Abstract
Background
Most data on the clinical presentation, diagnostics, and outcomes of patients with COVID-19 have been presented as case series without comparison to patients with other acute respiratory illnesses.
Methods
We examined emergency department patients between February 3 and March 31, 2020 with an acute respiratory illness who were tested for SARS-CoV-2. We determined COVID-19 status by PCR and metagenomic next generation sequencing (mNGS). We compared clinical presentation, diagnostics, treatment, and outcomes.
Findings
Among 316 patients, 33 tested positive for SARS-CoV-2; 31 without COVID-19 tested positive for another respiratory virus. Among patients with additional viral testing (27/33), no SARS-CoV-2 co-infections were identified. Compared to those who tested negative, patients with COVID-19 reported longer symptoms duration (median 7d vs. 3d, p < 0.001). Patients with COVID-19 were more often hospitalized (79% vs. 56%, p = 0.014). When hospitalized, patients with COVID-19 had longer hospitalizations (median 10.7d vs. 4.7d, p < 0.001) and more often developed ARDS (23% vs. 3%, p < 0.001). Most comorbidities, medications, symptoms, vital signs, laboratories, treatments, and outcomes did not differ by COVID-19 status.
Interpretation
While we found differences in clinical features of COVID-19 compared to other acute respiratory illnesses, there was significant overlap in presentation and comorbidities. Patients with COVID-19 were more likely to be admitted to the hospital, have longer hospitalizations and develop ARDS, and were unlikely to have co-existent viral infections.
Funding
National Center for Advancing Translational Sciences, National Heart Lung Blood Institute, National Institute of Allergy and Infectious Diseases, Chan Zuckerberg Biohub, Chan Zuckerberg Initiative.
Research in context.
Evidence before this study
Emerging data on the clinical presentation, diagnostics, and outcomes of patients with COVID-19 have commonly been presented as case series. Without control patients, it is not clear whether and how the clinical features, diagnostics, and outcomes differ from other respiratory infections.
Added value of this study
When compared to other patients with acute respiratory illness not caused by COVID-19, many of the clinical features and outcomes occur at similar rates. Notably different, patients with COVID-19 had a longer duration of symptoms, particularly fatigue, fever, and myalgias, were more likely to be admitted to the hospital and for a longer duration, and more likely to develop ARDS compared to those without COVID-19. Those infected with SARS-CoV-2 were unlikely to have co-existent viral infections when examined by PCR and metagenomic next generation sequencing.
Implications of all the available evidence
Given the considerable overlap in clinical features and outcomes, studies seeking to describe features unique to COVID-19 should employ a control group. Viral co-infection rates are variable and may be context specific.
Alt-text: Unlabelled box
1. Introduction
The severe acute respiratory coronavirus 2 (SARS-CoV-2) and its associated clinical disease, COVID-19, led to a global pandemic in early 2020, with more than 3 million cases and more than 200,000 deaths as of April 2020. [1] The initial published reports of COVID-19 describe the most common presenting symptoms as fever, cough, and dyspnea. [2], [3], [4], [5], [6] While many people recovered, reports from China, Italy, and the United States showed that approximately 5% of patients required intensive care, and 1.7 to 7.2% died. [1,7,8] The majority of clinical and outcomes data on COVID-19 have been from Asia and Europe, [4,6,7,[9], [10], [11], [12], [13], [14]] although data are now emerging from the United States. In particular, studies have reported the clinical features and outcomes of hospitalized patients in Seattle, New York City, and Northern California. [15], [16], [17], [18], [19] However, reports have predominantly focused on patients diagnosed with COVID-19 and have not described in detail the presentation of patients with acute respiratory illness who did not have COVID-19. Without control patients, it is uncertain whether COVID-19 presents differently from other respiratory infections.
The prevalence of viral co-infections in patients with COVID-19 appears to be low in most but not all studies. [[15], [16], [17], [18],[20], [21], [22], [23]] However, these studies used conventional microbiological techniques to evaluate for co-infections that are limited in their ability to diagnose respiratory infections. [24] Understanding the true scope of co-infections in patients with COVID-19 is critical to pursue appropriate diagnostics and management. Metagenomic next-generation sequencing (mNGS) offers a powerful alternative to test for viruses in a respiratory sample in an unbiased manner. [25]
Here we report the clinical characteristics, diagnostics, and outcomes of all patients presenting with respiratory illness to a tertiary academic medical center in San Francisco at the outset of the COVID-19 pandemic. We compare patients with COVID-19 disease to patients presenting during the same time period with an acute respiratory illness and report the prevalence of viral respiratory infections using both conventional microbiology and mNGS.
2. Methods
2.1. Setting and design
We conducted a retrospective cohort study to describe the characteristics, diagnostics, and outcomes of patients with respiratory illness presenting to the University of California, San Francisco (UCSF) Health Emergency Department (ED) during the COVID-19 outbreak, comparing patients with and without COVID-19 disease. We identified all patients 18 years or older who underwent testing for COVID-19 within 24 h of presentation to the ED between February 3 and March 31, 2020. Patients were tested for SARS-CoV-2 if they met U.S. Centers for Disease Control and Prevention (CDC) clinical testing criteria. [26]
Two physicians blinded to patients’ COVID-19 status, independently reviewed the documented clinical presentation of all patients and included only those who presented with acute respiratory symptoms (e.g., cough, dyspnea) or influenza-like illness symptoms (e.g., fever, myalgias). Discordant results were re-reviewed together and a consensus decision was reached on all cases (Appendix Fig. 1). If patients had multiple encounters during the time period, the first encounter was examined. Patients who were discharged and readmitted within 48 h were considered a single clinical encounter and outcomes ascertained throughout the encounter.
Fig. A1.
Cohort flow diagram.
2.2. Patient characteristics
Patient medical records were reviewed by trained physician chart reviewers and relevant data on initial presentation, radiology findings, and outcomes were abstracted using standardized case review forms. Additional information on patient demographics, vital signs, and laboratory results were obtained from the Epic-based electronic health record. We characterized patients’ comorbidities and their presenting signs and symptoms based on the admission History & Physical and Emergency Department documentation. If a specific comorbidity was not mentioned in the admission documentation, it was considered not present. Records were also reviewed to obtain results of laboratory tests and chest imaging reports within the first 24 h after admission.
2.3. Clinical microbiological testing
Clinician-ordered testing for COVID-19 was carried out at the UCSF Clinical Microbiology Laboratory using an in-house Clinical Laboratory Improvements Amendments (CLIA)-validated reverse transcriptase polymerase chain reaction (PCR) assay. This assay was performed for 290/316 (92%) of patients on RNA extracted from oropharyngeal and/or nasopharyngeal swab specimens using primers targeting two regions of the SARS-CoV-2 N gene. The analytical sensitivity/specificity of the in-house assay compared to the US CDC assay performed at the CDC was 97% and 100%, respectively. Twenty-six (8%) of the patients had SARS-CoV-2 PCR testing ordered at the study site but performed at the Centers for Disease Control or other institutions using their clinically validated assays. At the time of the study, PCR results were available at the earliest within 3 h, and the median time to result was 16 h.
Conventional PCR testing for other respiratory viruses was carried out at the discretion of treating clinicians for 270/316 (85%) of patients on pooled nasopharyngeal+oropharyngeal or nasopharyngeal swab specimens using two types of commercial assays as detailed in Appendix table 2. The first was a 12-target respiratory viral PCR assay (adenovirus, influenza AH1/AH3/B, human metapneumovirus, human rhinovirus, parainfluenza viruses 1–4, respiratory syncytial viruses A/B) manufactured by Luminex, Inc. The second was a 3-target (influenza A/B, respiratory syncytial virus) assay manufactured by Diasoren, Inc. Bacterial and fungal respiratory pathogens were assessed by semi-quantitative cultures. Patient blood cultures were performed via inoculation into BD Bactec Plus Aerobic and Lytic Anaerobic media (Becton Dickinson).
Table 2.
Laboratory and imaging findings within 24 h of presentation among 316 patients presenting with acute respiratory illness and tested for COVID-19.
| Lab normal values | COVID-19 positive (n = 33) | COVID-19 negative (n = 283) | P value | |
|---|---|---|---|---|
| Complete blood count | ||||
| White blood cell count | ||||
| Leukopenia* | 3.4–10.0 × 109/L | 3/33 (9%) | 10/279 (4%) | 0.148 |
| Leukocytosis✝ | 0/33 (0%) | 110/279 (39%) | <0.001 | |
| Neutrophil count | 1.8–6.8 × 109/L | |||
| Neutropenia* | 2/33 (6%) | 7/274 (3%) | 0.250 | |
| Neutrophilia✝ | 4/33 (12%) | 126/274 (46%) | <0.001 | |
| Lymphocyte count | 1.0–3.4 × 109/L | |||
| Lymphopenia* | 18/33 (55%) | 92/274 (34%) | 0.018 | |
| Lymphocytosis✝ | 0/33 (0%) | 15/274 (6%) | 0.384 | |
| Platelet count | 140–450 × 109/L | |||
| Thrombocytopenia* | 7/33 (21%) | 31/279 (11%) | 0.093 | |
| Thrombocytosis✝ | 0/33 (0%) | 14/279 (5%) | 0.377 | |
| Hemoglobin | 13.6–17.5 g/dL | |||
| Anemic* | 19/33 (58%) | 176/280 (63%) | 0.554 | |
| Chemistry | ||||
| Hyponatremia* | 135–145 mmol/L | 11/32 (34%) | 56/274 (20%) | 0.071 |
| Hypernatremia✝ | 1/32 (3%) | 12/274 (4%) | v | |
| Creatinine, elevated✝ (%) | 0.73–1.18 mg/dL | 11/32 (34%) | 71/274 (26%) | 0.306 |
| Aspartate transaminase, elevated✝ | 5–44 U/L | 10/28 (36%) | 38/217 (18%) | 0.022 |
| Alanine transaminase, elevated✝ | 10–61 U/L | 3/28 (11%) | 22/217 (10%) | 1.000 |
| Troponin I, elevated | <0.05 ug/L | 2/13 (15%) | 37/161 (23%) | 0.735 |
| Procalcitonin, elevated | <0.26 ug/L | 4/25 (16%) | 44/125 (35%) | 0.065 |
| Venous blood gas | ||||
| pH | 7.31–7.41 | |||
| Acidemic* | 0/29 (0%) | 28/192 (15%) | 0.031 | |
| Alkalemic✝ | 11/29 (38%) | 46/192 (24%) | 0.116 | |
| Hypercarbic✝ | 41–51 mmHg | 1/29 (4%) | 54/192 (28%) | 0.002 |
| Elevated lactate✝ | 0.5–2.0 mmol/L | 5/29 (17%) | 51/194 (26%) | 0.295 |
| Chest X-ray findings | ||||
| X-ray within first 24 h | 33/33 (100%) | 277/283 (98%) | 1.000 | |
| Patchy/hazy opacities | ||||
| Unilateral | 4/33 (12%) | 37/277 (13%) | 0.001 | |
| Bilateral | 18/33 (55%) | 67/277 (24%) 173/277 (63%) | ||
| Not present | 12/33 (33%) | 37/277 (13%) | ||
| Focal consolidation | ||||
| Unilateral | 1/33 (3%) | 29/277 (11%) | 0.368 | |
| Bilateral | 2/33 (6%) | 13/277 (5%) | ||
| Not Present | 30/33 (91%) | 235/277 (85%) | ||
| Interstitial abnormalities | ||||
| Unilateral | 0/33 (0%) | 7/277 (3%) | 0.561 | |
| Bilateral | 4/33 (12%) | 52/277 (19%) | ||
| Not Present | 29/33 (88%) | 218/277 (79%) | ||
| Pleural effusion | ||||
| Unilateral | 1/33 (3%) | 18/277 (7%) | 0.031 | |
| Bilateral | 0/33 (0%) | 18/277 (7%) | ||
| Not Present | 32/33 (97%) | 241/277 (87%) |
Legend
Results reflect lab tests and imaging tests performed within 24 h of presentation.
COVID-19 - Coronavirus Disease 2019.
lower than the lower limit of normal.
greater than the upper limit of normal.
2.4. Respiratory virus detection by metagenomic sequencing
To further screen for the presence of other respiratory viral pathogens, metagenomic next generation sequencing (mNGS) of RNA was performed on available residual RNA initially extracted for COVID-19 clinical PCR testing. At our institution, during the time period of the study, SARS-CoV-2 PCR was performed using in-house CLIA validated PCR tests for the majority of samples. This in-house PCR test involved first extracting RNA from patient swab samples and then carrying out reverse transcriptase PCR as described in the Methods. Of the 316 PCR tests performed, leftover RNA was available for mNGS analysis on 178 patients. To balance the need for timely turnaround with the desire to assess a sufficiently large fraction of the cohort, we performed mNGS on 60% (N = 107) of these 178 samples, which included as many SARS-CoV-2 positive samples as possible (mNGS data generated on 14) plus an arbitrary selection of SARS-CoV-2 negative samples. SARS-CoV-2 negative samples were distributed as evenly as possible throughout the study timeframe and were selected blinded to patient characteristics and outcomes.
After DNase treatment, human ribosomal RNA depletion was carried out using FastSelect (Qiagen). To control for background contamination, we included negative controls (water and HeLa cell RNA) as well as positive controls (spike-in dilution series of RNA standards from the External RNA Controls Consortium [ERCC]). [27] The latter enabled subsequent bioinformatic assessment of the total RNA mass input in each sample. [28]
RNA was then fragmented and subjected to a modified metagenomic spiked sequencing primer enrichment (MSSPE) library preparation method. [29] Briefly, a 1:1 mixture of the NEBNext Ultra II RNAseq Library Prep (New England Biolabs) random primer stock and a pool of SARS-CoV-2 primers at 100 µM was used at the first strand synthesis step of the standard RNAseq library preparation protocol to enrich for the recovery of reads spanning the length of the SARS-CoV-2 genome sequence in the context of mNGS analysis. [30] RNA-seq libraries underwent 146 nucleotide paired-end Illumina sequencing on an Illumina NovaSeq 6000.
2.5. mNGS bioinformatic and phylogenetic analysis
Following demultiplexing, reads were host- and quality-filtered and then subjected to viral reference based alignment at both the nucleotide and amino acid level against sequences in the National Center for Biotechnology Information (NCBI) nucleotide (NT) and non-redundant (NR) databases, followed by assembly using previously validated bioinformatics pipelines. [31,32] We used spike-in positive control ERCC RNA standards to bioinformatically calculate the input RNA for the mNGS assay. Ten samples had insufficient (<25 pg) input RNA for accurate analysis and so were considered invalid, leaving 97 subjects available for analysis.
Negative control (water and HeLa cell RNA) samples enabled estimating the number of background reads to each virus, which were normalized by input mass determined based on the ratio of sample reads to spike-in positive control ERCC RNA standards. [28] Viruses with sequencing reads significantly greater compared to negative controls (adjusted p value < 0.05 using a Holm-Bonferroni correction within each sample) were identified by modeling the number of background reads as a negative binomial distribution with mean and dispersion fitted on the negative controls. For phylogenetic analysis of SARS-CoV-2 viruses, we constructed genomes using minimap2 [33] to align reads to the reference MN908947.3 and iVar [34] to trim primers and call variants, then restricted to samples with at least 10-fold coverage of at least 97% (2929 kgbases) of the genome (n = 10), and utilized the Nextstrain [35] pipeline to build a phylogenetic tree using iqtree. [36] Viral genomic data is publicly accessible via gisaid.org (Global Initiative on Sharing All Influenza Data) [37] and Genbank (MT385414 - MT385497).
2.6. Treatment and outcomes
Clinical treatment and outcomes were ascertained through a combination of chart review and extraction of structured fields from the electronic health record. Medication records were reviewed to identify the administration of relevant antibiotics. We determined if patients required respiratory support at any point during their hospitalization: nasal cannula, high flow nasal cannula, noninvasive ventilation (bilevel or continuous positive airway pressure), or endotracheal intubation. Patients were considered to have new-onset cardiomyopathy if a treating physician documented the diagnosis. Acute respiratory distress syndrome (ARDS) was defined according to the Berlin definition by two physicians. [38] Acute kidney injury was defined using the Kidney Disease: Improving Global Outcomes definition. [39] Outcome ascertainment was censored on April 25, 2020.
2.7. Statistical analysis
We used descriptive statistics to characterize the features of patients grouped by COVID infection. Where clinically relevant, we dichotomized continuous variables. For normally distributed continuous variables, we calculated the mean and standard deviation and tested for differences using t-tests. For non-normally distributed continuous variables, we calculated the median and interquartile range and tested for differences using the Wilcoxon rank sum test. For categorical and dichotomous variables, we evaluated differences between groups using the chi-square test or Fisher's exact test. The analyses were not adjusted for multiple comparisons and should be interpreted as descriptive and exploratory. The Human Research Protection Program Institutional Review Board at the University of California, San Francisco, approved this study (IRB# 16–20,956). We used Stata version 14.2 (College Station, TX) and SAS version 9.4 (Cary, NC) to conduct all analyses.
2.8. Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
3. Results
3.1. Demographic characteristics and comorbidities
Out of 316 patients who presented with acute respiratory illness and underwent testing for COVID-19, 33 (10%) tested positive for SARS-CoV-2 by PCR. Patients with a positive COVID-19 test result were more likely to have traveled to an area of community transmission in the past 21 days or to have had contact with someone with COVID-19 (46% vs 11%, p < 0.001), to be married (64% vs. 36%, p = 0.02), or to identify as Asian (42% vs. 24%, p = 0.010) (Table 1). Patients who tested positive were also more likely to report never smoking tobacco (61% vs. 40%, p = 0.001) and to have undergone solid organ transplantation (12% vs. 3%, p = 0.027). The prevalence of hypertension and diabetes did not differ significantly between COVID-19 positive and negative patients. There was no significant difference by COVID-19 status of the proportion of patients taking an angiotensin-converting enzyme inhibitor or angiotensin II receptor blocker.
Table 1.
Characteristics of 316 patients presenting with acute respiratory illness and tested for COVID-19.
| COVID-19 positive (n = 33) | COVID-19 negative (n = 283) | P value | |
|---|---|---|---|
| Demographics | |||
| Age, median (IQR), yr | 63 (50, 75) | 62 (43, 72) | 0.243 |
| Female sex | 12 (36%) | 140 (50%) | 0.154 |
| Marital status | |||
| Married or partnered | 21 (64%) | 103 (36%) | 0.019 |
| Single | 7 (21%) | 136 (48%) | |
| Divorced | 2 (6%) | 18 (6%) | |
| Widowed | 2 (6%) | 19 (7%) | |
| Housing insecure | 1 (3%) | 44 (16%) | 0.063 |
| Race | |||
| White | 8 (24%) | 124 (44%) | 0.010 |
| Black or African American | 2 (6%) | 50 (18%) | |
| Asian | 14 (42%) | 69 (24%) | |
| Hispanic or Latino ethnicity | 5 (15%) | 21 (8%) | 0.128 |
| Required interpreter | 6 (18%) | 46 (16%) | 0.777 |
| Travel to an area with known community transmission in last 21 days or known COVID exposure | 15 (46%) | 31 (11%) | <0.001 |
| Comorbidities | |||
| Tobacco use | |||
| Current smoker | 0 (0%) | 52 (18%) | 0.001 |
| Former smoker | 9 (27%) | 47 (17%) | |
| Never smoker | 20 (61%) | 113 (40%) | |
| Unknown | 4 (12%) | 71 (25%) | |
| Hypertension | 16 (49%) | 119 (42%) | 0.479 |
| Coronary artery disease | 5 (15%) | 38 (13%) | 0.785 |
| Diabetes | 9 (27%) | 50 (18%) | 0.180 |
| Cancer, active (excluding non-melanoma skin cancer) | 5 (15%) | 42 (15%) | 0.962 |
| Cancer, in remission (excluding non-melanoma skin cancer) | 5 (15%) | 19 (7%) | 0.090 |
| Prior stroke | 0 (0%) | 25 (9%) | 0.090 |
| Chronic kidney disease | 7 (21%) | 28 (10%) | 0.049 |
| Liver disease | 0 (0%) | 13 (5%) | 0.375 |
| Human immunodeficiency virus | 0 (0%) | 15 (5%) | 0.382 |
| Chronic obstructive pulmonary disease/emphysema | 1 (3%) | 41 (15%) | 0.098 |
| Asthma | 4 (12%) | 38 (13%) | 1.000 |
| Chronic bronchitis | 0 (0%) | 5 (2%) | 1.000 |
| Congestive heart failure | 4 (12%) | 43 (15%) | 0.798 |
| Solid organ transplant | 4 (12%) | 8 (3%) | 0.027 |
| Other immunosuppressive condition | 5 (15%) | 33 (12%) | 0.560 |
| Home medications | |||
| Steroids | 5 (15%) | 26 (9%) | 0.275 |
| Immunosuppression medications (aside from steroids) | 6 (18%) | 35 (13%) | 0.347 |
| ACE inhibitors or ARB | 6 (18%) | 43 (15%) | 0.654 |
| Signs and Symptoms | |||
| Onset of symptoms relative to presentation, d (IQR) | 7 (5, 9) | 3 (2,7) | <0.001 |
| Fever, patient reported | 27 (82%) | 125 (44%) | <0.001 |
| Fatigue/malaise | 28 (85%) | 140 (50%) | <0.001 |
| Cough | 28 (85%) | 208 (74%) | 0.156 |
| Dry | 12 (43%) | 62 (30%) | 0.298 |
| Productive | 10 (36%) | 77 (37%) | |
| Unspecified | 6 (21%) | 69 (33%) | |
| Myalgia | 20 (61%) | 77 (27%) | <0.001 |
| Dyspnea | 23 (70%) | 171 (60%) | 0.301 |
| Chest pain | 5 (15%) | 81 (29%) | 0.100 |
| Sore throat | 9 (27%) | 73 (26%) | 0.855 |
| Congestion/Rhinorrhea | 10 (30%) | 74 (26%) | 0.610 |
| Diarrhea | 9 (27%) | 45 (16%) | 0.101 |
| Nausea | 8 (24%) | 48 (17%) | 0.300 |
| Vomiting | 5 (15%) | 28 (10%) | 0.350 |
| Abdominal pain | 4 (12%) | 26 (9%) | 0.535 |
| Headache | 7 (21%) | 47 (17%) | 0.506 |
| Altered mentation | 2 (6%) | 39 (14%) | 0.280 |
| Presenting vital signs | |||
| Tachycardia (HR > 100 beats/min) | 16 (49%) | 164 (58%) | 0.299 |
| Low mean arterial pressure (<60 mmHg) | 0 (0%) | 2 (1%) | 1.00 |
| Tachypnea (RR > 20 breaths/min) | 13 (39%) | 124 (44%) | 0.616 |
| Fever (Tmax ≥ 100.4°F) | 15 (46%) | 69 (24%) | 0.010 |
| Highest level of respiratory support in the first 24 h | |||
| Nasal cannula | 10 (30%) | 64 (23%) | 0.864 |
| High flow nasal cannula | 2 (6%) | 23 (8%) | |
| CPAP or BiPAP | 0 (0%) | 10 (4%) | |
| Mechanical ventilation | 1 (3%) | 12 (4%) |
Legend:
COVID-19 - Coronavirus Disease 2019; IQR - interquartile range; ACE - angiotensin-converting enzyme; ARB - Angiotensin II receptor blockers; HR - heart rate; CPAP - continuous positive airway pressure; BiPAP - bilevel positive airway pressure; RR - respiratory rate.
3.2. Signs, symptoms and vital signs
Patients with COVID-19 reported a longer duration of symptoms prior to ED presentation (median 7 vs. 3 days, p < 0.001) (Table 1). COVID-19 patients reported fever (82% vs. 44%, p < 0.001), fatigue (85% vs. 50%, p < 0.001), and myalgias (61% vs 27%, p < 0.001), at a higher rate than COVID-19 negative patients. The presence and characteristics of cough, dyspnea, and chest pain did not differ based on COVID-19 infection. Gastrointestinal symptoms – nausea, vomiting, diarrhea, and abdominal pain – were present at similar rates in the two groups. With respect to vital sign abnormalities, tachycardia, hypotension, oxygen requirement, and tachypnea did not differ by COVID-19 status. However, patients with COVID-19 were more likely to present with a measured fever (46% vs 24%, p = 0.010).
3.3. Laboratory studies and imaging upon presentation
Lymphopenia was more common in patients with COVID-19 at the time of presentation (55% vs 34%, p = 0.018) (Table 2). Aspartate transaminase but not alanine transaminase was more often elevated in patients with COVID-19 (36% vs. 18% p = 0.022 and 11% vs. 10% p = 1.000, respectively). Patients with COVID-19 were less often acidemic (0% vs. 15%, p = 0.031) and less often found to be hypercarbic (4% vs. 28%, p = 0.002) by venous blood gas. Of the patients tested on presentation, neither troponin nor procalcitonin elevation differed by COVID-19 status. Chest X-rays were performed on all but 6 patients. Radiographs from patients with COVID-19 were more likely to reveal bilateral patchy or hazy opacities (55% vs. 24%, p = 0.001). Focal consolidations, interstitial abnormalities, and pleural effusions were observed at similar proportions.
3.4. Pathogen diagnostics
Clinicians ordered Influenza/Respiratory syncytial virus PCR testing for 99/316 (31%) patients and 12-target respiratory virus PCR for 171/316 (54%) patients; testing rates did not differ by COVID-19 status (Table 3). Orthogonal mNGS analysis was performed on swab specimens from 97/316 (31%) of patients to provide additional broad range screening of both common and uncommon viral pathogens. By PCR, SARS-CoV-2 was the most prevalent respiratory virus detected in 33/316 patients (10%). No co-infections with SARS-CoV-2 and other viruses were identified. Other respiratory viruses were identified in 31/194 (16%) of patients without COVID-19. Independent mNGS analyses corroborated 13/14 (93%) of SARS-CoV-2 infections and 11/11 (100%) of other respiratory viral infections detected by clinical PCR assays. Respiratory bacterial co-infection was not more common in patients with COVID-19 (11% vs. 18%, p = 1.000) and no cases of ventilator associated pneumonia were identified in COVID-19 patients. Bacteremia or fungemia was also not more common in patients with COVID-19 disease (5% vs. 7%, p = 1.00).
Table 3.
Results of infectious disease testing among 316 patients presenting with acute respiratory illness and tested for COVID-19.
| COVID-19 positive (n = 33) | COVID-19 negative (n = 283) | P value | |
|---|---|---|---|
| Other viral testing performed | 82% (27/33) | 69% (194/283) | 0.116 |
| Influenza/Respiratory syncytial virus PCR | 27% (9/33) | 32% (90/283) | 0.596 |
| 12-target respiratory virus PCR panel | 55% (18/33) | 54% (153/283) | 0.958 |
| Metagenomic next generation sequencing | 42% (14/33) | 29% (83/283) | 0.123 |
| Positive identification of virus other than SARS-CoV-2* | 0% (0/27) | 16% (31/194) | 0.025 |
| I nfluenza A† | 0/27 | 5/194 | |
| Influenza B† | 0/27 | 2/194 | |
| Respiratory syncytial virus† | 0/27 | 3/194 | |
| Rhinovirus‡ | 0/26 | 9/188 | |
| Metapneumovirus‡ | 0/26 | 8/188 | |
| Parainfluenza‡ | 0/26 | 1/188 | |
| Coronavirus-229E§ | 0/14 | 2/83 | |
| Coronavirus-NL63§ | 0/14 | 1/83 | |
| Bocavirus§ | 0/14 | 1/83 | |
| Blood culture ordered | 19/33 (58%) | 139/283 (49%) | 0.358 |
| Blood culture positive | 1/19 (5%) | 10/139 (7%) | 1.000 |
| Enterococcus faecalis | 0/19 | 1/139 | |
| Enterococcus faecium | 1/19 | 1/139 | |
| E. coli | 0/19 | 1/139 | |
| Group A Streptococcus | 0/19 | 2/139 | |
| Group C Streptococcus | 0/19 | 1/139 | |
| Group G Streptococcus | 0/19 | 1/139 | |
| Klebsiella pneumoniae | 0/19 | 1/139 | |
| Staphylococcus aureus | 0/19 | 1/139 | |
| Candida glabrata | 0/19 | 1/139 | |
| Sputum or lower respiratory culture ordered | 9/33 (27%) | 33/283 (12%) | 0.012 |
| Sputum or lower respiratory culture positive| | 1/9 (11%) | 6/33 (18%) | 1.000 |
| Enterobacter cloacae complex | 0/9 | 1/33 | |
| H. parainfluenzae | 0/9 | 3/33 | |
| Staphylococcus aureus | 0/9 | 1/33 | |
| Pseudomonas aeruginosa | 0/9 | 2/33 | |
| Stenotrophomonas maltophilia | 1/9 | 0/33 |
Legend: COVID-19 - Coronavirus Disease 2019; PCR - polymerase chain reaction.
One case of viral co-infection identified (i.e., 32 pathogenic viruses in 31 patients).
ascertained by Influenza/RSV PCR or 12-target respiratory viral PCR panel or metagenomic next generation sequencing; 194 patients without COVID-19 and 27 with COVID-19 had any additional viral testing done.
ascertained by 12-target respiratory viral PCR panel or metagenomic next generation sequencing; 188 patients without COVID-19 and 26 with COVID-19 had either test performed.
ascertained by mNGS only; 83 patients without COVID-19 and 14 with COVID-19 had mNGS testing performed.
One case of multiple bacterial pathogens identified by sputum culture (i.e., 7 pathogenic bacteria in 6 patients).
3.5. Genomic epidemiology of SARS-CoV-2
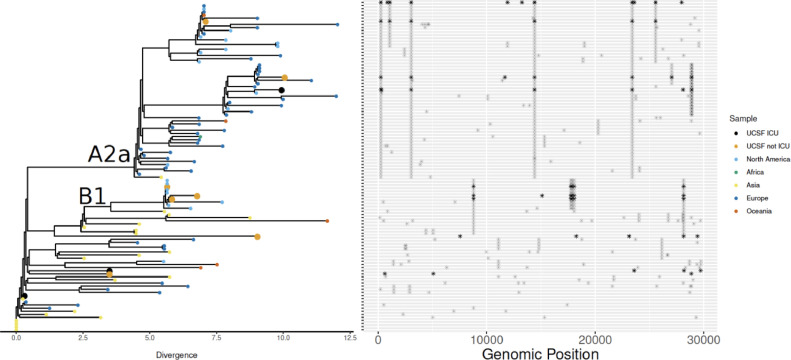
To understand the genomic epidemiology of SARS-CoV-2 in the cohort, phylogenetic analysis was performed. SARS-CoV-2 genomes with at least 97% coverage at 10-fold sequencing depth could be recovered from 10 of the 13 mNGS-positive subjects. These 10 genomes originate from several parts of the global SARS-CoV-2 phylogeny, with clades A2a (n = 3, widely prevalent in New York) and B1 (n = 3, detected in Washington State in February 2020) representing slightly more than half of the lineages we identified (Appendix Fig. 2). The SARS-CoV-2 isolated from patients who required ICU care were not associated with any single clade.
Fig. A2.
Genomic epidemiology of SARS-CoV-2 in study population. Phylogenetic analysis of 10 SARS-CoV-2 genomes from patients in the cohort indicated strains originating from a diversity of geographic locations. Single nucleotide polymorphisms are plotted in the panel adjacent to the phylogenetic tree. Most samples fell into the Nextstrain.org clades A2a (widely prevalent in New York) and B1 (detected in Washington State in February 2020). The SARS-CoV-2 from patients who required ICU care were not associated with any single clade.
3.6. Hospitalization treatment and outcomes
In all, 186 patients were hospitalized and patients with COVID-19 were more likely to be admitted (79% vs. 56%, p = 0.014) and have longer lengths of stay (median 10.7 vs. 4.7 days, p < 0.001). Among hospitalized patients, antibiotics and oseltamivir were used in similar proportions (Table 4). Hydroxychloroquine was more often used in patients with COVID-19 (22% vs. < 1%, p < 0.001); however, azithromycin and corticosteroids use did not differ by COVID-19 status. Six of 26 inpatients with COVID-19 were enrolled in a randomized trial of remdesivir. Respiratory support was provided in similar proportions of patients and, when respiratory support was needed, the level of support did not differ by COVID-19 status.
Table 4.
Treatment of 186 hospitalized patients with acute respiratory illness and tested for COVID-19.
| COVID-19 positive (n = 26) | COVID-19 negative (n = 160) | P value | |
|---|---|---|---|
| Antibiotics administered | 17/26 (65%) | 134/160 (84%) | 0.054 |
| Vancomycin | 8/26 (31%) | 72/160 (45%) | 0.126 |
| Piperacillin/tazobactam | 5/26 (19%) | 55/160 (35%) | 0.107 |
| Cefepime | 4/26 (15%) | 17/160 (11%) | 0.504 |
| Ceftriaxone` | 10/26 (39%) | 74/160 (46%) | 0.459 |
| Carbapenems | 3/26 (12%) | 19/160 (12%) | 1.000 |
| Azithromycin | 8/26 (31%) | 44/160 (28%) | 0.731 |
| Doxycycline | 7/26 (29%) | 70/160 (44%) | 0.106 |
| Fluoroquinolones | 4/26 (15%) | 32/160 (20%) | 0.581 |
| Other antibiotics | 4/26 (15%) | 43/160 (27%) | 0.329 |
| Oseltamivir | 3/26 (12%) | 15/160 (9%) | 0.729 |
| Remdesivir clinical trial* | 6/26 (23%) | 0/160 (0%) | <0.001 |
| Chloroquine | 0/26 (0%) | 0/160 (0%) | — |
| Hydroxychloroquine | 6/26 (22%) | 1/160 (<1%) | <0.001 |
| Steroids | 3/26 (12%) | 23/160 (14%) | 1.000 |
| No respiratory support | 6/26 (23%) | 55/160 (34%) | 0.255 |
| Respiratory support | |||
| Supplemental oxygen | 10/20 (50%) | 61/105 (58%) | 0.711 |
| High flow oxygen | 5/20 (25%) | 21/105 (20%) | |
| Noninvasive positive-pressure ventilation or invasive mechanical ventilation | 5/20 (25%) | 23/105 (22%) |
Legend
COVID-19 - Coronavirus Disease 2019.
Rows are not mutually exclusive, 1 patient received hydroxychloroquine and was enrolled in a blinded remdesivir trial.
Numerically, more patients with COVID-19 required ICU care compared to non-COVID-19 patients, although the difference was not statistically significant (42% vs. 26%, p = 0.092) (Table 5). When transferred to the ICU, there was no observed difference in the use of ICU interventions; however, patients with COVID-19 had a longer ICU length of stay (median 8.8 vs. 2.9 days, p = 0.005). Those diagnosed with COVID-19 were more likely to develop ARDS (23% vs. 4%, p < 0.001) but were no more likely to develop cardiomyopathy or acute kidney injury when compared to non-COVID-19 patients. Among those tested, patients diagnosed with COVID-19 were no more often observed to have abnormal coagulation tests or elevated troponin. Treatment administered to patients not admitted to the hospital are presented in Appendix Table 1.
Table 5.
Outcomes of 186 hospitalized patients with acute respiratory illness and tested for COVID-19.
| COVID-19 Positive (n = 26) | COVID-19 Negative (n = 160) | Difference in proportions (95% CI) | P value | |
|---|---|---|---|---|
| ICU admission | ||||
| ICU stay during hospitalization | 11/26 (42%) | 42/160 (26%) | 16% (4%, 36%) | 0.092 |
| Time to ICU, median days (IQR) | 3.1 (0.4, 4.77) | 0.3 (0.2, 0.4) | 0.027 | |
| ICU days, median days (IQR)* | 8.8 (2.7, 17.8) | 2.9 (1.6, 5.7) | 0.005 | |
| Intensive care unit interventions | ||||
| Endotracheal intubation | 6/11 (55%) | 21/42 (50%) | 5% (−28%, 38%) | 0.788 |
| Paralytics | 2/11 (18%) | 3/42 (7%) | 11% (−0.7%, 15%) | 0.275 |
| Prone positioning | 1/11 (9%) | 0/42 (0%) | 9% (−8%, 26%) | 0.208 |
| Vasopressors | 6/11 (55%) | 21/42 (50%) | 5% (−28%, 38%) | 0.788 |
| Extracorporeal membrane oxygenation | 0/11 (0%) | 0/42 (0%) | — | — |
| Renal replacement therapy | 1/11 (9%) | 5/42 (12%) | −3% (−23%, 17%) | 1.000 |
| Acute respiratory distress syndrome✝ | 6/26 (23%) | 7/160 (4%) | 20% (3%, 36%) | <0.001 |
| Acquired cardiomyopathy‡ | 0/26 (0%) | 5/160 (3%) | −3% (−6%, −0%) | 1.000 |
| Troponin tested | 14/26 (54%) | 113/160 (71%) | −17% (−37%, 3%) | 0.088 |
| Any troponin elevation | 5/14 (36%) | 37/113 (33%) | 3% (−24%, 30%) | 0.824 |
| Acute kidney injury§ | 10/26 (39%) | 56/160 (35%) | 4% (−16%, 24%) | 0.732 |
| AKI First day | 7/10 (70%) | 37/56 (66%) | 4% (−27%, 35%) | 0.808 |
| Abnormal coagulation test | ||||
| Elevated INR | 4/19 (21%) | 30/107 (28%) | −7% (−27%, 13%) | 0.779 |
| Elevated aPTT | 5/10 (50%) | 15/63 (24%) | 26% (−7%, 59%) | 0.085 |
| Elevated d-dimer | 4/4 (100%) | 14/16 (88%) | 12% (−4%, 28%) | 1.000 |
| Elevated fibrinogen | 8/9 (89%) | 12/20 (60%) | 29% (−0%, 59%) | 0.201 |
| Final diagnosis | ||||
| Pulmonary - infectious | 26/26 (100%) | 63/160 (39%) | 61% (53%, 69%) | <0.001 |
| Pulmonary - non-infectious | 0/26 (0%) | 27/160 (17%) | ||
| Other infectious | 0/26 (0%) | 24/160 (15%) | ||
| Cardiac | 0/26 (0%) | 19/160 (12%) | ||
| Malignancy | 0/26 (0%) | 6/160 (4%) | ||
| Renal | 0/26 (0%) | 3/160 (2%) | ||
| Other | 0/26 (0%) | 18/160 (11%) | ||
| Discharge disposition | 0.523 | |||
| Died | 1/26 (4%) | 16/160 (10%) | −6% (−15%, 3%) | |
| Home | 13/26 (50%) | 78/160 (49%) | 1% (−20%, 22%) | |
| Home hospice | 0/26 (0%) | 3/160 (2%) | −2% (−4%,0%) | |
| Home with services | 10/26 (39%) | 37/160 (23%) | 16% (−4%, 36%) | |
| Skilled nursing facility | 2/26 (8%) | 25/160 (16%) | 7% (−35%, 18%) | |
| Still admitted | 0/26 (0%) | 1/160 (1%) | −1% (−3%, 0%) | |
| Length of stay, median days (IQR)* | 10.7 (7.9, 22.7) | 4.7 (2.9, 7.0) | <0.001 |
Legend
All outcomes assessed through April 25, 2020.
COVID-19 - Coronavirus Disease 2019; ICU - intensive care unit; INR - international normalised ratio; aPTT - activated partial thromboplastin time,.
censored at April 25; length of stay for those still admitted, calculated.
ARDS defined using Berlin definition37.
based on treating physician diagnosis.
based on KDIGO definition38.
4. Discussion
While a number of studies describe the clinical features of patients with COVID-19, few have directly compared the clinical presentation and outcomes of COVID-19 to other respiratory illnesses. [23,[40], [41], [42], [43], [44]] Without a control group, and in settings of restricted COVID-19 test availability, we cannot ascertain whether COVID-19 presents differently from other forms of respiratory illnesses. In our study comparing acutely ill patients with and without COVID-19 presenting for emergency care, we found that patients with COVID-19 had a longer duration of symptoms, were more likely to be admitted to the hospital, had longer hospitalizations and were more likely to develop ARDS. Using standard laboratory PCR testing, and mNGS, we found a 16% prevalence of other respiratory viruses in the COVID-19 negative patients, and a lack of detectable viral co-infections in the COVID-19 positive patients.
Patients diagnosed with COVID-19 were more likely to be Asian (44%), which likely reflects differences in the dynamics of COVID-19 transmission early in the pandemic in San Francisco, where the proportion of people who self-identify as Asian is high (36%). [5] Although Asians were overrepresented in the initial COVID-19 cases at our institution, this is not indicative of the current situation in San Francisco, where Asians make up only 13% of the total number of COVID-19 cases. [45] COVID-19 patients were more likely to be never smokers, in line with other studies showing no link between tobacco use and increased COVID-19 risk. [4] [46,47] Largely similar comorbidity profiles were observed between COVID-19 positive and negative patients, aside from a higher proportion of chronic kidney disease and history of solid organ transplantation in COVID-19 patients.
Patients diagnosed with COVID-19 had a longer duration of symptoms prior to presentation and were more likely than control patients to report fever, fatigue and myalgias. It is notable, however, that 44% of COVID-19 negative patients reported fevers and systemic symptoms were common. In contrast to other reports, [4,6,7] COVID-19 positive patients in this cohort had relatively high rates of upper respiratory symptoms (21% with headache, 27% with sore throat, and 30% with congestion/rhinorrhea) and gastrointestinal symptoms. In terms of laboratory values, patients with COVID-19 were significantly more likely to have lymphopenia and no patient with COVID-19 had leukocytosis.
Determining rates of co-infection in patients with COVID-19 has significance given that SARS-CoV-2 testing may be deferred if an alternative respiratory pathogen is identified, especially in settings with limited test availability. In this cohort, no patients with COVID-19 had evidence of viral co-infection by either clinical PCR testing or by mNGS analysis. Only one COVID-19 positive patient had evidence of co-infection with a bacterial respiratory pathogen, and no difference in the prevalence of bacterial co-infection was identified based on COVID-19 status. These results are distinct from those reported in a recent study of COVID-positive patients that found a 21% rate of viral co-infections [23] but consistent with data from several other institutions demonstrating very low rates (≤6%) of viral or bacterial co-infection in hospitalized COVID-19 positive patients, including two recent large studies from New York City. [[15], [16], [17], [18],[20], [21], [22], [23]] Given the consistency in the low rate of co-infections across studies, it may be that there is an inherently low rate of viral and bacterial co-infection in COVID-19 patients. Alternatively, it is possible that early social distancing initiatives and school closures in San Francisco may have concomitantly reduced rates of other circulating respiratory viruses in our population. Similarly, the high rate of antibiotic use in our cohort may have contributed to a lower recovery rate of bacterial co-infection. Further investigation of co-infections in COVID-19 positive patients, and assessment of their potential impact on disease severity and outcomes is needed, especially if SARS-CoV-2 circulation extends to overlap with other highly prevalent seasonal respiratory pathogens.
Although patients with COVID-19 were more likely to be diagnosed with ARDS, there were no differences in their need for ICU care or mechanical ventilation. We also did not find significant differences in terms of acquired cardiomyopathy or troponin elevation during the hospitalization. Despite concerns for cardiac complications in COVID-19 positive patients, our findings highlight the importance of comparisons to control groups of hospitalized patients. [16,48,59] Large proportions of patients in both groups received broad-spectrum antibiotics, despite all of the COVID-19 positive patients having a confirmed viral etiology. This has important implications for antibiotic stewardship in the COVID-19 era and likely reflects clinical uncertainty about the true rate of bacterial co-infection early in the pandemic. COVID-19 was associated with longer hospital lengths of stay. While the duration of hospitalization may reflect the severity of illness, it could also be a marker of concern for late decompensation in these patients [50] or difficulties with hospital discharge due to requirements for isolation and infection control.
Prior studies describing the clinical presentation of patients with COVID-19, have for the most part, identified non-specific features that characterize respiratory infections in general. To our knowledge, this is the first U.S. study to identify characteristics distinguishing patients with COVID-19 from patients who underwent investigation for COVID-19 but were ultimately found to have an alternate diagnosis. Previous publications on this topic are primarily smaller in scope and are all outside of the US. [40,40,43] The clinical, laboratory, and imaging data we highlight have important implications for front line providers making decisions in real-time regarding the pre-test probability of COVID-19, especially in settings with limited access to rapid COVID-19 diagnostics.
In contrast to other areas in the United States, the Bay Area has not yet experienced a large surge in cases of COVID-19. The fact that resources were not strained may have affected the clinical course and outcomes observed. For example, while the sample size is not sufficient to evaluate differences in mortality, only one of the 33 with COVID-19 died (3%), which is lower than in other studies of hospitalized U.S. patients. [17,18] There is speculation that variations in circulating SARS-CoV-2 strains may affect pathogenicity and contribute to geographic differences in case fatality rates. [51,52] Exploratory phylogenetic analysis presented here demonstrated a diversity of strains among the COVID-19 patients requiring ICU care without a predominant clade; larger studies are needed to assess any potential relationship.
There are several limitations inherent to the study design and data available that should be considered when interpreting the results of this study. As a retrospective study based in a single academic medical center and focusing on patients presenting for emergency care, it may not generalize to other institutions with different patient populations or patients with milder forms of the disease. The study design relies on review of the medical record and thus variation in clinician assessment and documentation, particularly absent mention of symptoms and comorbidites, may result in misestimation of the prevalence of these clinical features. Although all patients in the COVID-19 negative group presented with respiratory complaints and/or influenza-like illness, only 56% of patients were given a final diagnosis of respiratory infection, which may affect the generalizability of our outcomes data. The low co-infection rate may have been influenced by incomplete testing for respiratory viral PCR and metagenomics though this is unlikely to have accounted for the full difference when compared to other cohorts. Additionally, as community-transmission increased, CDC clinical criteria for testing changed during the study period; this temporal change could bias the estimate of presenting clinical features. Finally, this study was undertaken at the end of the influenza season and during a period of social distancing, both of which likely impacted the prevalence of circulating viruses and the rate of co-infections.
In summary, while many clinical features of COVID-19 overlap with those of other acute respiratory illnesses, several unique characteristics were identified. Patients with COVID-19 had a longer duration of symptoms, particularly fatigue, fever, and myalgias, were more likely to be admitted to the hospital and for a longer duration, were unlikely to have co-existent viral infections, and were more likely to develop ARDS. Though this health system has not experienced a surge in COVID-19 cases, these key clinical characteristics may, in part, explain the observed differences in the propensity of COVID-19 to strain health systems. While we did find meaningful differences that may inform one's clinical suspicion for COVID-19, we did not find significant differences in cardiopulmonary comorbidities, ACE inhibitor/ARB use, or mortality rate. These findings enhance understanding of the clinical characteristics of COVID-19 in comparison to other acute respiratory illnesses.
Author contributions
Drs. Shah and Langelier had full access to all of the data and take responsibility for the integrity of the data and the accuracy of the data analysis.
Concept and design: Shah, Barish, Prasad, Kistler, Babik, Fang, Kangelaris, Langelier
Acquisition, analysis, or interpretation of data: Shah, Barish, Prasad, Kistler, Kamm, Li, Chiu, Babik, Fang, Kangelaris, Langelier, Abe-Jones, Alipanah, Alvarez, Botvinnik, Castaneda, The CZB CLIAhub Consortium, Dadasovich, Davis, Deng, Detweiler, Federman, Haliburton, Hao, Kerkhoff, Kumar, Malcolm, Mann, Martinez, Marya, Mick, Mwakibete, Najafi, Peluso, Phelps, Pisco, Ratnasiri, Rubio, Sellas, Sherwood, Spottiswoode, Tan, Yu
Drafting of the manuscript: Shah, Barish, Kistler, Kamm, Babik, Fang, Kangelaris, Langelier,
Critical revision of the manuscript for important intellectual content: All authors
Statistical analysis: Shah, Prasad, Li, Kamm, Hao, Martinez
Obtained funding: Shah, Chiu, Fang, Kangelaris, Langelier, DeRisi,
Supervision: Shah, Kistler, Chiu, Kangelaris, Langelier, DeRisi
Declaration of interests
Dr. Prasad reports personal fees from EpiExcellence, LLC, outside the submitted work. Dr. Chiu reports grants from National Institutes of Health/NHLBI, grants from National Institutes of Health/NIAID, during the conduct of the study. Dr. Peluso reports grants from Gilead Sciences, outside the submitted work. Dr. Deng has a patent 62/667,344 pending. All other authors have nothing to disclose.
Funding
This study was supported by the National Center for Advancing Translational Sciences (KL2TR001870), the National Heart Lung Blood Institute (1K23HL138461–01A1, R01-HL105704), National Institute of Allergy and Infectious Diseases (T32 AI060530, R33-AI120977), the Chan Zuckerberg Biohub, the Chan Zuckerberg Initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data sharing
Data used to complete this analysis used protected health elements. Researchers can contact the corresponding authors to request access to the study data.
Contributor Information
Sachin J. Shah, Email: sachin.shah@ucsf.edu.
Charles Langelier, Email: chaz.langelier@ucsf.edu.
Appendix
Tables A1 and A2, Figs. A1 and A2.
Table A1.
Treatment of Emergency department and observation patients with COVID19 infection.
| COVID positive (n = 7) | COVID negative (n = 123) | P Value | |
|---|---|---|---|
| Treatment | |||
| Doxycycline | 2/7 (29%) | 13/123 (11%) | 0.186 |
| Fluoroquinolones | 0/7 (0%) | 3/123 (2%) | 1.00 |
| Azithromycin | 2/7 (29%) | 4/123 (3%) | 0.033 |
| Cephalosporin | 1/7 (14%) | 4/123 (3%) | 0.245 |
| TMP-SMX | 0/7 (0%) | 2/123 (2%) | 1.00 |
| Oseltamivir | 0/7 (0%) | 4/123 (3%) | 1.00 |
| No antimicrobials given on dc | 3/7(43%) | 100/123 (80%) | 0.041 |
| Respiratory support | |||
| Supplemental oxygen | 0/7 (0%) | 3/123 (3%) | 1.00 |
| High Flow | 0/7 (0%) | 0/123(0%) | – |
| Crystalloid bolus volume within first 24 h (mean, SD) | 1000 (0) n = 3 | 1351.4 (716) n = 37 | 0.406 |
Table A2.
Complete microbiological test results for each patient.
| Patient | COVID-19 PCR | Respiratory Viral PCR | mNGS | Respiratory Culture Pathogen | Blood Culture Pathogen | Multiplex Viral PCR Ordered | RSV/Flu PCR Ordered |
|---|---|---|---|---|---|---|---|
| 1 | negative | n/a | yes | yes | |||
| 2 | negative | negative | yes | yes | |||
| 3 | negative | n/a | yes | yes | |||
| 4 | negative | n/a | yes | no | |||
| 5 | negative | negative | no | no | |||
| 6 | negative | negative | Candida glabrata | no | no | ||
| 7 | negative | n/a | yes | no | |||
| 8 | negative | n/a | yes | no | |||
| 9 | negative | n/a | no | no | |||
| 10 | negative | n/a | yes | no | |||
| 11 | negative | n/a | no | no | |||
| 12 | negative | Human Metapneumovirus | n/a | yes | yes | ||
| 13 | negative | Rhinovirus + RSV | n/a | yes | yes | ||
| 14 | negative | Rhinovirus | Rhinovirus A | yes | no | ||
| 15 | negative | negative | yes | yes | |||
| 16 | negative | n/a | yes | yes | |||
| 17 | negative | n/a | no | no | |||
| 18 | negative | n/a | no | no | |||
| 19 | negative | n/a | yes | no | |||
| 20 | negative | n/a | yes | no | |||
| 21 | SARS-CoV-2 | n/a | yes | yes | |||
| 22 | negative | n/a | yes | no | |||
| 23 | negative | n/a | no | no | |||
| 24 | negative | n/a | no | no | |||
| 25 | SARS-CoV-2 | SARS-CoV-2 | yes | no | |||
| 26 | negative | n/a | yes | yes | |||
| 27 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 28 | negative | n/a | no | no | |||
| 29 | SARS-CoV-2 | invalid | yes | yes | |||
| 30 | negative | Human metapneumovirus | n/a | yes | no | ||
| 31 | negative | n/a | no | no | |||
| 32 | negative | negative | yes | yes | |||
| 33 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 34 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 35 | negative | n/a | yes | no | |||
| 36 | negative | n/a | Staphylococcus aureus | no | no | ||
| 37 | negative | negative | yes | yes | |||
| 38 | negative | n/a | no | no | |||
| 39 | negative | negative | yes | yes | |||
| 40 | negative | negative | no | no | |||
| 41 | negative | n/a | yes | no | |||
| 42 | negative | n/a | yes | no | |||
| 43 | negative | n/a | Group A Streptococcus | yes | no | ||
| 44 | negative | n/a | yes | no | |||
| 45 | negative | n/a | yes | yes | |||
| 46 | negative | n/a | no | no | |||
| 47 | negative | n/a | yes | no | |||
| 48 | negative | Rhinovirus | Rhinovirus C | yes | no | ||
| 49 | negative | negative | yes | yes | |||
| 50 | negative | n/a | Group G Streptococcus | yes | no | ||
| 51 | negative | Human CoV 229E | yes | yes | |||
| 52 | negative | n/a | no | no | |||
| 53 | negative | n/a | yes | no | |||
| 54 | negative | negative | yes | yes | |||
| 55 | negative | RSV | RSV | yes | yes | ||
| 56 | negative | n/a | no | no | |||
| 57 | negative | n/a | yes | no | |||
| 58 | negative | n/a | yes | yes | |||
| 59 | negative | n/a | Klebsiella pneumoniae | yes | yes | ||
| 60 | negative | n/a | yes | no | |||
| 61 | negative | negative | yes | yes | |||
| 62 | negative | n/a | no | no | |||
| 63 | negative | n/a | no | no | |||
| 64 | negative | negative | no | no | |||
| 65 | negative | n/a | no | no | |||
| 66 | negative | n/a | no | no | |||
| 67 | SARS-CoV-2 | n/a | no | no | |||
| 68 | negative | n/a | no | no | |||
| 69 | negative | n/a | yes | no | |||
| 70 | SARS-CoV-2 | SARS-CoV-2 | yes | no | |||
| 71 | negative | n/a | no | no | |||
| 72 | negative | n/a | yes | yes | |||
| 73 | negative | negative | no | no | |||
| 74 | negative | n/a | no | no | |||
| 75 | negative | n/a | yes | no | |||
| 76 | negative | n/a | yes | no | |||
| 77 | negative | negative | yes | yes | |||
| 78 | negative | negative | yes | yes | |||
| 79 | negative | Influenza A | n/a | no | yes | ||
| 80 | negative | n/a | no | no | |||
| 81 | negative | n/a | yes | no | |||
| 82 | negative | invalid | no | no | |||
| 83 | negative | n/a | no | no | |||
| 84 | negative | n/a | yes | yes | |||
| 85 | negative | negative | yes | yes | |||
| 86 | SARS-CoV-2 | SARS-CoV-2 | yes | no | |||
| 87 | SARS-CoV-2 | SARS-CoV-2 | yes | no | |||
| 88 | negative | negative | no | no | |||
| 89 | negative | negative | H. parainfluenzae | no | no | ||
| 90 | negative | n/a | no | no | |||
| 91 | negative | n/a | no | no | |||
| 92 | negative | n/a | yes | yes | |||
| 93 | negative | n/a | no | no | |||
| 94 | SARS-CoV-2 | n/a | yes | yes | |||
| 95 | SARS-CoV-2 | n/a | yes | yes | |||
| 96 | SARS-CoV-2 | n/a | no | no | |||
| 97 | negative | negative | yes | no | |||
| 98 | negative | negative | no | no | |||
| 99 | negative | Human metapneumovirus | n/a | yes | yes | ||
| 100 | negative | n/a | no | no | |||
| 101 | negative | Human metapneumovirus | Human Metapneumovirus | yes | yes | ||
| 102 | negative | n/a | yes | no | |||
| 103 | negative | n/a | yes | no | |||
| 104 | negative | n/a | no | no | |||
| 105 | negative | n/a | yes | no | |||
| 106 | SARS-CoV-2 | n/a | yes | no | |||
| 107 | SARS-CoV-2 | n/a | no | yes | |||
| 108 | SARS-CoV-2 | invalid | yes | yes | |||
| 109 | negative | n/a | yes | yes | |||
| 110 | SARS-CoV-2 | SARS-CoV-2 | Stenotrophomonas maltophilia | Enterococcus faecium | yes | yes | |
| 111 | negative | n/a | yes | no | |||
| 112 | negative | RSV | RSV | yes | yes | ||
| 113 | negative | negative | yes | yes | |||
| 114 | negative | n/a | no | no | |||
| 115 | negative | negative | Staphylococcus aureus | yes | yes | ||
| 116 | negative | negative | no | yes | |||
| 117 | negative | Human CoV NL63 | no | no | |||
| 118 | negative | n/a | Group A Streptococcus | yes | no | ||
| 119 | negative | n/a | yes | yes | |||
| 120 | negative | n/a | yes | yes | |||
| 121 | negative | Rhinovirus | Rhinovirus A | yes | no | ||
| 122 | SARS-CoV-2 | n/a | no | no | |||
| 123 | negative | negative | yes | yes | |||
| 124 | negative | negative | no | yes | |||
| 125 | negative | negative | yes | yes | |||
| 126 | negative | n/a | yes | no | |||
| 127 | negative | n/a | no | no | |||
| 128 | negative | n/a | yes | yes | |||
| 129 | SARS-CoV-2 | n/a | yes | no | |||
| 130 | negative | n/a | yes | no | |||
| 131 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 132 | negative | negative | Enterobacter cloacae complex | yes | yes | ||
| 133 | negative | n/a | no | no | |||
| 134 | negative | n/a | no | no | |||
| 135 | negative | Influenza A virus | no | no | |||
| 136 | negative | n/a | no | no | |||
| 137 | negative | n/a | Group C Streptococcus | yes | yes | ||
| 138 | negative | n/a | yes | yes | |||
| 139 | negative | n/a | no | no | |||
| 140 | negative | negative | no | no | |||
| 141 | negative | Influenza B | n/a | no | yes | ||
| 142 | negative | n/a | no | no | |||
| 143 | negative | n/a | no | no | |||
| 144 | negative | n/a | no | yes | |||
| 145 | negative | negative | Enterococcus faecium | yes | no | ||
| 146 | negative | n/a | yes | no | |||
| 147 | negative | n/a | yes | no | |||
| 148 | negative | invalid | yes | yes | |||
| 149 | negative | n/a | no | yes | |||
| 150 | negative | n/a | yes | no | |||
| 151 | negative | n/a | no | no | |||
| 152 | negative | negative | no | yes | |||
| 153 | negative | negative | no | no | |||
| 154 | negative | n/a | yes | yes | |||
| 155 | negative | Human metapneumovirus | Human Metapneumovirus | yes | yes | ||
| 156 | negative | n/a | yes | yes | |||
| 157 | negative | negative | no | no | |||
| 158 | negative | invalid | no | yes | |||
| 159 | negative | n/a | yes | yes | |||
| 160 | negative | Parainfluenza virus 4 | no | no | |||
| 161 | negative | negative | no | no | |||
| 162 | negative | n/a | H. parainfluenzae | no | no | ||
| 163 | negative | n/a | no | no | |||
| 164 | negative | n/a | no | no | |||
| 165 | negative | n/a | yes | yes | |||
| 166 | negative | n/a | yes | no | |||
| 167 | negative | n/a | no | no | |||
| 168 | negative | Influenza A | Influenza A virus | yes | yes | ||
| 169 | negative | n/a | yes | yes | |||
| 170 | negative | negative | yes | yes | |||
| 171 | negative | n/a | Enterococcus faecalis | no | no | ||
| 172 | negative | n/a | yes | yes | |||
| 173 | SARS-CoV-2 | n/a | no | no | |||
| 174 | negative | n/a | yes | yes | |||
| 175 | SARS-CoV-2 | n/a | no | no | |||
| 176 | negative | Rhinovirus | Rhinovirus A | H. parainfluenzae | yes | no | |
| 177 | negative | n/a | no | no | |||
| 178 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 179 | negative | n/a | yes | no | |||
| 180 | negative | n/a | no | no | |||
| 181 | negative | n/a | no | no | |||
| 182 | negative | negative | no | no | |||
| 183 | negative | Human metapneumovirus | invalid | yes | no | ||
| 184 | negative | n/a | no | no | |||
| 185 | negative | negative | yes | no | |||
| 186 | negative | n/a | yes | yes | |||
| 187 | negative | n/a | no | no | |||
| 188 | negative | negative | yes | yes | |||
| 189 | negative | n/a | yes | no | |||
| 190 | negative | Human Metapneumovirus | no | no | |||
| 191 | negative | n/a | no | no | |||
| 192 | negative | n/a | no | no | |||
| 193 | negative | n/a | no | no | |||
| 194 | negative | negative | yes | no | |||
| 195 | negative | n/a | no | no | |||
| 196 | negative | invalid | yes | no | |||
| 197 | negative | n/a | yes | no | |||
| 198 | negative | n/a | no | no | |||
| 199 | negative | n/a | no | no | |||
| 200 | negative | n/a | no | no | |||
| 201 | SARS-CoV-2 | n/a | yes | no | |||
| 202 | negative | n/a | yes | no | |||
| 203 | negative | n/a | yes | no | |||
| 204 | negative | n/a | yes | yes | |||
| 205 | negative | negative | yes | yes | |||
| 206 | negative | negative | no | no | |||
| 207 | negative | n/a | no | no | |||
| 208 | negative | n/a | no | no | |||
| 209 | negative | negative | no | no | |||
| 210 | negative | n/a | no | no | |||
| 211 | negative | Human CoV 229E | no | no | |||
| 212 | negative | n/a | no | no | |||
| 213 | negative | n/a | no | no | |||
| 214 | negative | n/a | yes | yes | |||
| 215 | negative | negative | yes | yes | |||
| 216 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 217 | negative | n/a | no | no | |||
| 218 | negative | invalid | yes | no | |||
| 219 | negative | n/a | yes | no | |||
| 220 | negative | n/a | no | no | |||
| 221 | negative | negative | yes | yes | |||
| 222 | negative | n/a | no | no | |||
| 223 | negative | negative | no | yes | |||
| 224 | negative | n/a | no | no | |||
| 225 | negative | invalid | yes | no | |||
| 226 | negative | negative | no | no | |||
| 227 | negative | n/a | no | yes | |||
| 228 | negative | n/a | no | no | |||
| 229 | negative | negative | yes | yes | |||
| 230 | negative | Rhinovirus C | no | no | |||
| 231 | negative | n/a | no | no | |||
| 232 | negative | negative | yes | yes | |||
| 233 | negative | n/a | yes | no | |||
| 234 | negative | n/a | no | no | |||
| 235 | negative | n/a | yes | no | |||
| 236 | negative | n/a | no | no | |||
| 237 | negative | Influenza A virus | no | no | |||
| 238 | negative | Influenza A virus | no | no | |||
| 239 | negative | n/a | yes | yes | |||
| 240 | negative | negative | no | no | |||
| 241 | negative | n/a | yes | yes | |||
| 242 | negative | negative | yes | yes | |||
| 243 | negative | n/a | no | no | |||
| 244 | negative | n/a | yes | no | |||
| 245 | negative | Rhinovirus | Rhinovirus A | yes | no | ||
| 246 | negative | n/a | yes | no | |||
| 247 | negative | n/a | no | no | |||
| 248 | negative | n/a | yes | yes | |||
| 249 | negative | n/a | no | no | |||
| 250 | negative | n/a | no | no | |||
| 251 | negative | n/a | no | no | |||
| 252 | negative | n/a | yes | no | |||
| 253 | negative | Rhinovirus C | no | no | |||
| 254 | negative | negative | yes | no | |||
| 255 | negative | n/a | E. coli | yes | no | ||
| 256 | negative | n/a | yes | no | |||
| 257 | negative | negative | yes | yes | |||
| 258 | negative | n/a | yes | no | |||
| 259 | negative | n/a | no | no | |||
| 260 | negative | Human Bocavirus | yes | yes | |||
| 261 | SARS-CoV-2 | n/a | yes | no | |||
| 262 | negative | n/a | yes | no | |||
| 263 | SARS-CoV-2 | SARS-CoV-2 | no | no | |||
| 264 | SARS-CoV-2 | n/a | yes | no | |||
| 265 | negative | n/a | yes | no | |||
| 266 | negative | n/a | no | no | |||
| 267 | negative | n/a | no | no | |||
| 268 | negative | Human metapneumovirus | n/a | yes | yes | ||
| 269 | negative | n/a | yes | no | |||
| 270 | negative | n/a | yes | yes | |||
| 271 | negative | negative | Pseudomonas aeruginosa | yes | yes | ||
| 272 | negative | negative | yes | yes | |||
| 273 | negative | n/a | yes | yes | |||
| 274 | negative | Rhinovirus A | yes | yes | |||
| 275 | negative | negative | yes | yes | |||
| 276 | SARS-CoV-2 | n/a | no | no | |||
| 277 | negative | negative | yes | yes | |||
| 278 | negative | n/a | yes | yes | |||
| 279 | negative | Influenza B | n/a | yes | yes | ||
| 280 | negative | n/a | no | no | |||
| 281 | negative | n/a | no | no | |||
| 282 | negative | negative | no | no | |||
| 283 | negative | negative | no | no | |||
| 284 | negative | n/a | no | no | |||
| 285 | negative | n/a | no | no | |||
| 286 | negative | negative | no | no | |||
| 287 | negative | negative | yes | yes | |||
| 288 | negative | negative | no | no | |||
| 289 | negative | invalid | no | no | |||
| 290 | SARS-CoV-2 | n/a | yes | yes | |||
| 291 | negative | n/a | yes | no | |||
| 292 | negative | n/a | Pseudomonas aeruginosa | yes | no | ||
| 293 | negative | n/a | yes | no | |||
| 294 | negative | n/a | no | no | |||
| 295 | SARS-CoV-2 | negative | no | no | |||
| 296 | negative | n/a | no | no | |||
| 297 | negative | n/a | no | no | |||
| 298 | negative | n/a | yes | no | |||
| 299 | negative | n/a | no | no | |||
| 300 | negative | n/a | no | no | |||
| 301 | negative | negative | no | yes | |||
| 302 | negative | n/a | yes | no | |||
| 303 | negative | n/a | yes | yes | |||
| 304 | negative | n/a | yes | no | |||
| 305 | negative | n/a | yes | yes | |||
| 306 | negative | n/a | no | no | |||
| 307 | negative | n/a | yes | no | |||
| 308 | negative | n/a | no | no | |||
| 309 | SARS-CoV-2 | SARS-CoV-2 | yes | no | |||
| 310 | negative | n/a | yes | yes | |||
| 311 | negative | n/a | yes | no | |||
| 312 | negative | n/a | yes | no | |||
| 313 | negative | n/a | yes | no | |||
| 314 | SARS-CoV-2 | n/a | yes | yes | |||
| 315 | negative | n/a | no | no | |||
| 316 | negative | n/a | no | no |
Legend: Respiratory culture: sputum, endotracheal aspirate or bronchoalveolar lavage; negative: not detected; n/a = not applicable because RNA from patient sample unavailable for testing; invalid = sample unable to be analyzed by mNGS due to insufficient (<25 pg) RNA.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. February 2020 doi: 10.1016/S1473-3099(20)30120-1. Published onlineS1473309920301201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D., Hu B., Hu C. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. February 7, 2020 doi: 10.1001/jama.2020.1585. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet North Am Ed. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W., Ni Z., Hu Y. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. February 28, 2020 doi: 10.1056/NEJMoa2002032. Published onlineNEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet North Am Ed. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: asingle-centered, retrospective, observational study. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. 0(0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected Pneumonia in Wuhan, China. JAMA. February 7, 2020 doi: 10.1001/jama.2020.1585. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grasselli G., Zangrillo A., Zanella A. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA. April 6, 2020 doi: 10.1001/jama.2020.5394. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mo P., Xing Y., Xiao Y. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. March 24, 2020 [Google Scholar]
- 10.Onder G., Rezza G., Brusaferro S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA. March 23, 2020 doi: 10.1001/jama.2020.4683. Published online. [DOI] [PubMed] [Google Scholar]
- 11.Chen T., Wu D., Chen H. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020:368. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with Coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X.-.W., Wu X.-.X., Jiang X.-.G. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020:368. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young B.E., Ong S.W.X., Kalimuddin S. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. March 3, 2020 doi: 10.1001/jama.2020.3204. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatraju P.K., Ghassemieh B.J., Nichols M. Covid-19 in critically Ill patients in the Seattle region-case series. N Engl J Med. March 30, 2020 doi: 10.1056/NEJMoa2004500. Published onlineNEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arentz M., Yim E., Klaff L. Characteristics and outcomes of 21 critically Ill patients with COVID-19 in Washington State. JAMA. March 19, 2020 doi: 10.1001/jama.2020.4326. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal P., Choi J.J., Pinheiro L.C. Clinical characteristics of Covid-19 in New York City. N Engl J Med. April 17, 2020 doi: 10.1056/NEJMc2010419. Published onlineNEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson S., Hirsch J.S., Narasimhan M. Presenting characteristics, Comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. April 22, 2020 doi: 10.1001/jama.2020.6775. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers L.C., Parodi S.M., Escobar G.J., Liu V.X. Characteristics of hospitalized adults with COVID-19 in an integrated health care system in California. JAMA. April 24, 2020 doi: 10.1001/jama.2020.7202. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Liu J., Zhao X. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu Province: a Multicenter Descriptive Study. Clin Infect Dis. February 29, 2020 doi: 10.1093/cid/ciaa199. Published onlineciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellberg B., Haddix M., Lee R. Community Prevalence of SARS-CoV-2 among patients with influenzalike illnesses presenting to a Los Angeles medical center in March 2020. JAMA. March 31, 2020 doi: 10.1001/jama.2020.4958. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D., Liu L., Zhang M. Co-infections of SARS-CoV-2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. 2020;63(4):606–609. doi: 10.1007/s11427-020-1668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of Co-infection between SARS-CoV-2 and other respiratory Pathogens. JAMA. April 15, 2020 doi: 10.1001/jama.2020.6266. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain S., Self W.H., Wunderink R.G. Community-acquired Pneumonia requiring hospitalization among U.S. Adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langelier C., Kalantar K.L., Moazed F. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA. 2018;115(52):E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CDC. Coronavirus Disease 2019 (COVID-19) Centers for disease control and prevention. Published February. 2020;11 https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html [Google Scholar]
- 27.Lemire A., Lea K., Batten D. Development of ERCC RNA Spike-In control mixes. J Biomol Tech. 2011;22(Suppl):S46. [Google Scholar]
- 28.Mayday M.Y., Khan L.M., Chow E.D., Zinter M.S., DeRisi J.L. Miniaturization and optimization of 384-well compatible RNA sequencing library preparation. PLoS ONE. 2019;14(1) doi: 10.1371/journal.pone.0206194. Thomas T, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng X., Achari A., Federman S. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat Microbiol. 2020;5(3):443–454. doi: 10.1038/s41564-019-0637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning JE. SARS-CoV-2 Enrichment sequencing by spiked primer MSSPE method. Published onlineApril 7, 2020. doi: 10.17504/protocols.io.beshjeb6. [DOI]
- 31.Naccache S.N., Federman S., Veeraraghavan N. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24(7):1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha S., Ramesh A., Kalantar K. Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals Neuroinvasive Chikungunya Virus outbreak and other unrealized pathogens. MBio. 2019;10(6) doi: 10.1128/mBio.02877-19. Duggal N, editor. e02877-19, /mbio/10/6/mBio.02877-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. Birol I, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grubaugh N.D., Gangavarapu K., Quick J. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(1):8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadfield J., Megill C., Bell S.M. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. Kelso J, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minh B.Q., Schmidt H.A., Chernomor O. IQ-TREE 2: new Models and efficient methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. Teeling E, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GISAID - Next hCoV-19 App. Accessed 25 April 2020. https://www.gisaid.org/epiflu-applications/next-hcov-19-app/
- 38.Acute Respiratory Distress Syndrome The Berlin Definition. JAMA. 2012;307(23) doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 39.Notice. Kidney Int Suppl (2011) 2012;2(1):1. [Google Scholar]
- 40.Sun Y, Koh V, Marimuthu K, et al. Epidemiological and Clinical Predictors of COVID-19. 2020, 7. [DOI] [PMC free article] [PubMed]
- 41.Zhao D., Yao F., Wang L. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun H., Sun Z., Wu J., Tang A., Hu M., Xiang Z. Laboratory data analysis of novel coronavirus (COVID-19) screening in 2510 patients. Clin Chim Acta. 2020;507:94–97. doi: 10.1016/j.cca.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Tang Y., Mo Y. A diagnostic model for coronavirus disease 2019 (COVID-19) based on radiological semantic and clinical features: a multi-center study. Eur Radiol. April 16, 2020 doi: 10.1007/s00330-020-06829-2. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med (CCLM) 2020 doi: 10.1515/cclm-2020-0398. 0(0) [DOI] [PubMed] [Google Scholar]
- 45.San Francisco Open Data. DataSF. Accessed 29 May 2020. https://datasf.org/opendata/
- 46.Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8(4):e20. doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lippi G., Henry B.M. Active smoking is not associated with severity of coronavirus disease 2019 (COVID-19) Eur. J. Intern. Med. 2020;75:107–108. doi: 10.1016/j.ejim.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo T., Fan Y., Chen M. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. March 27, 2020 doi: 10.1001/jamacardio.2020.1017. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi S., Qin M., Shen B. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. March 25, 2020 doi: 10.1001/jamacardio.2020.0950. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet North Am Ed. 2020 doi: 10.1016/S0140-6736(20)30566-3. 0(0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brufsky A. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J Med Virol. 2020 doi: 10.1002/jmv.25902. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao H., Lu X., Chen Q. Patient-Derived mutations impact pathogenicity of SARS-CoV-2. Infectious Diseases (except HIV/AIDS) 2020 [Google Scholar]