Abstract
We engineered P. putida for the production of isobutanol from glucose by preventing product and precursor degradation, inactivation of the soluble transhydrogenase SthA, overexpression of the native ilvC and ilvD genes, and implementation of the feedback‐resistant acetolactate synthase AlsS from Bacillus subtilis, ketoacid decarboxylase KivD from Lactococcus lactis, and aldehyde dehydrogenase YqhD from Escherichia coli. The resulting strain P. putida Iso2 produced isobutanol with a substrate specific product yield (Y Iso/S) of 22 ± 2 mg per gram of glucose under aerobic conditions. Furthermore, we identified the ketoacid decarboxylase from Carnobacterium maltaromaticum to be a suitable alternative for isobutanol production, since replacement of kivD from L. lactis in P. putida Iso2 by the variant from C. maltaromaticum yielded an identical YIso/S. Although P. putida is regarded as obligate aerobic, we show that under oxygen deprivation conditions this bacterium does not grow, remains metabolically active, and that engineered producer strains secreted isobutanol also under the non‐growing conditions.
Keywords: isobutanol, ketoacid decarboxylase, metabolic engineering, microaerobic, Pseudomonas putida
Abbreviations
- 2‐KIV
2‐ketoisovalerate
- AlsS
acetolactate synthase
- BHI
brain–heart infusion
- KDC
ketoacid decarboxylase
- LB
Lysogeny broth
1. INTRODUCTION
Biofuel production from renewable feed stocks is of special importance because of the finite nature of the currently used crude oil derivatives and growing concerns about climate change 1. Isobutanol is an attractive alternative to the employed fossil fuels. It has several advantages such as a higher energy density, compatibility with existing engines, lower vapor pressure and volatility, as well as a lower corrosivity compared to bio‐ethanol 2, 3. Furthermore, isobutanol is used in the chemical industry and can be used to produce the gaseous alkene precursor isobutene 4.
Isobutanol can be synthesized via the branched‐chain amino acid biosynthesis and the so‐called Ehrlich pathway to convert pyruvate to isobutanol (Figure 1). The first step in this route is the conversion of two pyruvate molecules to 2‐acetolactate catalyzed by the acetolactate synthase (AlsS), which is usually feedback inhibited by the branched‐chain amino acids l‐valine, l‐leucine, and l‐isoleucine. However, AlsS from Bacillus subtilis has been shown to be feedback‐resistant and therefore has been applied for isobutanol production in several studies 5, 6. Then, 2‐acetolactate is reduced to 2,3‐dihydroxyisovalerate and subsequently converted to 2‐ketoisovalerate (2‐KIV) by the ketoacid reductoisomerase IlvC and dihydroxyacid dehydratase IlvD, respectively. Finally, isobutanol is synthesized from 2‐KIV in two more reaction steps of the Ehrlich pathway. The decarboxylation of 2‐KIV to isobutyraldehyde is catalyzed by ketoacid decarboxylases (KDCs) that are not widespread in nature. Especially KivD from Lactococcus lactis has been proved as an efficient variant in, e.g. E. coli and C. glutamicum 5, 7. The last step from isobutyraldehyde to isobutanol requires an aldehyde reductase or alcohol dehydrogenase. A number of NADH and NADPH dependent enzymes are available that catalyze this reaction 8.
Figure 1.
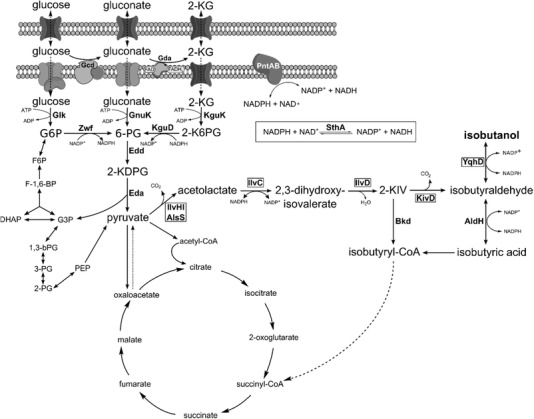
The central metabolism of P. putida KT2440 with the Ehrlich pathway. Abbreviations (coding genes are given in brackets): G6P: glucose‐6‐phosphate 2‐KG: 2‐ketogluconate, 2‐K6PG: 2‐keto‐6‐phosphogluconate, 6‐PG: 6‐phosphogluconate, 2‐KDPG: 2‐keto‐3‐deoxy‐6‐phosphogluconate, G3P: glyceraldehyde‐3‐phosphate, 1,3‐bPG: 1,3‐bisphosphoglycerate, 3‐PG: 3‐phosphoglycerate, 2‐PG: 2‐phosphoglycerate, PEP: phosphoenolpyruvate, DHAP: dihydroxyacetone‐phosphate, F‐1,6‐bP: fructose‐1,6‐bisphosphate, F6P: fructose‐6‐phosphate, CoA: co‐enzyme A, Gcd: glucose dehydrogenase (gcd), gad: gluconate 2‐dehdyrogenase (gad), PQQ: pyrroloquinoline quinone, Glk: glucokinase (glk), Zwf: glucose‐6‐phosphate 1‐dehydrogenase (zwf‐1, zwf‐2, zwf‐3), GnuK: gluconate kinase (gnuK), KguD: 2‐6‐phosphoketogluconate reductase (kguD), KguK: 2‐ketogluconate kinase (kguK), Edd: 6‐phosphogluconate dehydratase (edd), Eda: 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase (eda), IlvHI/AlsS: acetolactacte synthase (ilvHI/alsS), IlvC: ketolacid reductoisomerase (ilvC), IlvD: dihydroxyacid dehydratase (ilvD), KivD: ketoacid decarboxylase (kivD), Bkd: branched‐chain ketoacid dehydrogenase complex (bkd), Yqhd: aldehyde reductase (yqhD), AldH: aldehdye dehdyrogenases, PntAB: pyridine nucleotide transhydrogenase (membrane bound) (pntAB), SthA: pyridine nucleotide transhydrogenase (soluble) (sthA)
Several microorganisms have been engineered for isobutanol production such as E. coli, C. glutamicum, B. subtilis, and yeast such as Saccharomyces cerevisiae 5, 7, 9, 10. Although highly efficient E. coli and C. glutamicum strains have been constructed 6, 7, the relatively low tolerance of most microbial systems against isobutanol hampers commercialization of isobutanol production processes. In contrast, pseudomonads have an intrinsic tolerance against organic compounds and solvents 11, 12 making them promising candidates for isobutanol production.
Among them, Pseudomonas putida is a Gram‐negative, saprophytic soil bacterium with a genome size of 6.18 Mbp 13. It has been reported to promote plant growth, prevent plant diseases, and can efficiently remove organic soil pollutants and environmental contaminants 14. P. putida features a versatile metabolism using the Entner–Doudoroff pathway for glucose catabolism, shows resistance against oxidative stress conditions, and genetic engineering tools are readily available 15, 16, 17. The carbohydrate substrate spectrum is limited and confined to hexoses 18, however, P. putida has been recently engineered to concomitantly consume xylose, cellobiose, and glucose, which are the basic building blocks of the abundant polysaccharides cellulose and hemicellulose 19. As a result of these achievements, P. putida has emerged as a promising candidate for industrial biotechnology 20, 21. Recent works have engineered this bacterium for the production of polyhydroxyalkanoates, the nylon precursor cis,cis‐muconic acid 22 and aromatic compounds like p‐coumaric acid or trans‐cinnamate 23, 24. P. taiwanensis VLB120 has been applied for the production of phenol 25, 26.
PRACTICAL APPLICATION
The relatively low tolerance of most microbial systems against isobutanol hampers commercialization of isobutanol production processes. In contrast, pseudomonads have an intrinsic tolerance against organic compounds and solvents making them promising candidates for isobutanol production. Therefore, we engineered Pseudomonas putida KT2440 for the production of this alcohol by preventing product and precursor degradation and increasing the flux from pyruvate toward isobutanol. The achieved overall isobutanol yield is significantly higher compared to other engineered P. putida strains; however, rather low compared to tailored E. coli and C. glutamicum strains. Therefore, this study paths the way to construct more efficient P. putida strains for isobutanol production in future studies.
In this study, we engineered P. putida for the production of isobutanol from glucose by preventing product and precursor degradation and increasing the flux from pyruvate towards isobutanol. We identified KivD from Carnobacterium maltaromaticum as a suitable alternative to KivD from L. lactis to drive the decarboxylation of 2‐ketoisovalerate and finally we showed that isobutanol production can also be achieved under oxygen deprivation conditions with this obligate aerobic bacterium.
2. MATERIALS AND METHODS
2.1. Bacterial strains and plasmids
Bacterial strains, their respective genotype, plasmids, and oligonucleotides used in this study are listed in Table 1.
Table 1.
Overview of strains, plasmids and oligonucleotides used in this study
| Strain, plasmid or oligonucleotide | Relevant characteristic(s) or sequence (5′ → 3′) | Source, reference or purpose |
|---|---|---|
| Strains | ||
| Pseudomonas putida KT2440 | Wild type strain, DSM‐6125, ATCC47054 | 27 |
| Carnobacterium maltaromaticum LMA28 | 28 | |
| Lactococcus lactis subsp. cremoris MG1363 | 29 | |
| Corynebacterium glutamicum | Wild type strain ATCC13032 | American type culture collection |
| P. putida GN346 | P. putida KT2440 Δupp, ΔpedE, ΔpedI, ΔpedH, ΔaldB‐I | 30 |
| P. putida EP1 | P. putida GN346 ΔbkdAA | This work |
| P. putida EP2 | P. putida EP1 ΔsthA | This work |
| P. putida EP3 | P. putida EP2 Δgcd | This work |
| P. putida Iso1 | P. putida EP1 + pIP02 | This work |
| P. putida Iso2 | P. putida EP2 + pIP02 | This work |
| P. putida Iso3 | P. putida EP2 + pIP03 | This work |
| P. putida Iso4 | P. putida EP2 + pIP04 | This work |
| P. putida Iso5 | P. putida EP3 + pIP02 | This work |
| P. putida Iso6 | P. putida EP2 + pIP05 | This work |
| Plasmids | ||
| pBB1 | pACYC184/pBL1 derivative, chloramphenicol resistance, Ptac promoter and trpA terminator | 31 |
| pSA55 | Expression plasmid for adh2 of S. cerevisae and kivD of L. lactis | 5 |
| pBB1 yqhD | pBB1 Ptac yqhD | This work |
| pBB1 kivD yqhD | pBB1 Ptac kivD yqhD | This work |
| pNG413.1 | pBBR1MCS2 derivative, apramycin resistance, araC, PBAD, lacZ | 32 |
| pSEVA231 | pBBR1 derivative, kanamycin resistance, mobilizable (oriT) | 33 |
| pIP01 | pSEVA231Ptac kivD yqhD | This work |
| pIP02 | pNG413 araC PBAD kivD yqhD alsS ilvC ilvD | This work |
| pIP03 | pIP02, yqhD was changed for adhA from L. lactis | This work |
| pIP04 | pIP02, yqhD was changed for adhA from C. glutamicum | This work |
| pIP05 | pIP02, kivD was changed for kdcA from C. maltaromaticum | This work |
| pEMP04 | pSEVA231 Ptac kivD yqhD alsS ilvC ilvD | Ingenza Ltd. |
| pEMP012 | pEMP04, yqhD was changed for adhA from L. lactis | This work |
| pEMP013 | pEMP04, yqhD was changed for adhA from C. glutamicum | This work |
| pEMP014 | pEMP04, kivD was changed for kdcA from C. maltaromaticum | This work |
| Oligonucleotide | ||
| yqhd1 | AACTGCAGAACCAATGCATTGGAGGAGACACAACA TGAACAACTTTAATCTGCACACCCCAACC | Construction of pBB1yqhd, PstI site underlined |
| yqhd2 | CCGCTCGAGAAAGCTTAGCGGGCGGCTT CGTATATACG | Construction of pBB1yqhd, XhoI site underlined |
| kivd1 | TCCCCCCGGGAGGAGACACAACATGTATACAGTAGGAG ATTACCTAT | Construction of pBB1 kivd yqhd, XmaI site underlined |
| kivd2 | CCAATGCATTGGTTCTGCAGTTTTATGATTTATTTTGTTC AGCAAAT | Construction of pBB1 kivd yqhd, PstI site underlined |
| bkdaa1 | CTGGATCCCATTCAGACCTCCATGACC | Deletion of bkdAA |
| bkdaa2 | CGGCCGCTTCAGAGCTCACATGAGATGAACGA CCACAAC | Deletion of bkdAA |
| bkdaa3 | TGTTGTGGTCGTTCATCTCATGTGAGCTCTG AAGCGGC | Deletion of bkdAA |
| bkdaa4 | GCTTGTCGACCCGTCGTCACTGCCGTAG | Deletion of bkdAA |
| bkdaagc1 | GTACCGACGATGCCGCT | Verification of bkdAA deletion |
| bkdaagc2 | GCCGTGCCACTAAGATGTAG | Verification of bkdAA deletion |
| stha1 | GCCGCTTTGGTCCCGGATCCACAGCATCCAGTACGT CCGC | Deletion of sthA |
| stha2 | GTTGAAATCGGTCTCTCCGACCTGAACGCCGCGCACA TTAAC | Deletion of sthA |
| stha3 | GTTAATGTGCGCGGCGTTCAGGTCGGAGAGACCGATTT CAAC | Deletion of sthA |
| stha4 | TTGCATGCCTGCAGGTCGACTGGTTGGGCAAACCCTGC TTGG | Deletion of sthA |
| sthagc1 | ATGGCTATTCGACGCTGCTG | Verification of sthA deletion |
| sthagc2 | ACTATGGCTGCGAACTGCTG | Verification of sthA deletion |
| gcd1 | GCCGCTTTGGTCCCGGATCCTGACCTTGAGTTGTTCC TTG | Deletion of gcd |
| gcd2 | GACCTGACGGAGAACCTACATTAGCCGAGT AAGCGACAC | Deletion of gcd |
| gcd3 | GTGTCGCTTACTCGGCTAATGTAGGTTCTCCGTCA GGTC | Deletion of gcd |
| gcd4 | TTGCATGCCTGCAGGTCGACGACAACATCAGCAACG ACC | Deletion of gcd |
| gcdgc1 | GGGATGGGTTTCAATGGTTC | Verification of gcd deletion |
| gcdgc2 | GGCACAAGATGTTCTCAAGG | Verification of gcd deletion |
| png1 | AGCTCTAAGGAGGTTATAAAAACATATGTATACAGTAGG AGATTACC | Construction of pIP02, pIP03, pIP04 and pIP05 |
| png2 | GAGAATAGGAACTTCGAACTGCAGGTCGACTCAGAGG CCTTCCAGC | Construction of pIP02, pIP03 and pIP04 |
| png3 | AGCTCTAAGGAGGTTATAAAAACATATGTACACTGTT GGAAATTATTTGTTA | Construction of pIP05 |
| pbb1 | TCGGAGCTCCGCGAATTGCAAGCTGATCCG | Construction of pIP01, SacI site underlined |
| pbb2 | ATCGGATCCCTTAGCGGGCGGCTTCGTAT | Construction of pIP01, BamHI site underlined |
| kdca1 | TTGCTAAACAAAATTCATAAAACTGCAGAACCAATGC | Amplification of kdcA gene |
| kdca2 | AATGCATTGGTTCTGCAGTTTTATGAATTTTGTTTAGC AAAGACTTTC | Amplification of kdcA gene |
| p41 | TTGCTAAACAAAATTCATAAAACTGCAGAACCAATG CATTG | Construction of pEMP014 |
| p42 | TAATTTCCAACAGTGTACATGTTGTGTCTCCTCCCGG | Construction of pEMP014 |
| p43 | TCATTGATTTTACTAAATAAGCCAGGAGGACAGCTAT | Construction of pEMP012 |
| p44 | CGTACTACTGCTGCTTTCATGTTGTGTCTCCTCCAATGC | Construction of pEMP012 |
| p45 | GTGTGGCGATTCGTTTCTAAGCCAGGAGGACAGCTA TGAC | Construction of pEMP013 |
| p46 | TGGGGTGCAGCAGTGGTCATGTTGTGTCTCCTCCAA TGCATTG | Construction of pEMP013 |
| adha1 | GCATTGGAGGAGACACAACATGAAAGCAGCAGTA GTACG | Amplification of adhA gene from L. lactis |
| adha2 | GTCATAGCTGTCCTCCTGGCTTATTTAGTAAAATC AATGACCATCC | Amplification of adhA gene from L. lactis glutamicum |
| adha3 | TGCATTGGAGGAGACACAACATGACCACTGCTGCACC | Amplification of adhA gene from C. glutamicum |
| adha4 | GTCATAGCTGTCCTCCTGGCTTAGAAACGAATCG CCACACG | Amplification of adhA gene from C. glutamicum |
| alss1 | GAGGAAAGCGGCCGCGCTCTTCGGGGCGGAGCTTGTTG | Construction of pEMP04, NotI site underlined |
| alss2 | TTAGATCTCGAGGCTCTTCGGGCCTAGAGAGCTTTCG TTTTCATG | Construction of pEMP04, XhoI site underlined |
| ilvc1 | GAGGAAGCGGCCGCGCTCTTCGAAGAAAGTCGCCATCATC | Construction of pEMP04, NotI site underlined |
| ilvc2 | TTAGATCTCGAGGCTCTTCGGGCTTAGTTCTTGGTC TTGTCGAC | Construction of pEMP04, XhoI site underlined, |
| ilvd1 | GAGGAAGCGGCCGCGCTCTTCGCGGCGCCCGTG | Construction of pEMP04, NotI site underlined |
| ilvd2 | TTAGATCTCGAGGCTCTTCGGGCTCAGAGGCCTTCCAG | Construction of pEMP04, XhoI site underlined |
| pip011 | GAGGAAGCGGCCGCGCTCTTCGCGTGACTGGGAAAACCC TGGCGACTAGTCTTGGACTC | Construction of pEMP04, NotI site underlined |
| pip012 | TTAGATCTCGAGGCTCTTCGGGCTTAGCGGGCGGCTTCG TATATACGGCGGCTGA | Construction of pEMP04, XhoI site underlined |
2.2. Media and culture conditions
E. coli DH5α was grown aerobically in Lysogeny broth (LB) complex medium containing 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl 34 at 37ºC as 5 mL cultures in glass test tubes on a rotary shaker at 120 rpm (Infors AG, Bottmingen, Switzerland). C. maltaromaticum and L. lactis were grown in brain–heart infusion (BHI) broth (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) at 30°C on a rotary shaker at 120 rpm. For longtime storage, P. putida was kept as 30% (w/v) glycerol stock at −70°C and was streaked out for cultivation on LB solid medium with 15 g/L agar. The first preculture of P. putida was prepared by inoculation of 5 mL LB medium in a test tube with a single colony. The culture was cultivated at 30°C on a rotary shaker (Edmund Bühler GmbH, Bodelshausen, Germany) at 175 rpm overnight and used to inoculate, a second overnight preculture to an optical density at 600 nm (OD600) of 0.01–0.02 in 50 mL DeBont minimal medium (pH 7) 35, which was supplemented with 5.4 g/L glucose and 0.5 g/L yeast extract. Cells from the second preculture were harvested by centrifugation (4500 × g, 15 min, 4°C), resuspended in DeBont medium, and used to inoculate 50 mL DeBont medium, to an OD600 of about 0.1–0.2. The main culture was supplemented with 5.4 g/L glucose, 0.5 g/L isobutanol, or 2.9 g/L 2‐ketoisovalerate, respectively. The second pre‐ and main cultures were performed in 500 mL baffled Erlenmeyer flasks filled with 50 mL medium on a rotary shaker at 175 rpm at 30°C. Micro‐aerobic shaking flask cultivations were carried out in sealed 100 mL Müller‐Krempel bottles as 25 mL cultures that were inoculated to an OD600 of 15–20. To obtain sufficient biomass, the second preculture was performed in 100 mL LB medium in a 500 mL baffled Erlenmeyer flask that was cultivated on a rotary shaker (175 rpm) overnight at 30°C. Cells from the second preculture were harvested by centrifugation (4500 × g, 15 min, 4°C) and resuspended in 25 mL DeBont minimal medium (pH 7) supplemented with 5.4 g/L glucose and 15 g/L 3‐morpholino‐propanesulfonic acid. To induce plasmid‐based gene expression, 0.2% (w/v) l‐arabinose was supplemented. For plasmid‐bearing strains, 50 µg/mL kanamycin or 50 µg/mL apramycin were added to the medium.
2.3. Recombinant DNA work
Standardized cloning procedures such as PCR and DNA restrictions were carried out according to Sambrook and Russell, 2001. Plasmids were isolated from 5 mL liquid cultures using the E.Z.N.A.® Plasmid Mini Kit (Omega Bio‐tek, Inc., Norcross, USA) following manufacturer's instructions. PCR fragments were purified with the NucleoSpin® Gel and PCR Clean‐up Kit (Macherey‐Nagel GmbH & Co. KG, Düren, Germany) according to the manufacturer's instructions. Chromosomal DNA of E. coli MG1655, P. putida, C. maltaromaticum, and L. lactis was isolated using the Nucleospin® Microbial DNA Kit (Macherey‐Nagel GmbH & Co. KG, Düren, Germany) following the protocol of the manufacturer. Electrocompetent cells were prepared for E. coli and P. putida as described previously 53, 54. E. coli DH5α and P. putida strains were electroporated with an Eporator (Eppendorf AG, Hamburg, Germany) at 2.5 kV with 600 Ω resistance. All enzymes for recombinant DNA work were obtained from Thermo Fisher Scientific Inc. (Darmstadt, Germany) and oligonucleotides were synthesized by biomers.net GmbH (Ulm, Germany, listed in Table 3).
Table 3.
Overview of engineered P. putida strains cultivated under oxygen deprivation conditions
| Strain | YIso/S [mg/gGLC] | qs [g g−1 h−1] | Y2‐KG/S [mg/gGLC] |
|---|---|---|---|
| KT2440 | 0 | 0.11 ± 0.01 | 42 ± 32 |
| Iso2 | 9 ± 1 | 0.14 ± 0.01 | 120 ± 9 |
| Iso3 | 5 ± 2 | 0.12 ± 0.01 | 183 ± 8 |
| Iso4 | 4 ± 1 | 0.13 ± 0.01 | 193 ± 12 |
| Iso5 | 0 | 0.01 ± 0.00 | 0 |
| Iso6 | 19 ± 2 | 0.07 ± 0.01 | 397 ± 14 |
2.4. Plasmid construction
yqhD was amplified from genomic DNA of E. coli MG1655 using the primers yqhd1/yhqd2, digested with PstI/XhoI and ligated into PstI/XhoI‐digested pBB1 yielding pBB1 yqhD. kivD was subsequently added before yqhD, amplified from pSA55 with the primer pair kivd1/kivd2, digested with PstI/XmaI, and ligated into PstI/XmaI‐digested pBB1 yqhd creating plasmid pBB1 kivD yqhD. Ptac, kivD, and yqhD were amplified from plasmid pBB1 kivD yqhD using the primers pbb1/pbb2. The resulting PCR fragment was digested with BamHI/SacI and subsequently ligated into BamHI/SacI‐digested pSEVA231 to create plasmid pIP01. Plasmid pEMP04 was constructed using the inABLE DNA assembly method from Ingenza Ltd. The B. subtilis alsS and P. putida ilvC and ilvD genes were amplified using primer pairs alss1/alss2, ilvc1/ilvc2, and ilvd1/ilvd2, respectively. Additionally, a 5′ truncated version of pIP01 was amplified using primer pair pip011/pip012. The PCR products were digested using SapI and annealed oligonucleotides were ligated at each terminus. Ligation of the oligonucleotides results in the generation of 5′ and 3′ 16 nt single stranded overhangs that are complementary between fragments resulting in the DNA assembling in the predefined order. The genes of pEMP04 were amplified using the primers png1/png2 and cloned by Gibson Assembly 36 into NdeI/SalI‐digested pNG413.1 yielding plasmid pIP02. kdcA from C. maltaromaticum LMA28, adhA from L. lactis MG1363, and adhA from C. glutamicum was amplified using the respective genomic DNA with the primers kdca1/kdca2, adha1/adha2, and adha3/adha4 and cloned together via Gibson Assembly with a PCR fragment from pEMP04 that was amplified with the primers p41/p42, p43/p44, or p45/p46 to construct plasmid pEMP014, pEMP012, and pEMP013. To exchange Ptac with araC PBAD the genes of pEMP012, pEMP013 and pEMP014 were amplified using the primers png1/png2 for pEMP012/013 and png3/png2 for pEMP014 and cloned by Gibson Assembly into NdeI/SalI‐digested pNG413.1, constructing the plasmids pIP03, pIP04, and pIP05.
2.5. Determination of μ and Y X/S
Growth rates were determined by linear regression of ln(OD600) plotted against time (in hours) during the exponential growth phase. Biomass yields Y X/S (g/g) were calculated by linear regression of the biomass concentration cx (g/L) plotted against the respective glucose concentration (g/L) during the exponential growth phase.
2.6. Construction of P. putida deletion mutants
Chromosomal deletions in P. putida were carried out using the 5‐fluorouracil (5‐FU)/upp counterselection system 37. Deletions of the bkdAA gene (encoding the α‐subunit of the ketoacid dehydrogenase complex), the sthA gene (encoding soluble transhydrogenase) and the gcd gene (encoding glucose dehydrogenase) were performed using the integration vector pJOE6261.2. The flanking regions (about 500 bp) of each gene were amplified by PCR from chromosomal DNA of P. putida using the primer pairs bkdaa1/bkdaa2 and bkdaa3/bkdaa4, stha1/stha2, and stha3/stha4, gcd1/gcd2 and gcd3/gcd4. The two respective PCR fragments were purified and cloned into SalI/BamHI‐restricted pJOE6261.2 by Gibson Assembly. Finally, the assembly mix was used to transform P. putida by electroporation. The first selection was carried out on LB agar with 50 µg/L kanamycin and a kanamycin‐resistant clone was afterward grown in liquid LB medium for 24 h. The second recombination event was induced by plating cells on LB agar with 50 µg/L 5‐FU. Deletion mutants were identified by colony PCR using the primer pairs bkdaagc1/bkdaagc2, sthagc1/sthagc2, and gcdgc1/gcdgc2, respectively.
2.7. Analytics
Biomass formation was measured by determination of the OD600 (Ultrospec 10, GE Healthcare, USA) at specific time points. The cell dry weight (gCDW/L) was correlated to the OD600 in several independent cultivations with a correlation factor of 0.346 gCDW/L per OD (data not shown). Shaking flasks were sampled directly in the incubator using an injection syringe (100 Sterican®, 0.80 × 120 mm, B.Braun, Melsungen, Germany). For the determination of isobutanol, 2‐KIV, 2‐ketogluconate (2‐KG), and glucose concentrations, 2 mL of the main culture was harvested by centrifugation (12 100 × g, 5 min, room temperature (RT)) and the supernatant was analyzed via HPLC. Glucose concentrations were measured enzymatically with a test kit from r‐biopharm (r‐biopharm AG, Darmstadt, Germany).
2.8. HPLC metabolite quantification
Isobutanol, 2‐KIV and 2‐KG were measured with a Agilent 1200 series HPLC system equipped with a Rezex ROA organic acid H (8%) column (300 by 7.8 mm, 8 µm; Phenomenex) protected by a Phenomenex guard column carbo‐H (4 by 3.0 mm inside diameter) 38. Samples and standards were treated with a phosphate precipitation protocol before HPLC measurements. More precisely, 500 µL of sample volume was mixed with 45 µL 4 M NH3 and 50 µL 1.2 M MgSO4 followed by 5 min incubation at RT and centrifugation for 5 min at 7000 × g. Pellets were discarded and the supernatant was mixed with 500 µL 0.1 M H2SO4, incubated for 15 min at RT, and centrifuged for 15 min at 7000 × g. The resulting supernatant was used for HPLC injection with an injection volume of 10 µL. Separation was carried out under isocratic conditions at 50°C column temperature for 60 min with 5 mM H2SO4 as the mobile phase at a constant flow rate of 0.4 mL/min. Detection of isobutanol, 2‐KIV, and 2‐KG was achieved with a refractive index detector at 32°C. Quantification of all analytes was done with a 7‐point calibration curve for each component as an external reference standard.
3. RESULTS
3.1. Preventing product and precursor degradation
Pseudomonads are well‐known for their ability to degrade a variety of organic substances to utilize them as carbon and energy sources 21. Since the genomic repertoire provides annotated routes for the degradation of isobutanol and 2‐ketoisovalerate (Figure 1), we initially characterized growth on both compounds (Figure 2). P. putida showed exponential growth on isobutanol with a μ of 0.27 ± 0.01 h−1 as well as on 2‐KIV with a μ of 0.33 ± 0.01 h−1 that is 52% of the growth rate on glucose (Figure 2A,B). Recently, several enzymes involved in n‐butanol degradation were identified 39 and Simon et al. 30 constructed P. putida Δupp ΔpedE ΔpedI ΔpedH ΔaldB‐I (P. putida GN346) to inactivate two alcohol dehdyrogenases (PedE, PedH) and two aldehyde dehydrogenases (PedI, AldB‐I) and showed that the introduced deletions prevented n‐butanol consumption. Accordingly, P. putida GN346 was unable to utilize isobutanol as sole carbon and energy source (Figure 2A).
Figure 2.
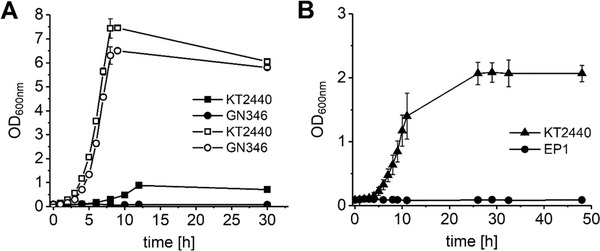
(A) Growth of P. putida KT2440 and P. putida GN346 in DeBont minimal medium containing 0.5 g/L isobutanol (filled symbols) or 5.4 g/L glucose (open symbols). (B) Growth of P. putida and P. putida EP1 in DeBont minimal medium containing 2.9 g/L 2‐ketoisovalerate. Experiments were performed in triplicates and error bars represent the corresponding standard deviation
P. putida possesses a branched chain ketoacid dehydrogenase (BCKDH) complex that converts 2‐ketoacids to the respective decarboxylated CoA‐derivatives 40, 41 which are, after further conversion steps, funneled into the TCA cycle. To prevent the consumption of the precursor 2‐KIV, we inactivated the α‐subunit of the BCKDH by deletion of the bkdAA gene in P. putida GN346. In contrast to the wild‐type, the resulting strain P. putida EP1 was unable to grow on 2‐KIV as carbon source (Fig. 2B), and therefore was used as basis for further strain engineering.
3.2. Engineering P. putida for isobutanol production
To drain the carbon from pyruvate to 2‐KIV, we constructed a plasmid harboring the alsS gene encoding the acetolactate synthase from Bacillus subtilis, which is not feedback inhibited by branched chain amino acids, and the native ilvCD genes encoding the ketolacid reductoisomerase and dihydroxyacid dehydratase (Figure 1). For the conversion of 2‐KIV to isobutanol, we additionally cloned kivD encoding the KDC from Lactococcus lactis and yqhD encoding an aldehyde reductase from E. coli (Fig. 1). AlsS, KivD, and YqhD were previously applied for isobutanol production in other hosts such as C. glutamicum and E. coli 5, 7. The resulting plasmid pIP02 expresses all cloned genes under control of the l‐arabinose inducible PBAD promoter and was used to transform P. putida EP1 yielding P. putida Iso1. In minimal medium with glucose, P. putida Iso1 showed a µ = 0.56 ± 0.02. Although the Y X/S was reduced by 25% compared to the wild‐type, no isobutanol was produced during the cultivation (Table 2).
Table 2.
Overview of growth, 2‐ketogluconate (2‐KG) and isobutanol production of P. putida and its engineered derivatives
| Strain | μ [h−1] | Y X/S [g/g] | Y 2‐KG/S [mg/gGLC] | Y Iso/S [mg/gGLC] |
|---|---|---|---|---|
| KT2440 | 0.62 ± 0.01 | 0.40 ± 0.01 | 0 | 0 |
| GN346 | 0.59 ± 0.01 | 0.39 ± 0.01 | 0 | 0 |
| Iso1 | 0.56 ± 0.02 | 0.30 ± 0.01 | 0 | 0 |
| Iso2 | 0.25 ± 0.01 | 0.13 ± 0.01 | 438 ± 20 | 22 ± 2 |
| Iso3 | 0.14 ± 0.01 | 0.06 ± 0.01 | 833 ± 100 | 13 ± 1 |
| Iso4 | 0.19 ± 0.01 | 0.09 ± 0.02 | 771 ± 15 | 14 ± 0.0 |
| Iso5 | 0.18 ± 0.02 | 0.28 ± 0.01 | 0 | 0 |
| Iso6 | 0.28 ± 0.01 | 0.10 ± 0.01 | 633 ± 39 | 21 ± 1 |
The synthesis of isobutanol from glucose requires 2 mol NAD(P)H per mol isobutanol. The reduction of acetolactate is catalyzed by NADPH‐dependent ketolacid reductoisomerase (IlvC), while the conversion of isobutyraldehyde to isobutanol can be catalyzed by NAD(P)H‐dependent aldehyde/alcohol dehydrogenases such as YqhD (Figure 1). Since YqhD is NADPH‐dependent, the engineered iosobutanol pathway should consume 2 mol NADPH per mol isobutanol. P. putida possesses a membrane‐bound and a soluble transhydrogenase. The latter is encoded by the sthA gene 42 and has in E. coli been reported to favor the re‐oxidation of NADPH to NADP+ under reduction of NAD+ to NADH 43, 44.
To test whether the inactivation of the soluble transhydrogenase is beneficial for isobutanol production, we deleted the sthA gene in P. putida EP1, yielding P. putida EP2 which was transformed with the plasmid pIP02. The resulting strain P. putida Iso2 showed in minimal medium containing 5.4 g/L glucose, a growth rate of 0.26 ± 0.01 h−1, a Y X/S of of 0.13 ± 0.01 g/g, and produced 438 ± 20 mg/gGLC 2‐KG and for the first time isobutanol with a Y Iso/S of 22 ± 2 mg/gGLC (Table 2, Figure 3). We also replaced the aldehyde reductase gene yqhD on the overexpression plasmid pIP02 with the adhA genes encoding NADH‐dependent alcohol dehydrogenase variants from Lactococcus lactis and Corynebacterium glutamicum, respectively. The plasmids pIP03 and pIP04 were used to transform P. putida EP2, yielding P. putida Iso3 and Iso4, which were characterized in minimal medium with glucose (Table 2). Both strains showed reduced growth rates and about 40% lower product yields compared to P. putida Iso2. All engineered strains with deletion of sthA converted 40% to 83% of the available glucose into 2‐KG that was secreted into the culture broth (Table 2). To avoid 2‐KG secretion and to improve isobutanol production, we constructed P. putida EP3 by deletion of the gcd gene encoding periplasmatic glucose dehydrogenase in P. putida EP2. To construct P. putida Iso5, P. putida EP3 was transformed with the plasmid pIP02. In fact, P. putida Iso5 did not secrete any 2‐KG, however, inactivation of GCD also abolished isobutanol production completely (Table 2).
Figure 3.
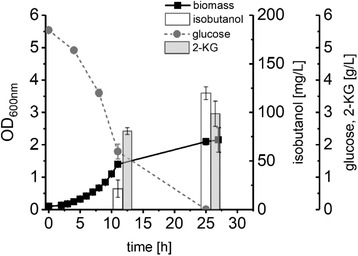
Growth (black circles), glucose consumption (grey circles), isobutanol production (white bars) and 2‐KG formation (grey bars) of P. putida Iso2 in DeBont minimal medium containing glucose. Experiments were performed in triplicates and error bars represent the corresponding standard deviation
3.3. Ketoacid decarboxylase from Carnobacterium maltaromaticum is suitable for isobutanol production
The key enzyme for isobutanol production via the Ehrlich pathway is ketoacid decarboxylase (KDC) converting 2‐KIV to isobutyraldehyde (Figure 1). So far, only KDC from L. lactis has been proven as suitable variant that efficiently catalyzes this reaction 2, 5. Recently, the genome sequence of Carnobacterium maltaromaticum LMA28 28 was published that harbors a gene encoding a putative KDC. KDC from C. maltaromaticum shows 54% identity to the KDC enzyme from L. lactis. To test the suitability of KDC from C. maltaromaticum for isobutanol production, we replaced the kivD gene on plasmid pIP02 with the respective gene from C. maltaromaticum. Plasmid pIP05 was transformed into P. putida EP2 and the resulting strain P. putida Iso6 was characterized. P. putida Iso6 showed a growth rate of 0.28 ± 0.01 h−1, a Y X/S of of 0.10 ± 0.01 g/g. and produced 633 ± 39 mg/gGLC 2‐KG. Furthermore, P. putida Iso6 secreted as much isobutanol as P. putida Iso2 with a Y Iso/S of 21 ± 1 mg/gGLC (Table 2) showing that KDC from C. maltaromaticum LMA28 is a useful alternative to KDC from L. lactis.
3.4. Microaerobic isobutanol production in P. putida
P. putida is regarded as an obligate aerobic bacterium 18. However, since the implementation of the synthetic isobutanol pathway theoretically enables a closed redox balance, we tested the capabilities of our engineered P. putida strains to produce isobutanol from glucose in a zero‐growth bioprocess under oxygen deprivation conditions 45. Therefore, we inoculated P. putida WT and Iso2–6 to an OD600 of 15–20 in closed bottles filled with minimal medium containing 5.4 g/L glucose and characterized substrate consumption and (by‐) product formation (Table 3). In the micro‐aerobic environment P. putida WT showed no growth, but remained metabolically active and consumed the glucose that was converted to 2‐KG. With the exception of P. putida Iso5, all other engineered strains consumed glucose and produced isobutanol. P. putida Iso6 showed the best performance under oxygen deprivation conditions. Compared to the WT the q S was reduced by 37% and P. putida Iso6 produced about 10% less isobutanol compared to the aerobic shaking flask experiments (Table 3).
4. DISCUSSION
P. putida is an emerging host for industrial biotechnology 46, 47, 48. However, this bacterium is also known to efficiently metabolize a broad range of substrates including amino and organic acids and alcohols 21. As shown here, P. putida grows rapidly on isobutanol as well as on its precursor 2‐KIV. Although P. putida KT2440 possesses four aldehyde dehydrogenases and about 10 alcohol dehydrogenases, Simon et al. 30 showed that deletion of the two alcohol dehydrogenase genes pedE and pedH and the two aldehyde dehydrogenases genes pedI and aldB‐I is sufficient to prevent n‐butanol degradation. Accordingly, we found that this strain background also prevents growth on the branched‐chain alcohol isobutanol. P. putida possesses a branched chain ketoacid dehydrogenase complex that converts 2‐ketoacids to the respective decarboxylated CoA‐derivatives 40, 41. As expected and also observed for P. taiwanensis VLB120 41, inactivation of the BCKDH abolished growth on 2‐ketoisovalerate. To avoid auxotrophies, we relinquished the inactivation of the l‐valine forming transaminase IlvE, the 2‐isopropylmalate synthase LeuA and the 2‐ketoisovalerate hydroxymethyltransferase PanB as has been applied to improve isobutyric acid production with P. taiwanensis strain VLB120 41.
Since AHAIR is usually NADPH‐dependent, the synthesis of one molecule of isobutanol either requires two molecules of NADPH or one NADH plus one NADPH molecule depending on the applied alcohol/aldehyde dehydrogenase variant for the reduction of isobutyraldehyde to isobutanol. Optimization of NAD(P)H availability has already been shown to be a crucial factor for isobutanol production with other hosts such as E. coli and C. glutamicum 6, 49. Recently, Nikel et al. 16 showed that P. putida cells growing on glucose exhibit a slight catabolic overproduction of reducing power and run a biochemical cycle that favors NADPH formation. Therefore, we applied in our experiments the broad‐substrate range NADPH‐dependent aldehyde reductase YqhD 50, which has also been successfully applied for isobutanol production with E. coli 8. However, expression of the synthetic pathway in P. putida Iso1 to channel pyruvate toward isobutanol did not result in isobutanol production from glucose. Similar to E. coli, P. putida possesses a membrane bound (PntAB) and a soluble transhydrogenase (SthA) to balance the overall redox state of the cell (Figure 1). SthA has in E. coli been reported to favor the oxidation of NADPH to NADP+, accompanied with the reduction of NAD+ to NADH 43, 44. To improve NADPH availability, we inactivated SthA that resulted in isobutanol formation in P. putida Iso2 under aerobic conditions. Accordingly, expression of two adhA genes encoding NADH‐dependent alcohol dehydrogenases from L. lactis and C. glutamicum, which have previously been shown to be suitable for isobutanol production 7, 8, instead of YqhD, led to significantly reduced isobutanol yields in the ΔsthA background (Table 2).
Inactivation of SthA resulted in isobutanol production, however, also in the secretion of significant amounts of 2‐KG. In P. putida a majority of the glucose is converted in the periplasm by glucose dehydrogenase (Gcd) to gluconate, which is transported to the cytoplasm and activated by the gluconate kinase to feed the Entner–Doudoroff pathway with 6‐phosphogluconate. Usually, only a small fraction of gluconate is converted in the periplasm by gluconate dehydrogenase to 2‐KG, which is subsequently transported into the cytoplasm to finally form 6‐phosphogluconate via 2‐KG kinase and 2‐ketogluconate‐6‐P reductase 16. Since deletion of gcd abolished 2‐KG production completely, the synthesis of this molecule occurs solely in the periplasm via the described route. The accumulation of 2‐KG in the culture broth indicates a transport inhibition of gluconate and/or 2‐KG from the periplasm to the cytoplasm by an unknown mechanism and/or an inhibition or limitation of the ATP‐dependent conversion to the phosphorylated derivatives. The latter might result as consequence of a perturbed redox state due to the inactivated transhydrogenase SthA.
P. putida is an obligate aerobic bacterium, however, in a bioelectrochemical system P. putida was metabolically active under anoxic conditions when an electron mediator was applied for redox balancing in a high‐yield 2‐KG production system 51, 52. Since isobutanol synthesis enables regeneration of NAD(P)+, we cultivated P. putida WT and the engineered derivatives under microaerobic conditions. All strains showed no growth (data not shown) but with the exception of P. putida Iso5, remained metabolically active and P. putida Iso2‐4 and 6 also secreted 2‐KG, isobutanol, and further unidentified products. However, according to the zero‐growth the q S values are low compared to aerobic conditions (e.g. for P. putida WT 0.11 vs. 1.55 g g−1 h−1) . The capability of P. putida to remain metabolically active opens the possibility to develop dual‐phase production processes that comprise an aerobic growth phase for rapid biomass formation and a micro‐aeobic or anaerobic production phase 45.
This study paths the way to construct more efficient P. putida strains for isobutanol production in future studies. The overall isobutanol yield is significantly higher compared to other engineered P. putida strains 41, however, rather low compared to tailored E. coli and C. glutamicum strains 49. Product and precursor degradation can be prevented by the presented deletions in this study, however, improving NAD(P)H and pyruvate availability 49 will be crucial to achieve high‐yield isobutanol production strains.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
ACKNOWLEDGEMENTS
We thank Mira Lenfers‐Lücker and Sven Göbel (Institute of Biochemical Engineering, University of Stuttgart, Germany) for assistance during HPLC analysis and experimental procedures. This work was funded by the European Commission H2020 project Empowerputida under the grant agreement No. 635536. This work was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Nitschel R, Ankenbauer A, Welsch I, et al. Engineering Pseudomonas putida KT2440 for the production of isobutanol. Eng Life Sci. 2020;20:148–159. 10.1002/elsc.201900151
REFERENCES
- 1. Tiffany, D. , Nelson, E. , Tilman, D. , Hill, J. , et al., Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. USA 2006, 103, 11206–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eikmanns, B.J. and Blombach, B. , Isobutanol, in: Bisaria, V. S., Kondo, A. (Eds.), Bioprocessing of Renewable Resources to Commodity Bioproducts, John Wiley & Sons, Inc., Weinheim: 2014, pp. 327–352. [Google Scholar]
- 3. Chen, C.‐T. and Liao, J.C. , Frontiers in microbial 1‐butanol and isobutanol production. FEMS Microbiol. Lett. 2016, 363, fnw020. [DOI] [PubMed] [Google Scholar]
- 4. Wilson, J. , Gering, S. , Pinard, J. , Lucas, R. , et al., Bio‐production of gaseous alkenes: ethylene, isoprene, isobutene. Biotechnol. Biofuels 2018, 11, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atsumi, S. , Hanai, T. , Liao, J.C. , Non‐fermentative pathways for synthesis of branched‐chain higher alcohols as biofuels. Nature 2008, 451, 86–89. [DOI] [PubMed] [Google Scholar]
- 6. Bastian, S. , Liu, X. , Meyerowitz, J.T. , Snow, C.D. , et al., Engineered ketol‐acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2‐methylpropan‐1‐ol production at theoretical yield in Escherichia coli . Metab. Eng. 2011, 13, 345–352. [DOI] [PubMed] [Google Scholar]
- 7. Blombach, B. , Riester, T. , Wieschalka, S. , Ziert, C. , et al., Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 2011, 77, 3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atsumi, S. , Wu, T.Y. , Eckl, E.M. , Hawkins, S.D. , et al., Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010, 85, 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li, S. , Huang, D. , Li, Y. , Wen, J. , et al., Rational improvement of the engineered isobutanol‐producing Bacillus subtilis by elementary mode analysis. Microb. Cell Fact. 2012, 11, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuda, F. , Ishii, J. , Kondo, T. , Ida, K. , et al., Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb. Cell Fact. 2013, 12, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kieboom, J. , Dennis, J.J. , de Bont, J.A.M. , Zijlstra, G.J. , Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 1998, 273, 85–91. [DOI] [PubMed] [Google Scholar]
- 12. Udaondo, Z. , Duque, E. , Fernández, M. , Molina, L. , et al., Analysis of solvent tolerance in Pseudomonas putida DOT‐T1E based on its genome sequence and a collection of mutants. FEBS Lett. 2012, 586, 2932–2938. [DOI] [PubMed] [Google Scholar]
- 13. Belda, E. , van Heck, R.G.A. , José Lopez‐Sanchez, M. , Cruveiller, S. , et al., The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016, 18, 3403–3424. [DOI] [PubMed] [Google Scholar]
- 14. Regenhardt, D. , Heuer, H. , Heim, S. , Fernandez, D.U. , et al., Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 2002, 4, 912–915. [DOI] [PubMed] [Google Scholar]
- 15. Nikel, P.I. , Martínez‐García, E. , de Lorenzo, V. , Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014, 12, 368–79. [DOI] [PubMed] [Google Scholar]
- 16. Nikel, P.I. , Chavarria, M. , Fuhrer, T. , Sauer, U. , et al., Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner‐Doudoroff, Embden‐Meyerhof‐Parnas, and pentose phosphate pathways. J. Biol. Chem. 2015, 290, 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Udaondo, Z. , Ramos, J.‐L. , Segura, A. , Krell, T. , et al., Regulation of carbohydrate degradation pathways in Pseudomonas involves a versatile set of transcriptional regulators. Microb. Biotechnol. 2018, 11, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. dos Santos, V.A.P.M. , Heim, S. , Moore, E.R.B. , Stratz, M. , et al., Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004, 6, 1264–1286. [DOI] [PubMed] [Google Scholar]
- 19. Dvořák, P. and de Lorenzo, V. , Refactoring the upper sugar metabolism of Pseudomonas putida for co‐utilization of cellobiose, xylose, and glucose. Metab. Eng. 2018, 48, 94–108. [DOI] [PubMed] [Google Scholar]
- 20. Poblete‐Castro, I. , Becker, J. , Dohnt, K. , dos Santos, V.M. , et al., Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [DOI] [PubMed] [Google Scholar]
- 21. Nikel, P.I. and de Lorenzo, V. , Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans‐metabolism. Metab. Eng. 2018, 50, 142–155. [DOI] [PubMed] [Google Scholar]
- 22. Kohlstedt, M. , Starck, S. , Barton, N. , Stolzenberger, J. , et al., From lignin to nylon: Cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida . Metab. Eng. 2018, 47, 279–293. [DOI] [PubMed] [Google Scholar]
- 23. Calero, P. , Jensen, S.I. , Nielsen, A.T. , Broad‐Host‐Range ProUSER vectors enable fast characterization of inducible promoters and optimization of p ‐coumaric acid production in Pseudomonas putida KT2440. ACS Synth. Biol. 2016, 5, 741–753. [DOI] [PubMed] [Google Scholar]
- 24. Molina‐Santiago, C. , Cordero, B.F. , Daddaoua, A. , Udaondo, Z. , et al., Pseudomonas putida as a platform for the synthesis of aromatic compounds. Microbiology 2016, 162, 1535–1543. [DOI] [PubMed] [Google Scholar]
- 25. Köhler, K.A.K. , Rückert, C. , Schatschneider, S. , Vorhölter, F.‐J. , et al., Complete genome sequence of Pseudomonas sp. strain VLB120 a solvent tolerant, styrene degrading bacterium, isolated from forest soil. J. Biotechnol. 2013, 168, 729–730. [DOI] [PubMed] [Google Scholar]
- 26. Wynands, B. , Lenzen, C. , Otto, M. , Koch, F. , et al., Metabolic engineering of Pseudomonas taiwanensis VLB120 with minimal genomic modifications for high‐yield phenol production. Metab. Eng. 2018, 47, 121–133. [DOI] [PubMed] [Google Scholar]
- 27. Bagdasarian, M. , Lurz, R. , Ruckert, B. , Franklin, F.C. , et al., Specific‐purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF 1010‐derived vectors, and a host‐vector system for gene cloning in Pseudomonas . Gene 1981, 16, 237–247. [DOI] [PubMed] [Google Scholar]
- 28. Cailliez‐Grimal, C. , Chaillou, S. , Anba‐Mondoloni, J. , Loux, V. , et al., Complete chromosome sequence of Carnobacterium maltaromaticum LMA 28. Genome Announc. 2013, 1, e00115‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wegmann, U. , O'Connell‐Motherway, M. , Zomer, A. , Buist, G. , et al., Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007, 189, 3256–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simon, O. , Klebensberger, J. , Mükschel, B. , Klaiber, I. , et al., Analysis of the molecular response of Pseudomonas putida KT2440 to the next‐generation biofuel n‐butanol. J. Proteomics 2015, 122, 11–25. [DOI] [PubMed] [Google Scholar]
- 31. Krause, F.S. , Blombach, B. , Eikmanns, B.J. , Metabolic engineering of Corynebacterium glutamicum for 2‐ketoisovalerate production. Appl. Environ. Microbiol. 2010, 76, 8053–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graf, N. and Altenbuchner, J. , Functional characterization and application of a tightly regulated MekR/P mekA expression system in Escherichia coli and Pseudomonas putida. Appl. Microbiol . Biotechnol. 2013, 97, 8239–8251. [DOI] [PubMed] [Google Scholar]
- 33. Silva‐Rocha, R. , Martínez‐García, E. , Calles, B.N. , Chavarría, M. , et al., The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2012, 41, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sambrook, J. and Russell, D.W. , Molecular cloning: a laboratory manual., 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor: 2001. [Google Scholar]
- 35. Hartmans, S. , Smits, J.P. , van der Werf, M.J. , Volkering, F. , et al., Metabolism of styrene oxide and 2‐phenylethanol in the styrene‐degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 1989, 55, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gibson, D.G. , Young, L. , Chuang, R.‐Y. , Venter, J.C. , et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- 37. Graf, N. and Altenbuchner, J. , Development of a method for markerless gene deletion in Pseudomonas putida. Appl. Environ . Microbiol. 2011, 77, 5549–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchholz, J. , Schwentner, A. , Brunnenkan, B. , Gabris, C. , et al., Platform Engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L‐lysine, L‐valine, and 2‐ketoisovalerate. Appl. Environ. Microbiol. 2013, 79, 5566–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vallon, T. , Simon, O. , Rendgen‐Heugle, B. , Frana, S. , et al., Applying systems biology tools to study n ‐butanol degradation in Pseudomonas putida KT2440. Eng. Life Sci. 2015, 15, 760–771. [Google Scholar]
- 40. Sokatch, J.R. , McCully, V. , Roberts, C.M. , Purification of a branched‐chain keto acid dehydrogenase from Pseudomonas putida . J. Bacteriol. 1981, 148, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lang, K. , Zierow, J. , Buehler, K. , Schmid, A. , Metabolic engineering of Pseudomonas sp. strain VLB120 as platform biocatalyst for the production of isobutyric acid and other secondary metabolites. Microb. Cell Fact. 2014, 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nikel, P.I. , Pérez‐Pantoja, D. , de Lorenzo, V. , Pyridine nucleotide transhydrogenases enable redox balance of Pseudomonas putida during biodegradation of aromatic compounds. Environ. Microbiol. 2016, 18, 3565–3582. [DOI] [PubMed] [Google Scholar]
- 43. Sauer, U. , Canonaco, F. , Heri, S. , Perrenoud, A. , et al., The soluble and membrane‐bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli*. J. Biol. Chem. 1998, 273, 85–91. [DOI] [PubMed] [Google Scholar]
- 44. Fuhrer, T. and Sauer, U. , Different biochemical mechanisms ensure network‐wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 2009, 191, 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lange, J. , Takors, R. , Blombach, B. , Zero‐growth bioprocesses: A challenge for microbial production strains and bioprocess engineering. Eng. Life Sci. 2017, 17, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikel, P.I. , Chavarría, M. , Danchin, A. , de Lorenzo, V. , From dirt to industrial applications: Pseudomonas putida as a Synthetic Biology chassis for hosting harsh biochemical reactions. Curr. Opin. Chem. Biol. 2016, 34, 20–29. [DOI] [PubMed] [Google Scholar]
- 47. Martínez‐García, E. and de Lorenzo, V. , Molecular tools and emerging strategies for deep genetic/genomic refactoring of Pseudomonas . Curr. Opin. Biotechnol. 2017, 47, 120–132. [DOI] [PubMed] [Google Scholar]
- 48. Martínez‐García, E. and de Lorenzo, V. , Pseudomonas putida in the quest of programmable chemistry. Curr. Opin. Biotechnol. 2019, 59, 111–121. [DOI] [PubMed] [Google Scholar]
- 49. Blombach, B. and Eikmanns, B.J. , Current knowledge on isobutanol production with Escherichia coli, Baciullus subtilis and Corynebacterium glutamicum. Bioeng . Bugs 2011, 2, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jarboe, L.R. , YqhD: a broad‐substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol. 2011, 89, 249–257. [DOI] [PubMed] [Google Scholar]
- 51. Lai, B. , Yu, S. , Bernhardt, P. V , Rabaey, K. , et al., Anoxic metabolism and biochemical production in Pseudomonas putida F1 driven by a bioelectrochemical system. Biotechnol. Biofuels 2016, 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu, S. , Lai, B. , Plan, M.R. , Hodson, M.P. , et al., Improved performance of Pseudomonas putida in a bioelectrochemical system through overexpression of periplasmic glucose dehydrogenase. Biotechnol. Bioeng. 2018, 115, 145–155. [DOI] [PubMed] [Google Scholar]
- 53. Dower, W. J. , Miller, J. F. and Ragsdale, C. W. , High efficiency transformation of E.coli by high voltage electroporation. Nucleic Acids Res. 1988, 16, 6127–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choi, K. H. , Kumar, A. and Schweizer, H. P. , A 10‐min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 2006, 64, 391–397. [DOI] [PubMed] [Google Scholar]


