Abstract
Radiomics is an emerging field using the extraction of quantitative features from medical images for tissue characterization. While MRI-based radiomics is still at an early stage, it showed some promising results in studies focusing on breast cancer patients in improving diagnoses and therapy response assessment. Nevertheless, the use of radiomics raises a number of issues regarding feature quantification and robustness. Therefore, our study aim was to determine the robustness of radiomics features extracted by two commonly used radiomics software with respect to variability in manual breast tumor segmentation on MRI. A total of 129 histologically confirmed breast tumors were segmented manually in three dimensions on the first post-contrast T1-weighted MR exam by four observers: a dedicated breast radiologist, a resident, a Ph.D. candidate, and a medical student. Robust features were assessed using the intraclass correlation coefficient (ICC > 0.9). The inter-observer variability was evaluated by the volumetric Dice Similarity Coefficient (DSC). The mean DSC for all tumors was 0.81 (range 0.19–0.96), indicating a good spatial overlap of the segmentations based on observers of varying expertise. In total, 41.6% (552/1328) and 32.8% (273/833) of all RadiomiX and Pyradiomics features, respectively, were identified as robust and were independent of inter-observer manual segmentation variability.
Subject terms: High-throughput screening, Data acquisition, Data processing, Breast cancer
Introduction
Radiomics is a technique that is used to extract large amounts of quantitative information from routine medical images that decode information about a region of interest (ROI). The majority of radiomics articles published concerns its application in the oncological field1–4. Here, radiomics bears the advantage of non-invasively quantifying the underlying phenotype of the entire tumor for multiple lesions simultaneously, in contrast to tissue biopsy, which samples only a small part of a single (often heterogeneous) tumor2,5. The ability to characterize the tumor and to establish links to the underlying biology6 and ultimately clinical outcomes, allows a more patient-tailored treatment7, enabling ‘precision medicine’8,9. Recently, several articles have outlined the potential clinical applicability of radiomics in the field of breast cancer for different purposes, e.g. diagnosis10,11, tumor response prediction12–14, prediction of molecular tumor subtype15,16, and prediction of axillary lymph node metastases17,18.
Although these results are promising, issues regarding features robustness as well as the comparability of results, including inter-observer segmentation variability, need to be addressed19–24. In order to extract clinically useful information from medical images and to use features as clinical biomarkers, it is important that extracted features are reproducible, standardized and robust25,26. All consecutive steps in the radiomics workflow induce potential uncertainties regarding feature robustness27,28. Since there used to be no gold standard or guideline for extraction of image features for radiomics use, an initiative –Image Biomarker Standardization Initiative (IBSI)- was launched as an effort to standardize the entire radiomics extraction process and encourage feature robustness29.
ROI segmentation is an important step after image acquisition in the radiomics workflow, and one of the largest bottlenecks30. Traditionally, the edges (2D) or surfaces (3D) of the ROI are segmented, thereby defining a region from which features will be extracted. Segmentation can be performed either manually, semi-automatically, or completely automatically. Both manual and semi-automatic segmentation are prone to inter- and intra-observer variabilities, with the degree of observer experience playing an important role31–33.
To the best of our knowledge, no articles have been published on the effect of manual inter-observer segmentation variability on MRI-based feature robustness in breast cancer patients. MRI is the most accurate modality for neoadjuvant systemic therapy response monitoring in breast cancer patients and as such much used in daily clinical practice34–37. In this article, we investigate the robustness of MR radiomics features, extracted using two commonly used radiomics software, with respect to variations in manual tumor segmentation of breast cancer patients.
Results
Study population
After the application of inclusion and exclusion criteria, 102 patients were included in the final analysis. Twenty-one of these patients were diagnosed with multifocal breast cancer, bringing the total number of tumors analyzed in this study to 129. Of these, 94 tumors (73%) were assigned ‘easy tumors’ and the remaining 35 tumors (27%) were assigned ‘challenging tumors’. The tumor volume between both groups was significant differently (5.3 vs 10.4 for ‘easy and challenging tumors’, respectively, p = 0.03).
Segmentation variability
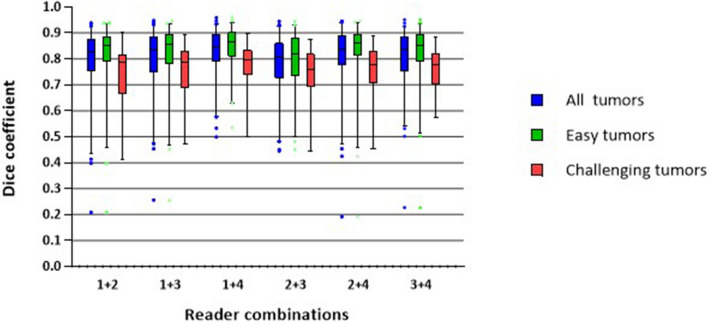
DSC distributions of all observer combinations are shown in Fig. 1. The mean DSC was 0.81 (range 0.19–0.96). The mean DSC was higher for the ‘easy tumors’ compared to the ‘challenging tumors’ (0.83 vs. 0.75, respectively, p < 0.001). The mean DSC for each observer combination separately, for all tumors, ranged between 0.78 and 0.83, where the segmentations of the breast radiologist and the medical student showed the highest overlap.
Figure 1.
Tumor segmentation variability for pairwise comparison of the different observers. (1) Dedicated breast radiologist, (2) Radiology resident, (3) Ph.D. candidate with a medical degree and (4) Medical student.
Pre-processing and feature extraction
The bin width for image discretization (calculated from the ROI greyscale range) was 0.1. Discretization of the scans with bins 0.1 wide resulted in a mean of 61 grayscale values per image (range 27–131). RadiomiX and Pyradiomics software extracted a total of 1328 and 833 features for each ROI, respectively. The extracted radiomics features included shape features, first-order statistical, intensity-histogram based, fractal, local intensity, and texture matrix-based features from both unfiltered and filtered images (wavelet decompositions). The RadiomiX software extracts more feature groups compared to the Pyradiomics software, namely intensity histogram (IH), fractal, local intensity, and gray level dependency zone matrix (GLDZM) features.
Radiomics feature robustness
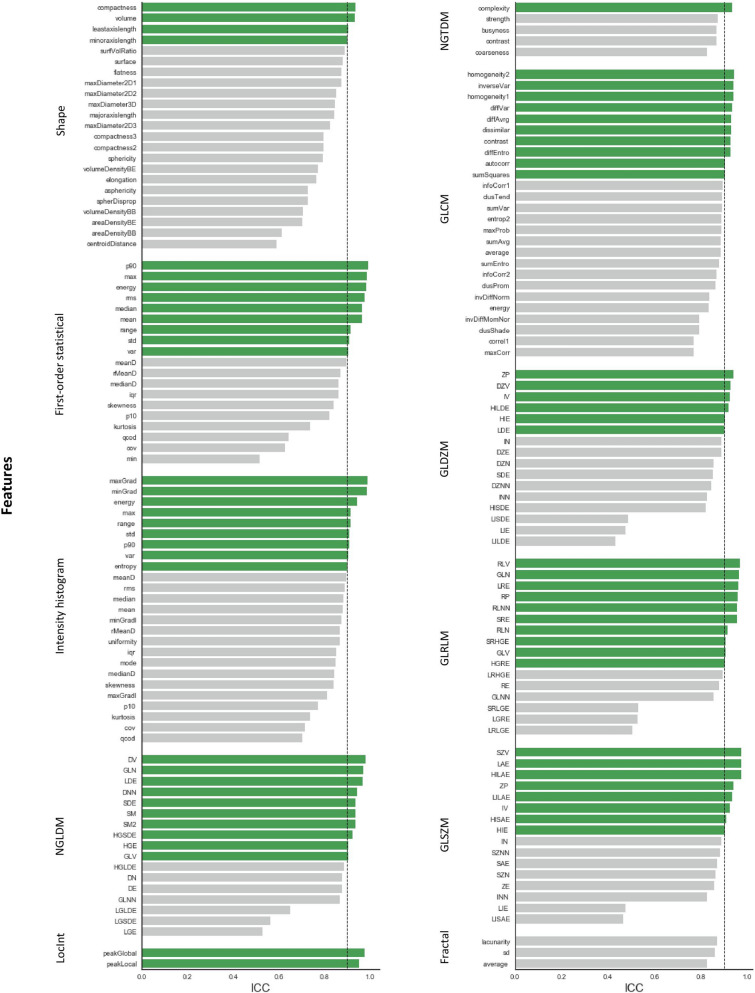
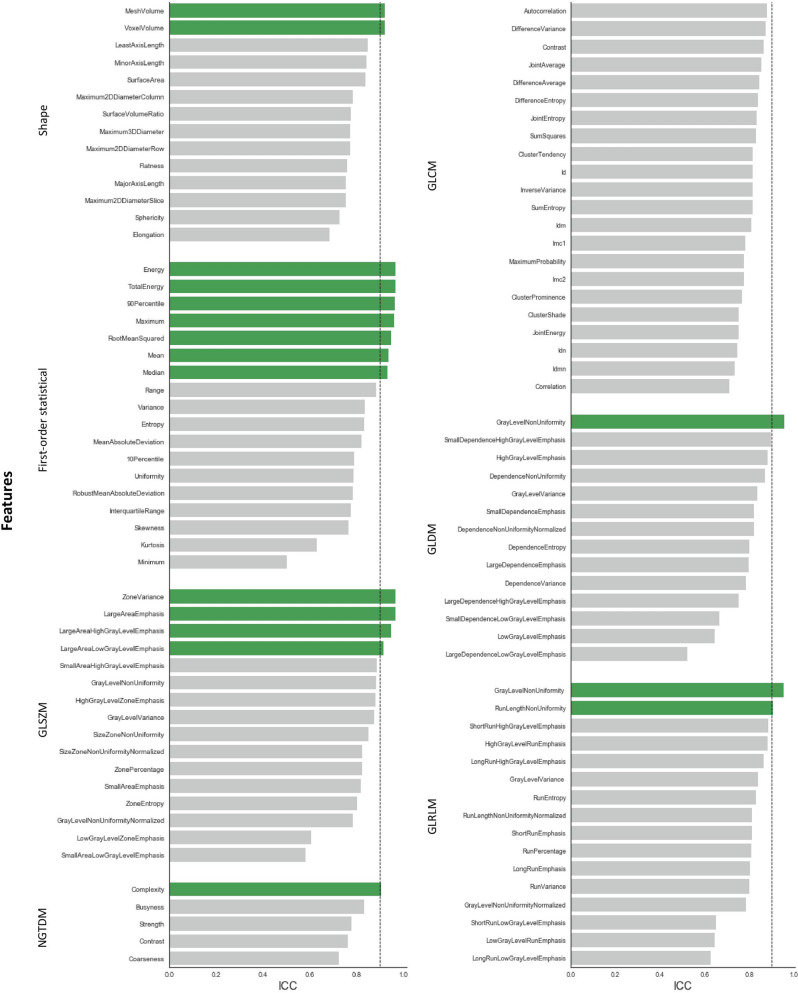
The average ICC for all RadiomiX features was 0.86 (95% CI 0.85–0.86) and for all Pyradiomics features 0.84 (95% CI 0.83–0.84). Table 1 presents the average ICC value per feature group for both software. The local intensity features scored the highest average ICC value for the RadiomiX features, and the first-order statistical features score the highest average ICC for the Pyradiomics features.
Table 1.
Average ICC values per feature group of the unfiltered and wavelet RadiomiX and Pyradiomics features.
| Feature group (n) | OncoRadiomiX | Pyradiomics | ||
|---|---|---|---|---|
| Mean ICC | Range | Mean ICC | Range | |
| Shape | 0.79 | 0.57–0.93 | 0.80 | 0.69–0.92 |
| Signal intensity | ||||
| First-order statistics | 0.85 | 0.51–0.99 | 0.84 | 0.50–0.97 |
| IH | 0.76 | 0.63–0.98 | – | – |
| Fractal | 0.81 | 0.79–0.83 | – | – |
| LocInt | 0.95 | 0.93–0.96 | – | – |
| GLCM | 0.76 | 0.49–0.88 | 0.80 | 0.71–0.88 |
| GLRLM | 0.79 | 0.56–0.96 | 0.81 | 0.63–0.95 |
| GLSZM | 0.80 | 0.55–0.98 | 0.84 | 0.58–0.97 |
| GLDZM | 0.76 | 0.50–0.92 | – | – |
| NGTDM | 0.78 | 0.57–0.85 | 0.80 | 0.72–0.91 |
| (N)GLDM | 0.83 | 0.55–0.96 | 0.79 | 0.52–0.96 |
| Wavelet | 0.81 | 0.01–0.99 | 0.81 | 0.12–0.99 |
The percentage of features that scored an ICC > 0.90, and thus were labeled by our pre-determined ICC cut-off as robust, was 41.6% (552/1328) for RadiomiX features and 32.8% (273/833) for Pyradiomics features. The unfiltered RadiomiX features (i.e., calculated on the unfiltered images) had an average ICC value of 0.79 (95% CI 0.77–0.81), of which 41.1% (69/168) were robust (Fig. 2). The unfiltered Pyradiomics features had an average ICC value of 0.81 (95% CI 0.79–0.83), of which 16.2% (17/105) were robust (Fig. 3). The results of the wavelet feature groups for both software are presented in the supplementary material 1 and 2.
Figure 2.
ICC values of all unfiltered RadiomiX features with robust features (ICC > 0.90) shown in green.
Figure 3.
ICC values of all unfiltered Pyradiomics features with robust features (ICC > 0.90) shown in green.
The percentage of robust RadiomiX features for the ‘easy tumors’ and the ‘challenging tumors’ was 57.5% (763/1328) and 17.2% (228/1328), respectively. When only considering the 168 unfiltered features, 50.0% (84/168) of the ‘easy tumors’ were robust and 20.2% (34/168) of the ‘challenging tumors’ (supplementary material 3). The percentage of robust Pyradiomics features for the ‘easy tumors’ and the ‘challenging tumors’ was 35.7% (297/833) and 28.6% (238/833), respectively. When only considering the 105 unfiltered features, 23.8% (25/105) of the ‘easy tumors’ were robust and 14.3% (15/105) of the ‘challenging tumors’ (supplementary material 4).
Discussion
In this study, our ultimate goal was to define a list of robust MRI radiomics features, independent of inter-observer segmentation variability, which could facilitate further breast MRI-based radiomics research. We successfully identified a subgroup of robust features for two commonly used radiomics software (41.6% of all RadiomiX features and 32.8% of all Pyradiomics features) in the presence of inter-observer segmentation variability (mean DSC of 0.81).
Although MRI feature robustness has already been investigated for different tumor sites (e.g., cervical cancer19 and glioblastoma23), the effect of inter-observer variability segmentation is most likely tumor-site specific38. The feature groups enclosing the most robust features in previous investigations (shape19 and, Intensity-histogram and GLCM23) are different from what we found to be the feature group enclosing the most robust features (local intensities and GLRLM). Most likely this could be explained that different tumor sites influence inter-observer variability. Although one must not forget that the differences in MRI sequences and, feature extraction software also influence this variability. Therefore, the MRI feature robustness cannot be generalized and must be examined for each specific tumor site, taking into account different MRI sequences and feature extraction software.
In addition, feature robustness for both radiomics software was identified for ‘easy tumors’ and ‘challenging tumors’. The number of robust features increased for ‘easy tumors’ and decreased for ‘challenging tumors’ in both software with significant differences between the mean DSC of the ‘easy’ and ‘challenging’ tumors (0.83 vs. 0.75, respectively, p < 0.001). The fact that the ‘challenging tumors’ were more irregular, often with spiculae, causes more segmentation variability and therefore less robust features. Furthermore, the significant difference in the DSC between easy and challenging tumors could be attributed to the sensitivity of the metric to tumor volume. Easy tumors were on average significantly smaller than challenging ones; therefore, a minor difference in segmentation of a small tumor would have a more profound effect on the DSC, compared to those with larger volumes.
A detailed comparison to previous studies is limited to one similar study. Saha et al.39 investigated the impact of breast MRI segmentation variability on radiomics feature robustness, whereby features were extracted using in-house software. Their reported mean ICC of 0.85 for all features, using semi-automatic breast tumor segmentation, is comparable to the average ICC reported in this study. Although the segmentations were performed by four fellow breast radiology trainees, the DSC results they report (range 0.506–0.740) were much lower than the DSC results in our analysis (range 0.783–0.827). We consciously opted for people with different segmentation expertise to ensure observer-independence of the robust features, consequently widening the applicability. Approximately 10% of the tumor features in their article were found to be robust, compared to 41.1% in this study. Solely 20 textural features (GLCM) were comparable between the studies, whereby the ICC of these features showed a substantial difference (average 0.26, range 0.09–0.51).
While we present the robust features for two different radiomics software, our aim is solely to facilitate future application of our findings. Both software have different pre-processing steps, and different groups of features, and comparing the software is beyond the scope of this study. A global initiative to standardize radiomic features extraction using different radiomics software–Imaging Biomarkers Standardization Initiative (IBSI)- was started to address these issues in a more comprehensive fashion40.
To overcome the problem of inter-observer variability with respect to ROI segmentation, promising steps towards (semi-)automatic segmentation have been taken in other tumor sites41–45. However, little work has been published on fully automatic segmentation software for DCE-MRI of the breast33,46–48. Most software, including semi-automatic segmentation, still require manual input or adjustments33,46,47, and would still be significantly slower than fully automated segmentation. Recent work on automatic MRI breast tissue segmentation reported encouraging results but was performed on only 30 patients48. The current lack of reliable, validated and widely available automatic segmentation software tools, and the need for manual input in semi-automated segmentation, demonstrate that manual segmentation remains important. The use of protocols or guidelines could encourage more reproducible manual segmentation results49,50. Furthermore, by providing precise instructions before the start of segmentation, inter-observer segmentation variability can be minimized.
There are some limitations to this study. Although an ICC threshold value of 0.90 was chosen to determine feature robustness, the significance of this threshold for radiomics models for patients’ outcome prediction is yet to be investigated. The inclusion of more patients and observers will allow better generalization of the results and development of robust radiomics signatures. Furthermore, we identified feature robustness to segmentation observer variability. However, due to the lack of data, we were not able to assess the robustness of radiomics features to differences in image acquisition, pre-processing and feature extraction, which are other major challenges in radiomics analysis. These are the aim of our current studies.
In conclusion, this study shows the intuitive notion that more complex, challenging tumors lead to less robust features. We identified radiomics features robust to inter-observer variations across two different radiomics software, which could be used for preselection of radiomics features in future radiomics analysis concerning MRI-based breast radiomics. Ultimately, this study identified a list of robust radiomics features, which is independent of inter-observer segmentation variability in breast MRI for two commonly used software.
Material and methods
Study population
In this single-center retrospective study, we collected data on 138 patients with histologically confirmed invasive breast cancer, who were planned for receiving NST and underwent a pretreatment DCE-MRI between January 2011 and December 2017 in Maastricht University Medical Center+. The institutional research board of the MUMC+ approved the study and waived the requirement for informed consent and the further need of guidelines. Exclusion criteria were: pathologically confirmed mastitis carcinomatosa, MR scan artifacts, or refusal of medical record usage by the patient. Furthermore, we excluded patients that underwent breast MRI exams with non-standard acquisition parameters, due to the use of a different MRI scanner. All histologically confirmed breast tumors were included in the analysis. The complete process is summarized in the flowchart presented in Fig. 4.
Figure 4.
Flowchart of the patient population in the study.
Imaging data
All images were acquired by two clinically interchangeable (i.e. provide qualitatively similar images) 1.5 T MRI scanners (Philips Intera and Philips Ingenia), using a dynamic contrast-enhanced T1-weighted (DCE-T1W) sequence with similar acquisition protocols (Table 2). The patients were scanned in prone position using a 16-channel dedicated breast coil. The DCE-T1W images were obtained before and after intravenous injection of gadolinium-based contrast Gadobutrol (Gadovist (EU)) with a volume of 15 cc and a flow rate of 2 ml/s. One pre-contrast image and five post-contrast images were obtained for each patient.
Table 2.
Imaging parameters for the breast DCE T1W sequence for both scanners.
| Scanner 1 Philips Ingenia (n) |
Scanner 2 Philips Intera (n) |
|
|---|---|---|
| Number of tumors | 100 | 29 |
| Field strength (T) | 1.5 | 1.5 |
| Slice thickness (mm) | 1.0 | 1.0 |
| Repetition time (ms) | 7.5 (88), 7.6 (12) | 7.4 (13), 7.5 (15), 7.6 (1) |
| Echo time (ms) | 3.4 | 3.4 |
| Flip angle (°) | 10 | 10 |
| Echo train length | 89* (range 62–175) | 80* (range 60–85) |
| Pixel spacing (mm) | 0.792 (3), 0.852 (1), 0.922 (2), 0.952 (47), 0.952 (47) | 0.852 (1), 0.942 (1), 0.972 (26), 0.992 (1) |
| Temporal resolution (s) | 95 | 98 |
*Average.
Tumor segmentation
The T1W images acquired two minutes post-contrast administration were used for the 3D tumor segmentation, as this is generally accepted to be the peak of enhancement of breast cancers51. Tumors were independently segmented by four observers with different degrees of experience in breast MR imaging: a dedicated breast radiologist with 11 years of clinical breast MRI experience (ML), a radiology resident with one year of breast MRI clinical experience (TvN), a Ph.D. candidate with a medical degree but no breast MRI clinical experience (RG) and a medical student with no experience whatsoever (NV) (Fig. 5). Segmentations were performed manually with Mirada RTx (v1.2.0.59, Mirada Medical, Oxford, UK). Agreements regarding segmentation procedures were made prior to tumor segmentation: (i) observers were allowed to adjust the image grayscale to optimize the visualization of the tumor; (ii) lymph nodes, pectoral muscle, and skin were excluded from segmentation; (iii) spiculae were only segmented if histologically confirmed. All observers had access to the radiology report during segmentation but were blinded to each other’s segmentations.
Figure 5.
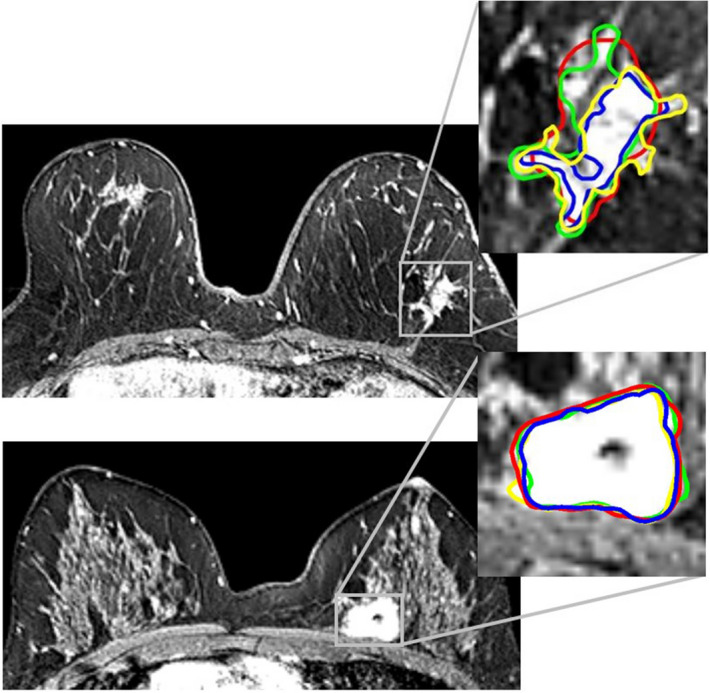
Two invasive breast tumors in the left breast on the 2-min post-contrast DCE-MRI with four single manual segmentations (colored margins: red, blue, green and yellow) fused. Upper: ‘challenging tumor’ with a mean DSC of 0.78 (range 0.71–0.82). Lower: ‘easy tumor’ with a mean DSC of 0.90 (range 0.89–0.91).
Image pre-processing and feature extraction
Radiomics feature extraction is generally performed after image pre-processing. Pre-processing is designed to increase data homogeneity, as well as to reduce image noise and computational requirements. Both radiomics software have the optionality to perform image normalization internally before feature extraction, which varies to an extent across the software. Pyradiomics centers the image around the mean and standard deviation based on all gray values of the image, while RadiomiX normalizes the images after removal of background data (non-breast voxels containing air). This transforms the voxel grayscale values to a more comparable range without changing image textures. Each image was discretized by resampling the grayscale values using a fixed bin width of 0.1 in order to reduce image noise and computational burden. The Pyradiomics community52 recommends the number of bins to be in range of 16–128. We calculated the optimal bin width by extracting the greyscale ranges within all the ROIs and choosing a width that maximizes the number of ROIs that fall in the abovementioned range of bins. Finally, voxel size was standardized across the cohorts to isotropic 1.0 mm3 voxels by means of linear interpolation. For each manually segmented ROI, features were extracted using two commonly used radiomics software: RadiomiX Discovery Toolbox software (OncoRadiomics SA, Liège, Belgium) and the open-source Pyradiomics software, version 2.1.252,53. A mathematical description of all RadiomiX features can be found in supplementary material 5. The Pyradiomics feature description can be found online54. Both software are IBSI compliant for most features, with a note being added in case of differences.
Data analyses
Segmentation variability analysis
Features with (near) zero variance across all tumors, i.e. features that have the same value across ninety-five percent or more of the observations, were excluded from the analysis as they carry no discriminative value. To evaluate the variability of the remaining features introduced by manual segmentation, the volumetric Dice Similarity Coefficient (DSC) was calculated for all pairs of observers. The DSC is a metric that quantifies the agreement (or ‘overlap’) between two segmentations55. A DSC of 1 indicates perfect spatial overlap of the segmentations, whereas 0 indicates no agreement, i.e. no spatial overlap of the segmentations, and a good overlap is considered with DSC > 0.7 as indicated by the literature56. The DSC was calculated as:
where A is the set of voxels contained in the first contour, B is the set of voxels contained in the second contour, || indicates the cardinality of the sets, and ∩ is the intersection between the first and second sets57. The DSC was calculated using Python (Version 3.6.3150.1013).
Radiomics feature robustness analysis
Feature robustness was assessed by evaluating the two-way random single measure intraclass correlation coefficient (ICC) (2,1). The two-way random model approach was chosen as it allows generalization of the results to any other rater with similar characteristics57. The ICC ranges between 0 and 1, with values closer to 1 representing stronger feature robustness to differences in segmentations. We chose a pre-defined ICC cut-off of > 0.9 to select highly stable features that are insensitive to segmentation variability57. Feature robustness was calculated for all RadiomiX and Pyradiomics features. The settings for image pre-processing (normalization, discretization, and resampling) in both radiomics software were checked for disparities. Calculations were performed in R studio (version 1.1.456, Vienna, Austria)58 using the IRR package version 0.8459.
Easy- versus challenging-to-segment tumors analysis
The differences in feature robustness and inter-observer tumor segmentation variability between ‘easy-to-segment’ and ‘challenging-to-segment’ tumors ones, hereinafter referred to as ‘easy tumors’ and ‘challenging tumors’, were assessed. This classification was unanimously determined by the dedicated breast radiologist (ML). ‘Easy tumors’ were defined as homogenous, round tumors with relatively sharp (albeit sometimes irregular) margins, without spiculae or areas of accompanying non-mass enhancement. Tumors not meeting these criteria were categorized as ‘challenging tumors’ (Fig. 5). To compare DSC results between ‘easy’ and ‘challenging’ tumors we used the independent samples t-test, performed in R studio using the IRR package.
Supplementary information
Acknowledgements
This work was supported by the Dutch Cancer Society (KWF Kankerbestrijding), Project number 12085/2018-2.
Author contributions
Hereby an original research manuscript is submitted to Scientific Reports journal. All authors have read the present manuscript and approved to submit. All authors have made a substantial contribution to the manuscript. R.G. and N.V., A.I wrote the manuscript, which was checked and revised several times by all authors. R.G., M.S., and T.v.N. contributed to the concept of the manuscript. Specific work on the design of the manuscript and figure preparation was performed by R.G., A.I., J.v.T., R.L. and H.W. Tumor segmentations were performed by R.G., M.L., T.v.N., and N.V. Data analysis and interpretation was performed by R.G., N.V., A.I and H.W.
Competing interests
Dr. Woodruff and Dr. Leijenaar have (minority) shares in the company OncoRadiomics. Dr. Smidt received a grant of the company Servier for microbiome research. The rest of authors declare no competing interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: N. M. H. Verbakel, A. Ibrahim, M. L. Smidt and H. C. Woodruff.
Supplementary information
is available for this paper at 10.1038/s41598-020-70940-z.
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer (Oxford, England: 1990) 2012;48(4):441–6. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillies R, Kinahan P, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogowicz M, Vuong D, Huellner MW, Pavic M, Andratschke N, Gabrys HS, et al. CT radiomics and PET radiomics: ready for clinical implementation? Q. J. Nucl. Med. Mol. Imaging. 2019;63:355–370. doi: 10.23736/S1824-4785.19.03192-3. [DOI] [PubMed] [Google Scholar]
- 5.Davnall F, Yip CS, Ljungqvist G, Selmi M, Ng F, Sanghera B, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3(6):573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossmann P, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:e23424. doi: 10.7554/eLife.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim A, Vallières M, Woodruff H, Primakov S, Beheshti M, Keek S, et al., editors. Radiomics Analysis for Clinical Decision Support in Nuclear Medicine. Seminars in Nuclear Medicine. New York: Elsevier; 2019. [DOI] [PubMed] [Google Scholar]
- 8.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 9.Walsh S, de Jong EE, van Timmeren JE, Ibrahim A, Compter I, Peerlings J, et al. Decision support systems in oncology. JCO Clin. Cancer Inform. 2019;3:1–9. doi: 10.1200/CCI.18.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milenkovic J, Dalmis MU, Zgajnar J, Platel B. Textural analysis of early-phase spatiotemporal changes in contrast enhancement of breast lesions imaged with an ultrafast DCE-MRI protocol. Med. Phys. 2017;44(9):4652–4664. doi: 10.1002/mp.12408. [DOI] [PubMed] [Google Scholar]
- 11.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ. Breast Cancer. 2017;3:43. doi: 10.1038/s41523-017-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Li Z, Qu J, Zhang R, Zhou X, Li L, et al. Radiomics of multi-parametric MRI for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin. Cancer Res. 2019;25:3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 13.Xiong Q, Zhou X, Liu Z, Lei C, Yang C, Yang M, et al. Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin. Transl. Oncol. 2019;22:50–59. doi: 10.1007/s12094-019-02109-8. [DOI] [PubMed] [Google Scholar]
- 14.Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res. Treat. 2018;173:455–463. doi: 10.1007/s10549-018-4990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur. Radiol. 2016;26(2):322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- 16.Monti S, Aiello M, Incoronato M, Grimaldi AM, Moscarino M, Mirabelli P, et al. DCE-MRI pharmacokinetic-based phenotyping of invasive ductal carcinoma: a radiomic study for prediction of histological outcomes. Contrast Media Mol. Imaging. 2018;2018:5076269. doi: 10.1155/2018/5076269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui X, Wang N, Zhao Y, Chen S, Li S, Xu M, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using radiomics features of DCE-MRI. Sci. Rep. 2019;9(1):2240. doi: 10.1038/s41598-019-38502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Wang T, Yang L, Wang Y, Li H, Zhou X, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using mammography-based radiomics method. Sci. Rep. 2019;9(1):4429. doi: 10.1038/s41598-019-40831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiset S, Welch ML, Weiss J, Pintilie M, Conway JL, Milosevic M, et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019;135:107–114. doi: 10.1016/j.radonc.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Q, Duan J, Duan Z, Meng X, Ma C, Zhu J, et al. Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: impact of tumor segmentation variability. Quant Imaging Med. Surg. 2019;9(3):453–464. doi: 10.21037/qims.2019.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belli ML, Mori M, Broggi S, Cattaneo GM, Bettinardi V, Dell'Oca I, et al. Quantifying the robustness of [(18)F]FDG-PET/CT radiomic features with respect to tumor delineation in head and neck and pancreatic cancer patients. Phys. Med. 2018;49:105–111. doi: 10.1016/j.ejmp.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Pavic M, Bogowicz M, Wurms X, Glatz S, Finazzi T, Riesterer O, et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018;57(8):1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 23.Tixier F, Um H, Young RJ, Veeraraghavan H. Reliability of tumor segmentation in glioblastoma: Impact on the robustness of MRI-radiomic features. Med. Phys. 2019;46:3582–3591. doi: 10.1002/mp.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traverso A, Kazmierski M, Shi Z, Kalendralis P, Welch M, Nissen HD, et al. Stability of radiomic features of apparent diffusion coefficient (ADC) maps for locally advanced rectal cancer in response to image pre-processing. Phys. Med. 2019;61:44–51. doi: 10.1016/j.ejmp.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: the facts and the challenges of image analysis. Eur. Radiol. Exp. 2018;2(1):36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018;102(4):1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br. J. Radiol. 2017;90(1070):20160665. doi: 10.1259/bjr.20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys. Med. Biol. 2016;61(13):R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwanenburg, A. L. S., Vallières, M. & Löck, S. Image biomarker standardisation initiative. arXiv preprint arXiv:161207003.
- 30.Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys. Med. Biol. 2016;61(17):6553–6569. doi: 10.1088/0031-9155/61/17/6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njeh CF. Tumor delineation: the weakest link in the search for accuracy in radiotherapy. J. Med. Phys. 2008;33(4):136–140. doi: 10.4103/0971-6203.44472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beresford MJ, Padhani AR, Taylor NJ, Ah-See ML, Stirling JJ, Makris A, et al. Inter- and intraobserver variability in the evaluation of dynamic breast cancer MRI. J. Magn. Reson. Imaging. 2006;24(6):1316–1325. doi: 10.1002/jmri.20768. [DOI] [PubMed] [Google Scholar]
- 33.Saha A, Grimm LJ, Harowicz M, Ghate SV, Kim C, Walsh R, et al. Interobserver variability in identification of breast tumors in MRI and its implications for prognostic biomarkers and radiogenomics. Med. Phys. 2016;43(8):4558. doi: 10.1118/1.4955435. [DOI] [PubMed] [Google Scholar]
- 34.Lobbes MB, Prevos R, Smidt M, Tjan-Heijnen VC, van Goethem M, Schipper R, et al. The role of magnetic resonance imaging in assessing residual disease and pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy: a systematic review. Insights Imaging. 2013;4(2):163–175. doi: 10.1007/s13244-013-0219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houssami N, Turner R, Morrow M. Preoperative magnetic resonance imaging in breast cancer: meta-analysis of surgical outcomes. Ann. Surg. 2013;257(2):249–255. doi: 10.1097/SLA.0b013e31827a8d17. [DOI] [PubMed] [Google Scholar]
- 36.Woolf DK, Padhani AR, Taylor NJ, Gogbashian A, Li SP, Beresford MJ, et al. Assessing response in breast cancer with dynamic contrast-enhanced magnetic resonance imaging: are signal intensity-time curves adequate? Breast Cancer Res. Treat. 2014;147(2):335–343. doi: 10.1007/s10549-014-3072-x. [DOI] [PubMed] [Google Scholar]
- 37.Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019;30:1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 38.van Timmeren JE, Leijenaar RTH, van Elmpt W, Wang J, Zhang Z, Dekker A, et al. Test-retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography. 2016;2(4):361–365. doi: 10.18383/j.tom.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha A, Harowicz MR, Mazurowski MA. Breast cancer MRI radiomics: an overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med. Phys. 2018;45(7):3076–3085. doi: 10.1002/mp.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwanenburg, A., Leger, S., Vallières, M. & Löck, S. Image biomarker standardisation initiative. arXiv preprint arXiv:161207003.
- 41.Hong J, Park BY, Lee MJ, Chung CS, Cha J, Park H. Two-step deep neural network for segmentation of deep white matter hyperintensities in migraineurs. Comput. Methods Programs Biomed. 2019;183:105065. doi: 10.1016/j.cmpb.2019.105065. [DOI] [PubMed] [Google Scholar]
- 42.Ghavami N, Hu Y, Gibson E, Bonmati E, Emberton M, Moore CM, et al. Automatic segmentation of prostate MRI using convolutional neural networks: Investigating the impact of network architecture on the accuracy of volume measurement and MRI-ultrasound registration. Med. Image Anal. 2019;58:101558. doi: 10.1016/j.media.2019.101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kugelman J, Alonso-Caneiro D, Read SA, Hamwood J, Vincent SJ, Chen FK, et al. Automatic choroidal segmentation in OCT images using supervised deep learning methods. Sci. Rep. 2019;9(1):13298. doi: 10.1038/s41598-019-49816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abernethy AP, Etheredge LM, Ganz PA, Wallace P, German RR, Neti C, et al. Rapid-learning system for cancer care. J. Clin. Oncol. 2010;28(27):4268. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heye T, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Feuerlein S, et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra-and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266(3):812–21. doi: 10.1148/radiol.12120255. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Giger ML, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad. Radiol. 2006;13(1):63–72. doi: 10.1016/j.acra.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 47.Lin MQ, Chen JH, Wang XY, Chan SW, Chen SP, Su MY. Template-based automatic breast segmentation on MRI by excluding the chest region. Med. Phys. J. Transl. Name Med. Phys. 2013;40(12):122301. doi: 10.1118/1.4828837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakran S, Chatterjee S, Singhal M, Gupta RK, Singh A. Automatic outer and inner breast tissue segmentation using multi-parametric MRI images of breast tumor patients. PLoS ONE. 2018;13(1):e0190348. doi: 10.1371/journal.pone.0190348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuller CD, Nijkamp J, Duppen JC, Rasch CR, Thomas CR, Jr, Wang SJ, et al. Prospective randomized double-blind pilot study of site-specific consensus atlas implementation for rectal cancer target volume delineation in the cooperative group setting. Int. J. Radiat. Oncol. Biol. Phys. 2011;79(2):481–489. doi: 10.1016/j.ijrobp.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell DM, Perry L, Smith S, Elliott T, Wylie JP, Cowan RA, et al. Assessing the effect of a contouring protocol on postprostatectomy radiotherapy clinical target volumes and interphysician variation. Int. J. Radiat. Oncol. Biol. Phys. 2009;75(4):990–993. doi: 10.1016/j.ijrobp.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 51.El Khouli RH, Macura KJ, Jacobs MA, Khalil TH, Kamel IR, Dwyer A, et al. Dynamic contrast-enhanced MRI of the breast: quantitative method for kinetic curve type assessment. AJR Am. J. Roentgenol. 2009;193(4):W295–300. doi: 10.2214/AJR.09.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pyradiomics feature description. Available from: https://pyradiomics.readthedocs.io/en/latest/features.html.
- 53.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramesh A, Kambhampati C, Monson JR, Drew P. Artificial intelligence in medicine. Ann. R. Coll. Surg. Engl. 2004;86(5):334. doi: 10.1308/147870804290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 56.Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans. Med. Imaging. 1994;13(4):716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- 57.Koo TK, Li MY. A Guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016;15(2):155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Racine JS. RStudio: a platform-independent IDE for R and Sweave. J. Appl. Econom. 2012;27(1):167–172. [Google Scholar]
- 59.Gamer, M., Lemon, J., Fellows, I. & Sing, P. Various Coefficients of Interrater Reliability and Agreement. IRR: R package version 084. 2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.