INTRODUCTION
The Type I interferon (IFN) response is a complex antiviral signaling pathway that is initiated in infected cells to transcriptionally reprogram neighboring cells to render them refractory against productive viral infection. The importance of this pathway in controlling vesicular stomatitis virus (VSV) infection in vivo is underscored by the observation that mice lacking IFN signaling components are highly susceptible to lethal VSV infection (1). In order to productively infect the host, wild-type (wt) VSV suppresses Type I IFN responses through one or more virus-encoded suppressors (2). The most prominent is the matrix (M) protein which is sufficient to suppress IFN gene expression in the absence of other viral components (3, 4) and shuts off multiple cellular biosynthesis pathways including host transcription (3, 5, 6), host nuclear-cytoplasmic RNA export (7–9) to block cellular protein production, and host translation (10–13). Position 51 of the M protein is crucial for these effects on cellular RNA (3, 5, 6) and protein production (8, 10, 11, 13–15), as well as many of the other cytotoxic effects associated with VSV infection including induction of cell rounding (16, 17) and regulation of apoptosis (18–21). This “shut-off” of cellular biosynthesis gives VSV the upper-hand in the virus vs. host battle and is a strategy used by many different RNA viruses (22).
While the exact mechanism utilized by VSV to suppress the IFN response remains to be elucidated, there is a strong correlation between a virus’s ability to block host gene expression and its ability to suppress IFN (10). For example, strain T1026R1 (R1) contains a M51R mutation in the M protein [M(M51R)] (3) and is delayed in its ability to inhibit host gene expression (23, 24) as well as being unable to suppress IFN gene expression (25). However, it is possible that VSV employs more than one mechanism to evade the IFN response. For example, Marcus compared the IFN-inducing capacity of 36 plaque-derived subpopulations of field isolate VSV-IN no. 22 and found that only isolate 22–20 was an excellent inducer of IFN, while its sister plaque isolates induced little to no IFN in primary chicken embryo cells. These results could be explained if, in addition to M-mediated global inhibition of host transcription, a second distinct viral function specifically suppresses an early step in IFN gene induction. While VSV mutant R1 may be defective in both functions, isolate 22–20 may be defective in only the one responsible for limiting IFN induction at an early step of transcription.
In support of this hypothesis, we recently reported that expression of the wt M protein by viral infection and via transfection inhibited activation of NF-κB, a key transcriptional regulator of the IFN response. We also reported that R1 and recombinant viruses expressing the M(M51R) mutation were unable to prevent NF-κB activation. Coinfection experiments revealed that viruses encoding a wt M protein blocked R1-mediated activation of NF-κB, however the recombinant virus encoding the M(M51R) mutation did not limit this activation (26). These findings suggest that this single mutation abrogates both inhibition of a step upstream of NF-κB activation and host gene expression.
The goal of this study was to characterize these viruses in mouse L929 cells and determine if 22–20 is indeed defective in an early step of IFN gene induction. We chose murine L929 cells as a model system as they are sensitive to VSV infection and are derived from peripheral organ tissues that can normally suppress VSV infection in vivo (27, 28).
To begin, we sequenced the genomes of 22–20 and 22–25 and identified a novel D52G mutation in the M protein of 22–20 that abrogates VSV mediated inhibition of NF-κB activation – confirming a role of VSV in specifically suppressing an early step in IFN gene induction. To further investigate this phenomenon, we characterized the effect of the D52G mutation in mouse L929 cells on (1) IFN mRNA and protein synthesis in virus infected cells and (2) luciferase protein expression driven from constitutive, NF-κB-dependent, and IFN-dependent promoters.
Our collective results support a model in which the M(D52G) mutation in 22–20 allows early activation of the IFN pathway in L929 cells via NF-κB activation; but nevertheless, the virus ultimately suppresses IFN gene expression. We conclude that inhibition of NF-κB activation and suppression of the IFN response during VSV infection are independent functions, and that the M protein is critical for both functions.
MATERIALS AND METHODS
Cells, viruses, and infections
Mouse L929 cells (ATCC CCL-1) were from the American Type Culture Collection and were grown as previously described (26, 29). The generation and maintenance of L929 cells stably expressing a luciferase reporter gene (pGL4.32 or pGL4.50 plasmid) or the CAT reporter gene (pTWU54 plasmid) were produced as previously described (26, 30). VSV field isolates 22–20 and 22–25 were generous gifts from Dr. Philip Marcus (University of Connecticut) and have been previously described (31, 32). The heat resistant (HR) strain of the Indiana serotype of VSV was used as the wt virus, and its mutant T1026R1, a temperature-stable revertant of T1026, was isolated by Stanners et al (24). All viruses were grown on Vero cells as previously described (24). Cells were infected with each virus at a multiplicity of infection (MOI) of 5 plaque forming units (PFU)/cell unless otherwise stated. Virus was adsorbed in MEM for 1 hour at 37 °C in the absence of serum, after which complete medium was added.
Plasmids
The pGL4.50 ([luc2/CMV/Hygro) plasmid contains the firefly luciferase reporter gene cloned behind the CMV promoter and the pGL4.32 (luc2P/NF-κB/Hygro) plasmid, which contains the same reporter gene cloned behind an inducible Nuclear Factor-κB (NF-κB) dependent promoter element, were obtained from Promega. Plasmid pCIN contains the luciferase reporter gene cloned behind the IFN-β promoter and was created by modification of pTWU54 (30). Plasmids were prepared using EndoFree Maxi Prep kits (Qiagen) according to the manufacturer’s instructions.
Luciferase Assay
L929 cells stably transfected with pGL4.32 or pGL4.50 were passed approximately 48 hours prior to infection. Cells were infected at an MOI of 25 and collected at the indicated time post-infection. Cells were washed with ice cold PBS and harvested in 1x Reporter Lysis Buffer (Promega) and luciferase activity was determined as previously described (26).
CAT Assay
CAT activity was determined by the method previously described (3). Briefly, L929 cells stably transfected with plasmid pTWU54 modified to contain the neomycin gene, were treated with Poly(I):poly(C) to induce the IFN-β promoter and infected with the indicated virus. At 3- and 6-hours post-infection (hpi), cells were harvested and CAT activity was measured.
Immunofluorescence
Immunofluorescence analysis was done as previously described (26) except that cells were blocked with UltraCruz® Blocking Reagent (sc-516214, Santa Cruz Biotechnology) for 30 minutes followed by incubation with an NF-κB antibody conjugated to Alexa Fluor® 488 (sc-8008 AF488, Santa Cruz Biotechnology) for 90 minutes at room temperature.
Quantitative RT-PCR (RT-qPCR)
IFN mRNA was quantitated by RT-qPCR as previously described (26). Briefly, total RNA was isolated from cells, reverse transcribed into cDNA, and the commercially available mouse IFN-β TaqMan expression assay (Mm00439546_s1, Applied Biosystems) was used for Real-Time PCR analysis of IFN-β mRNA production. Samples were run in triplicate and the HPRT endogenous control Taqman Gene Expression Assay (Mm00446968_m1) was used for relative quantification. All calculations were done using the ddCT method.
Mouse IFN-β ELISA
L929 cells were grown to 80–90% confluency in 24-well microtiter plates and infected at a MOI of 5 with the indicated virus. Culture media supernatants were collected and stored at −80°C until use. VeriKine-HS Mouse IFN Beta Serum ELISAs (Catalog No. 42410, PBL Assay Science) were run according to the manufacturer’s instructions.
Growth Curve Assay
L929 cells in 35-mm dishes were infected at an MOI of 5 PFU/cell with wt, R1, 22–20 and 22–25 VSV. After 1 hour of adsorption, the media was removed and cells were washed with PBS before being re-fed with 2 ml of EMEM containing 10% HS. At the indicated times post-infection, the supernatant was mixed gently by slowly pipetting up and down, and 1 ml of medium was removed from the dishes, aliquoted, and stored at −80°C. The yield of virus was determined by plaque assays on Vero cells and was expressed as PFU per cell.
Statistical Analysis
Statistical analysis throughout this paper was performed using the Student’s t test and an asterisk indicates significant reduction (P<0.05). Results were expressed as means and error bars indicate the ± standard error of the means (SEM).
RESULTS AND DISCUSSION
Genome sequencing reveals a novel mutation in the M protein of 22–20
The genomes of these viruses were sequenced and compared to determine if there was a second viral component involved in suppression of the IFN response (29). These near-complete genome sequences, methods, and accession numbers have been reported by Russell et al. (29).
Our sequence analysis revealed a novel mutation at position 52 of the 22–20 M protein. We verified this mutation in the original stock (acquired from Dr. Philip Marcus) and in two different virus stocks that were regrown from this original stock; therefore, we are confident that this is a true mutation. This mutation changed the highly conserved aspartic acid residue normally located at position 52 to glycine [M(D52G)]. Significantly, this newly identified mutation occurs one base away from the well-characterized M(M51R) mutation found in several VSV strains (ts082, R1) with a diminished ability to inhibit host gene expression and IFN response (3, 5, 6, 33). Furthermore, according to a structural analysis performed by Graham et al, the mutation at position 52 lies within a well-conserved region of amino acids that have been proposed to interact with the cellular protein Rae1 (34). Rae1 facilitates mRNA nuclear-cytoplasmic export (15, 34) and regulates host transcription initiation in VSV infected cells (35). Therefore, a mutation within this region has the potential to disrupt the virus’s ability to interfere with host gene expression.
Comparison of these genomes identified an additional amino acid difference in the N gene of these viruses. An aspartic acid (D) was located at position 371 of the 22–25 N protein, while a glycine (G) was identified at this location in 22–20. We aligned the sequences of the N gene of several strains of VSV Indiana including wt, R1, and the NCBI reference sequence strain, and found a glycine at this position. This suggests that glycine is conserved amongst several viruses of both IFN-inducing and IFN-suppressing phenotypes – reducing the likelihood that this mutation in the N protein is responsible for inhibiting the host IFN response.
Murine L929 cells are susceptible to 22–20 and 22–25 infection and produce infectious progeny with similar kinetics
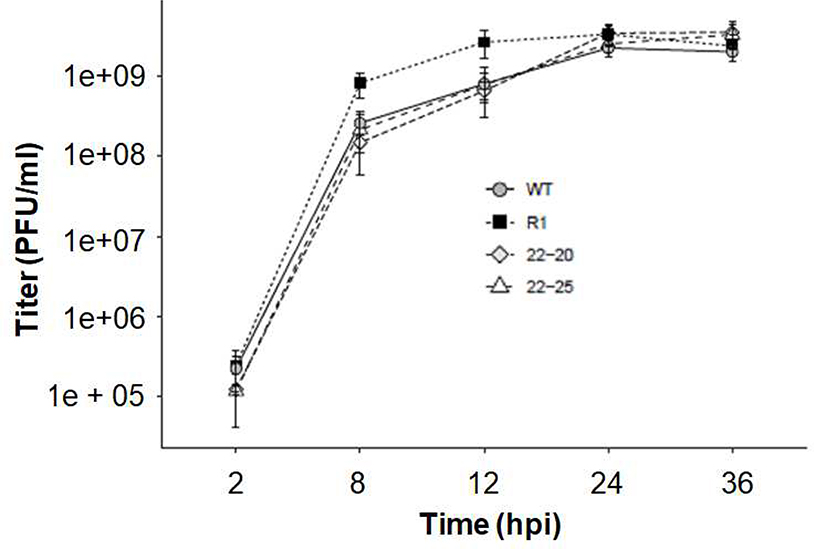
To test whether there were differences in the kinetics of infectious progeny release in murine L929 cells following 22–20 and 22–25 infection, single-cycle growth curve experiments were conducted (Fig. 1). L929 cells were infected with the indicated virus at a MOI of 5 and the viral titers were determined by Vero plaque assays at the indicated times post-infection. As expected, there was no significant difference in production of infectious virus between R1 and wt (18). Importantly, 22–20 and 22–25 grew to similar titers and produced similar levels of infectious progeny as wt and R1. Therefore, the M(D52G) mutation in 22–20 is not significantly detrimental to its ability to produce infectious viral progeny in mouse L929 cells.
Fig. 1.
22–20 and 22–25 have similar viral replication kinetics. L929 cells were infected with the indicated virus at a multiplicity of 5 PFU/cell. Following adsorption cells were washed and re-fed with fresh complete medium. At the indicated times post-infection, a small aliquot was removed and stored at −80°C. Supernatants were tested by plaque assay on Vero cells to determine the amount of progeny virus. A “mock” sample was taken at each time point and verified negative for virus by plaque assay. Data represent the mean of three independent experiments and the titer at each time point was determined in duplicate. All p-values > .05 per student’s t-test.
22–20 D52G mutation induces NF-κB activation and translocation in L929 cells
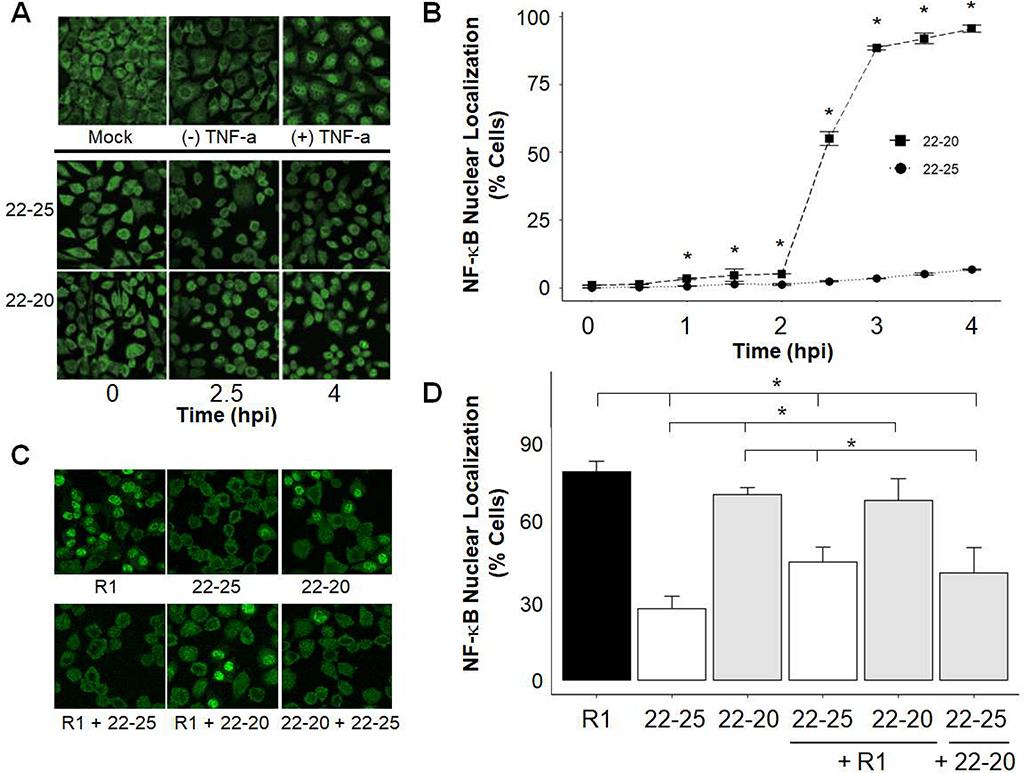
We previously determined that the methionine at position 51 (M51) of M is essential for inhibiting NF-κB activation, one of the first steps in IFN gene induction (26). The goal of this work was to determine if the novel D52G mutation located immediately adjacent to this residue similarly affects NF-κB induction. Immunofluorescence was performed to determine if the M(D52G) mutation in 22–20 M affected nuclear translocation of the p65 subunit NF-κB in L929 cells. As expected, NF-κB localized to the cytoplasm in mock-infected cells or untreated cells; however, treatment with TNF-α, a known activator of NF-κB (36), resulted in nuclear translocation of NF-κB (Fig. 2A). When examining virus-infected cells, little nuclear staining was observed during the 4-hour time course in cells infected with 22–25 (Fig. 2A and B). These findings confirm our previous report that the wt M protein inhibits a pathway that leads to nuclear localization of NF-κB in L929 cells (26). In contrast, NF-κB was detected in the nuclei of 1% of 22–20-infected cells by 2 hpi but increased rapidly to 55% and 90% by 2.5 and 3 hpi, respectively (Fig. 2A and B), therefore the D52G mutation in 22–20 abolished this function of the M protein. The kinetics of this activation were very similar to our findings with R1 and recombinant viruses encoding the M(M51R) mutation (26).
Fig. 2:
Nuclear Localization of NF-κB occurs rapidly in cells infected with viruses containing the M(D52G) and M(M51R) mutations. Cells were TNF-α treated, mock infected, or infected at an MOI of 10 for the indicated time (A, B) or coinfected at an MOI of 25 for each virus for 5 hours (C, D). The p65 subunit of NF-κB was visualized by immunofluorescence and confocal microscopy. Four or five images of each sample were taken, the total number of cells per image was counted, and the percentage of cells with nuclear NF-κB staining was determined. Data represent the mean values from three independent experiments in panel B and four independent experiments in panel D. Error bars indicate the SEM. Representative images are shown in A and C. * p-value <.05 per student’s t-test.
Coinfection assays were performed to determine if viruses encoding mutant M proteins were able to inhibit virus-mediated activation of NF-κB. Similar to Fig. 2A and 2B, minimal nuclear NF-κB staining was detected in 22–25-infected cells, while NF-κB was activated in cells infected with R1 or 22–20 (Fig. 2C and D). Reduced nuclear localization was detected in cells coinfected with R1 and 22–25, compared to cells infected with R1 alone. In contrast, 22–20 was not able to significantly reduce NF-κB activation and translocation in the nucleus of R1 infected cells. These findings were consistent when different virus stocks were tested and when cells were infected at a MOI of 5. Therefore, the functional M protein encoded by 22–25 was able to block viral activation of NF-κB; however, the M(D52G) mutant protein in 22–20 could not. These findings indicate that the wt M protein is essential for suppression of activated NF-κB and that the M(D52G) mutation abrogates this function. Taken together, these findings indicate that the methionine and aspartic acid residues at position 51 and 52, respectively, of the M protein are essential for blocking activation of NF-κB in L929 cells.
22–20 suppresses IFN-β mRNA and protein expression in mouse L929 cells
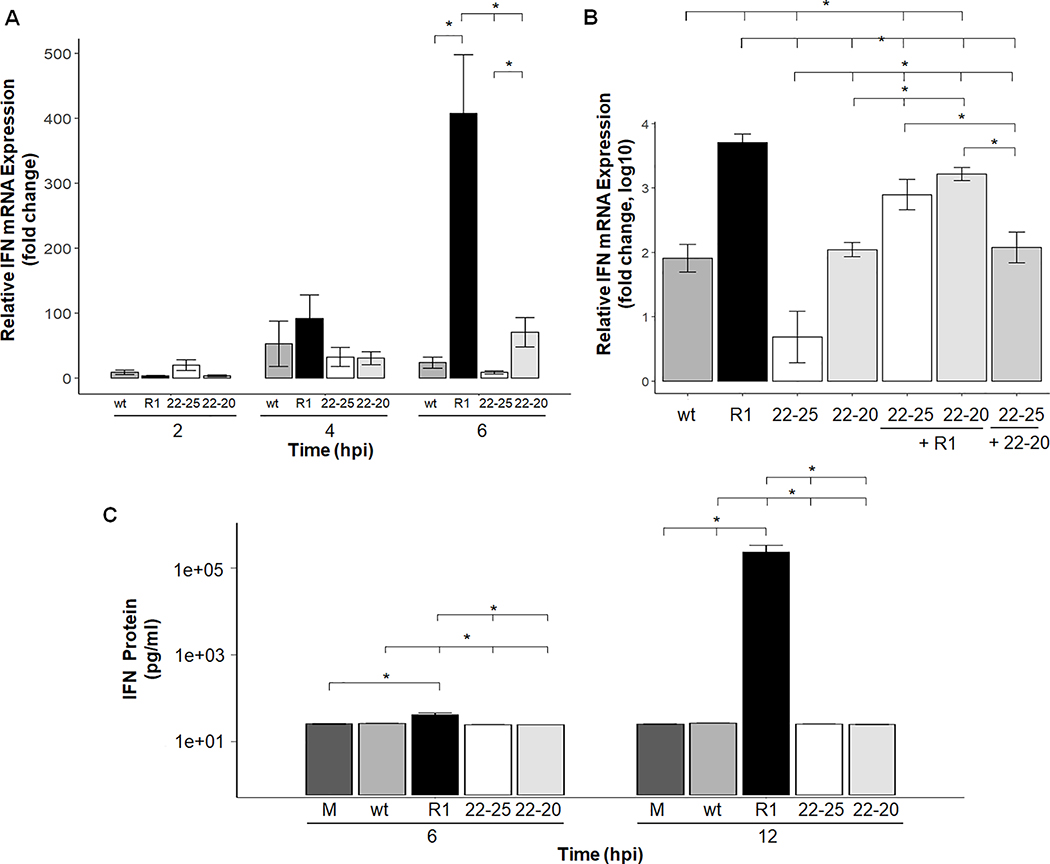
Our previous findings indicate a tight correlation between NF-κB activation and subsequent expression of IFN mRNA in virus infected cells. For example, viruses encoding wt M proteins do not activate NF-κB and produce very little IFN mRNA, whereas recombinant viruses encoding the M51R mutation potently activate NF-κB and induce IFN mRNA expression. Since the D52G mutation induces NF-κB in mouse L929 cells, we measured IFN mRNA and protein production in response to 22–20 and 22–25 infection. Total RNA was isolated at 2, 4 and 6 hpi and analyzed by Real-Time PCR analysis. As expected, R1 infection with the M(M51R) mutation induced much more IFN mRNA expression than wt and 22–25 in L929 cells by 4- and 6-hours post-infection (Fig. 3A). 22–20 infected cells expressed significantly less IFN mRNA compared to R1 infected cells, despite viral mediated NF-κB activation (Fig. 3A). These findings indicate that while the M(M51R) mutation is detrimental to VSV’s ability to suppress IFN mRNA production, the M(D52G) mutation does not perturb this function of the M protein. It is worth noting that while we observed very little IFN mRNA production in 22–20 infected L929 cells, the original studies conducted with 22–20 in chicken embryo cells potently induced expression. This indicates that suppression of the IFN response may be mediated in a cell-type specific manner and represents a future avenue of investigation.
Fig. 3.
IFN-β mRNA expression and protein production are suppressed by the wt and M(D52G) strain of VSV but not by the M(M51R) virus in mouse L929 cells. L929 cells were infected at an MOI of 5 for the indicated time (A) or coinfected at an MOI of 5 for each virus for 6 hours (B). Total RNA was isolated, reverse transcribed and IFN-β mRNA quantitated by real-time PCR. Four independent experiments were performed and each sample was run in triplicate. Samples were normalized to HPRT gene expression which was stable over the time course tested. Data is represented as fold change relative to mock-infected cells. Error bar = mean +/−SEM. (C) IFN-β concentrations in VSV-infected L929 cells. Monolayers were infected with the indicated virus at an MOI of 5. At 6 and 12 hpi media from each well was collected and the concentration of IFN-β protein was quantitated by an ELISA assay. A mock sample at each time point was also collected and verified negative for IFN-β protein production. Each sample was tested in triplicate and the data shown represents the average of three separate assays. * p-value <.05 per student’s t-test.
Next, coinfection experiments were conducted to determine if 22–20 was able to limit viral induction of IFN mRNA. As shown in Figure 3B, coinfection of IFN-inducing R1 with both 22–20 and 22–25 led to a statistically significant reduction in IFN mRNA expression compared to single infection with R1, indicating that 22–20 can limit R1-mediated induction of IFN mRNA. However, coinfection with 22–20 and 22–25 yielded IFN mRNA levels comparable to infection with 22–20 alone. Next, we examined how much IFN protein was produced in VSV-infected cells. Cells were infected with the indicated virus at a MOI of 5 and 1 ml of media was collected after 6 or 12 hours of infection. The amount of IFN protein contained in the media was determined by an ELISA assay. Consistent with the mRNA results discussed above, significant amounts of IFN protein (over 1400 pg/ml) were detected in R1-infected cells, however little of this protein was made in cells infected with wt, 22–20, or 22–25 (Fig. 3C). Collectively, this data indicates that 22–20 does not induce a strong Type I IFN response in mouse L929 cells.
Both 22–20 and 22–25 limit gene expression from the IFN-β, NF-κB-dependent, and CMV promoter in L929 cells
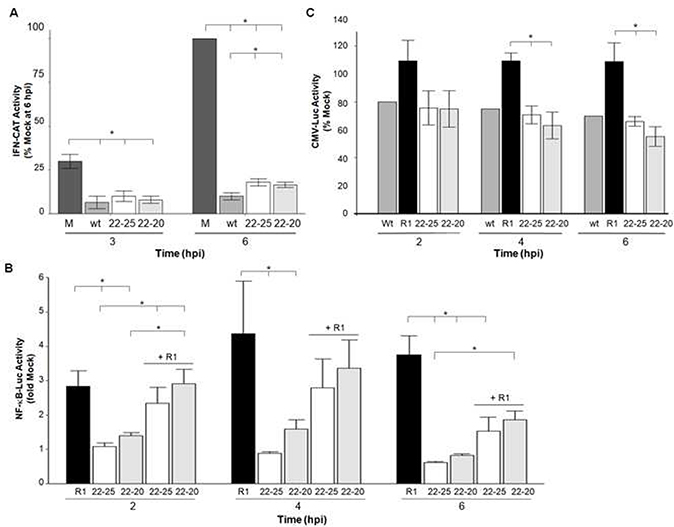
Despite NF-κB activation and translocation into the nucleus, infection with 22–20 does not result in Type I IFN gene expression in L929 cells. One possible explanation for this is that the 22–20 M protein may retain the ability to inhibit host transcription – thus rendering NF-κB availability in the nucleus a moot point. Therefore, reporter assays were conducted to evaluate the effect of the M(D52G) mutation on the ability of 22–20 to inhibit host gene expression from constitutive, NF-κB-dependent, and IFN-dependent promoter constructs in L929 cells. As expected, viruses that produce little IFN protein (Fig. 3C) similarly limit gene expression from an IFN-β driven promoter (Fig. 4A). Cells that were stably transfected with the pCIN plasmid were treated with poly(I):poly(C) to induce expression of CAT from the IFN-β promoter and infected with 22–20 or 22–25 at a MOI of 5. Poly(I):poly(C) treatment did induce expression of CAT in mock infected cells. However, infection with 22–20 or 22–25 severely limited this induction, indicating that these viruses were able to limit poly(I):poly(C)-mediated induction of the IFN-β promoter. We next measured the effect of this mutation on gene expression from an inducible NF-κB-dependent promoter (Fig. 4B). L929 cells stably transfected with the pGL4.32 plasmid, which contains the Luc2P gene under the control of an inducible NF-κB-dependent promoter, were infected with VSV for 2, 4, or 6 hours and luciferase activity was measured. In preliminary experiments (data not shown), we determined that a higher MOI of 25 was necessary to limit luciferase expression, because the stably transfected L929 cells expressed luciferase from a strong (SV40) promoter. This was true even for the wt virus, which is well-known to inhibit host gene expression. To keep our methods consistent, and allow comparison of the results, the immunofluorescence coinfections (Fig. 2C and 2D) were also done at a MOI of 25.
Fig. 4.
The 22–20 virus encoding the M(D25G) mutation limits reporter gene expression from various promoters similarly to viruses containing a wt M protein. (A) To induce the IFN-β promoter, L929 cells stably transfected with an IFN-β CAT reporter construct (modified pTWU54) were treated with poly(I):poly(C) and then infected with the indicated virus at an MOI of 5. Lysates were collected at 3 and 6 hpi and the amount of CAT activity was determined. Data was calculated as percent of mock at 6 hpi. The average of two independent experiments is shown and error bars represent the SEM. (B) L929 cells stably transfected with a NF-κB-dependent luciferase reporter construct (pGL4.32) were infected at a MOI of 25 or coinfected (MOI of 25 for each virus) with the indicated virus for 2, 4, or 6 hours. Lysates were collected and the amount of luciferase present was determined. Samples were run in triplicate and data shown is the mean of three independent luciferase experiments. (C) L929 cells stably transfected with the constitutive CMV (pGL4.50) were infected at a MOI of 25 with the indicated virus for 2, 4, or 6 hours. Lysates were collected and the amount of luciferase present was determined. Samples were run in triplicate and the data shown is the mean of three independent experiments. * p-value <.05 per student’s t-test.
As expected, NF-κB dependent-luciferase expression increased over the times tested in cells infected with R1; however, 22–20 and 22–25 infection limited gene expression as the cells produced less luciferase than mock-infected cells. This suggests that 22–20 suppresses host cell transcription from this promoter despite our finding that this virus permits NF-κB translocation into the nucleus. Coinfection experiments were done to determine if 22–20 could limit R1-mediated induction of the NF-κB promoter. We found slightly significant statistical evidence that coinfection with R1 and 22–20 reduced luciferase expression from the NF-κB-dependent promoter relative to R1 infection alone (p=0.058). Luciferase expression was significantly reduced in R1 and 22–25 coinfected cells relative to R1 infection alone. Finally, we measured the effects of VSV infection on transcription of a reporter gene controlled by a constitutive CMV promoter (Fig. 4C). As expected, wt infection limited luciferase expression from this promoter compared to mock infection over a 6-hour time course. The wt virus was only tested once in this assay as it has been previously reported that the wt M protein limits CMV-dependent expression of a reporter gene (3, 5). Both 22–20 and 22–25 limited transcription from the CMV promoter in comparison to infection with R1. These findings indicate that the M(D52G) mutation in 22–20 did not perturb the virus’s ability to inhibit host gene expression as both 22–20 and 22–25 inhibit host transcription governed by multiple independent promoters (as demonstrated by reporter assays). Therefore, the aspartic acid at position 52 is not essential for this function of M.
In summary, both wt and 22–25 prevent NF-κB activation, and do not produce IFN mRNA or protein. In R1-infected cells, NF-κB is activated, IFN mRNA is induced and IFN protein is produced because M(M51R) abrogates viral inhibition of both NF-κB activation and host gene expression. In contrast, even though 22–20 activates NF-κB at early times post-infection, IFN mRNA and protein production are suppressed because this virus retains its ability to limit host transcription and translation. We propose that the M(M51R) in R1 disrupts two independent functions, inhibition of NF-κB activation and inhibition of host gene expression, while M(D52G) selectively disrupts viral inhibition of NF-κB. We conclude that suppression of NF-κB and suppression of the IFN response are independent, genetically separable functions of the VSV M protein and have experimental evidence to suggest that the IFN response is suppressed via M-mediated inhibition of host cell transcription. The precise NF-κB-independent mechanism by which 22–20 suppresses the IFN response is the subject of ongoing work.
Highlights.
Characterization of a novel VSV M protein mutation [M(D52G)] in the 22–20 strain
The M(D52G) mutation perturbs M-mediated inhibition of NF-κB activation
22–20 inhibits IFN gene expression in L929 cells
Despite this mutation, 22–20 inhibits host transcription from multiple promoters
Inhibition of NF-κB activation is not necessary for IFN suppression by VSV
ACKNOWLEDGMENTS
We thank Hyla Sweet for her support and insightful comments on the manuscript. We thank the Rochester Institute of Technology (RIT) College of Science (COS) for helping to support this work and the COS Summer Undergraduate Research Program and the RIT Honors Program for supporting many of the undergraduates who contributed to this work. This research was supported by the NIH/NIAID grant R15 AI058969 (M. F.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Data availability
Raw Illumina reads are available from the Sequence Read Archive at BioProject number PRJNA508804. Genome sequences were deposited in GenBank with accession numbers: MH919396 for 22–20, MH919397 for 22–25, MH919398 for T1026R21, and MH919399 for HR.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–21. [DOI] [PubMed] [Google Scholar]
- 2.Marcus PI, Sekellick MJ. 1987. Interferon induction by viruses. XV. Biological characteristics of interferon induction-suppressing particles of vesicular stomatitis virus. J Interferon Res 7:269–84. [DOI] [PubMed] [Google Scholar]
- 3.Ferran MC, Lucas-Lenard JM. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol 71:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J Virol 77:4646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black BL, Lyles DS. 1992. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol 66:4058–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik SY, Banerjea AC, Harmison GG, Chen CJ, Schubert M. 1995. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J Virol 69:3529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Her LS, Lund E, Dahlberg JE. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845–8. [DOI] [PubMed] [Google Scholar]
- 8.Petersen JM, Her LS, Varvel V, Lund E, Dahlberg JE. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol Cell Biol 20:8590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell 6:1243–52. [DOI] [PubMed] [Google Scholar]
- 10.Connor JH, Lyles DS. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J Virol 76:10177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor JH, Lyles DS. 2005. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J Biol Chem 280:13512–9. [DOI] [PubMed] [Google Scholar]
- 12.Whitlow ZW, Connor JH, Lyles DS. 2006. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. J Virol 80:11733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitlow ZW, Connor JH, Lyles DS. 2008. New mRNAs are preferentially translated during vesicular stomatitis virus infection. J Virol 82:2286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen JM, Her LS, Dahlberg JE. 2001. Multiple vesiculoviral matrix proteins inhibit both nuclear export and import. Proc Natl Acad Sci U S A 98:8590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17:93–102. [DOI] [PubMed] [Google Scholar]
- 16.Lyles DS, McKenzie MO. 1997. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology 229:77–89. [DOI] [PubMed] [Google Scholar]
- 17.Simon KO, Whitaker-Dowling PA, Youngner JS, Widnell CC. 1990. Sequential disassembly of the cytoskeleton in BHK21 cells infected with vesicular stomatitis virus. Virology 177:289–97. [DOI] [PubMed] [Google Scholar]
- 18.Desforges M, Despars G, Bérard S, Gosselin M, McKenzie MO, Lyles DS, Talbot PJ, Poliquin L. 2002. Matrix protein mutations contribute to inefficient induction of apoptosis leading to persistent infection of human neural cells by vesicular stomatitis virus. Virology 295:63–73. [DOI] [PubMed] [Google Scholar]
- 19.Kopecky SA, Willingham MC, Lyles DS. 2001. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J Virol 75:12169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecky SA, Lyles DS. 2003. Contrasting effects of matrix protein on apoptosis in HeLa and BHK cells infected with vesicular stomatitis virus are due to inhibition of host gene expression. J Virol 77:4658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopecky SA, Lyles DS. 2003. The cell-rounding activity of the vesicular stomatitis virus matrix protein is due to the induction of cell death. J Virol 77:5524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyles DS. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev 64:709–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunigan DD, Lucas-Lenard JM. 1983. Two transcription products of the vesicular stomatitis virus genome may control L-cell protein synthesis. J Virol 45:618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanners CP, Francoeur AM, Lam T. 1977. Analysis of VSV mutant with attenuated cytopathogenicity: mutation in viral function, P, for inhibition of protein synthesis. Cell 11:273–81. [DOI] [PubMed] [Google Scholar]
- 25.Marcus PI, Sekellick MJ. 1980. Interferon induction by viruses. III. Vesicular stomatitis virus: interferon-inducing particle activity requires partial transcription of gene N. J Gen Virol 47:89–96. [DOI] [PubMed] [Google Scholar]
- 26.Varble AJ, Ried CD, Hammond WJ, Marquis KA, Woodruff MC, Ferran MC. 2016. The vesicular stomatitis virus matrix protein inhibits NF-κB activation in mouse L929 cells. Virology 499:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trottier MD, Lyles DS, Reiss CS. 2007. Peripheral, but not central nervous system, type I interferon expression in mice in response to intranasal vesicular stomatitis virus infection. J Neurovirol 13:433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 8:e1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell TM, Santo EE, Golebiewski L, Haseley NS, Ferran MC. 2019. Near-Complete Genome Sequences of Vesicular Stomatitis Virus Indiana Laboratory Strains HR and T1026R1 and Plaque Isolates 22–20 and 22–25. Microbiol Resour Announc 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulares AH, Ferran MC, Lucas-Lenard J. 1996. NF-kappaB activation Is delayed in mouse L929 cells infected with interferon suppressing, but not inducing, vesicular stomatitis virus strains. Virology 218:71–80. [DOI] [PubMed] [Google Scholar]
- 31.Marcus PI, Sekellick MJ, Spiropoulou CF, Nichol ST. 1993. Interferon induction by viruses. XXII. Vesicular stomatitis virus-Indiana: M-protein and leader RNA do not regulate interferon induction in chicken embryo cells. J Interferon Res 13:413–8. [DOI] [PubMed] [Google Scholar]
- 32.Marcus PI, Sekellick MJ. 1994. Interferon induction: regulation by both virus and cell. Hokkaido Igaku Zasshi 69:1320–31. [PubMed] [Google Scholar]
- 33.Coulon P, Deutsch V, Lafay F, Martinet-Edelist C, Wyers F, Herman RC, Flamand A. 1990. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol 71 ( Pt 4):991–6. [DOI] [PubMed] [Google Scholar]
- 34.Graham SC, Assenberg R, Delmas O, Verma A, Gholami A, Talbi C, Owens RJ, Stuart DI, Grimes JM, Bourhy H. 2008. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog 4:e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajani KR, Pettit Kneller EL, McKenzie MO, Horita DA, Chou JW, Lyles DS. 2012. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog 8:e1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hohmann HP, Brockhaus M, Baeuerle PA, Remy R, Kolbeck R, van Loon AP. 1990. Expression of the types A and B tumor necrosis factor (TNF) receptors is independently regulated, and both receptors mediate activation of the transcription factor NF-kappa B. TNF alpha is not needed for induction of a biological effect via TNF receptors. J Biol Chem 265:22409–17. [PubMed] [Google Scholar]