Abstract
Human cytomegalovirus (HCMV) is the most common congenital infection. A gB subunit vaccine (gB/MF59) is the most efficacious clinically tested to date, having achieved 50% protection against primary infection of HCMV-seronegative women. We previously identified that gB/MF59 vaccination primarily elicits non-neutralizing antibody responses, with variable binding to gB genotypes, and protection associated with binding to membrane-associated gB. We hypothesized that gB-specific non-neutralizing antibody binding breadth and function are dependent on epitope and genotype specificity, and ability to interact with membrane-associated gB. We mapped twenty-four gB-specific monoclonal antibodies (mAbs) from naturally HCMV-infected individuals for gB domain specificity, genotype preference, and ability to mediate phagocytosis or NK cell activation. gB-specific mAbs were primarily specific for Domain II and demonstrated variable binding to gB genotypes. Two mAbs facilitated phagocytosis with binding specificities of Domain II and AD2. This investigation provides novel understanding on the relationship between gB domain specificity and antigenic variability on gB-specific antibody effector functions.
Introduction
Each year, an estimated 40,000 children in the U.S. are born with congenital human cytomegalovirus (HCMV) infection (cCMV), with roughly 8,000 afflicted children developing long term sequelae of disease such as sensorineural hearing loss, neurodevelopmental delay, visual impairment, and psychomotor disability [1, 2]. While mothers with primary HCMV infection during pregnancy have rates of vertical transmission up to 40%, mothers with HCMV reactivation or reinfection can also transmit the virus to their developing infant but at a rate in the range of <1–4%, suggesting partial protection from preexisting immunity [3–5]. Additionally, HCMV infection post-transplantation and consequent graft rejection, remain significant complications for solid-organ transplant patients, especially for those HCMV naïve recipients with transplants from HCMV seropositive donors [6, 7]. Although the protective correlates of immunity against HCMV infection have not been fully elucidated, there is ample evidence that immune factors elicited by natural infection can confer some degree of protection for mothers, infants, and immunosuppressed individuals against HCMV reinfection or reactivation [8, 9] Thus, understanding how natural immunity against HCMV mediates protection may offer the basis for development of an effective HCMV vaccine.
Numerous HCMV vaccine candidates have been tested clinically, including live-attenuated virus, viral glycoprotein subunit formulations, viral vectors, and single/bivalent DNA plasmids, yet few have demonstrated sufficient protection as compared to natural immunity to warrant late stage clinical development [10]. Interestingly, the glycoprotein B (gB/MF59) subunit vaccine achieved partial efficacy in preventing HCMV primary infection in distinct cohorts of HCMV seronegative postpartum women (50%) and adolescent girls (43%) [11, 12]. Moreover, in HCMV seronegative patients receiving liver or kidney transplants from HCMV seropositive donors, the gB/MF59 vaccine was shown to reduce the magnitude and duration of posttransplant HCMV viremia [13]. gB is a viral fusion protein, essential for viral infection of host cells, and is a target of both neutralizing and non-neutralizing antibodies [14]. Counterintuitively, in the gB/MF59 phase II trials, vaccination elicited negligible neutralizing responses when compared with HCMV seropositive individuals [15, 16]; yet, gB/MF59 vaccination elicited effector-cell mediated antibody responses, including antibody dependent cellular phagocytosis (ADCP). This observation prompted a hypothesis that non-neutralizing antibodies against gB may contribute to protection against HCMV acquisition [15, 16]. Moreover, effector antibody functions mediated by NK cells may be crucial for targeting HCMV-infected cells, further underlying the significance of non-neutralizing antibodies in anti-HCMV immunity [17, 18].
gB is a homotrimeric class III fusion protein that contains 5 defined antigenic domains, namely AD1 - AD5, with AD2 site 1, AD4 (Domain II), and AD5 (Domain I) as the target of neutralizing antibodies, and AD2 site 2 and AD1 as reported primary targets of non-neutralizing antibodies [14, 19]. The transmembrane and cytosolic domains of gB (AD3), are thought to be obscured from antibody recognition in the membrane-associated portion. Yet, the epitope-specific antibody responses stimulated by natural HCMV infection and gB/MF59 vaccination demonstrate AD3 immunodominance, with 76% of total linear gB-specific antibody responses directed at this region in naturally HCMV infected vs 32% in gB/MF59 vaccinated individuals [15]. Variation in epitope-specific antibody binding may also stem from gB genotype-specific differences amongst clinical HCMV strains [19]. In fact, differences amongst HCMV gB genotypes correlate with cell tropism during HCMV infection [20]. Interestingly, it was recently reported that the ability of gB/MF59 vaccine sera to bind to membrane associated gB, via a transfected cell binding assay, predicts risk of HCMV acquisition [21]. Furthermore, like HSV gB and VSV protein G, HCMV gB undergoes significant conformational transition from prefusion to postfusion states to mediate viral and host-cell membrane fusion [22, 23]. Thus, the characterization of antibodies that bind distinctly to soluble postfusion gB constructs and cell-associated gB, with the latter possibility representing a distinct prefusion-like structure, is needed to guide vaccine development.
Because there are stark differences in the gB epitope binding profiles of naturally-infected individuals and gB/MF59 vaccinees [15, 24], defining immune-protection offered by prior exposure to natural HCMV infection and vaccination will require an understanding of how gB as an antigen induces potentially protective effector antibody responses. The aim of this study is to characterize the non-neutralizing gB-specific antibody epitope-specificity and functions in natural HCMV infection for a panel of gB-specific mAbs isolated by memory B cell cultures from three naturally infected individuals [25]. A panel of 24 mAbs with no HCMV neutralizing or weakly neutralizing activity were analyzed for their specificity for gB domains and genotypes, and effector functions including ADCC and ADCP. Understanding gB diversity, and how gB-specific antibodies bind and mediate antiviral functions across genotypes to different antigenic domains, will be critical if we hope to optimize anti-gB antibody responses in future iterations of an HCMV vaccine.
Methods
gB-specific monoclonal antibodies (mAbs)
gB-specific monoclonal antibodies (mAbs) were isolated from healthy adult volunteers with previous natural HCMV infection. Subjects were consented for blood sampling in accordance with NIH guidelines [25]. B cell isolation and recombinant monoclonal antibody isolation and production using a human IgG1 backbone was performed as previously described [25]. Briefly, total RNA from isolated memory B cells, which screened positive for binding activity against gB, was converted to cDNA using a reverse transcription kit (Invitrogen), and IgG genes were identified by PCR primers. After cloning the variable regions of gB-specific mAbs from B cells from three donors the VH and VL sequences were cloned into a human IgG1 vector and recombinantly expressed in HEK293 cells in Zhiqiang An’s laboratory at the University of Texas Health Science Center at Houston. This panel of mAbs was previously screened by Xia et al. for neutralization, and deemed non-neutralizing with a neutralization IC50 titer above 1μg/mL for HCMV strain AD169r in ARPE-19 cells [25].
Mapping gB-specific mAb domain specificity by binding antibody multiplex assay (BAMA)
Antibody responses against soluble full length gB protein, lacking the transmembrane domain and the hydrophobic portion of the membrane proximal region (FLgB(−)TMD) (generously provided by Sanofi), soluble gB ectodomain, lacking the cytosolic domains, and individual gB domains were measured as previously described [26]. Carboxylated fluorescent beads (Luminex) were covalently coupled to purified HCMV antigens and incubated with monoclonal antibodies in assay diluent (phosphate-buffered saline, 5% normal goat serum, 0.05% Tween 20, and 1% Blotto milk). The gB domain antigen panel, included FLgB(−)TMD, gB ectodomain, gB domain I, gB domain II, gB AD-1 (myBiosource), and biotinylated linear gB AD-2 site 2 (biotin-AHSRSGSVQRVTSS), and biotinylated linear gB AD-2 site 1 (biotin-NETIYNTTLKYGD) are previously described [15]. All gB proteins are based on the Towne strain gB (genotype 1). HCMV glycoprotein–specific antibody binding was detected with phycoerythrin-conjugated goat anti-human IgG (2 μg/mL, Southern Biotech). Beads were washed and acquired on a Bio-Plex 200 instrument (Bio-Rad), and results were expressed as mean fluorescence intensity. A panel HCMV seronegative plasma samples (n=30) were included to determine nonspecific baseline levels of binding. Minimal background activity was observed, so the threshold for positivity for each antigen was set at the mean value (100 MFI) of negative control sera to each antigen + two standard deviations. Blank beads were used in all assays to account for nonspecific binding. All assays included tracking of CMV immunoglobulin (Cytogam, generously gifted by CSL Behring) standard by Levy-Jennings charts. The preset assay criteria for sample reporting were coefficient of variation per duplicate values of ≤20% for each sample and ≥100 beads counted per sample. All mAbs were analyzed at a concentration of 30 μg/mL for each antigen: FLgB(−)TMD, gB ectodomain, gB Domain I, gB Domain II, gB AD-1 and gB AD-2 sites 1 and 2. This concentration was predetermined to be within the linear range of binding based on testing serial dilutions of a small subset of gB specific mAbs (1–155, 1–235, 2–43, 1–189, 3–54, and 3–74).
gB-specific mAb binding strength measured via ELISA
gB-specific monoclonal antibody binding responses were measured against FLgB(−)TMD, gB regional epitopes (Ectodomain, Domain I), or gB peptides (AD2 site 1, AD2 site 2). All gB constructs were solubilized in 0.1 M NaHCO3, with proteins plated at a concentration of 3 μg/mL and peptides at 10 μg/mL respectively. Plates were washed then blocked before adding gB-specific mAbs at either 5 μg/mL for conformational protein ELISAs or 100 μg/mL for peptide ELISAs. After incubation with mAbs, plates were washed and incubated with a 1:5000 goat-anti human HRP conjugated IgG secondary (Jackson ImmunoResearch). ELISAs were developed with SeraCare ELISA kit and read at 450 nm.
Assessment of gB-specific mAb affinity via surface plasmon resonance (SPR)
Postfusion trimeric gB ectodomain was captured on an NTA sensor chip to ~500 response units (RUs) per cycle using a Biacore X100 (GE Healthcare). The chip was doubly regenerated using 0.35 M EDTA and 0.1 M NaOH followed by 0.5 mM NiCl2. Three samples containing only buffer were injected over both ligand and reference flow cells, followed by single injections of each mAb at a concentration of 25 nM. Samples that did not initially result in interpretable sensorgrams were repeated using a concentration of 250 nM. The resulting data were double-reference subtracted and fit to a 1:1 binding model using the Biacore X100 Evaluation software.
gB genotype-specific transfected cell binding
HEK293T cells were cultured overnight to ~50% confluency in a T25 flask, and then co-transfected using TransIT-mRNA Transfection Kit (Mirus Bio) with a GFP-expressing mRNA plasmid (Miltenyi Biotec) and a second plasmid encoding either gB from genotypes 1, 2, 3, 4, or 5 (UPenn, Drew Weissman). Transfected cells were incubated at 37°C and 5% CO2 for 24 hours, washed with PBS once, and detached using 0.05% trypsin + EDTA (Thermo Fisher Scientific). Cells were re-suspended in DMEM complete medium and counted using Countess Automated Cell Counter (Invitrogen). 100,000 cells were placed in tubes and stained with LIVE/DEAD Aqua Dead Cell Staining Kit (Thermo Fisher Scientific) diluted 1:1000 at room temperature for 20 minutes. 50,000 live cells were placed in each well of a 96-well V-bottom plate (Corning). The plates were centrifuged at 1,200 × g for 5 minutes and the supernatants were aspirated. Cells were incubated with monoclonal antibodies, which were diluted to 5 μg/ml in duplicate in DMEM complete medium, at 37°C and 5% CO2 for 2 hours. After washing with wash buffer (PBS + 1% FBS) twice, cells were incubated with PE-conjugated mouse anti-human IgG Fc (Southern Biotech) diluted 1:200 at 4°C for 30 minutes. Following two additional wash steps, cells in tubes and plates were resuspended and fixed in PBS + 10% formalin for 10 minutes at room temperature. Fixed cells were washed once and resuspended in wash buffer for flow cytometry. Events were acquired on LSR II machine (BD biosciences) using high-throughput sampler (HTS). Data were analyzed with Flowjo software (Tree Star, Inc.), and the PE+ population was identified from the live GFP+ cell population for each sample. Non-specific binding of PE-conjugated mouse anti-human IgG Fc was corrected in the analysis. gB-genotype preference for gB-specific mAbs defined if % PE positive population of a single transfected gB genotype was greater than 5 times the % PE positive population of the lowest bound cell-associated gB genotype by that mAb.
Natural killer (NK) cell CD107a degranulation assay
Cell-surface expression of CD107a was used as a marker for NK cell degranulation [27, 28]. MRC5 cells were plated at 5×104 cells/well in a 96-well flat-bottom tissue culture plate and allowed to incubate for 24 hours at 37°C. Cells were infected with AD169r-GFP at an MOI of 1.0, then incubated a further 48 hours at 37°C. Following incubation, supernatant was removed and the infected cell monolayers were washed once with RPMI 1640 containing 10% FBS, HEPES, Pen-Strep-L-Glut, Gentamicin (R10 media) before addition of NK cells. Primary human NK cells were isolated from peripheral blood mononuclear cells (PBMC) after overnight rest in R10 media with 10ng/mL IL-15 (Miltenyi Biotech) by depletion of magnetically labeled cells (Human NK cell isolation kit, Miltenyi Biotech). 5×104 live NK cells were added to each well containing HCMV-infected MRC5 cell monolayers. mAbs were diluted in R10 and added to the cells at a final dilution of 25μg/mL in duplicate. Brefeldin A (GolgiPlug, 1 μl/ml, BD Biosciences), monensin (GolgiStop, 4μl/6mL, BD Biosciences), and CD107a-FITC (BD Biosciences, clone H4A3) were added to each well and the plates were incubated for 6 hours at 37ºC in a humidified 5% CO2 incubator. NK cells were then gently resuspended, taking care not to disturb the MRC5 cell monolayer, and the NK containing supernatant was collected and transferred to 96-well V-bottom plates. The recovered NK cells were washed with PBS, and stained with LIVE/DEAD Aqua Dead Cell Stain at a 1:1000 dilution for 20 minutes at room temperature. The cells were then washed with 1%FBS PBS and stained for 20 minutes at room temperature with the following panel of fluorescently conjugated antibodies diluted in 1%FBS PBS: CD56-PECy7 (BD Biosciences, clone NCAM16.2), CD16-PacBlue (BD Biosciences, clone 3G8), and CD69-BV785 (BioLegend, Clone FN50). The cells were then washed twice and re-suspended in 1% paraformaldehyde fixative for flow cytometric analysis. Data analysis was performed using FlowJo software (v9.9.6). Data is reported as the % of CD107a+ live NK cells (singlets, lymphocytes, aqua blue-, CD56+ and/or CD16+, CD107a+) and NK cell CD16 downregulation index (DRI) [29]. CD16 DRI represents the ratio between NK cell CD16 MFI measured for control samples containing NK cells incubated with target cells alone, and NK cell CD16 MFI measured for experimental samples containing NK cells, target cells, and test antibodies, according the following equation: CD16 DRI = (MFI CD16 control sample)/(MFI CD16 experimental sample). All final data represent specific activity, determine by subtraction of non-specific activity observed in assays performed with mock-infected cells.
Antibody dependent cellular phagocytosis (ADCP)
Approximately 3.5×106 PFU of concentrated, sucrose gradient-purified AD169r-GFP virus was transferred to a 100,000 kDa Amicon filter (Millipore), then buffer exchanged with 1× PBS, concentrated down to approximately 100 μL, and transferred to a microcentrifuge tube. Next, 10 μg of AF647 NHS ester (Invitrogen) reconstituted in DMSO was added to the concentrated, purified virus for direct fluorescent conjugation, then this reaction mixture was incubated at room temperature for 1 hour with constant agitation. The reaction was quenched with 80 μL of 1 M Tris-HCl, pH 8.0, then the fluorophore-labelled virus was diluted 25× in wash buffer (PBS + 0.1% FBS). mAbs were diluted to 0.1 mg/mL in wash buffer, then 10 μL of each diluted mAb was combined with 10 μL of diluted, fluorophore-conjugated virus in a round-bottom, 96-well plate and allowed to incubate at 37°C for 2 hours. Following this incubation step, 50,000 THP-1 cells were added to each well, suspended in 200 μL primary growth media. Plates were centrifuged at 1200× g and 4°C for 1 hour in a spinoculation step, then incubated at 37°C for an additional hour. Cells were re-suspended and transferred to a 96-well V-bottom plate, then washed twice prior to fixation in 100 μL DPBS + 1% formalin. Events were acquired on LSR II machine (BD biosciences) using the HTS. The % AF647+ cells was calculated from the full THP-1 cell population and reported for each sample. A cutoff for a sample mediating ADCP was defined as > 99% AF647+ signal from THP1 cells incubated with fluorophore conjugated virus and either an HCMV seronegative control or non-HCMV-specific mAb.
Results
Mapping the domain-specificity of non-neutralizing gB-specific mAbs from HCMV seropositive individuals
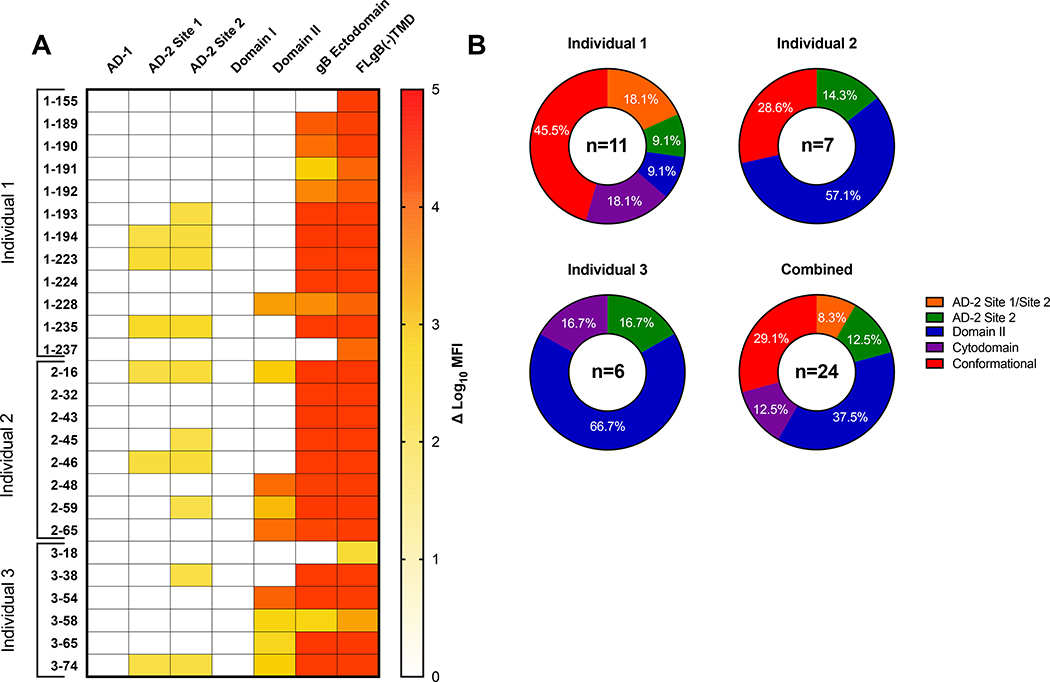
We first mapped the binding specificity of the panel of gB-specific mAbs for defined neutralizing and non-neutralizing antigenic domains of gB by binding antibody multiplex assay (BAMA). The most frequent binding specificity was against Domain II (37.5%), as well as AD2 site 2 (12.5%) and cytodomain (12.5%) (Figure 1A). While 4 mAbs demonstrated detectable binding to both linear AD2 site 1 and site 2, these mAbs may be AD2 site 2 specific, given that previously described AD2 site 1 mAbs have neutralizing activity [30, 31]. Interestingly, there were three mAbs, 1–155, 1–237, and 3–18, which bound FLgB(−)TMD, but did not bind a gB ectodomain that excludes the membrane proximal region (MPER) and the cytodomain containing AD3, suggesting specificity for the cytosolic domain which includes one of these regions. There are two groups of clonal populations of mAbs found in Individual 1, including mAbs 1–189, 1–190, 1–191, 1–192 which bind soluble FLgB(−)TMD and gB ectodomain, but not to a defined epitope (termed conformational binding), and mAbs 1–223 and 1–224 to AD2 and conformationally gB respectively. Antigenic domain-specificity of gB-specific mAbs was heterogenous in Individual 1, but was dominated by Domain II in individuals 2 (57.1%) and 3 (66.7%) (Figure 1B). The binding profile for the combined panel of 24 gB-specific mAbs notably excludes any AD1 or AD5 (Domain I) binding mAbs, despite the polyclonal CMV-IgG preparation Cytogam measurably binding to all antigens (data not shown).
Figure 1. gB domain-specific mAb binding determined by BAMA.
(A) Heat map of gB domain-specific binding strength for 0gB-specific mAbs represented as log10 mean fluorescent intensity (MFI). (B) Pie charts representing the percentage of total gB-specific mAbs binding to each domain specificity assessed by BAMA per each naturally HCMV infected individual and all individuals combined. AD-2 Site 1/Site 2: Binding to both the AD-2 Site 1 and Site 2 peptide, AD-2 site 2: binding to the site 2 peptide only, Domain II: binding to Domain II only, Cytodomain: binding to full length gB, but not gB ectodomain, Conformational: binding to full length gB and gB ectodomain, but no other domain.
mAb binding kinetics to FLgB(−)TMD and trimeric post-fusion gB ectodomain
Next, we sought to describe the binding kinetics of each gB-specific mAb to the FLgB(−)TMD protein vs a trimeric post-fusion gB ectodomain construct. By understanding discrepancies in binding between FLgB(−)TMD and the post-fusion gB ectodomain, we aimed to highlight novel HCMV cytodomain-specific mAbs and their binding kinetics. Multiple mAbs (1–155, 1–189, 1–191, 1–192, 1–237) exhibited poor binding strength to the post-fusion gB ectodomain, but retained robust binding to FLgB(−)TMD. MAbs 3–18 and 3–58 exhibited poor gB binding to both FLgB(−)TMD and post fusion gB ectodomain (Table 1).
Table 1.
gB-specific mAb immunogenetics, binding strength, and avidity to full length gB and gB ectodomain
| Monoclonal Antibody | Heavy Chain a | Light Chain a | CDRH3 Length a | Neutralization IC50 (μg/mL) a | Epitope Specificity | Full length gB binding EC50 (μg/mL)b | gB ectodomain binding EC50 (μg/mL) b | gB ectodomain apparent avdity KD (nM) c |
|---|---|---|---|---|---|---|---|---|
| 1-155 | IGHV1-69*01 | IGLV2-11*01 | 14 | 0 | Cytodomain | 0.002 | 6.903 | 6.2 |
| 1-189 | IGHV1-69*01 | IGKV4-1*01 | 11 | 0 | Conformational | 0.007 | 2.562 | 702 |
| 1-190 | IGHV1-69*01 | IGKV4-1*01 | 11 | 0 | Conformational | 0.019 | 3.716 | 1.88 |
| 1-191 | IGHV1-69*01 | IGKV4-1*01 | 11 | 0 | Conformational | 0.009 | 6.163 | 10.8 |
| 1-192 | IGHV1-69*06 | IGKV4-1*01 | 11 | 0 | Conformational | 0.010 | > 100 | 116 |
| 1-193 | IGHV1-18*01 | IGKV2-30*01 | 22 | 1 | AD2 Site 2 | 0.003 | 0.004 | 0.0003 |
| 1-223 | IGHV1-69*03 | IGLV2-23*01 | 22 | 8.3 | AD2 Site 1/Site 2 | 0.009 | 0.013 | 0.301 |
| 1-224 | IGHV1-69*03 | IGKV3-20*01 | 22 | 6.4 | Conformational | 0.006 | 0.011 | 0.116 |
| 1-228 | IGHV3-13*01 | IGKV1-33*01 | 10 | 0 | Domain II | 0.001 | 8.014 | N.D. |
| 1-235 | IGHV4-31*03 | IGKV4-1*01 | 11 | 0 | AD2 Site 1/Site 2 | 0.001 | 0.002 | 0.019 |
| 1-237 | IGHV1-69*01 | IGKV4-1*01 | 12 | 0 | Cytodomain | 0.003 | 2.656 | 5.27 |
| 2-16 | IGHV1-69*01 | IGKV3-15*01 | 15 | 11 | Domain II | 0.002 | 0.004 | 0.0004 |
| 2-32 | IGHV4-59*01 | IGKV4-1*01 | 12 | 0 | Conformational | 0.003 | 0.006 | N.D. |
| 2-43 | IGHV6-1*01 | IGKV4-1*01 | 14 | 15 | Conformational | 0.003 | 0.004 | 0.0034 |
| 2-45 | IGHV4-34*02 | IGKV3-15*01 | 24 | 9.8 | AD2 Site 2 | 0.003 | 0.006 | 0.0004 |
| 2-48 | IGHV1-2*02 | IGKV3-20*01 | 14 | 14.6 | Domain II | 0.003 | 0.006 | 0.113 |
| 2-59 | IGHV4-34*02 | IGLV1-47*01 | 18 | 2.3 | Domain II | 0.006 | 0.010 | 0.0016 |
| 2-65 | IGHV1-2*02 | IGKV3-20*01 | 14 | 6.4 | Domain II | 0.007 | 0.018 | 0.081 |
| 3-18 | IGHV3-21*01 | IGKV1D-8*01 | 26 | 0 | Cytodomain | 16.370 | > 100 | 22.5 |
| 3-38 | IGHV1-18*01 | IGKV4-1*01 | 14 | 0 | AD2 Site 2 | 0.006 | 0.008 | 0.0003 |
| 3-54 | IGHV1-46*01 | IGKV1-5*03 | 22 | 1 | Domain II | 0.006 | 0.011 | 0.474 |
| 3-58 | IGHV3-30*03 | IGLV3-10*01 | 34 | 0 | Domain II | 5.733 | 20.560 | 1.82 |
| 3-65 | IGHV3-33*01 | IGKV4-1*01 | 16 | 0 | Domain II | 0.003 | 0.003 | 0.036 |
| 3-74 | IGHV3-30-3*01 | IGLV3-25*03 | 28 | 0 | Domain II | 0.601 | 0.723 | 1.52 |
Heavy chain and light chain genes, as well as CDRH3 length listed as described in (Xia et al 2017).
Neutralization previously defined as an IC50 < 1 (μg/mL) by (Xia et al 2017), ”> xx” no neutralization at the highest dilution at which the mAb was tested
binding to full length gB and post fusion gB ectodomain EC50 (μg/mL) measured by ELISA.
binding kinetics to post fusion gB ectodomain measured by SPR
Three isolated gB-specific mAbs with binding to both the linear AD2 site 1 and site 2 regions, regardless of individual donor, demonstrated both high binding strength and avidity for FLgB(−)TMD (EC50 0.001 – 0.009 μg/mL) and gB ectodomain (0.002 – 0.013 μg/mL) (Table 1). gB mAbs with AD2 site 2 only specificity also had robust binding to gB-TMD (EC50 0.003 – 0.006 μg/mL) as well as gB ectodomain (0.004 – 0.008 μg/mL) (Table 1). Of the mAbs which bound Domain II, all except three, 1–228, 3–58 and 3–74, showed strong binding to both FLgB(−)TMD and gB ectodomain (EC50 < 0.020 μg/mL) (Table 1). Specifically, mAb 3–74, which binds Domain II predominantly, demonstrated poor binding and avidity to both FLgB(−)TMD and gB ectodomain by ELISA and SPR. Two Domain II-specific mAbs bound with an interesting pattern to gB ectodomain. Domain II-specific mAb 1–228 had strong binding to gB-TMD (EC50 0.001 μg/mL) but very poor binding to gB ectodomain by ELISA (EC50 8.014 μg/mL) and no detectable (N.D.) avidity by SPR (Table 1). Further, Domain II-specific mAb 3–58 had poor binding strength to both FLgB(−)TMD (EC50 5.73 μg/mL) and gB ectodomain (EC50 20.56 μg/mL) (Table 1).
MAbs 1–155 and 1–237 which bound to the FLgB(−)TMD and not the ectodomain in the gB domain mapping, and therefore potentially bind within the cytodomain, followed predictable patterns of binding with strong binding to FLgB(−)TMD (EC50 < 0.004 μg/mL) and negligible binding strength and avidity to gB ectodomain (Figure 1A, Table 1). MAb 3–18, also determined to be cytodomain-specific by BAMA, demonstrated poor binding measured by both ELISA (EC50 > 100 μg/mL) and SPR (KD 22.5 nM) to the ectodomain as well. Finally, those mAbs which didn’t have a definable epitope specificity by gB domain mapping, and termed “conformational”, had highly variable binding strength to gB ectodomain, but fairly strong binding to FLgB(−)TMD. Interestingly, mAb 2–32, which bound both FLgB(−)TMD (EC50 0.003 μg/mL) and gB ectodomain (EC50 0.006 μg/mL) well via ELISA, as well as has binding detected to both the gB-TMD and gB ectodomain antigens via BAMA (Figure 1A), demonstrated undetectable binding to gB ectodomain by SPR (N.D., not detectable).
Cell associated gB DNA transfected-cell binding
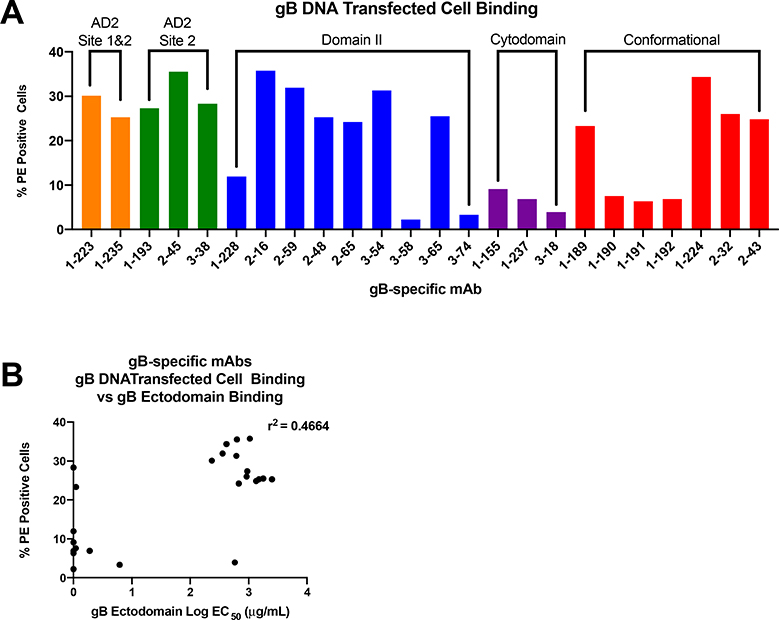
We have recently discovered that the magnitude of binding to gB DNA transfected cells is a correlate of protection against primary HCMV acquisition in postpartum and adolescent women vaccinated with gB/MF59 in phase II clinical trials [21]. Next, we investigated whether binding to cell associated gB is dependent upon gB-specific mAb antigenic site specificity. gB-specific mAbs with AD-2 specificity demonstrated high magnitude binding to cell associated gB more consistently than mAbs from any other specificity represented in our panel (Figure 2A). Cytodomain-specific mAbs bind poorly to cell associated gB, consistent with epitope location in the transmembrane and cytosolic compartment [14]. Notably, binding to cell associated gB was not correlated to strength of binding to soluble gB measured by ELISA (Figure 2B).
Figure 2. gB DNA transfected cell binding.
(A) gB-specific mAb binding to cell-associated gB on the surface of gB DNA transfected cells. (B) gB-specific mAbs generally bind with high magnitude to cell-associated gB, but this binding strength is not highly correlated to strength of binding (EC50) to soluble gB ectodomain measured by ELISA.
Cell associated gB genotype-specific mAb binding
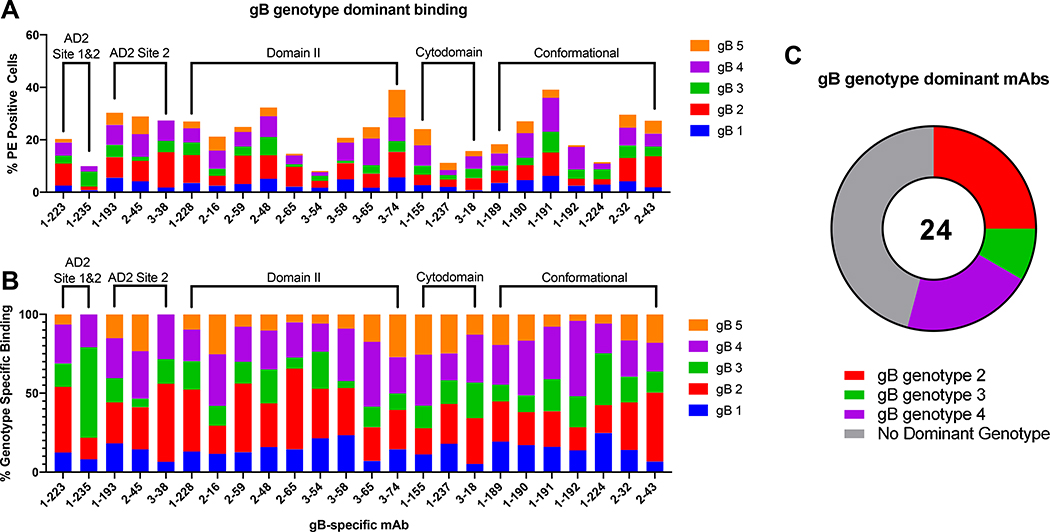
We previously reported that gB/MF59 vaccinees may have had reduced acquisition of HCMV strains with gB1 genotype, the genotype matched to the vaccine construct, suggesting the possibility of strain-specific protection [32]. A s such, we next explored how non-neutralizing gB-specific mAbs differentially recognize gB genotypes 1–5, as expressed on the surface of a cell. Comparison of total, and genotype specific, gB transfected cell bound populations of each gB-specific mAb (Figure 3A) highlights variability for cell-associated gB binding across epitope specificities. While a majority of gB-specific mAbs bound all 5 genotypes, there was unequal binding magnitude across genotypes for the same mAb. To compare genotype preference amongst gB-specific mAbs (Figure 3B), we next assessed the percent representation of each genotype in the sum total gB genotype binding for each mAb. Here, there emerged a trend toward preference for gB-specific mAbs isolated from these naturally-infected donors for gB genotypes 2 and 4, representing the cumulative genotype preference for 11 of 14 gB-specific mAbs with gB genotype specific binding (Figure 3C). To understand trends in mAb genotype binding preference, mAbs which bound a single gB genotype with a % PE positive population greater than 5X the % PE positive population of the lowest bound gB genotype were considered to have preferred binding to that genotype. These findings are consistent across epitope specificities. Interestingly, mAbs which were unable to bind gB ectodomain measured by ELISA or BAMA, but bound FLgB(−)TMD and classified as cytodomain-specific, demonstrated equivalent cell associated gB binding across all tested gB genotypes (Figure 3A). Because the cytodomain, including AD3, is thought to be inaccessible for mAb binding in the known post-fusion structure, these data implicate differences in epitope presentation of soluble gB and cell-associated gB.
Figure 3. Cell associated gB genotype-specific mAb binding.
(A) Percentage of gB genotype-specific transfected cells bound by gB-specific mAbs, grouped by domain specificity. (B) Percentage of gB genotype-specific transfected cells bound by the total population of gB transfected cells, demonstrating genotype preference for each gB-specific mAbs, listed by domain specificity. (C) gB-genotype preference for gB-specific mAbs defined if % PE positive population of a single transfected gB genotype was greater than 5 times the % PE positive population of the lowest bound cell-associated gB genotype by that mAb.
Antibody dependent cellular cytotoxicity (ADCC)
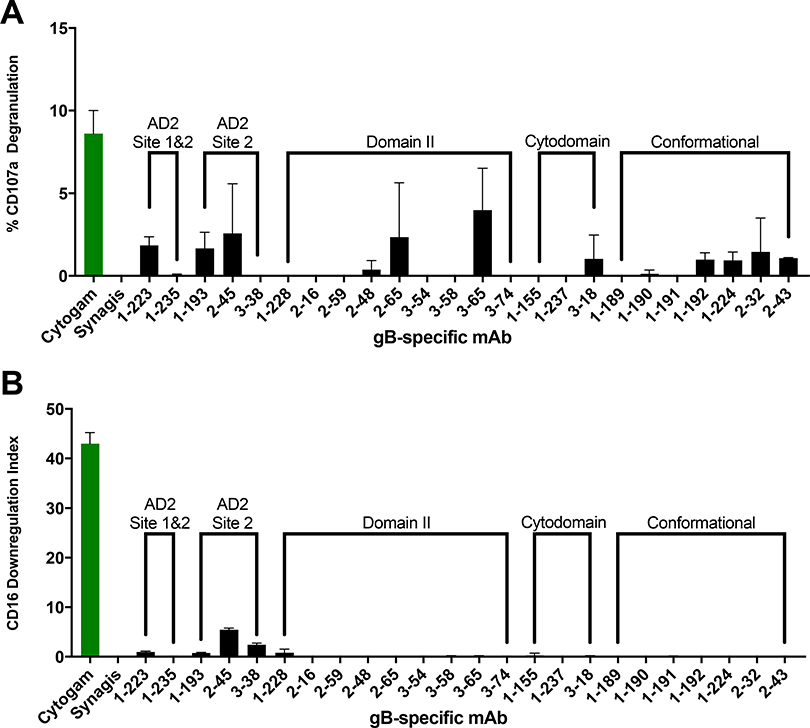
Antibody-mediated NK cell cytotoxicity has been demonstrated as a crucial mechanism for controlling HCMV infection [33, 34]. To distinguish the domain specificity of gB-specific mAbs which activate NK cells, we next screened our panel of mAbs for ability to mediate two well defined NK phenotypes of cytotoxic killing, cell-surface expression of CD107a [27, 28], indicative of degranulation, (Figure 4A) and CD16 downregulation, indicative of NK cell activation [35] (Figure 4B) in the presence of HCMV AD169r infected cell targets. While a number of mAbs across multiple domains demonstrated measurable CD107a degranulation, all responses were modest. Next, an additional marker of NK cell activation, CD16 downregulation calculated into a downregulation index (DRI) [29],was measured for each gB-specific mAb. Low levels of CD16 DRI were observed for AD2 site 2 specific mAbs, however activity was markedly lower than that observed for HCMV hyperimmunoglobulin (Cytogam). Overall, none of the gB-specific mAbs strongly activated NK cells in these assays.
Figure 4. gB-specific mAbs mediate negligible NK activation when compared to polyclonal CMV-specific antibody preparation (Cytogam).
gB-specific mAbs were screened for mediation of NK cell (A) CD107a degranulation and (B) CD16 downregulation (DRI) as measures of antibody-mediated NK cell activation. mAbs were compared against polyclonal CMV-specific antibody preparation (Cytogam) and monoclonal RSV-specific antibody preparation (Synagis). Data are presented as mAb specific response against AD169r-GFP infected ARPE19 cells minus mock infected negative control cells.
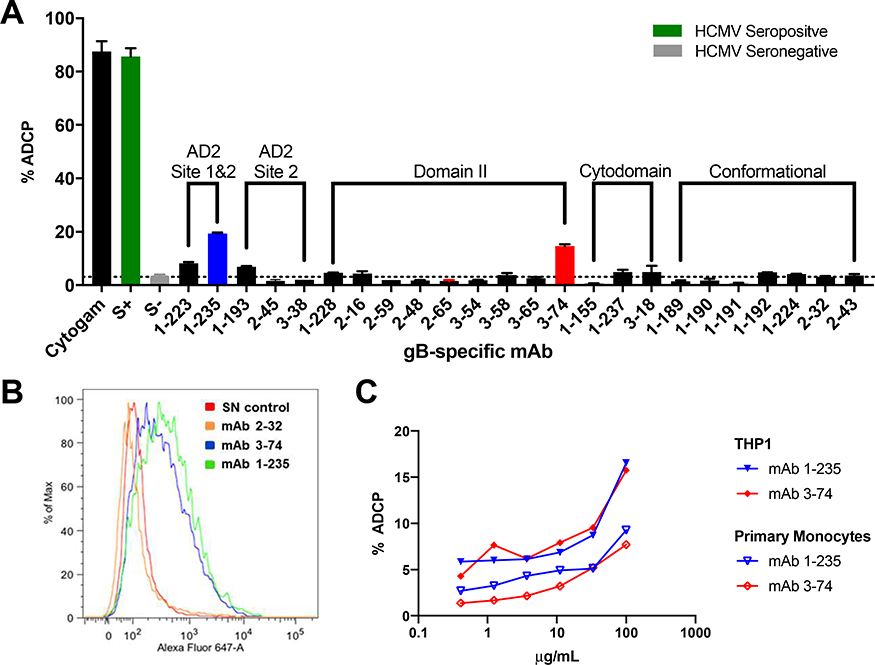
gB AD2 site 1/site 2 and Domain II-specific mAbs 1–235 and 3–74 mediate ADCP
In a study of functional antibody responses to gB/MF59 vaccination, vaccinees were found to demonstrate limited neutralization responses, but robust ADCP [15]. To identify the domain specificity of gB-specific mAbs which can mediate ADCP, we screened our panel of gB-specific mAbs for ability to mediate whole virion phagocytosis (Figure 5A). Two gB-specific mAbs, 1–235 and 3–74, mediated phagocytosis of AD169r-GFP virions, which exceeded the baseline ADCP level of non-specific mAbs and seronegative control plasma by 19.4% and 14.7% respectively. However, the magnitude of ADCP was modest when compared to HCMV hyperimmunoglobulin (cytogam). To better assess their ADCP potency with both THP-1 cells and primary monocytes isolated from healthy donors (Figure 5B), mAbs 1–235 and 3–74 were titrated from a concentration of 100 μg/mL. ADCP increased in a dose-dependent manner for 1–235 and 3–74 in both THP1 cells and primary monocytes. Importantly, the gB epitope specificity of ADCP mediating mAb 1–235 was AD2 Site 1 and Site 2, while 3–74 specificity was Domain II (Figure 1A).
Figure 5. gB AD2 and Domain-II specific mAbs 1–235 and 3–74 mediate antibody dependent cellular phagocytosis (ADCP).
(A) gB-specific mAbs were screened for ADCP activity. (B) A representative histogram for ADCP mediating mAbs as compared to a HCMV seronegative control. The magnitude of ADCP was titrated over a dilution series for the two ADCP mediating mAbs using (C) THP1 cells (solid symbols) and primary monocytes (open symbols).
Discussion
The dominant non-neutralizing antibody responses elicited by the partially-protective gB/MF59 vaccine has suggested a shift in the paradigm of immunogenicity endpoints for consideration when designing the next generation of HCMV vaccines [15]. While protective immunity through virus neutralization to prevent primary infection is considered the “gold standard” of HCMV vaccine development, next generation vaccine regimens will need to consider antibody effector functions beyond virus neutralization. Indeed, strategies such as cell-to-cell spread enable immunologically-covert spread of infection even in the presence of neutralizing antibodies [36]. In fact, prophylactic passive immunization with both neutralizing and non-neutralizing mAbs were considered equally protective in a murine cytomegalovirus challenge model [37]. Accordingly, non-neutralizing antibodies which mediate effector functions like ADCP, like those described in this study, may have potential importance as immunological endpoints of a protective HCMV vaccine.
A majority (> 90%) of gB-specific antibodies from B cell clones have no neutralizing activity [38]. Indeed, the partially-protective gB/MF59 vaccine elicited limited heterologous neutralization [15]. While the specificity and characteristics of neutralizing gB-specific mAbs have been well described, the types of naturally-elicited gB-specific mAbs that mediate non-neutralizing effector functions which may be critical for protection against HCMV acquisition remain to be fully characterized.
In this study, 24 non-neutralizing gB-specific mAbs isolated from naturally HCMV-infected individuals were assessed for epitope binding specificity and affinity, gB genotype preference, and Fc-mediated effector functions. These non-neutralizing mAbs bound predominantly to Domain II, or to one or both of the binding sites of AD2. Domain II is largely the target of neutralizing antibodies, with one study identifying Domain II-specific binding by neutralizing antibodies in greater than 90% of HCMV seropositive subjects tested. Interestingly, three mAbs from the panel demonstrated robust binding to the FLgB(−)TMD protein but failed to bind a gB ectodomain, suggesting specificity within the AD3 or MPER region, a portion of the gB construct located in the cytodomain of membrane-associated gB [14]. This potentially hidden region on an intact virion or infected cell could certainly be exposed through protein shedding from cell lysis or disruption of the HCMV virion [39], similarly to other viral structural antigens such as pp150 and pp28. However, it also raises questions regarding the structure of this membrane-associated region of the protein. Interestingly, none of the non-neutralizing gB-specific mAbs in this panel had appreciable binding to AD1 or Domain I. AD1 is traditionally an immunodominant domain and is targeted by both neutralizing and non-neutralizing mAbs [38, 40]. Furthermore, The lack of reactivity with AD1 may be the result of differences between the AD1 construct used in our BAMA studies versus the native protein epitope. As such, the lack of Domain I and AD1 binding from this representative panel is surprising and could be the result of sampling bias from only three individuals [25], by selection bias of excluding mAbs which bind to traditionally neutralizing domains, or non-optimal conformation of the protein used for B cell sorting to isolate these mAbs. Indeed, one fundamental challenge of this study, with only 3 individual donors, is limited in its ability to claim generalizability of specific ratios of total antibody binding to certain domains.
Antibodies mediating neutralization after gB/MF59 vaccination and natural infection can be strain specific [15, 41] and may not provide equivalent degrees of protection against all strains. It has been reported that breakthrough HCMV infections in the vaccine subjects receiving gB/MF59, which is a gB genotype 1 based vaccine, were more likely to be infected with HCMV strains expressing gB genotypes 3 or 5 than strains 1, 2, or 4 [32]; suggesting gB/MF59 induced antibodies were limited due to its strain-specific neutralization or antibody effector functions. Our study offers the first evidence that critical gB-specific non-neutralizing antibodies have different strain specific gB recognition when displayed on the surface of a cell. This panel of non-neutralizing gB-specific mAbs generally bound with greater preference to gB genotypes 2 and 4. These findings are in concordance with phylogenetic clustering based on next generation sequencing of the full length gB open reading frame of the 5 clinically significant HCMV gB genotypes 1/2/4 and 3/5 into two supergroups [32]. The lack of knowledge regarding which HCMV gB genotype or genotypes infected the individuals from which the mAbs were isolated limits the assertions we can make about how gB genotype specific antibodies may be raised by natural infection. Increased binding to gB genotypes 2 and 4 may be an artifact of the small number of individuals (3) included in this study. Clinically, prevalence of HCMV gB variants may be influenced by geography, immune status, and prior infection with HCMV of other gB variants [42–44]. Future efforts might address the question of gB variant specific recognition and antibody function by utilizing mAbs isolated from individuals that have been tested for endogenous viral strains or have known exposure to a specific gB genotype variant.
Interestingly, mAbs which were classified as AD2 site 2-specific, an epitope considered highly variable between HCMV strains, were able to bind cell associated gB regardless of genotype. Preserved gB genotype 2/4 preferential binding by AD2 site 2 specific mAbs 2–45 and 3–38 may be explained by the difference in antigens used for respective binding assays. Binding to a linear peptide fragment for classifying epitope specificity may capture a broad range of AD2 site 2-specific mAbs versus cell associated gB which may be more restrictive for conformation specific binding [19].
A notable finding from studying gB-specific mAbs that are cytodomain-specific, is their breadth of membrane associated gB binding. These FLgB(−)TMD-binding mAbs, 1–155, 1–237, and 3–18, demonstrate negligible gB ectodomain and postfusion gB trimer binding by both ELISA and SPR, yet yield comparable signals for transfected cell binding to other gB ectodomain binding mAbs. With AD3 thought to be buried in the cytodomain [14], these findings raise the question: how do presentation of gB epitopes differ as a soluble protein versus a membrane-associated protein? As a viral fusogen, gB is thought to undergo transformation from a prefusion conformation to a postfusion conformation to facilitate entry into a host cell [23, 45]. While this study does not define a specific prefusion structure, it does highlight how cell associated gB may expose different epitopes than soluble postfusion gB, potentially accounting for improved membrane associated gB binding of cytodomain-specific mAbs. Notably, it was recently reported that the ability of gB/MF59 vaccine sera to bind to gB transfected cells predicts risk of HCMV acquisition [21]. Taken together, these findings warrant further investigation of gB conformation-specific antibody binding, to parse out the epitope binding specificity and effector functions of gB vaccine-elicited Abs.
This study demonstrates the first effort to identify gB domain specificity of antibodies that mediate Fc receptor functions like ADCP and NK cell activation elicited by natural HCMV infection. While little is known about gB-specific non-neutralizing functions in naturally-infected individuals, investigations of gB/MF59 vaccinees has shown this class of antibody may indeed be a desirable target of vaccines that aim to protect against primary HCMV acquisition [15]. It is now evident that non-neutralizing mAbs are not limited to traditional “non-neutralizing” epitopes [14, 19]. Even more, these mAbs, while preferentially binding to certain gB genotypes, retain the ability to bind across a spectrum of gB genotype variants. Ultimately this work contributes to the field of HCMV vaccinology by emphasizing the impact of epitope binding specificity, binding strength, and genotype breadth amongst functional non-neutralizing gB-specific mAbs. By improving our knowledge of gB immunogenicity elicited by natural infection, and the specificity of antibody responses that may mediate key functions other than traditional neutralization, this work informs rational design of new HCMV vaccines that aim to reduce the devastating burden of HCMV disease in both congenital infection and transplant settings.
Supplementary Material
Acknowledgements
The gB-specific monoclonal antibodies were kindly provided by Dai Wang at Merck through a collaboration with Zhiqiang An’s laboratory at the University of Texas Health Science Center at Houston. This work was supported by grants from Merck Research Labs and the Welch Foundation (AU-0042–20030616 to Z.A), NIH/ National Institute of Allergy and Infectious Disease R21 (R21AI136556 to S.R.P). Dr. Permar consults for Merck, Pfizer, Sanofi, and Moderna vaccine programs and has sponsored programs from Merck and Moderna around CMV vaccine immunity. A patent application covering the mAbs described in the article has been submitted by Merck &Co., Inc. and University of Texas. D.W. is a current employee and stockholders of Merck & Co., Inc.
Footnotes
Declaration of interests
☐ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☒The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
S.P. consults for Merck, Pfizer, Sanofi, and Moderna vaccine programs and has sponsored programs from Merck and Moderna around CMV vaccine immunity.
A patent application covering the mabs described in the article has been submitted by Merck &Co., Inc. and University of Texas. D.W. is a current employee and stockholders of Merck & Co., Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kenneson A and Cannon MJ, Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol, 2007. 17(4): p. 253–76. [DOI] [PubMed] [Google Scholar]
- 2.Boppana SB, Ross SA, and Fowler KB, Congenital cytomegalovirus infection: clinical outcome. Clin Infect Dis, 2013. 57 Suppl 4: p. S178–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boppana SR, Lisa; Folwer Karen; Mach Michael; Britt William, Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. The New England Jouranal of Medicine, 2001. 344(18): p. 1366–1371. [DOI] [PubMed] [Google Scholar]
- 4.Manicklal S, et al. , The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev, 2013. 26(1): p. 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana SB and Britt WJ, Antiviral Antibody Responses and Intrauterine Transmission after Primary Maternal Cytomegalovirus Infection. The Journal of Infectious Diseases, 1995. 171: p. 1115–1121. [DOI] [PubMed] [Google Scholar]
- 6.Kotton C, Management of Cytomegalovirus infection in solid organ transplantation. Nature Reviews Nephrology, 2010. 12: p. 711–721. [DOI] [PubMed] [Google Scholar]
- 7.Streblow DN, Orloff SL, and Nelson JA, Acceleration of allograft failure by cytomegalovirus. Curr Opin Immunol, 2007. 19(5): p. 577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adler SP, et al. , Immunity Induced By Primary Human Cytomegalovirus Infection Protects Against Secondary Infection Among Women Of Childbearing Age. The Journal of Infectious Diseases, 1995. 171(1): p. 26–32. [DOI] [PubMed] [Google Scholar]
- 9.Smiley ML, et al. , The role of pretransplant immunity in protection from cytomegalovirus disease following renal transplantation. Transplantation, 1985. 40(2): p. 157–161. [DOI] [PubMed] [Google Scholar]
- 10.Bialas KM and Permar SR, The March towards a Vaccine for Congenital CMV: Rationale and Models. PLoS Pathog, 2016. 12(2): p. e1005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein DI, et al. , Safety and efficacy of a cytomegalovirus glycoprotein B (gB) vaccine in adolescent girls: A randomized clinical trial. Vaccine, 2016. 34(3): p. 313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pass RF, et al. , Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med, 2009. 360(12): p. 1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths PD, et al. , Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. The Lancet, 2011. 377(9773): p. 1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke HG and Heldwein EE, Crystal Structure of the Human Cytomegalovirus Glycoprotein B. PLoS Pathog, 2015. 11(10): p. e1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson CS, et al. , HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc Natl Acad Sci U S A, 2018. 115(24): p. 6267–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baraniak I, et al. , Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A, 2018. 115(24): p. 6273–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuijpers T, et al. , Human NK cells can control CMV infection in the absence of T cells. Blood, 2008. 112(3): p. 914–915. [DOI] [PubMed] [Google Scholar]
- 18.Mitrovic M, et al. , Innate immunity regulates adaptive immune response: lessons learned from studying the interplay between NK and CD8+ T cells during MCMV infection. Med Microbiol Immunol, 2012. 201(4): p. 487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer H, et al. , Glycoprotein gp116 of human cytomegalovirus contains epitopes for strain-common and strain-specific antibodies. Journal of General Virology, 1992. 73(9): p. 2375–2383. [DOI] [PubMed] [Google Scholar]
- 20.Tarrago D, Quereda C, and Tenorio A, Different Cytomegalovirus Glycoprotein B Genotype Distribution in Serum and Cerebrospinal Fluid Specimens Determined by a Novel Multiplex Nested PCR. Journal of Clinical Microbiology, 2003. 41(7): p. 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenks JA, et al. , Antibody binding to native cytomegalovirus glycoprotein B predicts vaccine efficacy. medRxiv, 2020: p. 2020.02.27.20028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rey FA, et al. , Different functional states of fusion protein gB revealed on human cytomegalovirus by cryo electron tomography with Volta phase plate. PLOS Pathogens, 2018. 14(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heldwein EE, et al. , Crystal Structure of Glycoprotein B from Herpes Simplex Virus 1. Science, 2006. 313(5784): p. 217–220. [DOI] [PubMed] [Google Scholar]
- 24.Baraniak I, et al. , Epitope-Specific Humoral Responses to Human Cytomegalovirus Glycoprotein-B Vaccine With MF59: Anti-AD2 Levels Correlate With Protection From Viremia. J Infect Dis, 2018. 217(12): p. 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia L, et al. , Active evolution of memory B-cells specific to viral gH/gL/pUL128/130/131 pentameric complex in healthy subjects with silent human cytomegalovirus infection. Oncotarget, 2017. 8(43): p. 73654–73669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bialas KM, et al. , Maternal Antibody Responses and N onprimary Congenital Cytomegalovirus Infection of HIV-1-Exposed Infants. J Infect Dis, 2016. 214(12): p. 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alter G, Malenfant JM, and Altfeld M, CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods, 2004. 294(1–2): p. 15–22. [DOI] [PubMed] [Google Scholar]
- 28.Betts MR, et al. , Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of Immunological Methods, 2003. 281(1–2): p. 65–78. [DOI] [PubMed] [Google Scholar]
- 29.Legris T, et al. , Antibody-Dependent NK Cell Activation Is Associated with Late Kidney Allograft Dysfunction and the Complement-Independent Alloreactive Potential of Donor-Specific Antibodies. Front Immunol, 2016. 7: p. 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McVoy MM, Tenorio E, and Kauvar LM, A Native Human Monoclonal Antibody Targeting HCMV gB (AD-2 Site I). Int J Mol Sci, 2018. 19(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauvar LM, et al. , A high-affinity native human antibody neutralizes human cytomegalovirus infection of diverse cell types. Antimicrob Agents Chemother, 2015. 59(3): p. 1558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson CS, et al. , Intrahost Dynamics of Human Cytomegalovirus Variants Acquired by Seronegative Glycoprotein B Vaccinees. Journal of Virology, 2019. 93(5): p. e01695–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z, et al. , Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J Virol, 2013. 87(13): p. 7717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa-Garcia M, et al. , Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J Immunol, 2015. 194(6): p. 2715–24. [DOI] [PubMed] [Google Scholar]
- 35.Bowles JA and Weiner GJ, CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J Immunol Methods, 2005. 304(1–2): p. 88–99. [DOI] [PubMed] [Google Scholar]
- 36.Jacob CL, et al. , Neutralizing antibodies are unable to inhibit direct viral cell-to-cell spread of human cytomegalovirus. Virology, 2013. 444(1–2): p. 140–7. [DOI] [PubMed] [Google Scholar]
- 37.Bootz A, et al. , Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog, 2017. 13(8): p. e1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potzsch S, et al. , B cell repertoire analysis identifies new antigenic domains on glycoprotein B of human cytomegalovirus which are target of neutralizing antibodies. PLoS Pathog, 2011. 7(8): p. e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimpel GR, Immune Defenses, in Medical Microbiology B. S, Editor. 1996, University of Texas Medical Branch at Galveston: Galveston (TX). [PubMed] [Google Scholar]
- 40.Speckner A, et al. , Antigenic domain 1 of human cytomegalovirus glycoprotein B induces a multitude of different antibodies which, when combined, results in incomplete virus neutralization. Journal of General Virology, 1999. 80(8): p. 2183–2191. [DOI] [PubMed] [Google Scholar]
- 41.Klein M, et al. , Strain-Specific Neutralization of Human Cytomegalovirus Isolates by Human Sera. Journal of Virology, 1999. 73(2): p. 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coaquette A, et al. , Mixed Cytomegalovirus Glycoprotein B Genotypes in Immunocompromised Patients. Clinical Infectious Diseases, 2004. 39(2): p. 155–161. [DOI] [PubMed] [Google Scholar]
- 43.Ishibashi K, et al. , Strain-specific seroepidemiology and reinfection of cytomegalovirus. Microbes Infect, 2008. 10(12–13): p. 1363–9. [DOI] [PubMed] [Google Scholar]
- 44.Gorzer I, et al. , Human cytomegalovirus (HCMV) genotype populations in immunocompetent individuals during primary HCMV infection. J Clin Virol, 2010. 48(2): p. 100–3. [DOI] [PubMed] [Google Scholar]
- 45.Si Z, et al. , Different functional states of fusion protein gB revealed on human cytomegalovirus by cryo electron tomography with Volta phase plate. PLOS Pathogens, 2018. 14(12): p. e1007452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.