Figure 1.
CD1c+ DC Heterogeneity Is Evident in Human BM
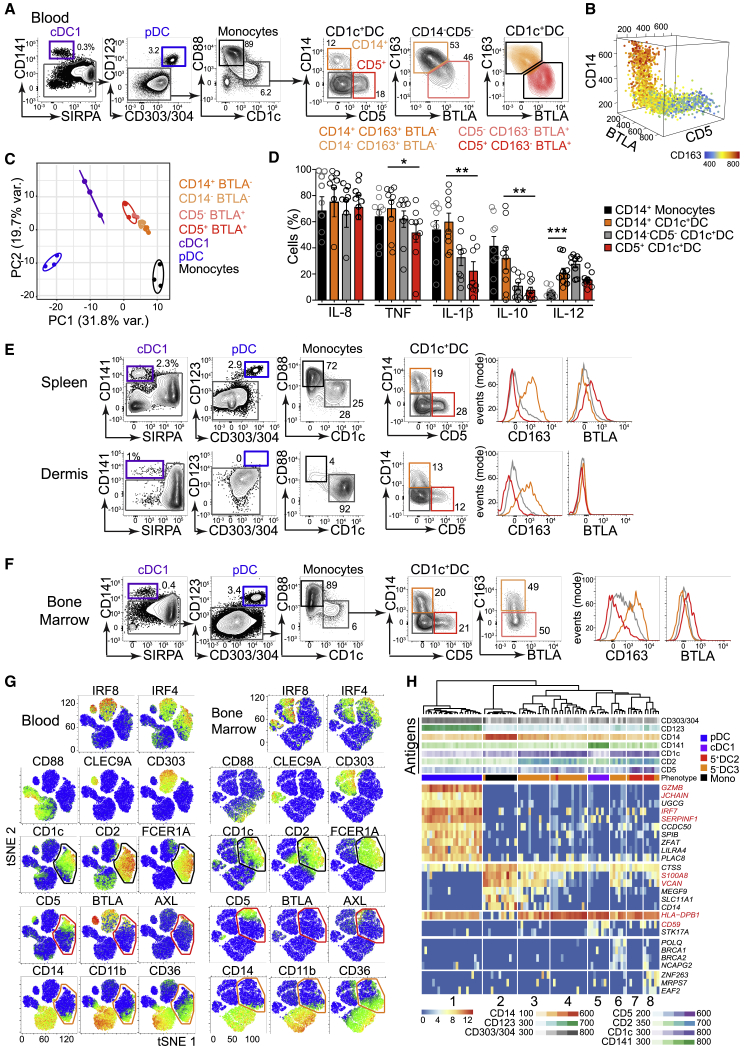
(A) Flow phenotyping of CD1c+ DCs from HC PB mononuclear cells (PBMCs) (representative example of n = 22), distinct from SIRPA−CD141+ cDC1s, CD123+CD303/4+ pDCs, and CD88+monocytes (Mono). CD14+CD163+BTLA− (orange), CD14−CD163+BTLA− (light orange), CD163−BTLA+CD5− (light red), and CD163−BTLA+CD5+ (red) CD1c+ DC subsets are indicated.
(B) 3D representation of CD14, CD5, and BTLA expression (flow cytometry) across the CD1c+ DC population. Heatmap shows expression of CD163.
(C) PCA of NanoString gene expression profiling of fluorescence-activated cell sorting (FACS)-purified DC subsets from n = 3 HC PBMCs. CD1c+ DCs were purified based on their expression of CD14, CD5, and BTLA (A).
(D) Intracellular flow analysis of in vitro cytokine elaboration (percentage of positive cells) by PB monocytes (black) and CD1c+DC subsets CD14+ (orange), CD14−CD5− (gray), and CD5+ (red) from n = 9 HC donors in response to 14-h stimulation with TLR agonists (CpG, poly(I:C), CL075, and lipopolysaccharide [LPS]). p values were derived from paired two-tailed t tests; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005. Bars show mean ± SEM, and circles represent individual donors.
(E and F) Representative examples of the flow profiling of DC subsets in human spleen (n = 3), dermis (n = 3) (E) and BM (n = 13) (F), gated as in (A). Histograms show CD163 and BTLA expression on CD14+ (orange), CD5+ (red) and CD14−CD5− (gray) CD1c+ DCs.
(G) tSNE visualization of the expression of TFs and surface markers across HC PB and BM lineage(lin, CD3,19,20,56,161)-HLA-DR+ cells by CyTOF analysis. Black gates indicate the CD1c+DC population distinct from CD88+monocytes, CLEC9A+cDC1 and CD303+pDC. Red and orange gates indicate expression of lymphocyte- or monocyte-associated antigens, respectively.
(H) Hierarchical clustering of single-cell transcriptomes of mature DCs from BM using all protein-coding, non-cell-cycle genes. Marker genes were identified within SC3 with parameters p < 0.01, area under the receiver operating characteristic curve (AUROC) > 0.85; cluster 1, pDCs (GZMB, JCHAIN); cluster 2, monocytes (S100A8, VCAN); cluster 3, CD14+ DC3s (HLA-DPB1); cluster 5, cDC1s (CD59). The top rows show fluorescence intensity of surface antigens (“Antigens”) from index-sorted cells, and “Phenotype” denotes their classification defined by surface markers.
See also Figure S1.