 | 7.9 | 11.9 | |
a12 blood spots on filter paper have not been properly preserved. The same individuals were involved during the transversal parasitological study including the three methods of malaria diagnostic
Notes: In Ambohitromby, a total of 173, 79 and 99 samples were analysed for T1, T2 and T3 respectively. In Miarinarivo, a total of 122, 65 and 52 samples were analysed for T1, T2 and T3, respectively, except at T3 were only 49 smears were read as 3 slides were unreadable
Abbreviations: n, sample size
Among the 70 positive samples identified by real-time PCR, 84.3% carried P. falciparum, 5.7% carried P. vivax, 1.4% carried P. malariae and 8.6% carried mixed infections always involving P. falciparum (Table 2). All mixed infections were observed in Ambohitromby.
Table 2.
Plasmodium species detected by real-time PCR in asymptomatic individuals at the three time points (T1-T3)
| Ambohitromby | Miarinarivo | Total by species (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | Total | T1 | T2 | T3 | Total | ||
| Sample size | 173 | 79 | 99 | 351 | 122 | 65 | 52 | 239 | 590 |
| P. falciparum | 17 | 9 | 7 | 33 | 13 | 11 | 2 | 26 | 59 (84.3) |
| P. vivax | 1 | 1 | 1 | 3 | 0 | 1 | 0 | 1 | 4 (5.7) |
| P. malariae | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 (1.4) |
| Mixed infectiona | 2 | 2 | 2 | 6 | 0 | 0 | 0 | 0 | 6 (8.6) |
| Total by village and time point (%) | 20 (11.6) | 12 (15.2) | 10 (10.1) | 42 (12.0) | 14 (11.5) | 12 (18.5) | 2 (3.8) | 28 (11.7) | 70 (100) |
aThe 6 mixed infections implicate P. falciparum with P. vivax (4), P. malariae (1) and P. ovale (1)
Mosquito species and behaviour
Abundance and diversity
In total, 2407 mosquitoes were collected during 72 HNs in Ambohitromby and Miarinarivo. As presented in Table 3, Anopheles was the most abundant mosquito genus collected (68.55%, n = 1650) followed by Culex (26.87%, n = 647), Mansonia (3.49%, n = 84), Aedes (0.99%, n = 24) and Coquillettidia (0.08%, n = 2). Among Anopheles, An. coustani was by far the most abundant representing 52.99% (751/1417) of the known potential malaria vectors in Madagascar, followed by An. arabiensis (28.93%, 410/1417), An. funestus (12.84%, 182/1417) and An. mascarensis (4.66%, 66/1417); An. gambiae was barely represented (0.56%, 8/1417). Detailed data covering all collected species are presented in Additional file 1: Table S3.
Table 3.
Mosquitoes collected by HLCs in Ambohitromby and Miarinarivo at the three time points T1-T3)
| Mosquito species | Ambohitromby | Miarinarivo | Total by species | Proportion (in %) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | Total | T1 | T2 | T3 | Total | |||
| Anopheles coustani a | 75 | 42 | 162 | 279 | 66 | 97 | 309 | 472 | 751 | 31.20 |
| Anopheles arabiensis a,b | 64 | 207 | 10 | 281 | 16 | 81 | 32 | 129 | 410 | 17.03 |
| Anopheles funestus a | 34 | 26 | 40 | 100 | 9 | 14 | 59 | 82 | 182 | 7.56 |
| Anopheles squamosus/cydippis | 13 | 43 | 29 | 85 | 8 | 40 | 15 | 63 | 148 | 6.15 |
| Anopheles mascarensis a | 24 | 13 | 12 | 49 | 5 | 2 | 10 | 17 | 66 | 2.74 |
| Anopheles rufipes | 9 | 10 | 0 | 19 | 5 | 11 | 9 | 25 | 44 | 1.83 |
| Anopheles maculipalpis | 7 | 9 | 0 | 16 | 10 | 9 | 5 | 24 | 40 | 1.66 |
| Anopheles gambiae a,b | 0 | 4 | 0 | 4 | 1 | 2 | 1 | 4 | 8 | 0.33 |
| Anopheles pretoriensis | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0.04 |
| Total Anopheles | 226 | 354 | 253 | 833 | 120 | 256 | 441 | 817 | 1650 | 68.55 |
| Culex antennatus | 30 | 258 | 4 | 292 | 49 | 99 | 25 | 173 | 465 | 19.32 |
| Culex quinquefasciatus | 5 | 103 | 0 | 108 | 15 | 23 | 19 | 57 | 165 | 6.86 |
| Culex giganteus | 0 | 0 | 0 | 0 | 2 | 4 | 2 | 8 | 8 | 0.33 |
| Culex univittatus | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 4 | 0.17 |
| Culex decens | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 3 | 0.12 |
| Culex bitaeniorhyncus | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 0.08 |
| Total Culex | 40 | 361 | 4 | 405 | 69 | 127 | 46 | 242 | 647 | 26.88 |
| Mansonia uniformis | 8 | 9 | 3 | 20 | 5 | 25 | 34 | 64 | 84 | 3.49 |
| Total Mansonia | 8 | 9 | 3 | 20 | 5 | 25 | 34 | 64 | 84 | 3.49 |
| Aedes tiptoni | 1 | 0 | 0 | 1 | 8 | 0 | 1 | 9 | 10 | 0.42 |
| Aedes skusea | 5 | 0 | 0 | 5 | 3 | 0 | 0 | 3 | 8 | 0.33 |
| Aedes albopictus | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | 0.12 |
| Aedes vittatus | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0.08 |
| Aedes circumlateolus | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0.04 |
| Total Aedes | 7 | 2 | 0 | 9 | 12 | 1 | 2 | 15 | 24 | 1.00 |
| Coquillettidia grandidieri | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 0.08 |
| Total Coquillettidia | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 0.08 |
| Total by village and time point | 281 | 726 | 260 | 1267 | 206 | 409 | 525 | 1140 | 2407 | _ |
aKnown potential malaria vectors in Madagascar
bAn. arabiensis and An. gambiae were identified by TaqMan assay among all An. gambiae (s.l.) collected (see Methods section)
Note: The proportion is equal to the total by species divided by the total of all species collected (n = 2407)
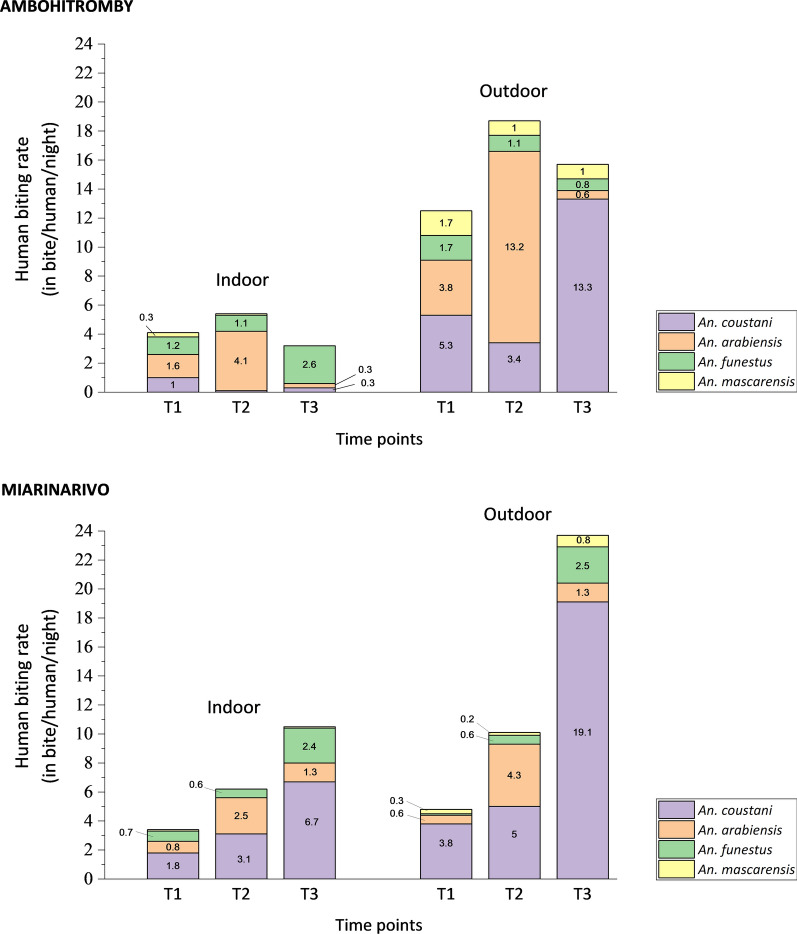
Analysis of total Anopheles caught by HLC revealed significant associations with time point and an interaction with village. The number of Anopheles was higher at T2 ( = 64.8, P < 0.001), especially in Ambohitromby, whereas at T1 and T3, numbers were higher in Miarinarivo (interaction term = 14.92, P < 0.001). The human-biting rate (HBR), corresponding to the number of collected mosquitoes per number of human-nights, was determined for the known potential malaria vectors, but An. gambiae due to the low number of captured mosquitoes (n = 8). Results over time and for each village are depicted in Fig. 3. Overall Anopheles HBR varies in each village over the time course of the survey, from 8 bites per human and per night (b/h/n) to 34.3 b/h/n. However, the bite frequency was higher at T2 in Ambohitromby, while being the highest at T3 in Miarinarivo. Strikingly, An. arabiensis was the malaria vector species with the highest HBR in Ambohitromby, with 17.3 b/h/n at T2, while in Miarinarivo, it was An. coustani with 25.9 b/h/n at T3. Nevertheless, An. coustani exhibited the highest biting frequency in both villages at T3, which corresponds to the time where the rice reached full maturity providing high shade on the padding fields suitable for An. coustani larval development.
Fig. 3.
Indoor and outdoor human-biting rate of malaria vectors at the three time points in Ambohitromby and Miarinarivo. Light numbers within the graphs indicate the mean bite per human and per night for each of the four Anopheles species
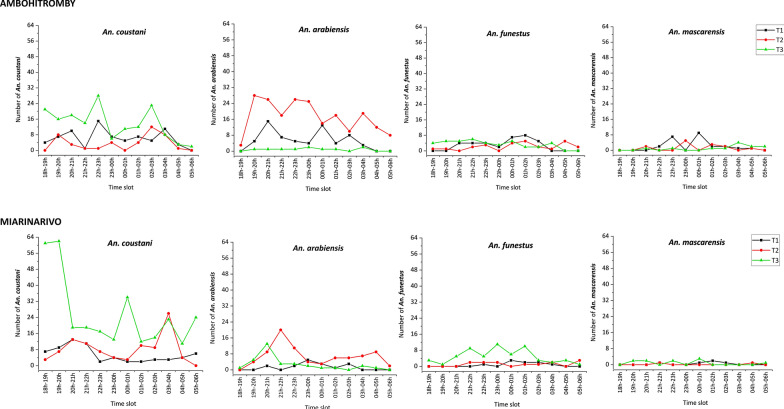
Looking at the hourly biting rate of the four potential malaria vectors across time-points in the two villages, the four mosquito species mostly bit all night long (Fig. 4). There were however variations in the biting pattern across the night and according to the time point, especially for An. coustani and An. arabiensis. Considering An. coustani, the HBR in the two villages was higher at T3 compared to T1 and T2. The highest HBR peak at T1 was between 22:00–23:00 h in Ambohitromby and between18:00–20:00 h in Miarinarivo. The HBR peak at T2 was higher in Miarinarivo compared to Ambohitromby and occurred between 03:00–04:00 h. At T3, the biting pattern over the night was overall similar between the two villages. The An. arabiensis HBR in the two villages was higher at T2 compared to T1 and T3. The highest HBR peak occurred between 19:00–20:00 h in Ambohitromby and between 21:00–22:00 h in Miarinarivo. An. funestus and An. mascarensis showed the lowest HBR peak in the two villages with the same biting pattern along the night.
Fig. 4.
Hourly biting rate of malaria vectors at the three time points in Ambohitromby and Miarinarivo. Data represent both indoor and outdoor HLCs collected mosquitoes
Endophagy rate of malaria vectors
Endophagy rates, representing the proportion of mosquitoes collected indoors over the total number of mosquitoes collected indoors and outdoors by HLCs, are summarized in Table 4 for the four more abundant potential malaria vector species, according to the time-course of the survey and for each village. Anopheles funestus exhibited the highest endophagy rate in both villages (58.00 ± 4.94% in Ambohitromby and 53.66 ± 5.51% in Miarinarivo). The lowest endophagy rate in Ambohitromby was exhibited by An. coustani (5.73 ± 1.39%), while it was An. mascarensis that exhibited the lowest endophagy rate in Miarinarivo (11.76 ± 7.81%). Anopheles arabiensis and An. coustani, known as zoo-anthropophilic species, exhibited higher endophagy rates in Miarinarivo (42.64 ± 4.35% and 29.24 ± 2.09%, respectively) compared to Ambohitromby (25.27 ± 2.59% and 5.73 ± 1.39%, respectively). Comparing the endophagy rate by mosquito species, village and time point revealed a number of associations. All Anopheles species, except for An. funestus which did not differ significantly between indoors and outdoors, were collected in higher numbers outdoors, while Anopheles arabiensis and An. coustani were collected in higher numbers at T2 and T3, respectively. From 17 PSCs, a total of 70 mosquitoes were collected resting indoors (Additional file 1: Table S4). Among those 42.10% were An. funestus, 31.57% An. arabiensis, 15.78% An. coustani and 10.52% An. mascarensis in Ambohitromby, while only An. funestus (96.15%) and An. mascarensis (3.84%) were collected resting indoors in Miarinarivo.
Table 4.
Proportion (in %) of the malaria vectors collected biting indoor by HLCs (endophagic rate)
| Species | Ambohitromby | Miarinarivo | ||||||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | Average proportion ± SE | T1 | T2 | T3 | Average proportion ± SE | |
| An. coustani | 16.00 (12/75) | 2.38 (1/42) | 1.85 (3/162) | 5.73 ± 1.39 | 31.82 (21/66) | 38.14 (37/97) | 25.89 (80/309) | 29.24 ± 2.09 |
| An. arabiensis | 29.69 (19/64) | 23.67 (49/207) | 30.00 (3/10) | 25.27 ± 2.59 | 56.25 (9/16) | 37.04 (30/81) | 50.00 (16/32) | 42.64 ± 4.35 |
| An. funestus | 41.18 (14/34) | 50.00 (13/26) | 77.50 (31/40) | 58.00 ± 4.94 | 88.89 (8/9) | 50.00 (7/14) | 49.15 (29/59) | 53.66 ± 5.51 |
| An. mascarensis | 16.67 (4/24) | 7.69 (1/13) | 0 (0/12) | 10.20 ± 4.32 | 20.00 (1/5) | 0 (0/2) | 10.00 (1/10) | 11.76 ± 7.81 |
Notes: Numbers in parenthesis represent the number of mosquitoes collected indoor over the total number of mosquitoes collected indoor and outdoor. Anopheles gambiae was not taken into account in this table due to its low number
Abbreviation: SE, standard error = sqrt (p(1 – p)/n)
Plasmodium carriage in Anopheles mosquitoes and entomological inoculation rate
Among 1715 anopheline mosquitoes captured by HLCs (n = 1650) and PSCs (n = 65), 1550 were tested for the presence of Plasmodium sporozoites by TaqMan and SYBR Green assays. As described in the methods section, DNA was first extracted from the head-thorax of individual mosquitoes and its quality assessed by amplification of the S7 gene. Using an equal volume of gDNA from at most 6 mosquitoes of the same species, 261 pools were assembled and tested for the presence of Plasmodium DNA, using the Plasmodium TaqMan assay. A total of 23 pools were found positive for Plasmodium DNA. Deconvolution of each positive pool to individual mosquito revealed that 28 mosquitoes carried Plasmodium DNA in their head-thorax, all of which had been captured by HLCs only. However, the SYBR Green assay for P. falciparum/P. vivax species detection was conclusive only for 13 (out 28) mosquitoes carrying either P. falciparum or P. vivax parasite (Table 5). The Plasmodium species of the 15 remaining TaqMan Plasmodium-positive mosquitoes could not be identified possibly due to the less effective SYBR Green assay used. It nevertheless cannot be excluded that these mosquitoes were infected with P. malariae or P. ovale or even lemur parasites as the TaqMan assay was targeting a region of the 18S gene highly conserved among Plasmodium species. Overall, 9 mosquitoes were positive for P. falciparum and 4 for P. vivax. These mosquitoes belong to three anopheline species: An. funestus; An. arabiensis; and An. coustani (with the latter species being the more frequently infected one) (Table 5). Based on the species-specific assay (SYBR Green), the sporozoite rate (SR) varied from 0 to 1.4% according to the Anopheles species. Including all Anopheles species and all mosquitoes captured by HLCs and PSCs the overall SR was 0.84%. Of note, An. rufipes, an anopheline species which is not known being a malaria vector in Madagascar, was found positive by the TaqMan Plasmodium assay (2/40). As there are increased reports on its role in malaria transmission in other countries [26–28], it might be worth to include this species for Plasmodium sporozoite carriage in future surveillance programs.
Table 5.
Plasmodium carriage in Anopheles mosquitoes analysed in pools and individually
| Species | Total screened | Pools analysed | Positive pools | Positive mosquitoes | Positive in species screening | Plasmodium species (n) | Sporozoite rate (SR) (%) |
|---|---|---|---|---|---|---|---|
| An. coustani | 714 | 122 | 10 | 14 | 7 | Pf (6); Pv (1) | Pf (0.84); Pv (0.14) |
| An. gambiae (s.l.) | 374 | 60 | 8 | 9 | 3 | Pf (1); Pv (2) | Pf (0.27); Pv (0.54) |
| An. funestus | 212 | 36 | 3 | 3 | 3 | Pf (2); Pv (1) | Pf (0.94); Pv (0.47) |
| An. squamosus | 116 | 20 | 0 | 0 | 0 | _ | 0 |
| An. mascarensis | 59 | 10 | 1 | 0 | 0 | _ | 0 |
| An. rufipes | 40 | 7 | 1 | 2 | 0 | _ | 0 |
| An. maculipalpis | 35 | 6 | 0 | 0 | 0 | _ | 0 |
| Total | 1550 | 261 | 23 | 28 | 13 | Pf (9); Pv (4) | Pf (0.58); Pv (0.26) |
Abbreviations: n, number of samples positive to Plasmodium species-specific; Pf, P. falciparum; Pv, P. vivax
Notes: Mosquitoes analysed include those collected by both HLCs and PSCs
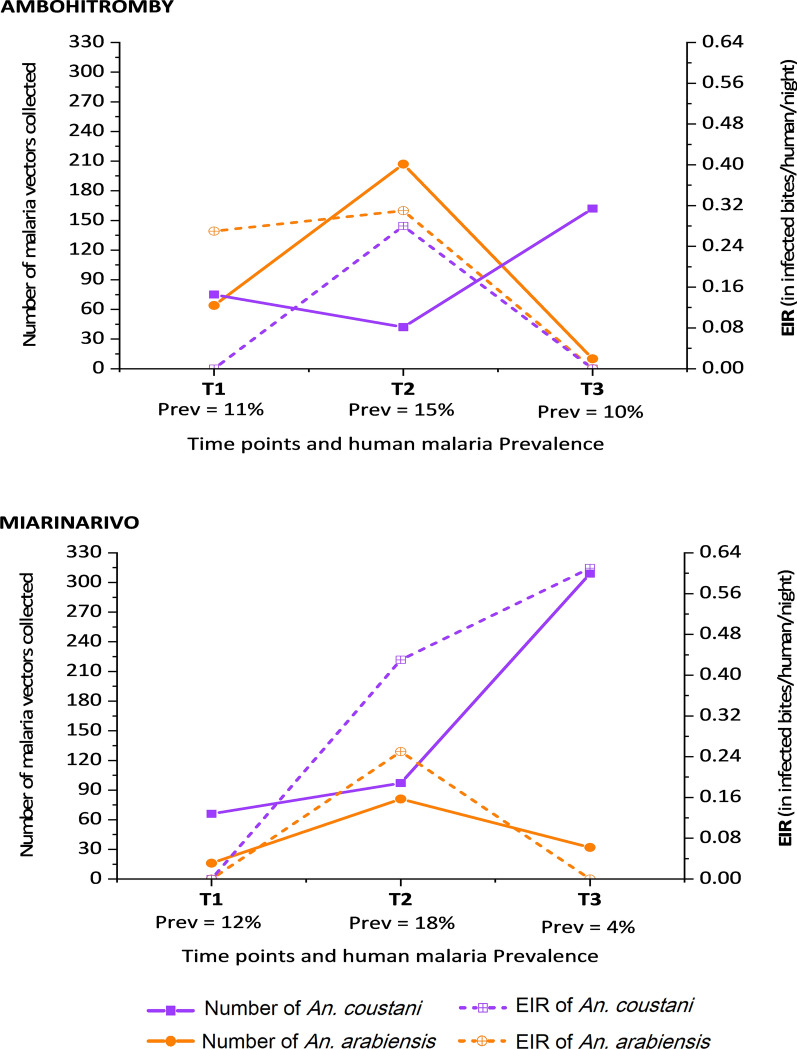
When looking at the entomological inoculation rate (EIR) as a proxy for malaria transmission, it appears that An. arabiensis and An. coustani contribute most to malaria transmission, but with striking differences between the two villages and over time (Table 6). Indeed, in Ambohitromby, An. arabiensis was the main vector at the beginning (T1) and the mid-term (T2) of transmission season, with EIRs of 0.27 and 0.31 ib/h/n respectively, followed by An. coustani at T2 (0.28 ib/h/n). By contrast, in Miarinarivo, An. coustani was the main vector at T2 and T3 (EIRs of 0.43 and 0.61 ib/h/n respectively), followed by An. arabiensis that played a vector role at T2 only (EIR of 0.25 ib/h/n). Plotting the number of An. arabiensis and An. coustani, and their respective EIR, show that the EIRs are not proportional to the number of captured mosquitoes and that this is evidenced in the two villages over the transmission season (Fig. 5). In Ambohitromby, An. arabiensis and An. coustani showed comparable densities at T1 with different EIRs (0.27 and 0 ib/h/n respectively); while their density at T2 and T3 was greatly different, but the EIR was similar for both species. In Miarinarivo, despite similar density of An. arabiensis and An. coustani at T2, An. coustani contributed most to malaria transmission and maintained this role at T3 with increased EIR possibly associated to its higher density.
Table 6.
Entomological indices of malaria vectors at the three time points in the two villages
| Species | Ambohitromby | Miarinarivo | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time point | HBR | n | (+) | SR (%) | EIR | Time point | HBR | n | (+) | SR (%) | EIR | |
| Anopheles coustani | T1 | 6.25 | 68 | 0 | 0 | 0 | T1 | 5.50 | 65 | 0 | 0 | 0 |
| T2 | 3.50 | 25 | 2 | 8.00 | 0.28 | T2 | 8.08 | 95 | 5 | 5.26 | 0.43 | |
| T3 | 13.50 | 159 | 0 | 0 | 0 | T3 | 25.75 | 297 | 7 | 2.36 | 0.61 | |
| Total | 7.75 | 252 | 2 | 0.40 | 0.03 | Total | 13.11 | 457 | 12 | 2.63 | 0.34 | |
| Anopheles arabiensis | T1 | 5.33 | 58 | 3 | 5.08 | 0.27 | T1 | 1.33 | 16 | 0 | 0 | 0 |
| T2 | 17.25 | 166 | 3 | 1.78 | 0.31 | T2 | 6.75 | 81 | 3 | 3.70 | 0.25 | |
| T3 | 0.83 | 10 | 0 | 0 | 0 | T3 | 2.67 | 32 | 0 | 0 | 0 | |
| Total | 7.81 | 234 | 6 | 2.52 | 0.20 | Total | 3.58 | 129 | 3 | 2.34 | 0.08 | |
| Anopheles funestus | T1 | 2.83 | 35 | 1 | 2.86 | 0.08 | T1 | 0.75 | 14 | 0 | 0 | 0 |
| T2 | 2.17 | 28 | 0 | 0 | 0 | T2 | 1.17 | 16 | 1 | 6.25 | 0.07 | |
| T3 | 3.33 | 42 | 0 | 0 | 0 | T3 | 4.92 | 77 | 1 | 1.30 | 0.06 | |
| Total | 2.78 | 105 | 1 | 0.95 | 0.03 | Total | 2.38 | 107 | 2 | 1.87 | 0.04 | |
| All 3 species | T1 | 14.42 | 163 | 4 | 2.45 | 0.35 | T1 | 7.58 | 94 | 0 | 0 | 0 |
| T2 | 22.92 | 222 | 5 | 2.25 | 0.52 | T2 | 16.00 | 192 | 9 | 4.69 | 0.75 | |
| T3 | 17.67 | 215 | 0 | 0 | 0 | T3 | 33.33 | 406 | 8 | 1.97 | 0.66 | |
| Total | 18.33 | 600 | 9 | 1.50 | 0.27 | Total | 18.97 | 692 | 17 | 2.46 | 0.47 | |
Abbreviations: HBR, human-biting rate (the number of collected mosquitoes per number of human-night (12 at each time point in each village)). The HBR is expressed in bite/human/night (b/h/n); n, the number of anopheline mosquitoes collected by HLCs and analysed by real time PCR. The DNA was extracted from individual head-thorax; (+) corresponds to the number of Plasmodium positive samples confirmed by the TaqMan 18S; SR, sporozoite rate (the number of positive samples divided by the number of analysed samples (n), in %); EIR, entomological inoculation rate (EIR = HBR × SR). It is expressed as infective bite/human/night (ib/h/n)
Fig. 5.
Variation of the density and EIR of An. arabiensis and An. coustani over time in Ambohitromby and Miarinarivo. Prev: human malaria prevalence
Discussion
The objective of this study, conducted in two neighbouring villages in a region of Madagascar where malaria is still a high public health problem, was to estimate the level of malaria transmission and to identify the mosquito vector species involved. Indeed, in that region, no such study had ever been conducted despite the high number of patients diagnosed with malaria at the local health centres. To our knowledge, the only study carried out at the same site, just provided entomological data and goes back to 1992 [29].
Similarity in human malaria prevalence in the two villages
Parasitological data in the asymptomatic villagers revealed malaria infection cases mainly due to P. falciparum, in addition to low levels of P. vivax and P. malariae. The overall malaria prevalence throughout the study period was 11.9% as determined by PCR, and 4.8% by microscopy. These values are similar to the ones reported in the Tsiroanomandidy study performed in March 2014 [13]. Like the Tsiroanomandidy study, the results of this study highlight the high prevalence of sub-microscopic Plasmodium carriage which represents 50–75% of the investigated cases (negative by microscopy, positive by PCR).
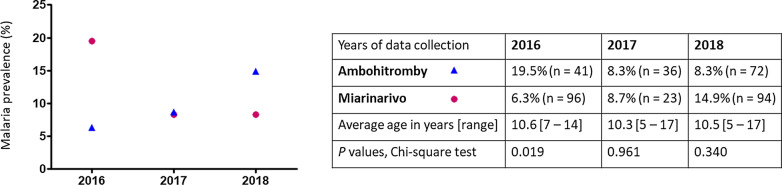
Data from this study show a similar malaria prevalence in the two villages. This is in sharp contrast to the significant difference in prevalence that was observed in 2016, between the school-age children of Ambohitromby and Miarinarivo (our unpublished data). Indeed, in 2016 a significant difference (P = 0.019, Chi-square test) in malaria RDT prevalence was observed between Ambohitromby (19.5%, n = 41) and Miarinarivo (6.3%, n = 96), but not in 2017 (this study) nor 2018 (Bourgouin et. al. unpublished data, Fig. 6). Such variation in malaria prevalence across years in the two villages, might result from better mosquito net coverage of the populations or climatic and ecological changes impacting Anopheles density [30, 31]. However, no mosquito net distribution was done between 2016 and 2018 in Andriba. Therefore, it might be possible that, in Ambohitromby 2016, the local conditions facilitated the development of an increased number of mosquito breeding sites leading to increased Anopheles vector population size and subsequent increased transmission. These results reflect the dynamic in malaria transmission in these two villages and the need to adapt locally vector control strategies.
Fig. 6.
Malaria prevalence in Ambohitromby and Miarinarivo in 2016, 2017 and 2018. Data were collected from asymptomatic school-aged children tested with RDT in March each year (at T2). P values < 0.05 are significant; n: sample size
Variation in the number of malaria vector species is associated with time point and village
Entomological data from HLCs and PSCs, showed that Anopheles species diversity was similar between the two villages surveyed (Tables 3, 4). However, the analysis of total Anopheles caught by HLCs across the malaria transmission season revealed significant associations with time point and an interaction with village, especially for An. arabiensis and An. coustani. Anopheles arabiensis was the species with the highest HBR at the mid-term of the transmission season (February) in Ambohitromby, while An. coustani was the one with the highest HBR at the onset (December) and at the late-term (April) in both villages. This change in HBR over the malaria transmission season could be explained by changes in the ecological environment, precisely the rice fields which constitute the main Anopheles larval habitats in the two villages. Indeed, the growing phases of the rice determine important changes in the characteristics of the Anopheles breeding sites [32]. Fields with rice in the early stages of growth and with young short plants, offer sunny breeding sites favourable for the development of An. gambiae (s.l.) larvae. As the rice plants grow, they shade the water of the rice fields which become less favourable for An. gambiae (s.l.) larvae, giving way to An. coustani larvae that prefer shaded breeding sites. This transition from An. gambiae (s.l.) to An. coustani in breeding sites by increasing vegetation cover, was also demonstrated for borrow pits in Ethiopia [33].
Through the analysis of the Anopheles vector feeding behaviour, only An. funestus showed a strong tendency for both biting and resting indoor, not departing from its known behaviour in Andriba [29] and in other African regions including Madagascar [5, 17, 34–37]. Both An. arabiensis and An. coustani exhibited a significant outdoor biting preference in each village towards increased endophagy in Miarinarivo (Table 4). This observation is suggestive of the presence of different populations for both vector species in each village. However, it cannot be excluded though that the local environment such as distance between houses and breeding sites, their number and size, and the structure of the villages itself contribute to the observed differences in An. arabiensis and An. coustani endophagy between the two villages. The absence of indoor resting An. arabiensis and An. coustani in Miarinarivo despite their abundance, might also advocate for the presence of different populations for both species in each village.
Different contribution of Anopheles species to malaria transmission between the two villages
Among 1550 mosquitoes tested, 28 were found positive for bearing Plasmodium sporozoites by TaqMan assay. The SYBR Green PCR assay allowed to identify either P. falciparum or P. vivax in 13 out of those 28 mosquito samples. Given the fact that the prevalence of P. malariae and P. ovale is very low in the studied community, this result suggests that the SYBR Green assay had a poor performance under our experimental condition, possibly linked to low sporozoite loads in the mosquito samples. As a consequence, it is difficult to discuss species specific rates (SR and EIR). All mosquitoes positive for Plasmodium spp were collected by HLCs, with 20/28 collected outdoors. Among the 28 Plasmodium-positive mosquitoes, 14 were An. coustani, 9 An. arabiensis, 3 An. funestus and 2 An. rufipes. Whereas An. funestus and An. arabiensis are well known malaria vectors in Madagascar, the contribution of An. coustani to malaria transmission has been suspected on several occasions due to its high density and propensity to anthropophily [38]. It was only recently that some An. coustani samples were detected CSP-positive by ELISA [9]. Data from this study, using a robust TaqMan assay, clearly demonstrated the vector role of An. coustani in malaria transmission in Andriba. Anopheles coustani is also known to be a malaria vector in continental Africa: in Cameroon [39]; Zambia [40, 41]; and Kenya [42]. This work also revealed that two out of 40 An. rufipes analysed by the TaqMan assay were found possibly carrying Plasmodium sporozoites. To date, An. rufipes has never been reported naturally infected with Plasmodium in Madagascar. In continental Africa, it was found naturally infected with P. falciparum in Burkina Faso [26, 43] and more recently in Cameroon [28]. Lastly, none of the An. mascarensis samples (n = 59) were found positive for Plasmodium although it is known as a malaria vector in other Malagasy areas [8, 10, 44, 45].
Surprisingly, this study revealed that An. coustani, was mainly responsible for malaria transmission in Miarinarivo, despite the presence of An. funestus and An. arabiensis. In that village, people were exposed to 61.2 ib/h infected bites per human (ib/h) from An. coustani during the study period (6 months), compared to only 5.4 ib/h in Ambohitromby, despite its relatively high abundance in this latter village. Plasmodium infected An. coustani (n = 12) were captured equally outdoor and indoor in Miarinarivo. By contrast, An. arabiensis was mainly responsible for malaria transmission in Ambohitromby, with 36 ib/h during the same study period, while it was responsible for 14.4 ib/h in Miarinarivo. The majority of infected An. arabiensis (9/10) were found outdoors. Anopheles funestus contributed to a minor extent to malaria transmission in both villages, being responsible for 5.4 and 7.2 ib/h during the whole survey in Ambohitromby and Miarinarivo respectively. Infected An. funestus (n = 3) were captured either indoors or outdoors. Overall, these results are similar to those observed in a study conducted in the Taveta district in Kenya, where malaria transmission, due to An. coustani, An. arabiensis and An. funestus occurred both indoors and outdoors [42]. The different contribution of malaria vector species might result from a different layout of the houses in each village and their distance from the rice fields as previously argued [45]. Indeed, the satellite view of the two villages shows that the houses in Miarinarivo are more numerous and very close to each other compared to houses in Ambohitromby, and that Miarinarivo is surrounded by more and closer rice fields, which is particularly favourable to the large number of An. coustani recorded in Miarinarivo.
In summary, the results of this study show that in neighbouring villages with a similar malaria prevalence in the human population, malaria transmission was driven by two different mosquito species and notably involved An. coustani as the major vector in Miarinarivo. Detailed analysis of the EIR over time (Table 6) shows that most malaria transmission occurred at the beginning and middle of the malaria transmission in Ambohitromby due to An. arabiensis, while occurring at the mid-course and vanishing of the malaria transmission season in Miarinarivo, due to An. coustani. Overall, the population in Ambohitromby was expected to receive 48.6 ib/h over the malaria season (November-April) compared to 84.6 ib/h in Miarinarivo.
Conclusions
Overall, this study demonstrates the variability of vector biology dynamics between two neighbouring villages with similar ecological settings. This is the first time that An. coustani has been clearly demonstrated as playing a major contribution in malaria transmission in an area of Madagascar, despite the presence of An. arabiensis and An. funestus known as major malaria vectors in the country. This finding was quite surprising as An. coustani is being known as a zoophilic and exophilic species in most areas of Madagascar where it was found. The results of this study can be used to better describe the epidemiology and transmission of malaria in Madagascar and to provide relevant information as guidance for adapted malaria vector control strategies. In an epidemiological context such as Madagascar, marked by the presence of both P. falciparum and P. vivax in combination with presence of several vector species, understanding the vector-specific contributions to the transmission of these two main Plasmodium species constitutes a challenge for malaria elimination.
Supplementary information
Additional file 1: Table S1. Sequences of the primers and TaqMan probes used for the morphological identification of An. gambiae/An. arabiensis and for Plasmodium detection in Anopheles mosquitoes. Table S2. Human population that participated in the study categorised by age group and sex. Table S3. Mosquitoes collected by HLCs inside and outside houses, in Ambohitromby and Miarinarivo at the three time points. Table S4. Number of mosquitoes collected resting indoor by PSC.
Acknowledgements
We thank the laboratory technicians from the Institut Pasteur de Madagascar who helped in data collection and experimentations: Mandaniaina R. Andriamiarimanana, Emma Rakotomalala, Rado L. Rakotoarison, Rakotoniaina M. Tanjona, Mamy D. Andrianatoandro, Faniry Randrianarisoa and Maminirina F. Ambinintsoa. We thank Miriam K. Laufer from the University of Maryland School of Medicine for critical reading of the manuscript. We also thank Juliette Paireau from the Institut Pasteur for helping with finding satellite images compatible with CC-BY license.
Abbreviations
- ACT
artemisinin-based combination therapy
- CSP
circumsporozoite protein
- DNA
deoxyribonucleic acid
- EIR
entomological inoculation rate
- ELISA
enzyme-linked immunosorbent assay
- GLMM
generalized linear mixed model
- GPS
global positioning system
- HLC
human landing catch
- HN
human-night
- ib/h/n
infective bite per human per night
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PSC
pyrethrum spray catch
- RDT
rapid diagnostic test
- RNA
ribonucleic acid
- SE
standard error
- SR
sporozoite rate
Authors’ contributions
MON, IVW and CB designed the study. JGY coordinated the field study, performed the experiments. NP and CB developed the mosquito TaqMan assays. TR supervised the mosquito field collections. RG provided the entomology expertise. RP performed the statistical analyses. IP, RP and MON critically read the manuscript. JGY and CB analysed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Institut Pasteur International Network to JGY as a doctoral Calmette-Yersin fellowship (award DI/EC/MAM/No479/14), to MON (award IPIN/G4 GROUP-02); by the Institut Pasteur de Madagascar to IVW (award IPM/IPal-VivaxDuffy) and to CB (award 007/IPM/DIR/PR/16); by French Agence Nationale de la Recherche grant to CB (award ANR-10-LABX-62-IBEID). The funders had no role in the design of the study and collection, analysis, interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
This study followed ethical principles according to the Helsinki Declaration and was approved by the Malagasy Ethical Committee of the Ministry of Health (agreements No. 122-MSANP/CE–2015 and No. 141-MSANP/CE–2014). Prior to carrying out study procedures, individual written informed consent was obtained from all study participants, or their parents or legal guardians in the case of minors.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jessy Goupeyou-Youmsi, Email: goupeyou.youmsi@gmail.com.
Tsiriniaina Rakotondranaivo, Email: tsiryrakoto22@gmail.com.
Nicolas Puchot, Email: nicolas.puchot@pasteur.fr.
Ingrid Peterson, Email: IPeterson@som.umaryland.edu.
Romain Girod, Email: rgirod@pasteur.mg.
Inès Vigan-Womas, Email: ivigan@pasteur.fr.
Richard Paul, Email: richard.paul@pasteur.fr.
Mamadou Ousmane Ndiath, Email: ousmane.ndiath@gmail.com.
Catherine Bourgouin, Email: catherine.bourgouin@pasteur.fr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04282-0.
References
- 1.World Health Organization. World malaria report 2019. Geneva: World Health Organization; 2019. https://www.who.int/publications-detail/world-malaria-report-2019. Accessed 9 Mar 2020.
- 2.Mouchet J, Blanchy S, Rakotonjanabelo A, Ranaivoson G, Rajaonarivelo E, Laventure S, et al. Epidemiological stratification of malaria in Madagascar. Arch Inst Pasteur Madagascar. 1993;60:50–59. [PubMed] [Google Scholar]
- 3.Howes RE, Mioramalala SA, Ramiranirina B, Franchard T, Rakotorahalahy AJ, Bisanzio D, et al. Contemporary epidemiological overview of malaria in Madagascar: operational utility of reported routine case data for malaria control planning. Malar J. 2016;15:502. doi: 10.1186/s12936-016-1556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang SY, Battle KE, Gibson HS, Ratsimbasoa A, Randrianarivelojosia M, Ramboarina S, et al. Spatio-temporal mapping of Madagascar’s Malaria Indicator Survey results to assess Plasmodium falciparum endemicity trends between 2011 and 2016. BMC Med. 2018;16:71. doi: 10.1186/s12916-018-1060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tantely ML, Le Goff G, Boyer S, Fontenille D. An updated checklist of mosquito species (Diptera: Culicidae) from Madagascar. Parasite. 2016;23:20. doi: 10.1051/parasite/2016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pock Tsy JM, Duchemin JB, Marrama L, Rabarison P, Le Goff G, Rajaonarivelo V, et al. Distribution of the species of the Anopheles gambiae complex and first evidence of Anopheles merus as a malaria vector in Madagascar. Malar J. 2003;2:33. doi: 10.1186/1475-2875-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratovonjato J, Randrianarivelojosia M, Rakotondrainibe ME, Raharimanga V, Andrianaivolambo L, Le Goff G, et al. Entomological and parasitological impacts of indoor residual spraying with DDT, alphacypermethrin and deltamethrin in the western foothill area of Madagascar. Malar J. 2014;13:21. doi: 10.1186/1475-2875-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrama L, Laventure S, Rabarison P, Roux J. Anopheles mascarensis (De Meillon, 1947): main vector of malaria in the region of Fort-Dauphin (south-east of Madagascar)] Bull Soc Pathol Exot. 1999;92:136–138. [PubMed] [Google Scholar]
- 9.Nepomichene TNJJ, Tata E, Boyer S. Malaria case in Madagascar, probable implication of a new vector, Anopheles coustani. Malar J. 2015;14:475. doi: 10.1186/s12936-015-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontenille D, Lepers JP, Coluzzi M, Campbell GH, Rakotoarivony I, Coulanges P. Malaria transmission and vector biology on Sainte Marie Island, Madagascar. J Med Entomol. 1992;29:197–202. doi: 10.1093/jmedent/29.2.197. [DOI] [PubMed] [Google Scholar]
- 11.Ihantamalala FA, Rakotoarimanana FMJ, Ramiadantsoa T, Rakotondramanga JM, Pennober G, Rakotomanana F, et al. Spatial and temporal dynamics of malaria in Madagascar. Malar J. 2018;17:58. doi: 10.1186/s12936-018-2206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tedrow RE, Rakotomanga T, Nepomichene T, Howes RE, Ratovonjato J, Ratsimbasoa AC, et al. Anopheles mosquito surveillance in Madagascar reveals multiple blood feeding behavior and Plasmodium infection. PLoS Negl Trop Dis. 2019;13:e0007176. doi: 10.1371/journal.pntd.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howes RE, Franchard T, Rakotomanga TA, Ramiranirina B, Zikursh M, Cramer EY, et al. Risk factors for Malaria infection in central Madagascar: insights from a cross-sectional population survey. Am J Trop Med Hyg. 2018;99:995–1002. doi: 10.4269/ajtmh.18-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Climate-Data.org. Climat Andriba. 2018. https://fr.climate-data.org/location/472281/. Accessed 12 Jun 2018.
- 15.Canier L, Khim N, Kim S, Sluydts V, Heng S, Dourng D, et al. An innovative tool for moving malaria PCR detection of parasite reservoir into the field. Malar J. 2013;12:405. doi: 10.1186/1475-2875-12-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. Module de formation à la lutte contre le paludisme: Entomologie du paludisme et lutte antivectorielle. Guide du participant. Geneva: World Health Organization; 2014. http://apps.who.int/iris/bitstream/handle/10665/136477/9789242505818_fre.pdf?sequence=1.
- 17.Grjebine A. Insectes diptères Culicidae Anophelinae. Paris: ORSTOM; 1966. http://www.documentation.ird.fr/hor/fdi:11315. Accessed 12 Jun 2018.
- 18.De Meillon B. The anophelini of the Ethiopian geographical region. Johannesburg: South African Institute of Medical Research; 1947. [Google Scholar]
- 19.Walker ED, Thibault AR, Thelen AP, Bullard BA, Huang J, Odiere MR, et al. Identification of field caught Anopheles gambiae s.s. and Anopheles arabiensis by TaqMan single nucleotide polymorphism genotyping. Malar J. 2007;6:23. [DOI] [PMC free article] [PubMed]
- 20.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 21.Tahar R. Immune response of Anopheles gambiae to the early sporogonic stages of the human malaria parasite Plasmodium falciparum. EMBO J. 2002;21:6673–6680. doi: 10.1093/emboj/cdf664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor SM, Juliano JJ, Trottman PA, Griffin JB, Landis SH, Kitsa P, et al. High-throughput pooling and real-time PCR-based strategy for malaria detection. J Clin Microbiol. 2010;48:512–519. doi: 10.1128/JCM.01800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandeu MM, Moussiliou A, Moiroux N, Padonou GG, Massougbodji A, Corbel V, et al. Optimized pan-species and speciation duplex real-time PCR assays for Plasmodium parasites detection in malaria vectors. PLoS ONE. 2012;7:e52719. doi: 10.1371/journal.pone.0052719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rath A, Prusty MR, Barik SK, Das M, Tripathy HK, Mahapatra N, et al. Development, standardization and validation of molecular techniques for malaria vector species identification, trophic preferences, and detection of Plasmodium falciparum. J Vector Borne Dis. 2017;54:25–34. [PubMed] [Google Scholar]
- 26.Diabaté DD, Mouline K, Lefèvre T, Awono-Ambene H, Ouédraogo J, Dabiré K. Anopheles rufipes remains a potential malaria vector after the first detection of infected specimens in 1960 in Burkina Faso. J Infect Dis Ther. 2013;1:3. [Google Scholar]
- 27.Awono-Ambene PH, Etang J, Antonio-Nkondjio C, Ndo C, Eyisap WE, Piameu MC, et al. The bionomics of the malaria vector Anopheles rufipes Gough, 1910 and its susceptibility to deltamethrin insecticide in North Cameroon. Parasit Vectors. 2018;11:253. doi: 10.1186/s13071-018-2809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabue RN, Awono-Ambene P, Etang J, Atangana J, Antonio-Nkondjio C, Toto JC, et al. Role of Anopheles (Cellia) rufipes (Gough, 1910) and other local anophelines in human malaria transmission in the northern savannah of Cameroon: a cross-sectional survey. Parasit Vectors. 2017;10:22. doi: 10.1186/s13071-016-1933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontenille D. Hétérogénéité de la transmission des paludismes à Madagascar. Mém Société R Belge Entomol. 1992;35:129–132. [Google Scholar]
- 30.Craig MH, Snow RW, le Sueur D. A climate-based distribution model of malaria transmission in Sub-Saharan Africa. Parasitol Today. 1999;15:105–111. doi: 10.1016/S0169-4758(99)01396-4. [DOI] [PubMed] [Google Scholar]
- 31.Kesteman T, Rafalimanantsoa SA, Razafimandimby H, Rasamimanana HH, Raharimanga V, Ramarosandratana B, et al. Multiple causes of an unexpected malaria outbreak in a high-transmission area in Madagascar. Malar J. 2016;15:57. doi: 10.1186/s12936-016-1113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravoniharimelina B, Romi R, Sabatinelli G. Etude longitudinale sur le gîtes larvaires d’Anopheles gambiae s.l. dans un canton de la province d’Antananarivo (Hautes terres centrale de Madagascar). Ann Parasitol Hum Comp. 1992;67:26–30.
- 33.Kiszewski AE, Teffera Z, Wondafrash M, Ravesi M, Pollack RJ. Ecological succession and its impact on malaria vectors and their predators in borrow pits in western Ethiopia. J Vector Ecol. 2014;39:414–423. doi: 10.1111/jvec.12117. [DOI] [PubMed] [Google Scholar]
- 34.Rajaonarivelo V, Le Goff G, Cot M, Brutus L. Les anophèles et la transmission du paludisme à Ambohimena, Village de la marge occidentale des hautes-terres malgaches. Parasites. 2004;11:75–82. doi: 10.1051/parasite/200411175. [DOI] [PubMed] [Google Scholar]
- 35.Antonio-Nkondjio C, Hinzoumbe Kerah C, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, et al. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 2006;43:1215–1221. doi: 10.1093/jmedent/43.6.1215. [DOI] [PubMed] [Google Scholar]
- 36.Oyewole IO, Awolola TS, Ibidapo CA, Oduola AO, Okwa OO, Obansa JA. Behaviour and population dynamics of the major anopheline vectors in a malaria endemic area in southern Nigeria. J Vector Borne Dis. 2007;44:56–64. [PubMed] [Google Scholar]
- 37.Cohuet A, Simard F, Wondji CS, Antonio-Nkondjio C, Awono-Ambene P, Fontenille D. High malaria transmission intensity due to Anopheles funestus (Diptera: Culicidae) in a village of savannah-forest transition area in Cameroon. J Med Entomol. 2004;41:901–905. doi: 10.1603/0022-2585-41.5.901. [DOI] [PubMed] [Google Scholar]
- 38.Fontenille D, Rakotoarivony I, Rajaonarivelo E, Lepers JP. Study of Culicidae in Firaisam-Pokontany of Ambohimanjaka in the environs of Tananarive: results of a longitudinal study, in particular on the vectorial transmission of malaria. Arch Inst Pasteur Madagascar. 1988;54:231–242. [PubMed] [Google Scholar]
- 39.Fornadel CM, Norris LC, Franco V, Norris DE. Unexpected anthropophily in the potential secondary malaria vectors Anopheles coustani s.l. and Anopheles squamosus in Macha, Zambia. Vector Borne Zoonotic Dis. 2011;11:1173–9. [DOI] [PMC free article] [PubMed]
- 40.Stevenson JC, Simubali L, Mbambara S, Musonda M, Mweetwa S, Mudenda T, et al. Detection of Plasmodium falciparum infection in Anopheles squamosus (Diptera: Culicidae) in an area targeted for malaria elimination, southern Zambia. J Med Entomol. 2016;53:1482–1487. doi: 10.1093/jme/tjw091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mwangangi JM, Muturi EJ, Muriu SM, Nzovu J, Midega JT, Mbogo C. The role of Anopheles arabiensis and Anopheles coustani in indoor and outdoor malaria transmission in Taveta District. Kenya. Parasit Vectors. 2013;6:114. doi: 10.1186/1756-3305-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holstein M. Un nouveau vecteur du paludisme en AOF: Anopheles rufipes Gough 1910. Off Rech Sci Tech O-m. 1949. https://www.documentation.ird.fr/hor/fdi:13035. Accessed 25 Mar 2020.
- 43.Le Goff G, Randimby FM, Rajaonarivelo V, Laganier R, Duchemin JB, Robert V, et al. Anopheles mascarensis of Meillon 1947, a malaria vector in the middle west of Madagascar? Arch Inst Pasteur Madagascar. 2003;69:57–62. [PubMed] [Google Scholar]
- 44.Marrama L, Jambou R, Rakotoarivony I, Leong Pock Tsi JM, Duchemin JB, Laventure S, et al. Malaria transmission in southern Madagascar: influence of the environment and hydro-agricultural works in sub-arid and humid regions: Part 1. Entomological investigations. Acta Trop. 2004;89:193–203. [DOI] [PubMed]
- 45.Endo N, Eltahir EAB. Environmental determinants of malaria transmission in African villages. Malar J. 2016;15:578. doi: 10.1186/s12936-016-1633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequences of the primers and TaqMan probes used for the morphological identification of An. gambiae/An. arabiensis and for Plasmodium detection in Anopheles mosquitoes. Table S2. Human population that participated in the study categorised by age group and sex. Table S3. Mosquitoes collected by HLCs inside and outside houses, in Ambohitromby and Miarinarivo at the three time points. Table S4. Number of mosquitoes collected resting indoor by PSC.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional files.