Abstract
BVES is a transmembrane protein, our previous work demonstrated that single nucleotide mutations of BVES in tetralogy of fallot (TOF) patients cause a downregulation of BVES transcription. However, the relationship between BVES and the pathogenesis of TOF has not been determined. Here we reported our research results about the relationship between BVES and the right ventricular outflow tract (RVOT) stenosis. BVES expression was significantly downregulated in most TOF samples compared with controls. The expression of the second heart field (SHF) regulatory network genes, including NKX2.5, GATA4 and HAND2, was also decreased in the TOF samples. In zebrafish, bves knockdown resulted in looping defects and ventricular outflow tract (VOT) stenosis, which was mostly rescued by injecting bves mRNA. bves knockdown in zebrafish also decreased the expression of SHF genes, such as nkx2.5, gata4 and hand2, consistent with the TOF samples` results. The dual-fluorescence reporter system analysis showed that BVES positively regulated the transcriptional activity of GATA4, NKX2.5 and HAND2 promoters. In zebrafish, nkx2.5 mRNA partially rescued VOT stenosis caused by bves knockdown. These results indicate that BVES downregulation may be associated with RVOT stenosis of non-syndromic TOF, and bves is probably involved in the development of VOT in zebrafish.
Subject terms: Embryology, Disease model
Introduction
Congenital heart disease (CHD) is the most common birth defect, exhibiting a mortality rate of more than 29%1. Within this group of conditions, the incidence of TOF is 7–10% in the USA2, 13.4% in Nigeria3, and 16% in India4. However, the aetiology of TOF has not been fully elucidated.
Anatomically, TOF has four distinct structural defects, namely, ventricular septal defect, overriding of the aorta, RVOT/pulmonary artery (PA) stenosis and right ventricular hypertrophy5. However, embryologically, the defects are thought to be caused by a single developmental error, involving the outflow tract funnel septum shifting left to right or forward, leading to a poor contraposition ventricular septal defect, overriding of the aorta and RVOT funnel stenosis5–11. RVOT stenosis eventually leads to right ventricular hypertrophy10. The degree of RVOT stenosis is a key clinical factor for the diagnosis of TOF10. However, the molecular mechanism behind RVOT/PA stenosis remains under investigation.
The development of the cardiac outflow tract has two major cell sources: one is neural crest cells, which provide cells for the distal development of the great artery by migration, while releasing signals to the SHF7; the other is anterior SHF12, and at the late stage of cardiac looping, cells from the anterior SHF are added to the outflow tract area, which promotes outflow tract elongation and the correct fusion of the outflow tract myocardial wall and ventricular septum13. Ablation of SHF may result in TOF due to an abnormal outflow tract8. In animal models, the knockout or knockdown of SHF regulatory network genes, such as Gata414, nkx2.512,15, Tbx116, Tbx2017, and Hand218, leads to abnormal development of the outflow tract. Functional mutations of SHF genes, such as NKX2.519–21, TBX122,23, GATA424,25, TBX2026, and HAND227, were found in TOF patients. The expression of these genes, including NKX2.528, GATA428, and TBX129, was significantly decreased in the cardiac tissue samples of TOF, indicating that the downregulation of SHF genes is associated with outflow tract stenosis.
BVES, a blood vessel epicardial substance, also known as POPDC1, belongs to the Popeye domain containing (POPDC) gene family, encodes a novel class of cyclic adenosine monophosphate (cAMP) effector proteins, is a highly evolutionarily conserved membrane protein and is highly expressed in adult heart and skeletal muscle in vertebrates30–32. In mice, Bves is expressed in the development of cardiomyocytes33 and coronary endothelial cells34 and is highly expressed in the adult conduction system35. Knockout of Bves in mice was shown to cause sinus bradycardia under stress in an age-dependent manner35. In humans, BVES was found to be more highly expressed in ventricles than in atria36. Functional mutations of the BVES gene were detected in a family affected by hereditary muscular dystrophy with arrhythmia37, and in non-syndromic TOF38,39. It has also been shown that the expression of BVES was downregulated in patients with heart failure36 and non-syndromic TOF39. However, the relationship between BVES downregulation in non-syndromic TOF and outflow tract stenosis has not been determined.
Zebrafish have been used as an animal model for the development of VOT. In zebrafish embryos, SHF cells are derived from mesodermal nkx2.5- and gata4-positive cells15. Zebrafish bves expression began at the zygote transcript stage (1 h post fertilization, 1 hpf) and persisted through the developmental stages of the heart, including the SHF area and other cardiac tissues40,41. It was reported that the expression of the Aggrecan and Cyp26 genes was changed in cardiac tissues with aortic valve disease and TOF42,43, and was also associated with the development of the cardiac outflow tract in zebrafish42,44. However, whether bves is involved in the development of VOT has not been determined.
In this study, BVES downregulation was detected in most of the 83 tissue samples from TOF patients together with the downregulation of the key SHF genes related to TOF. bves downregulation in zebrafish led to abnormal cardiac looping and VOT stenosis and subsequently led to downregulation of the key SHF genes. In addition, our findings suggested that bves and nkx2.5 mRNA may partially rescue the VOT phenotype caused by the downregulation of bves to varying degrees.
Materials and methods
All methods were performed in accordance with the relevant guidelines and regulations.
Samples
This study was approved by the Ethics Committee of Hunan Normal University (NO. 014050), Guangdong General Hospital and the Institutional Ethics Committee of Guangdong Academy of Medical Sciences (GDREC2016186A). Written informed consent was obtained from each subject or their guardian.
TOF tissue samples were obtained from the hypertrophic muscle tissue of RVOT during open-heart surgery for TOF. Normal control samples were obtained from the RVOT tissue of individuals who died in accidents and agreed beforehand for their organs to be donated. Samples from cases with congenital heart-related diseases were excluded. Information about patients and normal controls is shown in Tables 1 and 2, respectively. After obtaining the tissue, it was quickly cut into small, 4–5 mm pieces with surgical scissors, placed into an Eppendorf tube, snap-frozen in liquid nitrogen and stored at − 80 °C for analysis. Zebrafish samples were selected under a microscope for embryonic development, while a embryo was selected as a sample for the extraction of RNA.
Table 1.
Baseline information on the TOF samples.
| Variables | Statistics (n = 83) |
|---|---|
| Male gender (%) | 46 (54%) |
| Female gender (%) | 38 (46%) |
| Age (years) | 3.7 ± 8.9 |
| Patent foramen ovale | 56 (67%) |
| Atrial septal defect (%) | 7 (8%) |
| Patent ductus arteriosus (%) | 18 (22%) |
| Dextrocardia of aortic arch | 1 (1%) |
| Congenital right aorta | 4 (5%) |
| Venous anomalies (%) | 6 (7%) |
| Coronary artery anomalies | 3 (3%) |
| Endocardial cushion defect (%) | 15 (18%) |
| Mesocardiac | 1 (1%) |
| Other body defects | 5 (6%) |
Data are expressed as means and standard deviations, number or percentage. Venous anomalies contain persistent superior vena cava, inferior arch of innominate vein, persistent left superior vena cava into left atrium; Endocardial cushion defect contain tricuspid regurgitation, tricuspid insufficiency, mitral regurgitation, pulmonary regurgitation, pulmonary valve biology, pulmonary valve stenosis, absence of pulmonary valve.
Table 2.
Baseline information of the control samples.
| Gender | Age | Cause of death | Other defects | |
|---|---|---|---|---|
| CT1 | Male | 2 years | Drug abuse | No |
| CT2 | Female | 74 days | Encephalatrophy | No |
| CT3 | Male | 60 years | Encephalorrhagia | Hypertension |
Zebrafish lines
The AB strain of zebrafish was purchased from the Institute of Hydrobiology, Chinese Academy of Sciences. The transgenic line Tg (cmlc2:dsRed) was received as a gift from Didier Stainier, Max Planck Institute of Cardiovascular Research, Germany45, and Tg (flia:eGFP) from Qingshun Zhao, Nanjing Model Animal Research Center46. Adult zebrafish were raised and maintained under standard laboratory conditions47,48. The animal experimental protocol was approved by the Ethics Committee of Hunan Normal University (NO. 014050), and performed according to the relevant guidelines and regulations.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total cDNA was prepared from whole embryos or tissues and qRT-PCR was performed as previously described39. In patient samples, the data of Ct values were normalized to GAPHD, and the fold change between normal control samples and TOF tissue samples was quantified using the 2−ΔΔCT Livak Method. The significance was analyzed by Student’s t-test. In the same plate, the expression in patient tissue that was lower than the average expression of normal samples was defined as ‘downregulated’, while the expression in patient tissue was higher than the average expression of normal samples was defined as ‘upregulated’, and the data are presented as scatter points and histogram that were generated by GraphPad Prism 5. In zebrafish samples, the control group and knockdown group included at least three samples for each time point. The data of Ct values were normalized to gapdh, and the fold change between control group and knockdown group was quantified using the 2−ΔΔCT Livak Method. The significance was analyzed by Student’s t-test. The data are presented in the form of a histogram that were generated by GraphPad Prism 5. All primer sequences are shown in Table S1.
Plasmid construction
The construction of overexpression plasmid with pCMV-BVES and luciferase reporter plasmids with the human NKX2.5, GATA4 and MEF2C promoters were performed as previously described39. Promotors of TBX1, TBX20 and HAND2 were amplified by primer 1 (forward, ATCGGTACCGAATTCagaatgtccaacacaacatcc; reverse, GTGATATCAGATCTCccatcaggcccagtctgagg, the capital letters are homologous arms on Vector), primer 2 (forward, ATCGGTACCGAATTCagaatgtccaacacaacatcc, reverse, GTGATATCAGATCTCccatcaggcccagtctgagg, the capital letters are homologous arms on Vector), and primer 3 (forward, ATCGGTACCGAATTCacacgagtaaggccggtttt, reverse, GTGATATCAGATCTCcggttagagctgtttggggt, the capital letters are homologous arms on Vector), repectively. pGL3-Bias plasmid were digested by restriction enzyme, XhoI, to linearize the plasmid. Promotor sequences were ligated into vectors using ClonExpress Ultra One Step Cloning Kit (Vazyme).
Luciferase reporter assays
The luciferase reporter assays were analysesd as previously described39. HEK293T cell line was used, and the levels of firefly luciferase were standardised relative to that of Renilla luciferase.
Morpholinos and mRNA injections
Zebrafish embryos were injected as reported previously40, and the sequences of the bves morpholino oligos was designed intended to block the translation of bves (5′-GATGTTGTGTTGGACATTCTGAGGC-3′, synthesised by GeneTools). pXT7-bves and pXT7-nkx2.5 were linearized and used for in vitro transcription (Ribo m7G Cap Analogue, RiboMAX Large Scale RNA Production Systerm-T7; Promega). A total of 150 ng and 80 ng of capped mRNA of bves and nkx2.5 was coinjected with bves morpholino, respectively.
Phenotypic analyses of zebrafish
Embryos were incubated at 28.5 °C in petri dishes containing fresh water and maintained as described48. 6% methylcellulose was used to restrict the movement of zebrafish and Axiocam of Zeiss Company was used to photograph and analyse the morphology of juvenile fish at 48 hpf and 72 hpf. The heart phenotype of the juvenile fish at these two stages were analysed as previously described48. The range of motion of the heart was clearly seen, and the approximate position of VOT was observed. Under normal beating conditions of zebrafish heart, continuous photographs were taken with green fluorescence. Finally, the diastolic and systolic images were selected for analysis. Zeiss AxioVision 3.0.6 software and Adobe Photoshop were used to process images. We measured the width of ventricle outflow tract at the end-systolic stage using Digimizer, which normalised the scales added by Zeiss AxioVision 3.0.6 software automatically.
Western blot
Total protein samples were prepared in radioimmunoprecipitation assay (RIPA) buffer, and the protein concentration was determined by BCA assay (Beyotime). Protein isolation was carried out in a 12% SDS polyacrylamide gel. Then, the protein was transferred to nitrocellulose membranes, blocked with 8% skim milk, and incubated with anti-BVES antibody (1:1,500 dilution; Absin) and anti-β-ACTIN antibody (1:3,000 dilution; Proteintech). The signal densities of BVES protein bands were quantified and normalised to β-ACTIN using ImageJ.
Statistical analysis
The scatter points and histograms were generated by GraphPad Prism 5. To assess whether the experimental data of two groups and three groups were significantly different from each other, we applied Student’s t-test and one-way Anova, respectively. A p value of < 0.05 was considered statistically significant.
Results
Expression pattern of human BVES was related to pulmonary artery development
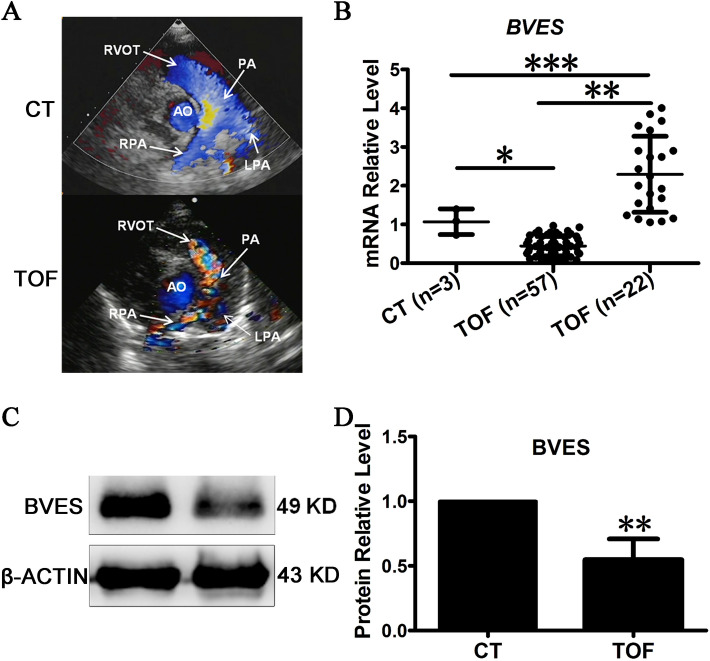
The RVOT in TOF patients showed a phenotype of RVOT stenosis in all samples examined (Fig. 1A). The parasternal short axis view of cardiac colour Doppler echocardiography is important to assess the size of, for instance, the RVOT, PV, main pulmonary artery, and ascending aorta49. In normal people, the blood flow of RVOT and PA was antegrade and presented a single colour, while in TOF patients, it was aliasing and turbulent because of stenosis and presented multiple colours50 (Fig. 1A). The expression pattern of BVES in RVOT tissues with TOF was examined by qRT-PCR, and 79 of the 83 cases were valid for qRT-PCR analysis. Among them, the downregulation of BVES was detected in 57 cases, accounting for 72.2% of the total, while there was upregulation of BVES in the remaining 22 cases (Fig. 1B). Western blot analysis showed that the level of BVES protein expression in the three samples with BVES downregulation was approximately half that in the control (Fig. 1C,D). These results indicate that BVES downregulation could be related to TOF.
Figure 1.
Relationship between the expression pattern of BVES and TOF. (A) The phenotype of RVOT stenosis in TOF diagnosed by echocardiography. RVOT, right ventricular outflow tract; AO, aorta; PA, pulmonary artery; RPA, right pulmonary artery; LPA, left pulmonary artery. CT, Control, the normal phenotype; TOF, tetralogy of fallot, the mal-phenotype of TOF patients. (B) qRT-PCR detected the expression of BVES. (C) Western blot detected the expression of BVES in downregulated samples. (D) Quantification of BVES by greyscale analysis. CT, control, RVOT tissue of normal controls; TOF, tetralogy of fallot, hypertrophic RVOT tissue of patients; n: number of samples. *p < 0.05; **p < 0.01; ***p < 0.001. The error bar shows the mean and SD.
Expression pattern of a set of SHF genes in RVOT stenosis with TOF
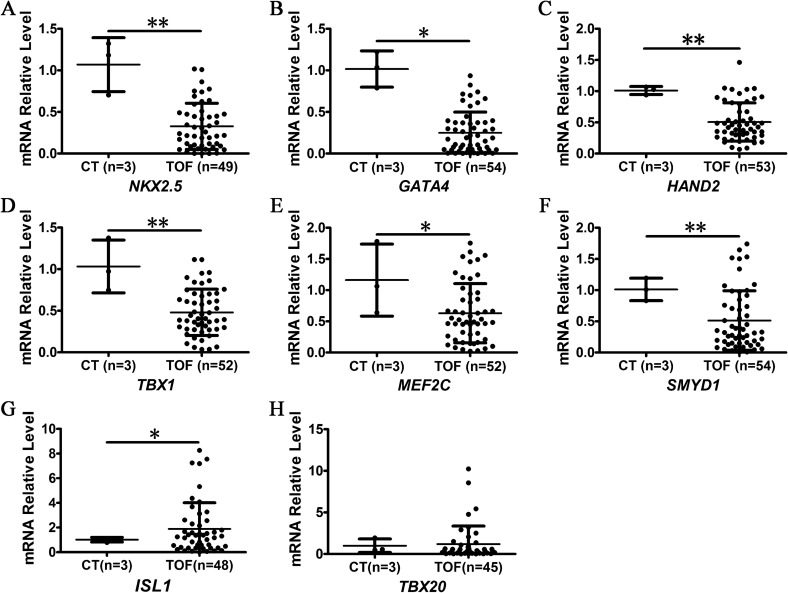
It has been shown that the genes in the SHF regulatory network are required for the development of the cardiac outflow tract2,5,6,9,10,51. To examine the expression pattern of the SHF genes in RVOT stenosis tissue, the gene expression patterns in the RVOT tissues of TOF with BVES downregulation were analysed by qRT-PCR. The results showed that the expression of human GATA4, NKX2.5, TBX1, HAND2, SMYD1 and MEF2C was significantly downregulated compared with that in the control (Fig. 2A–F), which is similar to the cases for GATA4, NKX2.5 and TBX1 observed in other studies28,29. Compared with the control samples, the mean expression of ISL1 was higher than that of the control group, which was significant (Fig. 2G), although approximately 41.7% of the RVOT stenosis samples (20/48) were downregulated. However, the expression of TBX20 was unexchanged (Fig. 2H). These results further indicate that the downregulation of the SHF genes is related to ROVT stenosis in TOF.
Figure 2.
Detection of SHF gene expression. CT, control, RVOT tissue of normal controls; TOF, tetralogy of fallot, hypertrophic RVOT tissue of patients; n, Number of samples. *p < 0.05; **p < 0.01. The error bar shows the mean and SD.
bves knockdown in zebrafish led to abnormal heart looping
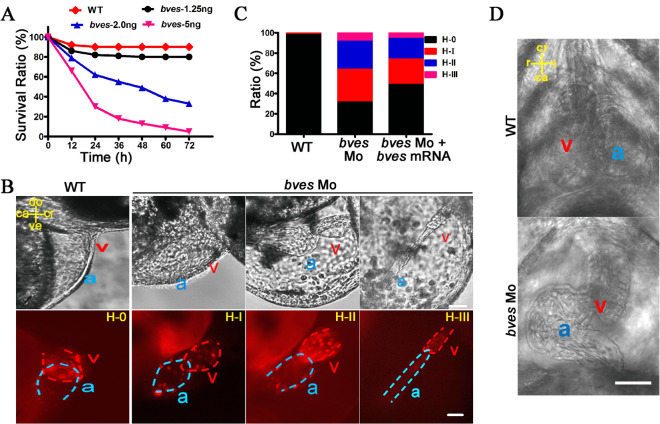
Previous studies showed that abnormal looping of the heart (the change of outer or inner curvature) disrupts the alignment between the ventricle and artery, leading to congenital heart phenotypes, such as TOF8,52,53. To explore the causal relationship in vivo between BVES downregulation in ROVT stenosis and the development of heart looping, a zebrafish bves knockdown model with bves morpholino was used. We chose a bves morpholino blocking bves translation that was reported in a previous report40. Knockdown efficiency was verified by analysing the survival rate of the morphants at 72 hpf. The results showed that the morphants with 2.0 ng were suitable for analysing the cardiac phenotype, which is consistent with a previous report40, while the morphants with 1.25 ng were similar to the wild type, and the morphants with 5 ng with a death occurred at 5 dpf (Fig. 3A). Therefore, a bves knockdown model, the morphants injected with 2.0 ng of morpholino were used in the subsequent analyses, we called it bves Mo.
Figure 3.
bves knockdown led to abnormal cardiac looping in zebrafish. (A) Survival ratio of wild type and morphants with 1.25, 2.0 and 5.0 ng of bves Mo at 72 hpf. WT, wild type; bves-1.25 ng, bves morphants with injection of 1.25 ng of bves Mo; bves-2.0 ng, bves morphants with injection of 2.0 ng of bves Mo; bves-5.0 ng, bves morphants with injection of 5.0 ng of bves Mo. (B) bves morphants showed cardiac defects at 48 hpf for the cmlc2:dsRed transgenic fish (a: atrium; v: ventricle), which were divided into four types based on the degree of cardiac dysplasia. H-0, the normal phenotypes; H-I, the moderate phenotypes; H-II, the strong phenotypes; H-III, the severe phenotypes. The position of the heart is marked by compass lines. ca, caudal; cr, cranial; do, dorsal; ve, ventral. Scale bar: 50 μm. (C) Data statistics of the different cardiac phenotypes in (B). (D) The morphants at 72 hpf with a left-looping heart in the ventral view. ca, caudal; cr, cranial; r, right; l, left. Scale bar: 50 μm. WT: wild type; bves Mo: bves morphants; bves Mo + bves mRNA, coinject bves morpholino and bves mRNA.
To determine the effect of bves knockdown on cardiac development, the embryos of Tg (cmlc2:dsRed) zebrafish were used for bves morpholino injection, in which RFP was localized at the cardiomyocytes, facilitating live imaging of heart morphology. At 48 hpf, the morphants showed looping defects, cardiac dysplasia, and cardiac oedema (Fig. 3B). Classification into four phenotypes could be performed according to the degree of cardiac abnormality48, and approximately 70% of bves morphants showed defects of various degrees of severity, and this phenotype was partially rescued by coinjection with bves morpholino and bves mRNA (Fig. 3C). At 72 hpf, approximately 6.7% (3/45) of the morphants showed a left-looping heart in the ventral view (Fig. 3D), and the morphant with left-looping heart was not found in the WT or coinjection with bves morpholino and bves mRNA. These results indicated that knockdown of bves led to abnormal cardiac looping.
bves knockdown in zebrafish causes VOT stenosis
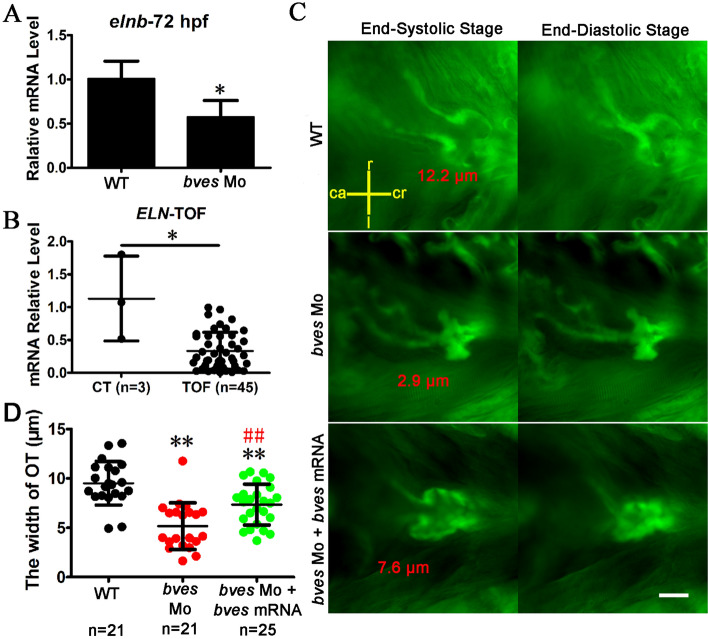
The RVOT is the site of circulation between the right ventricular and the pulmonary artery5,54. elnb, a marker of the outflow tract in zebrafish, showed high expression at 72 hpf55. Compared with the wild type, the expression of elnb was downregulated in bves morphants (Fig. 4A), consistent with TOF patient samples (Fig. 4B). The results showed that, in the bves morphants, the VOT was abnormal. Stenosis of the PA or the RVOT is an indicator of TOF. To study the role of bves in the development of the outflow tract, the embryos of Tg (flia:eGFP) zebrafish were used for the bves morpholino injection, in which GFP is expressed in all endothelial cells46. In the wild type (Fig. 4C, top row), the VOT at 72 hpf was open at the end-systolic stage, while the VOT was closed at the end-diastolic stage (Fig. 4C). In the bves morphants (Fig. 4C, middle row), the VOT was smaller and narrower than that in the wild type at the end-systolic stage. The width of VOT at the end-systolic stage was reduced approximately 50% (WT, 9.5 μm; bves Mo, 5.0 μm, p < 0.01) (Fig. 4D). The narrow phenotype was mostly rescued by bves mRNA (bves Mo + bves mRNA, 7.5 μm, p < 0.01 compared with WT; p < 0.01 compared with bves Mo) (Fig. 4C, bottom row; Fig. 4D). These results suggest that bves knockdown affected the development of outflow tract, leading to the phenotype of VOT stenosis.
Figure 4.
bves knockdown led to the phenotype of outflow tract stenosis in zebrafish with flia:eGFP. (A) qRT-PCR detected the expression of elnb in zebrafish at 72 hpf. WT, wild type; bves Mo, bves morphants. *p < 0.05. (B) qRT-PCR detected the expression of ELN in human tissue exapmles. CT, control, RVOT tissue of normal controls; TOF, tetralogy of fallot, hypertrophic RVOT tissue of patients. *p < 0.05. (C) bves morphants showed outflow tract defects at 72 hpf in Tg (flia:eGFP) zebrafish. The left column shows the outflow tract at the cardiac end-systolic stage, and the right column shows the end-diastolic stage. The words that marked by red color is the width of outflow tract. ca, caudal; cr, cranial; r, right; l, left. Scale bar = 15 μm. (D) Statistics about the width of outflow at the end-systolic stage in (C). Compared with the WT, the width of the outflow tract in bves knockdown mutants decreased by approximately 50% (WT, 9.5 μm; bves Mo, 5.0 μm, p < 0.01 compared with WT; bves Mo + bves mRNA, 7.5 μm, p < 0.01 compared with WT; p < 0.01 compared with bves Mo). OT, outflow tract; *Significance analysis with WT; #Significance analysis with bves Mo. **p < 0.01; ##p < 0.01. n, number of samples. The error bar shows the mean and SD. bves Mo: bves morphants; WT: wild type; bves Mo + bves mRNA, coinject bves morpholino and bves mRNA.
bves regulated VOT development via SHF genes
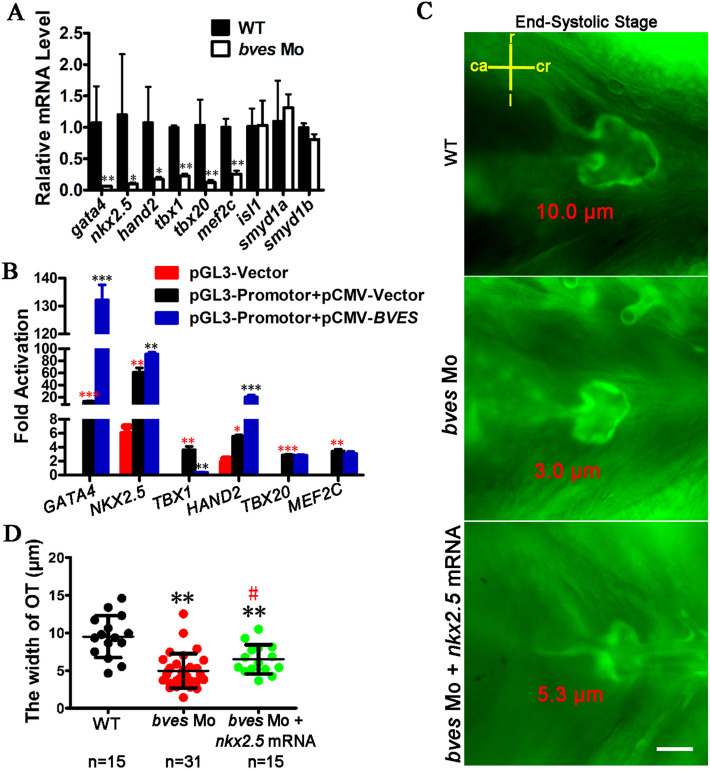
The development of VOT at the late stage of cardiac looping is closely related to the SHF genes2,5,6,9,10,51. To detect the effects of SHF genes on the development of outflow tract, their expression in the bves morphants at 48 hpf was analysed by qRT-PCR. As seen in Fig. 5A, the expression of gata4, nkx2.5, hand2, tbx1, tbx20 and mef2c was significantly downregulated, which is consistent with the downregulation detected in the RVOT tissues of TOF (Fig. 2), except for tbx20. The expression of smayd1a, smayd1b and isl1 was unchanged (Fig. 5A). To study the regulatory relationship between bves and the SHF genes, GATA4, NKX2.5, TBX1, HAND2, TBX20 and MEF2C were chosen for dual-fluorescence reporter system analysis. The results showed that the overexpression of BVES in the HEK293T cell line significantly increased the transcriptional activities of the promoters of GATA4, NKX2.5 and HAND2 by tenfold, 1.5-fold and threefold, respectively (Fig. 5B). For other transcriptional activities, TBX1 was decreased by ninefold, and TBX20 and MEF2C were unchanged. These results suggest that genes in the SHF regulatory network, such as GATA4, NKX2.5 and HAND2, are potential factors involved downstream of BVES.
Figure 5.
bves regulates the development of the outflow tract via SHF. (A) SHF gene expression was detected in bves morphants by qRT-PCR at 48 hpf. (B) Overexpression of BVES led to upregulation of transcriptional activity at the promoters of GATA4, NKX2.5 and HAND2, downregulation of transcriptional activity at the promoters of TBX1, and no change in transcriptional activity at the promoters of TBX20 and MEF2C. pCMV-Vector, empty vector of pCMV-Myc; pCMV-BVES, the vector overexpressing BVES; pGL3-Vector, empty vector of pGL3-Bias; pGL3-Promotor, vector containing the promotor of gene. Red *: significance analysis with pGL3-Vector group; Black *: significance analysis with pGL3-Promotor + pCMV-Vector group. *p < 0.05; **p < 0.01; ***p < 0.001. (C) Outflow tract phenotype of the wild type, bves Mo injected, bves Mo and nkx2.5 mRNA coinjected in end-systolic at 72 hpf in Tg (flia:eGFP). The words that marked by red color is the width of outflow tract. ca, caudal; cr, cranial; r, right; l, left. Scale bar = 15 μm. (D) The width of the outflow tract at the end-systolic stage in (C). Compared with the WT and bves Mo, the width of the outflow tract was partially rescued by nkx2.5 mRNA (WT, 9.5 μm, bves Mo, 5.0 μm, p < 0.01 compared with WT; bves Mo + nkx2.5 mRNA, 6.3 μm, p < 0.01 compared with WT, p < 0.05 compared with bves Mo). n, number of samples. OT, outflow tract. *Significance analysis with WT; #Significance analysis with bves Mo. #p < 0.01; **p < 0.01. The error bar shows the mean and SD. WT: wild type; bves Mo: bves morphants; bves Mo + nkx2.5 mRNA, coinject bves morpholino and nkx2.5 mRNA.
To further confirm that bves regulates VOT development through the SHF genes, we coinjected bves morpholino and nkx2.5 mRNA into Tg (flia:EGFP) transgenic zebrafish and analysed the development of VOT at 72 hpf. The results showed that the width of VOT was partially rescued by nkx2.5 mRNA (Fig. 5C,D) (WT, 9.5 μm, bves Mo, 5.0 μm, p < 0.01 compared with WT; bves Mo + Nkx2.5 mRNA, 6.3 μm, p < 0.01 compared with WT, p < 0.05 compared with bves Mo). These results indicated that bves regulated the development of VOT via its downstream genes, such as nkx2.5, gata4 and hand2. However, the results need to be further studied.
Discussion
Our previous report showed that BVES allelic variants are associated with RVOT stenosis in TOF patients, which leads to the downregulation of BVES itself at the transcriptional and protein levels in the tissues of cases of RVOT stenosis with TOF containing allelic variants38,39. These results suggest that the downregulation caused by the BVES allelic variants was related to the phenotype of RVOT stenosis, consistent with the finding that gene downregulation is associated with RVOT stenosis, as demonstrated by other investigators28,29,38. However, the mechanism underlying the association between BVES downregulation and RVOT stenosis has not been elucidated. In this study, BVES downregulation was detected in approximately 72.2% of the samples (57/79) from cases of RVOT stenosis with TOF with decreases of both mRNA and protein levels by half. The results presented in this paper are consistent with a previous report39, suggesting that the downregulation of the BVES gene in RVOT stenosis with TOF is an aetiology, and not only the aetiology that lead to the phenotype of TOF.
It has been shown that, at the late stage of cardiac looping, cells from the anterior SHF are added to the outflow tract area for outflow tract elongation and the correct fusion of the outflow tract myocardial wall and ventricular septum6,7,13. Ablation of SHF may result in TOF due to an abnormal outflow tract8. In this study, compared with the normal RVOT samples, the expression of genes in the SHF regulatory network, such as NKX2.5 and GATA4, HAND2, TBX1, MEF2C and SMYD1, were significantly decreased in the TOF samples, with BVES being mostly downregulation (Fig. 2) and occasionally upregulated (data unpublished). In animal models, knockout or knockdown of SHF regulatory network genes, such as Gata414 and nkx2.512,15, leads to abnormal outflow tract development. The expression of NKX2.5 and GATA4 in the open-heart surgery of CHD tissues is downregulated28, which is similar to that in the myocardial tissue at the earlier stage of embryonic development in the nkx2.5 and gata4 knockdown animal models12,15. These results suggest that the downregulation of gene expression detected in the tissues obtained during surgery may be similar to the gene downregulation in embryonic development. In our zebrafish model, knockdown of bves significantly decreased the expression of the five genes in the SHF regulatory network, including nkx2.5, gata4, hand2, mef2c and tbx1 (Fig. 5A), suggesting that bves downregulation may be the cause of the maldevelopment of heart looping and VOT. The results with a dual-fluorescence reporter system confirmed that BVES positively regulated the transcriptional activity of GATA4, NKX2.5 and HAND2 and negatively regulated the transcriptional activity of TBX1 promoters (Fig. 5B). The above results suggest that GATA4, NKX2.5 and HAND2 are potential downstream genes of the BVES gene.
Nkx2.5 and Gata4, as marker of cardiac progenitors, together regulate the arrangement of ventricle outflow14. In Gata4+/− mice, a small number of heterozygote mutants were double outlet right ventricle (DORV), and in Gata4+/−;Gata5+/− double heterozygote mutants, almost all embryos were DORV, some of which had further aortic stenosis. Hand2, a heart and neural crest derivative, is important for the development of the heart, especially in the outflow tract56. In the mutant mice of Hand2f/f;Mef2c-Anf-Cre, which specifically deleted Hand2 in the SHF progenitors, the OFT lumen was narrowed, and the OFT wall was thickened57. In other mutant mice of Hand2f/-;Wnt1-Cre cKO, which specifically deleted Hand2 in the neural crest, a variety of arterial malformations, including pulmonary stenosis, were observed18. In this paper, nkx2.5 mRNA only partially rescued the OFT malformation caused by bves Mo, which indicated that bves caused OFT abnormalities by widely regulating the heart development gene, not one of the heart development genes. The cells of outflow tract are different from anterior lateral plate mesoderm (ALPM), where SHF progenitors are specified in higher vertebrates. The descendants of gata4+ or nkx2.5+ cell at ALPM are crucial for the development of outflow tract by compromised progenitor cell proliferation15,58,59. Nkx2.5 and Gata4 play an important role in the outflow tract by regulating the SHF genes, but they also play a vital role in early specialization of cardiomyocyte progenitor cells60,61. In our bves knockdown model in zebrafish, the early heart development was affected (Fig. 3), so bves leading to the abnormal phenotypes of outflow tract may be affected both by the outflow tract development via regulating SHF genes and the indirect consequence of early defects via regulating early cardiac progenitor specification. However, the molecular regulatory mechanism between bves and nkx2.5, gata4 and hand2 requires further study.
BVES, encodes a transmembrane protein, that contains three transmembrane domains and a Popeye domain. The Popeye domain makes up a large part of the cytoplasmic portion of the protein and functions as a cAMP-binding domain62,63. In both mice and zebrafish, Bves act as effector proteins of cAMP, which control the development of the conduction system by combining ion channel genes35,37. Furthermore, BVES has a functional mutant in AV-block patients37,64. However, in the process of carcinogenesis, BVES inhibits epithelial–mesenchymal transition (EMT) and cell adhesion of cancer cells to prevent the formation of cancer via GEFT/Rho signalling and WNT signalling65. In our previous study, BVES also had functional polymorphisms in TOF patients39, but whether BVES is related to the maldevelopment of OFT has not been studied to date. In this paper, we illustrate that bves is related to the development of OFT in zebrafish, and the potential target genes of bves are nkx2.5, gata4 and hand2. Whether bves acts as an effector protein of cAMP or as a signal transduction member, such as GEFT/Rho signalling and WNT signalling, to regulate the expression of nkx2.5, gata4 and hand2 needs further study.
Zebrafish, as an animal model, has been used to study the pathogenesis of many diseases due to its simple developmental structure of the organs studied. In terms of studies on cardiac diseases, zebrafish have been used in research on heart failure66, congenital heart defects67,68, and outflow tract disease42,69. In this paper, we used morpholinos to downregulate bves expression in zebrafish. The downregulation of bves in zebrafish caused looping defects, cardiac dysplasia, cardiac oedema (Fig. 3B), stenosis of the VOT (Fig. 4C) and several heart rate abnormalities (data unpublished). Abnormal heart rhythm leads to abnormal ventricular contraction, which makes the cilia of the developing heart and outflow tract feel different shear stress, thereby affecting the development of the heart and outflow tract70,71. In our bves knockdown model, we found that some of the mutants had abnormal rhythms. Because the phenotype of outflow tract stenosis also appears in the mutants with normal heart development and rhythm, we think that the difference in outflow tract dynamics caused by ventricular contract was considerably less than outflow tract malformation. However, further studies need to be performed to support this hypothesis. In addition, the abnormal phenotypes of VOT and heart looping were partially rescued by bves mRNA (Figs. 3D, 4C), supporting the hypothesis that the phenotypes were partially caused by the downregulation of bves, and several might be caused by the toxicity of the morpholino. To further explore the molecular mechanism by which bves induced developmental abnormalities in the heart and VOT, it is necessary to establish a bves knockout zebrafish line.
In summary, we studied the relationship between BVES downregulation and the occurrence of RVOT stenosis in TOF using human RVOT stenosis samples and a zebrafish model of bves downregulation. Our results show that bves is required for the development of VOT in zebrafish, suggesting that BVES downregulation is associated with the occurrence of RVOT stenosis in non-syndromic TOF patients.
Supplementary information
Acknowledgements
This study was supported in part by grants from the National Natural Science Foundation of China (Nos.: 81670290, 81470449, 81370451, 31572349, 81670288, 81570279, 81600320, 81800289, 81974019, 81700338, 81470377, 81970324, 31872315), National Key Research and Development Program of China (2018YFA0108700, 2018YFC1002600, 2017YFA0105602), NSFC Projects of International Cooperation and Exchanges (81720108004), Science and Technology Planning Projects of Guangdong Province of China (2017A070701013, 2017B090904034, 2017B030314109, 2018B090944002, 2019B020230003), The Research Team Project of Natural Science Foundation of Guangdong Province of China (2017A030312007), The key program of Guangzhou science research plan (201904020047), the Special Project of Dengfeng Program of Guangdong Provincial People's Hospital (DFJH201802, DFJH201812, KJ012019119, KJ012019423), Hunan Provincial Natural Science Foundation of China (Nos. 2019JJ50394) and Guangdong Provincial Key Laboratory of South China Structural Heart Disease.
Author contributions
Y.S., X.W., W.Y., P.Z. and J.Z., designed and performed the experiments, analyzed and interpreted the data, wrote the paper; Y.L., Y.W. and X.F. did analyses and data interpretation, modified the paper; Y.C., H.W., S.Y. and Y.S. performed the experiments, such as luciferase reporter assays and so on; Z.J., Z.Z. and W.C. did zebrafish experiment; X.X. provided photos (Fig. 1A) and analysis for clinical indicators of TOF; J.C., H.Y., G.W., X.L. and X.Z. collected samples and information of TOF patients; Y.W., G.D., F.L., X.M. and X.Y. analyzed the data and designed figure layout. All authors reviewed the manuscript, and have no objection to this ranking.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yan Shi, Yongqing Li, Yuequn Wang and Ping Zhu.
Contributor Information
Jian Zhuang, Email: Zhuangjian5413@tom.com.
Xiushan Wu, Email: xiushanwu@yahoo.com.
Wuzhou Yuan, Email: yuanwuzhou@aliyun.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-70806-4.
References
- 1.Lloyd-Jones D, et al. Heart disease and stroke statistics—2009 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;3:480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Morgenthau A, Frishman WH. Genetic origins of tetralogy of fallot. Cardiol. Rev. 2018;2:86–92. doi: 10.1097/CRD.0000000000000170. [DOI] [PubMed] [Google Scholar]
- 3.Ekure EN, et al. Clinical epidemiology of congenital heart disease in Nigerian children, 2012–2017. Birth Defects Res. 2018;110:1233–1240. doi: 10.1002/bdr2.1361. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj R, et al. Epidemiology of congenital heart disease in India. Congenit. Heart Dis. 2015;5:437–446. doi: 10.1111/chd.12220. [DOI] [PubMed] [Google Scholar]
- 5.Apitz C, Webb GD, Redington AN. Tetralogy of Fallot. Lancet. 2009;9699:1462–1471. doi: 10.1016/S0140-6736(09)60657-7. [DOI] [PubMed] [Google Scholar]
- 6.Schleich JM, Abdulla T, Summers R, Houyel L. An overview of cardiac morphogenesis. Arch. Cardiovasc. Dis. 2013;11:612–623. doi: 10.1016/j.acvd.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Bajolle F, et al. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ. Res. 2006;3:421–428. doi: 10.1161/01.RES.0000202800.85341.6e. [DOI] [PubMed] [Google Scholar]
- 8.Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev. Biol. 2005;1:72–83. doi: 10.1016/j.ydbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Waldo KL, et al. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 2005;1:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Sommer RJ, Hijazi ZM, Rhodes JF. Pathophysiology of congenital heart disease in the adult: Part III: Complex congenital heart disease. Circulation. 2008;10:1340–1350. doi: 10.1161/CIRCULATIONAHA.107.714428. [DOI] [PubMed] [Google Scholar]
- 11.Anderson RH, Mori S, Spicer DE, Brown NA, Mohun TJ. Development and morphology of the ventricular outflow tracts. World J. Pediatr. Congenit. Heart Surg. 2016;5:561–577. doi: 10.1177/2150135116651114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;11:2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson RH, et al. Normal and abnormal development of the intrapericardial arterial trunks in humans and mice. Cardiovasc. Res. 2012;1:108–115. doi: 10.1093/cvr/cvs147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laforest B, Nemer M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev. Biol. 2011;2:368–378. doi: 10.1016/j.ydbio.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 15.Guner-Ataman B, et al. Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development. 2013;6:1353–1363. doi: 10.1242/dev.088351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;13:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi JK, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;10:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ. Res. 2008;12:1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- 19.Cao Y, et al. Single nucleotide polymorphism of NKX2-5 gene with sporadic congenital heart disease in Chinese Bai population. Int. J. Clin. Exp. Pathol. 2015;11:14917–14924. [PMC free article] [PubMed] [Google Scholar]
- 20.Gioli-Pereira L, et al. NKX25 mutations in patients with non-syndromic congenital heart disease. Int. J. Cardiol. 2010;3:261–265. doi: 10.1016/j.ijcard.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Yin J, et al. Search of somatic mutations of NKX2–5 and GATA4 genes in Chinese patients with sporadic congenital heart disease. Pediatr. Cardiol. 2018;40:17–22. doi: 10.1007/s00246-018-1955-z. [DOI] [PubMed] [Google Scholar]
- 22.Conti E, et al. DiGeorge subtypes of nonsyndromic conotruncal defects: Evidence against a major role of TBX1 gene. Eur. J. Hum. Genet. 2003;4:349–351. doi: 10.1038/sj.ejhg.5200956. [DOI] [PubMed] [Google Scholar]
- 23.Xu YJ, et al. Detecting 22q11.2 deletion in Chinese children with conotruncal heart defects and single nucleotide polymorphisms in the haploid TBX1 locus. BMC Med Genet. 2011;12:169. doi: 10.1186/1471-2350-12-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng T, Wang L, Zhou SF, Li X. Mutations of the GATA4 and NKX25 genes in Chinese pediatric patients with non-familial congenital heart disease. Genetica. 2010;11–12:1231–1240. doi: 10.1007/s10709-010-9522-4. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida A, et al. Genetic mutation analysis in Japanese patients with non-syndromic congenital heart disease. J. Hum. Genet. 2016;2:157–162. doi: 10.1038/jhg.2015.126. [DOI] [PubMed] [Google Scholar]
- 26.Huang RT, et al. TBX20 loss-of-function mutation responsible for familial tetralogy of Fallot or sporadic persistent truncus arteriosus. Int. J. Med. Sci. 2017;4:323–332. doi: 10.7150/ijms.17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu CX, et al. A novel HAND2 loss-of-function mutation responsible for tetralogy of Fallot. Int. J. Mol. Med. 2016;2:445–451. doi: 10.3892/ijmm.2015.2436. [DOI] [PubMed] [Google Scholar]
- 28.Tong YF. Mutations of NKX25 and GATA4 genes in the development of congenital heart disease. Gene. 2016;1:86–94. doi: 10.1016/j.gene.2016.04.061. [DOI] [PubMed] [Google Scholar]
- 29.Aguayo-Gomez A, et al. Identification of copy number variations in isolated tetralogy of Fallot. Pediatr. Cardiol. 2015;8:1642–1646. doi: 10.1007/s00246-015-1210-9. [DOI] [PubMed] [Google Scholar]
- 30.Reese DE, Zavaljevski M, Streiff NL, Bader D. bves: A novel gene expressed during coronary blood vessel development. Dev. Biol. 1999;1:159–171. doi: 10.1006/dbio.1999.9246. [DOI] [PubMed] [Google Scholar]
- 31.Smith TK, et al. Bves directly interacts with GEFT, and controls cell shape and movement through regulation of Rac1/Cdc42 activity. Proc. Natl. Acad. Sci. USA. 2008;24:8298–8303. doi: 10.1073/pnas.0802345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amunjela JN, Swan AH, Brand T. The role of the popeye domain containing gene family in organ homeostasis. Cells. 2019;8(12):1594. doi: 10.3390/cells8121594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andree B, Fleige A, Arnold HH, Brand T. Mouse Pop1 Is required for muscle regeneration in adult skeletal muscle. Mol. Cell. Biol. 2002;5:1504–1512. doi: 10.1128/mcb.22.5.1504-1512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith TK, Bader DM. Characterization of Bves expression during mouse development using newly generated immunoreagents. Dev. Dyn. 2006;6:1701–1708. doi: 10.1002/dvdy.20739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froese A, et al. Popeye domain containing proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J. Clin. Invest. 2012;3:1119–1130. doi: 10.1172/JCI59410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gingold-Belfer R, et al. Popeye domain-containing 1 is down-regulated in failing human hearts. Int. J. Mol. Med. 2011;1:25–31. doi: 10.3892/ijmm.2010.558. [DOI] [PubMed] [Google Scholar]
- 37.Schindler RF, et al. POPDC1(S201F) causes muscular dystrophy and arrhythmia by affecting protein trafficking. J. Clin. Invest. 2016;1:239–253. doi: 10.1172/JCI79562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, et al. Mutational and functional analysis of the BVES gene coding region in Chinese patients with non-syndromic tetralogy of Fallot. Int. J. Mol. Med. 2013;4:899–903. doi: 10.3892/ijmm.2013.1275. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, et al. The functional polymorphism R129W in the BVES gene is associated with sporadic tetralogy of Fallot in the Han Chinese population. Genet. Test Mol. Biomarkers. 2019;9:601–609. doi: 10.1089/gtmb.2019.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu YC, et al. Blood vessel epicardial substance (Bves) regulates epidermal tight junction integrity through atypical protein kinase C. J. Biol Chem. 2012;47:39887–39897. doi: 10.1074/jbc.M112.372078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu YC, et al. Knockdown of zebrafish blood vessel epicardial substance results in incomplete retinal lamination. ScientificWorldJournal. 2014;2014:803718. doi: 10.1155/2014/803718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambeau P, et al. Reduced aggrecan expression affects cardiac outflow tract development in zebrafish and is associated with bicuspid aortic valve disease in humans. Int. J. Cardiol. 2017;249:340–343. doi: 10.1016/j.ijcard.2017.09.174. [DOI] [PubMed] [Google Scholar]
- 43.Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge syndrome in the chick. Hum. Mol. Genet. 2006;23:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- 44.Rydeen AB, Waxman JS. Cyp26 enzymes facilitate second heart field progenitor addition and maintenance of ventricular integrity. PLoS Biol. 2016;11:e2000504. doi: 10.1371/journal.pbio.2000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;7288:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jing Y, et al. Antiangiogenic effects of AA-PMe on HUVECs in vitro and zebrafish in vivo. Onco Targets Ther. 2018;11:1871–1884. doi: 10.2147/OTT.S157747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mm W. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) Rerio. Eugene: University of Oregon Press; 1994. [Google Scholar]
- 48.Peng X, et al. CXXC5 is required for cardiac looping relating to TGFbeta signaling pathway in zebrafish. Int. J. Cardiol. 2016;214:246–253. doi: 10.1016/j.ijcard.2016.03.201. [DOI] [PubMed] [Google Scholar]
- 49.Swamy P, Bharadwaj A, Varadarajan P, Pai RG. Echocardiographic evaluation of tetralogy of Fallot. Echocardiography. 2015;32:S148–156. doi: 10.1111/echo.12437. [DOI] [PubMed] [Google Scholar]
- 50.Saraste M, et al. Transthoracic Doppler echocardiography as a noninvasive tool to assess coronary artery stenoses—A comparison with quantitative coronary angiography. J. Am. Soc. Echocardiogr. 2005;6:679–685. doi: 10.1016/j.echo.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 51.Topf A, et al. Functionally significant, rare transcription factor variants in tetralogy of Fallot. PLoS ONE. 2014;8:e95453. doi: 10.1371/journal.pone.0095453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yelbuz TM, et al. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation. 2002;4:504–510. doi: 10.1161/01.cir.0000023044.44974.8a. [DOI] [PubMed] [Google Scholar]
- 53.Abu-Issa R, Kirby ML. Patterning of the heart field in the chick. Dev. Biol. 2008;2:223–233. doi: 10.1016/j.ydbio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Felice V, Zummo G. Tetralogy of fallot as a model to study cardiac progenitor cell migration and differentiation during heart development. Trends Cardiovasc. Med. 2009;4:130–135. doi: 10.1016/j.tcm.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 55.Miao M, Bruce AE, Bhanji T, Davis EC, Keeley FW. Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 2007;2:115–124. doi: 10.1016/j.matbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 56.George RM, Firulli AB. Hand factors in cardiac development. Anat. Rec. (Hoboken) 2019;1:101–107. doi: 10.1002/ar.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia M, Luo W, Jin H, Yang Z. HAND2-mediated epithelial maintenance and integrity in cardiac outflow tract morphogenesis. Development. 2019;13:dev177477. doi: 10.1242/dev.177477. [DOI] [PubMed] [Google Scholar]
- 58.Colombo S, et al. Nkx genes establish second heart field cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression. Development. 2018;3:dev161497. doi: 10.1242/dev.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paffett-Lugassy N, et al. Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development. 2017;24:4616–4624. doi: 10.1242/dev.153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.George V, Colombo S, Targoff KL. An early requirement for nkx25 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol. 2015;1:10–22. doi: 10.1016/j.ydbio.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heicklen-Klein A, McReynolds LJ, Evans T. Using the zebrafish model to study GATA transcription factors. Semin. Cell Dev. Biol. 2005;1:95–106. doi: 10.1016/j.semcdb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Brand T. The Popeye domain containing genes and their function as cAMP effector proteins in striated muscle. J. Cardiovasc. Dev. Dis. 2018;5(1):18. doi: 10.3390/jcdd5010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindler RF, Poon KL, Simrick S, Brand T. The Popeye domain containing genes: Essential elements in heart rate control. Cardiovasc. Diagn. Ther. 2012;4:308–319. doi: 10.3978/j.issn.2223-3652.2012.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson I, et al. Novel recessive splice site mutation in POPDC1 (BVES) is associated with first-degree atrioventricular block and muscular dystrophy. Neuromuscul. Disord. 2017;27:S139–S140. [Google Scholar]
- 65.Han P, et al. Ten years of research on the role of BVES/POPDC1 in human disease: A review. Onco Targets Ther. 2019;12:1279–1291. doi: 10.2147/OTT.S192364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu XY, et al. A Zebrafish heart failure model for assessing therapeutic agents. Zebrafish. 2018;3:243–253. doi: 10.1089/zeb.2017.1546. [DOI] [PubMed] [Google Scholar]
- 67.Bournele D, Beis D. Zebrafish models of cardiovascular disease. Heart Fail. Rev. 2016;6:803–813. doi: 10.1007/s10741-016-9579-y. [DOI] [PubMed] [Google Scholar]
- 68.Grant MG, Patterson VL, Grimes DT, Burdine RD. Modeling syndromic congenital heart defects in Zebrafish. Curr. Top. Dev. Biol. 2017;124:1–40. doi: 10.1016/bs.ctdb.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Zeng XX, Yelon D. Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 2014;4:951–960. doi: 10.1016/j.celrep.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, et al. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. Elife. 2016;5:e10557. doi: 10.7554/eLife.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Courchaine K, Rykiel G, Rugonyi S. Influence of blood flow on cardiac development. Prog. Biophys. Mol. Biol. 2018;137:95–110. doi: 10.1016/j.pbiomolbio.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.