Abstract
Enteroaggregative haemorrhagic Escherichia coli (E. Coli, EAHEC) has been identified as the agent responsible for one of the largest outbreaks of gastroenteritis and Haemolytic-uremic syndrome (HUS) that is transmitted through food in Germany in 2011. The hypervirulent pathotype has a unique combination of two pathogens namely enterohemorrhagic E.coli strain (EHEC) which produces shiga/verotoxin and enteroaggregative E.coli toxins (EAEC) which produces toxins similar to ST and hemolysin. The toxin produced by the EAHEC strain is a hybrid pathotype that combines the virulence potential of the EAEC and EHEC strains that will damage the microcirculation, cause vasculitis and other toxic effects. The purpose of this study was to determine the percentage of samples infected with enteroaggregative hemorrhagic E. coli bacteria (EAHEC) in pediatric diarrhea patients at DR. Soetomo Hospital, Surabaya, Indonesia, 2015. This study used PCR (Polymerase Chain Reaction) method to detect enteroaggregative E. coli strains (CVD432 and aaic genes) and enterohemorrhagic E.coli (eae gene).The results showed that 33 out of 40 (82,5%) stool samples examined were detected enteroaggregative E. coli (EAEC), 4 out of 40 (10%) enterohemorrhagic E. coli (EHEC) and 3 out of 40 (7,5%) enteroaggregative haemorrhagic E. coli bacteria (EAHEC), which caused diarrhea in pediatric diarrhea patients at Dr. Soetomo General Hospital. The unique combination of genomic features of the Surabaya outbreak strain, containing characteristics from pathotypes EAEC and EHEC, suggested that it represents a new pathotype enteroaggregative haemorrhagic E. coli (EAHEC). It is expected that development of specific primer design and sequencing are needed to continue in this research.
Key words: E. coli, EAHEC, pediatric diarrhea patients, Dr. Soetomo General Hospital
Introduction
Diarrhea is a common cause of death in developing countries and the second leading cause of death in infants throughout the world. Same infectious diarrheal diseases are caused by viruses, bacteria, and parasites. Bacteria causes about 15% of cases in children, with the most common type being E. coli. Most strains of E. coli are harmless, but some pathogenic serotypes can cause serious food poisoning and diarrhea in humans.1
E. coli is classified based on its virulence characteristics, each group causes disease through different mechanisms. Virotypes or pathotype classification are based on the presence of certain virulence factors and their interactions with mammalian cells or tissues such as adhesion, and or invasion of mammalian cells, and toxin production. Pathogenetic E. coli are classified into five virotypes namely (1) Enterotoxigenic E. coli (ETEC),which produces heat-resistant exotoxin (LT) and heat-resistant enterotoxin (ST). (2) Enteropathogenic E. coli (EPEC), which adheres very well to epithelial cells, attaches adhesions or removes wounds and irritates but does not release toxins. (3) Enterohemorrhagic E. coli (EHEC), which produces shiga/verotoxin, named for its cytotoxic effect on vero cells, an African monkey kidney. (4) Enteroagregative E. coli (EAEC) is characterized by a pattern of attachment that is typical in humans and produces toxins similar to ST and hemolysin. (5) Enteroinvasive E. coli (EIEC) does not ferment lactose and produces disease by invading intestinal mucosal epithelial cells.2
In mid-May 2011 there was an outbreak of diarrhea in Germany. Until June 2, 2011, Germany found 830 cases of Hemolytic Uraemic Syndrome (HUS) with 46 deaths. The same cases were found in 11 other countries: Austria, Czech Republic, Denmark, France, the Netherlands, Norway, Spain, Sweden, Switzerland, the United Kingdom, and the United States. Symptoms of this disease in the form of abdominal pain are cramps and diarrhea. In some cases, it can even cause bloody diarrhea (haemorrhagic colitis). Fever and vomiting can also occur.3
Preliminary research on the genetic analysis shows, this bacterial strain is a mutant form of two bacteria E. coli, EAEC and EHEC called EAHEC. When these two bacteria combine, it is dangerous to humans. One of the bacteria will take toxic substances from other bacteria and produce toxins that are more dangerous because it causes severe diarrhea, even damaging the tissues, including the kidneys. This case of E. coli outbreaks has caused rare and life threatening kidney failure. A normal E. coli infection is also life-threatening, but generally only in the group of infants, children, and people who have weak immunity.4
In 2010, diarrheal diseases were ranked as the first inpatient in Indonesia with 71,889 cases and 1,289 of them died. As for outpatient care, there were 105,279 new cases of diarrhea. The number of diarrhea patients in East Java Province in 2006 was 970,554, with sufferers in toddlers 448,677. In 2008 in Surabaya reported 66,841 cases of diarrhea or diarrhea morbidity rate of 2,355 per 100,000 population and the number of cases of diarrhea in children under five was reported as 24,571 cases or 36.76% of all cases of diarrhea.5
Based on the fact of the disease incidence by E.coli and its impact on human life, people need more information about E.coli aspects as well as its role in the spread of diseases caused by food (Foodborne Diseases). Until now, the percentage of the diarrhea caused by EAHEC in children under five years old in the Dr. Soetomo Hospital Surabaya still unknown.
Materials and Methods
Forty faecal samples from diarrhea patients under five years old in Dr. Soetomo General Hospital who have not taken antibiotics yet were collected. Fresh faecal samples were cultured in MacConkey agar, then incubated at 37°C for 18-24 hours. All the colonies were collected and suspended in test tube containing 2 ml of sterile distilled water. The cell suspension was boiledat 100°C for 15 minutes, thencentrifugated at 10,000 rpm for 10 minutes. Supernatant fluid was transferred into a new sterile tube, DNA target is ready to use.
Amplification was performed using PCR Master Mix; AAICf primers, ATTGTCCTCAGGCATTTCAC and AAICr, ACGACACCCCTGATAAACAA and CVD432f primers, CTGGCGAAAGA CTGTATCAT and CVD432r primers, CAATGTATAGAAATCCGCTGTT (EAEC); EAEf primers, CCCAATT CGGCACAAGCATAAGC and EAEr, CCCGGATCCGTCTCGCCAGTATTCG (EHEC).6
A total of 25 μL PCR mixture consisting of PCR Mix 12.5 μL, 1 μL of each primer (AAIC and CVD432) forward primer, reverse primer 1 μL, 4.5 μL H2O, and 6 mL of the target DNA. PCR was carried out in a Bio-rad PCRsystem T100TM Thermal Cycler (BioRad). PCR stages are set as denaturation at 94°C for 20 seconds; primer annealing at 57°C for 20 seconds; primer extension at 72°C for 60 seconds; as many as 35 cycles; and the final stage is final extension at 72°C for10 minutes.
Total 5 μL of DNA amplification product by PCR analyzed with electrophoresis gel containing 2% agarose in 0.5× TBE buffer. Insert samples, positive controls, and DNA markers were loaded in the gel wells and provide an electrical of 100v until the indicator color reaches 1 cm from the bottom edge of the gel. The gel was shaken in liquid ethidium bromide. The DNA bands were observed using UV Transilluminator. DNA bands in 215, 630, and 881 base pairs meant samples showed positive for E. coli strain EAHEC.
This study used Polymerase Chain Reaction (PCR) for the detection of EAEC using AAIC and CVD primers.The AAIC primers will produce amplicons of 215 base pairs, CVD primers will produce amplicons of 630 base pairs. Positive results on testing with AAIC primers are shown by the emergence of 215 bp DNA bands seen in Figure 1 and CVD primers are shown by the emergence of 630 bp DNA bands in Figure 2.6 EHEC detection used EAE primers for PCR. The EAE primers will produce amplicons of 881 bp. Positive results in testing with EAE primer shown by the emergence of 881 bp DNA band as shown in the Figure 3.
Figure 1.
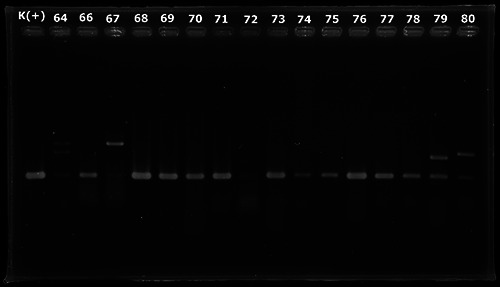
PCR amplification using primer AAIC.
Figure 2.
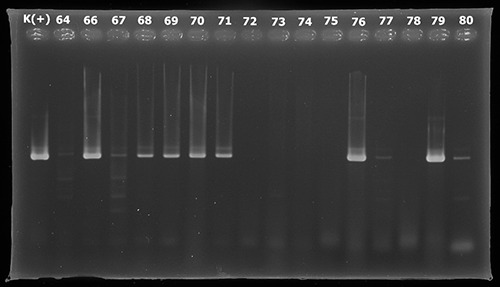
PCR amplification using primer CVD.
Figure 3.

PCR amplification using primer EAE.
Results
Samples with DNA fragments of 215 and 630 bp indicate that the samples are positive containing EAEC. Positive results can be detected at samples coded 4, 18, 32, 33, 35, 37, 38, 46, 49, 50, 51, 52, 53, 54, 57, 60, 62, 64, 66, 67, 68, 69, 70, 71, 76, 77, 79, 80, 82, 84, 86, 92 and 93.Samples with DNA fragments of 881 bp indicate that the sample is positive containing EHEC. Possitive results can be detected at samples with codes 13, 19, 35, 69, 86, 91, 91 and 97.
The results showed that 33(82.5%) out of 40 stool samples examined were detected enteroaggregative E. coli (EAEC), 4 (10%) out of 40 stool samples examined were detected enterohemorrhagic E. coli (EHEC), and 3 (7.5%) out of 40 stool samples examined were detected enteroaggregative haemorrhagic E. coli bacteria (EAHEC) which caused diarrhea in pediatric diarrhea patients at Dr. Soetomo General Hospital, Surabaya, Indonesia.
Discussion
Samples with DNA fragments at 215, 630, and 881 bp indicate that the sample is positive containing EAHEC. Possitive results can be detected at samples with codes35, 69, and 86. Based on the results of this study, it can be concluded that the EAHEC as one of the causes of diarrhea in children under five years old in Surabaya. There is 7.5% diarrhea caused by Enteroagregative Hemorrhagic E. Coli (EAHEC) in children under five years old at Dr. Soetomo Hospital Surabaya.
Conclusions
The finding of EAHEC, which is combination of EAEC and EHEC in Surabaya outbreak in 2015 suggested that it represents a new pathotype EAHEC in Indonesia. It is expected that sequencing and development of specific primer set are needed to continue and confirm in the next research
Acknowledgements
This research was supported by the Japan Initiative for Global Research Network on Infectious Diseases (JGRID); Institute of Tropical Disease (ITD) andDepartment of Pediatrics, Faculty of Medicine, Universitas Airlangga; Dr. Soetomo General Hospital, Surabaya, Indonesia. This article has been presented in International Conference of Infectious Diseases, Biothreats, and Military Medicine (INSBIOMM), Surabaya 2019.
Funding Statement
Funding: This work is supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID).
References
- 1.Weintraub A. Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J Med Microbiol. 2007, 56:4-8. [DOI] [PubMed] [Google Scholar]
- 2.Harrington SM, Dudley EG, Nataro JP. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett 2006; 254:12-18. [DOI] [PubMed] [Google Scholar]
- 3.Mellmann A, Harmsen D, Cummings CA, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE, 2011; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzuszkiewicz E, Thurmer A, Schuldes J, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero- Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 2011; 193:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kementrian Kesehatan Republik Indonesia,. 2011, Profil Kesehatan Indonesia 2010. http://www.depkes.go.id/resources/download/pusdatin/profilkesehatan-indonesia/profil-kesehatanindonesia-2010.pdf [Google Scholar]
- 6.Nguyen Tung Vu P, Le Van, et al. Detection and Characterization of Diarrheagenic Escherichia coli from Young Children in Hanoi, Vietnam. J Clin Microbiol 2005; 43, 755-760. [DOI] [PMC free article] [PubMed] [Google Scholar]


