Abstract
The “canonical” function of Pax7+ muscle stem cells (satellite cells) during hypertrophic growth of adult muscle fibers is myonuclear donation via fusion to support increased transcriptional output. In recent years, however, emerging evidence suggests that satellite cells play an important secretory role in promoting load-mediated growth. Utilizing genetically modified mouse models of delayed satellite cell fusion and in vivo extracellular vesicle (EV) tracking, we provide evidence for satellite cell communication to muscle fibers during hypertrophy. Myogenic progenitor cell-EV-mediated communication to myotubes in vitro influences extracellular matrix (ECM)-related gene expression, which is congruent with in vivo overload experiments involving satellite cell depletion, as well as in silico analyses. Satellite cell-derived EVs can transfer a Cre-induced, cytoplasmic-localized fluorescent reporter to muscle cells as well as microRNAs that regulate ECM genes such as matrix metalloproteinase 9 (Mmp9), which may facilitate growth. Delayed satellite cell fusion did not limit long-term load-induced muscle hypertrophy indicating that early fusion-independent communication from satellite cells to muscle fibers is an underappreciated aspect of satellite cell biology. We cannot exclude the possibility that satellite cell-mediated myonuclear accretion is necessary to maintain prolonged growth, specifically in the later phases of adaptation, but these data collectively highlight how EV delivery from satellite cells can directly contribute to mechanical load-induced muscle fiber hypertrophy, independent of cell fusion to the fiber.
Keywords: extracellular vesicles, Mmp9, Nr4a1, tdTomato, miRNA, Pax7-DTA
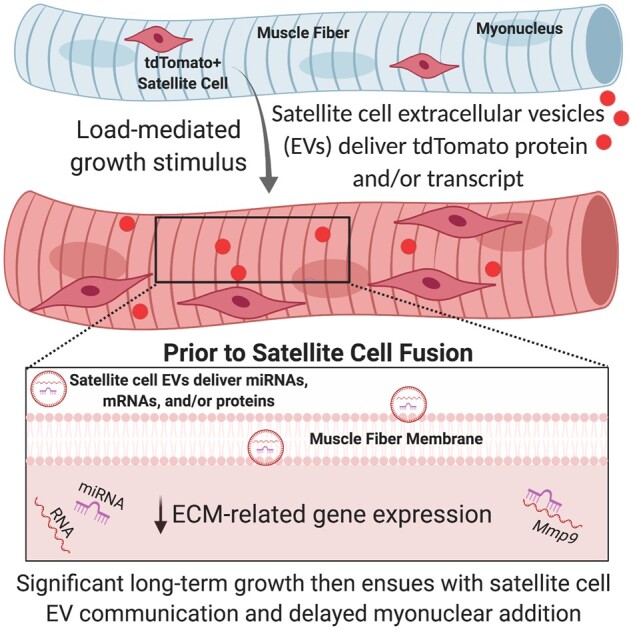
Graphical Abstract
Graphical Abstract.
Introduction
The primary muscle stem cell population is Pax7+ satellite cells.1 Throughout postnatal development, these cells fuse to growing muscle fibers and contribute new myonuclei.2 When satellite cells are depleted using a genetic approach in prepubertal murine muscle, muscle fibers are smaller by adulthood,3 and when depleted in young adulthood, cannot mount an effective hypertrophic response to mechanical loading.4–6 Conversely, adult (>4 months old) murine muscle can sustain short-term load-mediated hypertrophy (≤2 weeks) in the absence of satellite cells,6–10 although long-term hypertrophy (≥8 weeks) is attenuated.7,8,11 Some evidence suggests that the addition of new myonuclei to muscle fibers undergoing hypertrophy is necessary before growth ensues12,13 or is required beyond a certain threshold of growth to control the “myonuclear domain,”14–16 which is the maximal area that each myonucleus can transcriptionally govern, although this limit is equivocal and may be muscle and/or fiber type specific.9,14,17–19
Aside from myonuclear addition, another avenue whereby satellite cells could directly support muscle fiber hypertrophy is via secreted factors, which is plausible since satellite cells reside in very close proximity to muscle fibers, increase dramatically in abundance with mechanical overload (MOV), and are known to communicate with mononuclear cells in muscle to facilitate adaptation.7,8,20–22 Recently, our laboratory described how satellite cells deliver miRNA to fibrogenic cells via extracellular vesicles (EVs) early during mechanical loading to support long-term hypertrophic muscle growth.7 We hypothesized that satellite cells may also be communicating with muscle fibers, the largest cells by volume in muscle, via EVs early during loading to potentiate the long-term growth response. To test this hypothesis, we developed murine genetic models of delayed satellite cell fusion and in vivo EV tracking, complemented with in vivo satellite cell depletion experiments, in vitro primary myogenic progenitor cell (MPC) culture, unbiased transcriptome profiling, and in silico analyses. Our experiments revealed that satellite cells communicate with muscle fibers via EVs and contribute to load-mediated muscle hypertrophy in adult animals and that this involves the repression of matrix metalloproteinase 9 (Mmp9). Furthermore, delayed satellite cell fusion did not inhibit long-term muscle hypertrophy, which may in part be due to satellite cell EV communication to muscle fibers. Our findings, for the first time, reveal EV-mediated intercellular communication between satellite cells and muscle fibers during hypertrophy and point to new therapeutic frontiers for targeting these cells to preserve or enhance muscle mass.
Results
Primary MPC EVs Can Package tdTomato Protein and Transcript
Numerous investigations provide evidence for nonfusion-mediated transfer of genetically-driven cytoplasm-localized fluorescent reporters in vivo.23–27 Most recently, using a Cre/Lox genetic inducible model, cytoplasmic tdTomato (tdT) fluorescent protein was shown to be packaged in EVs from adipocytes.28 In context with our previous work showing EV communication from satellite cells to fibrogenic cells during hypertrophy,7 we hypothesized that tdT protein and/or transcript could be packaged in satellite cell EVs. We generated satellite cell-specific tamoxifen-inducible tdT mice (Pax7CreERT-tdTfl/+ or Pax7-tdT), as previously described,29–32 then isolated satellite cells from vehicle and tamoxifen-treated mice via fluorescent-activated cell sorting (FACS).33In vitro, EVs (characterized via nanoparticle tracking analysis) from proliferating tdT+ primary MPCs contained tdT protein (determined via single vesicle fluorescent correlational spectroscopy (FCS), high-magnification fluorescent imaging, and western blot) and messenger RNA (mRNA, determined via quantitative polymerase chain reaction, qPCR) (Figure S1). We isolated EVs via a polymer-based method (ExoQuick) as described by our laboratory,7 a polymer-based method combined with density gradient ultracentrifugation (DGUC), as well as ultracentrifugation (UC), and consistently found tdT transcript (Figure S1). Collectively, these data agree with evidence for cytoplasmic-localized reporter protein and/or mRNA being packaged in EVs23,28,34,35 and show that MPC EVs contain tdT protein and transcript.
Fusion-Independent Satellite Cell Communication to Muscle Fibers during MOV
Neural Wiskott–Aldrich syndrome protein (N-WASp) regulates actin polymerization and is reportedly essential for myoblast fusion throughout development and in vitro.36 To prevent satellite cell fusion during synergist ablation-induced MOV of the plantaris muscle, we developed Pax7CreERT-N-WASpfl/fl-tdTfl/+ mice (termed N-WASp/tdT onward) that, upon tamoxifen-induced recombination, demonstrate a loss of N-WASp gene expression simultaneous with tdT expression specifically in satellite cells (Figure 1A and B). We hypothesized that fusion-incompetent N-WASp-depleted satellite cells would communicate with muscle fibers via EVs during MOV, which could be detected by the appearance of tdT in muscle fibers. Following vehicle and tamoxifen administration in vivo and a minimum 2-week washout, we isolated N-WASp/tdT MPCs via FACS and confirmed tdT expression and N-WASp knockdown (P < 0.05, Figure 1C and D) with tamoxifen. The loss of N-WASp did not affect EV biogenesis or release as N-WASp−/tdT+ MPCs generated a similar amount of EVs as MPCs isolated from vehicle-treated N-WASp+/tdT− mice (Figure 1E). Furthermore, N-WASp−/tdT+ EVs contained tdT mRNA (Figure 1F). N-WASp depletion did not affect MPC proliferation (Figure 1G), but reduced myosin heavy chain (MyHC) area and myotube formation in vitro (P < 0.05, Figure 1H–J). We then subjected N-WASp+/tdT− and N-WASp−/tdT+ plantaris muscles to MOV for 7 days; sham-operated mice served as controls (Figure 1K). Satellite cell proliferation after 7 days of MOV was similar between N-WASp+/tdT− and N-WASp−/tdT+ mice (Figure 1L), but myonuclear accretion was abolished in the N-WASp−/tdT+ condition (P < 0.05, Figure 1M and N). In the absence of satellite cell fusion and myonuclear accretion, muscle fibers in N-WASp−/tdT+ mice displayed tdT fluorescence, pointing to fusion-independent delivery of tdT (Figure 1O). To validate this finding, we modeled tdT transfer by EVs in vitro by incubating myotubes generated from wild type C57BL/6J MPCs with EVs collected from proliferating N-WASp−/tdT+ MPCs (Figure 1P). We observed tdT puncta in myotubes that appeared similar to cytoplasmic tdT transferred via EVs reported in other cell culture models35 (Figure 1Q); tdT puncta in myotubes were not apparent when cultured with EVs from vehicle-treated N-WASp+/tdT− MPCs (not shown). These findings align with previous work showing mRNA and protein transfer via EVs between myogenic cells in vitro,37 as well as cytoplasmic reporter transfer between myogenic cells.34 Collectively, these data indicate that satellite cells communicate to muscle fibers before fusing during MOV, which is in part mediated by EVs.
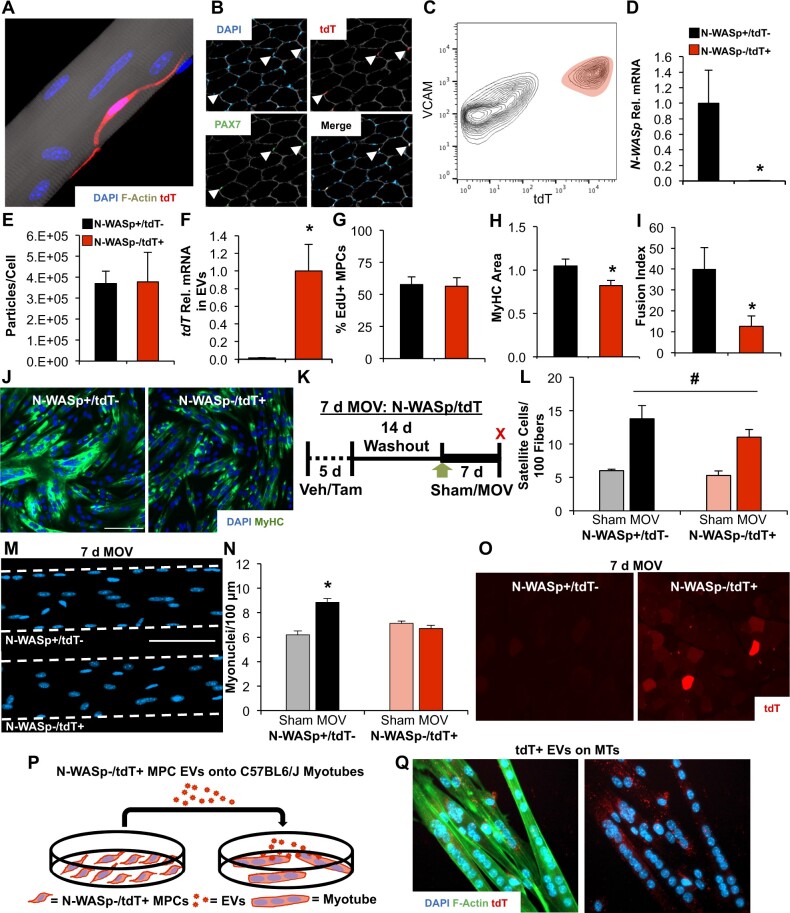
Figure 1.
Delayed satellite cell fusion and EV-dependent communication to muscle fibers during 7 days of MOV. (A) Confocal image of a tdT-expressing satellite cell (red) overlaid with DAPI (blue) on a muscle fiber (stained for F-actin, gray), illustrating Pax7Cre-mediated recombination of the floxed tdT transgene in the presence of tamoxifen. (B) Histochemical image depicting Pax7 immunostaining (green) that coincided with tdT fluorescence (red) in the correct anatomical location (beneath laminin, gray, and overlaid with DAPI, blue) after tamoxifen treatment. (C) Flow cytometry plot illustrating tdT+ cells coexpressing Vcam, which was used to identify and purify satellite cells via FACS in tamoxifen-treated mice; Vcam+/Cd31−/Cd45−/Sca1− cells were purified in vehicle-treated mice, as described previously,33 and tdT recombination was minimal (not shown). (D) Gene expression to confirm N-WASp recombination from in vivo-treated vehicle (N-WASp+/tdT−) versus tamoxifen-treated (N-WASp−/tdT+) primary MPCs purified via FACS, n = 2 biological replicates per condition. (E) Nanoparticle tracking analysis showing EV particle abundance normalized to cell count from N-WASp+/tdT− and N-WASp−/tdT+ MPCs, n = 2 biological replicates per condition. (F) Relative tdT mRNA abundance in EVs isolated from N-WASp+/tdT− (n = 2 biological replicates) and N-WASp−/tdT+ MPCs (n = 3 biological replicates). (G) Proliferation in N-WASp−/tdT+ and N-WASp+/tdT− MPCs (n = 3 biological replicates per condition). (H) MyHC area after 30 h of differentiation (n = 3 biological replicates per condition). (I) Fusion index (n = 3 biological replicates per condition). (J) Representative images of MPC fusion in N-WASp+/tdT− versus N-WASp−/tdT+ conditions; green is MyHC and blue is DAPI. (K) Study design schematic illustrating vehicle (Veh) and tamoxifen (Tam) treatment followed by sham or 7 days of MOV in N-WASp/tdT mice. (L) Satellite cells normalized to muscle fiber count in N-WASp+/tdT− (n = 5 sham and 5 MOV) versus N-WASp−/tdT+ (n = 6 sham and 7 MOV) mice. (M) Representative images of myonuclei in isolated single muscle fibers from MOV mice. (N) Myonuclear content analysis on isolated single muscle fibers. (O) Representative image of tdT fluorescence in MOV N-WASp−/tdT+ muscle fibers despite the prevention of satellite cell-mediated myonuclear accretion. (P) Study design schematic illustrating the treatment of wild type C57BL/6J myotubes with EVs from N-WASp−/tdT+ MPCs. (Q) Representative image showing tdT puncta (red) in C57BL/6J myotubes (stained for F-actin, green, and DAPI, blue) incubated with N-WASp−/tdT+ MPC EVs. *P < 0.05, #P < 0.05 effect of MOV, scale bars = 100 µm, all data are presented as mean ± SE.
MPC EVs Alter Muscle Cell Transcriptional Profiles in vitro in a Fashion Consistent with Satellite Cell-Dependency in vivo
Effectively blocking EV release in a cell type-specific fashion in vivo is complicated and presents a variety of technical challenges.38 In order to search for candidate muscle fiber genes whose expression is potentially affected by satellite cell EVs during hypertrophy in vivo, we turned to our previously published murine model of inducible satellite cell depletion, the Pax7-Diptheria Toxin A (Pax7-DTA) model.6,8,10 Specifically, using an unbiased approach, we aimed to identify genes that were similarly expressed between satellite cell replete and deplete muscle at rest, but were lower in the presence of satellite cells following MOV, which could indicate EV-mediated delivery of repressive microRNAs (miRNAs) to target cells by activated satellite cells.39 Plantaris muscles of adult mice with and without satellite cells underwent sham surgery or 7 days of MOV, and then muscles were profiled via microarray (Figure 2A). Mmp9 was the most differentially expressed gene in the microarray dataset that was lower in the presence of satellite cells after MOV (+41% in satellite cell replete and +80% in depleted muscle), fitting our above criteria (Figure 2B, Table S1). Mmp9 is synthesized and secreted by myotubes,40 regulates extracellular matrix (ECM) quality and turnover,41 and is robustly upregulated in bioengineered myofibers after mechanical loading,42 indicating it is mechanosensitive and expressed by muscle fibers during hypertrophy; Mmp9 is also highly-induced in myotubes by the presence of inflammatory cues.43 Worth noting is that Mmp9 is enriched in activated satellite cells,44–46 so lower transcript levels in the presence of satellite cells during MOV points to the presence of a repressive satellite cell-mediated mechanism (e.g., EV delivery of miRNA to target cells). We then profiled MPC EVs via miRNA microarray to identify miRNAs that may inhibit Mmp9. We recently showed that miR-206 is transferred by satellite cell EVs to fibrogenic cells during MOV to regulate collagen deposition,7 and confirm here that miR-206 is the most abundant miRNA in MPC EVs (Figure 2C). Expanding on our earlier work, we found numerous miRNAs enriched in MPC EVs (Figure 2C) that are experimentally shown to lower Mmp9 expression, such as miR-24,47–49 miR-149,50–55 and miR-486,56,57 as well as miRNAs that are validated to target the 3′-UTR of Mmp9 mRNA and reduce transcript levels, including Let-7e,58 miR-133a and -133b,59,60 and miR-32061 (Figure 2D). In silico predicted miRNA-mRNA target analysis62 further revealed that ECM remodeling is the most regulated process by MPC EV miRNAs (Figure 2E, Tables S2 and S3). Although the in vivo results from satellite cell-depleted mice could be explained by dysregulated communication from other cell types throughout muscle, we speculate that satellite cell-derived EVs contribute to Mmp9 repression specifically in muscle fibers during MOV.
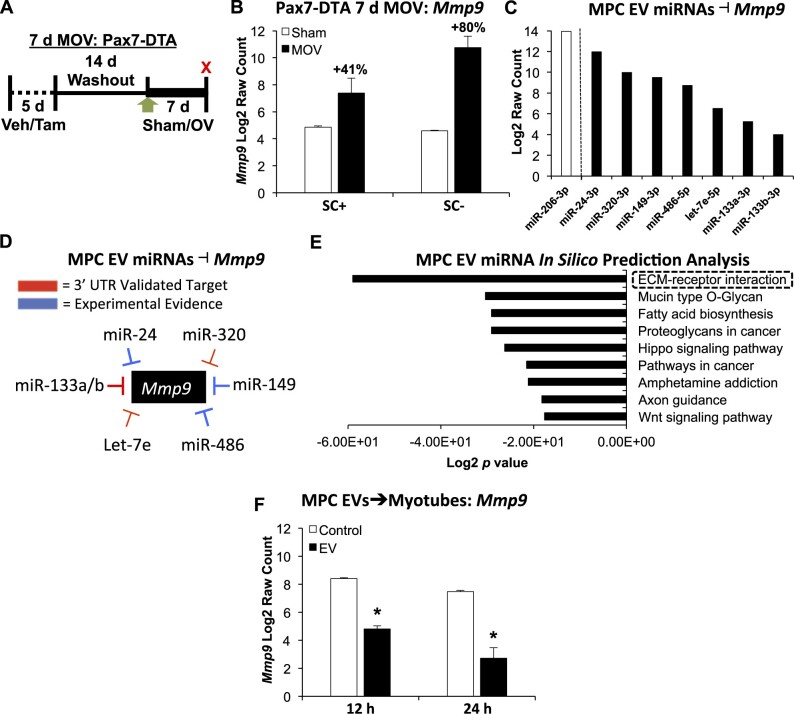
Figure 2.
Evidence for the impact of EV-mediated communication to muscle fibers in vitro and in vivo. (A) Study design schematic illustrating vehicle (Veh, SC+) and tamoxifen (Tam, SC−) treatment followed by sham or 7 days of MOV in Pax7-DTA mice. (B) Mmp9 mRNA levels in sham versus MOV in the presence and absence of satellite cells; n = 6 mice per group, two separate pools of three plantaris muscles for each group resulting in n = 2 pools per group. (C) miRNAs enriched in MPC EVs that influence Mmp9 levels in different experimental models; miR-206 was the most abundant miRNA measured. (D) Summary of evidence for miRNAs that are enriched in MPC EVs that affect Mmp9 via direct 3′-UTR targeting or indirectly via experimental manipulation using miRNA mimics and/or antagomirs (see “Results” section for specific studies). (E) DIANA miRPath analysis of miRNAs enriched in MPC EVs using the top 100 miRNAs. (F) Mmp9 mRNA levels in C57BL/6J myotubes incubated with MPC EVs for 12 or 24 h; one primary cell line was used to generate myotubes and was incubated with MPC EVs from two separate cell lines at each time point (n = 2 per time point: 12 and 24 h control, 12 and 24 h EV treatment). *Adjusted P < 0.05, all data are presented as mean ± SE.
We sought to experimentally validate the effects of satellite cell communication to muscle fibers via EVs using an in vitro approach coupled with unbiased RNA sequencing. We incubated myotubes with MPC EVs for 12 and 24 h and found that Mmp9 was robustly downregulated at both time points (adj. P < 0.05, Figure 2F). A variety of other transcripts associated with the Reactome term “ECM Organization”63 was also repressed at 12 or 24 h (Bmp2, Col8a1, Col12a1, Ltpb2, P4ha3, Scube3, Smoc2, Timp3, adj. P < 0.05, Table S4). We further identified mRNAs whose expression was higher in myotubes incubated with MPC EVs. Nr4a1, which is strongly associated with exercise adaptation,64 was elevated at both 12 and 24 h (adj. P < 0.05). The complete list of up- and downregulated genes in myotubes incubated with MPC EVs is found in Table S4. It is possible that EVs from other cell types in muscle could mediate the effects observed here, but collectively, these data show that MPC EVs elicit marked transcriptional changes in myotubes that simulate the effect of satellite cell EVs on muscle fibers during hypertrophy.
Delayed Satellite Cell Fusion Does Not Inhibit Long-Term Muscle Hypertrophy
Satellite cell fusion was prevented during the first week of MOV in N-WASp−/tdT+ muscle (see Figure 1N), which aligned with our in vitro data showing impaired MPC fusion (see Figure 1H–J). Recent evidence from our laboratory suggests that the presence of satellite cells and modest myonuclear accretion during the first week of MOV is sufficient to support robust long-term growth (up to 8 weeks) without continued myonuclear addition,7 but whether there is an early “critical window” for satellite cell fusion that is required to sustain prolonged hypertrophy is not clear.7,12,13,65 To determine the necessity of satellite cell fusion for maximal long-term hypertrophy, we subjected N-WASp+/tdT− and N-WASp−/tdT+ mice to 8 weeks of MOV; we also overloaded vehicle and tamoxifen-treated Pax7CreERT-N-WASpfl/fl mice (N-WASp,) for 2 weeks, and sham-operated mice served as controls (Figure 3A). Myonuclear accretion was attenuated at 2 weeks in the absence of N-WASp (P < 0.05), consistent with our 7-day experiments; however, myonuclear number was similar between conditions by 8 weeks (Figure 3B). Satellite cell abundance (Figure 3C), muscle hypertrophy (Figure 3D–G), and ECM content (3H and I) were not different between groups after 8 weeks. In our hands, N-WASp−/tdT+ MPCs eventually fuse to form large myotubes with enough time in culture (not shown), congruent with the in vivo data. Although we were not able to test the necessity of myonuclear accretion for maximal growth using this model, our results collectively suggest that delayed satellite cell fusion during MOV does not inhibit long-term skeletal muscle hypertrophy or result in excess ECM accumulation, and that the delivery of satellite cell secretory products to muscle fibers, specifically early during loading via EVs, may help promote prolonged muscle growth.
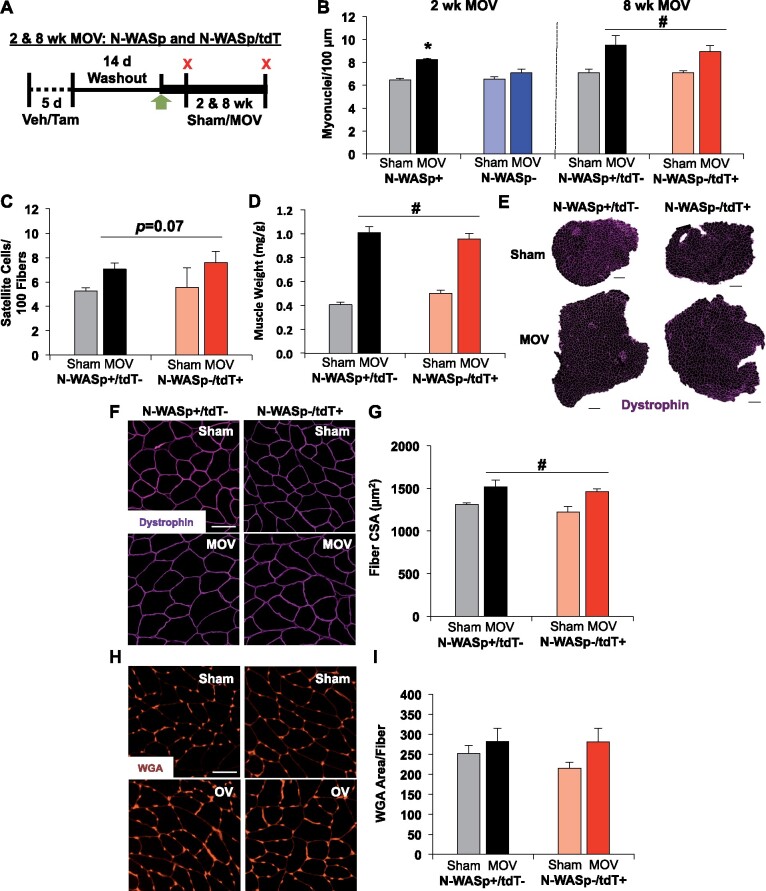
Figure 3.
Delayed satellite cell fusion does not inhibit long-term muscle hypertrophy. (A) Study design schematic illustrating vehicle (Veh) and tamoxifen (Tam) treatment followed by sham, 2 weeks, or 8 weeks of MOV in N-WASp (2 weeks) and N-WASp/tdT (8 weeks) mice. (B) Myonuclear content analysis on isolated single muscle fibers at 2 weeks (n = 5 sham and 8 MOV N-WASp+, n = 7 sham and 11 MOV N-WASp−) and 8 weeks (n = 4 sham and 4 MOV N-WASp+/tdT−, n = 4 sham and 5 MOV N-WASp−/tdT+). (C) Satellite cells normalized to muscle fiber count after 8 weeks of MOV. (D) Plantaris muscle wet weight normalized to body weight. (E) Representative plantaris muscle cross-sections labeled with dystrophin (pink), scale bar = 500 µm. (F) Magnified representative images of dystrophin, scale bar = 50 µm. (G) Average muscle fiber cross-sectional area (CSA). (H) Representative images of ECM glycosaminoglycan area evaluated by WGA staining, scale bar = 50 µm. (I) WGA area normalized to muscle fiber count. *P<0.05, #P < 0.05 effect of MOV, all data are presented as mean ± SE.
Discussion
Our laboratory and others report that adult muscle fibers can grow in the absence of satellite cell fusion and myonuclear addition, indicating flexibility within the myonuclear domain.18,19 Hypertrophy without increased myonuclear number has been demonstrated with mechanical,6–8,10,11,66 pharmacological,13,67–69 and genetic approaches,68,70–74 although this finding is not universal,4,12,13,75–79 which could be explained by a variety of factors.5,19 In recent years, a deeper appreciation for the secretory function of muscle stem cells in general,80–84 and satellite cells specifically7,8,21,22 has emerged in the context of skeletal muscle remodeling, with evidence for EV communication between mononuclear cells.7 It is becoming clear that the importance of satellite cells for promoting muscle hypertrophy extends beyond fusion and myonuclear addition, and the findings from this investigation provide the first evidence of EV-mediated communication to muscle fibers during mechanical loading, independent from satellite cell fusion. EV communication to muscle fibers early during hypertrophy may influence ECM-related signaling within muscle fibers, specifically targeting Mmp9. Furthermore, we show that early satellite cell fusion during loading is not necessary to produce maximal long-term growth.
We attribute the appearance of tdT fluorescence in muscle fibers of N-WASp−/tdT+ mice following 7 days of MOV at least in part to EV-mediated transfer of tdT protein and/or transcript from satellite cells, which is supported by our cell culture work. Other investigations in different experimental systems document the transfer of genetic cytoplasm-localized reporter fluorescence between cells,23–27,34,35 as well as to distant tissues specifically via EVs.23 Mesenchymal stem cells have also been shown to deliver transgene mRNAs to recipient cells via EVs that then elicit a biological effect.85 Recently, Flaherty et al. used an adipocyte-specific Cre-inducible cytoplasmic tdT model to show the packaging of tdT protein into EVs,28 consistent with what we find in our satellite cell-specific tdT model. The appearance of tdT in muscle fibers is likely not due to recombination in resident myonuclei since the Pax7-tdT cytoplasmic reporter is highly-specific to satellite cells.29–32 and Cre-inducible reporters using other lineage-specific promoters have consistently shown reporter gene expression is restricted to the cell that is expressing Cre, even when Cre-mediated “leakiness” does occur.86–89 Another intriguing explanation for tdT transfer from satellite cells to myofibers is tunneling nanotubes,90 that are capable of material transfer between myogenic progenitors and muscle fibers in culture,91 deserving further investigation. We ultimately conclude that reporter fluorescence in satellite cells can be transmitted to muscle fibers in the absence of fusion, specifically during times of adaptive remodeling.
MPC EVs lowered gene expression related to ECM regulation in myotubes, which we used to model the influence of satellite cell-derived EVs on muscle fibers in vivo. Myogenic cell-derived EVs promote effective regeneration in vivo,92 which could in part be mediated by the effect of EVs on growing de novo muscle fibers. In our data, Mmp9 was repressed at 12 and 24 h in EV-treated myotubes determined via RNA-seq, as well as in the presence of satellite cells during MOV determined via microarray. Mmp9 regulates ECM turnover and its inhibition improves muscle regeneration, controls fibrosis,93 and is protective of muscle mass in the face of pathological inflammation and muscular dystrophy,43,94 specifically in the early phase of dystrophic disease progression.95 Somewhat paradoxically, lifelong genetic depletion of Mmp9 is deleterious to muscle mass96 while constitutive muscle-specific overexpression promotes growth.97Mmp9 is pleiotropic95,98,99 and its regulation by muscle fibers during loading is not entirely clear.42 Mmp9 may affect myogenic and hypertrophic signaling in muscle,95,97 but it is also known to cleave collagen IV and laminin, and high levels of Mmp9 induce muscle membrane damage.44,94,97 Satellite cells could, therefore, mediate a “goldilocks” effect by fine-tuning muscle fiber Mmp9 levels through EV delivery of miRNAs early during MOV to manage ECM integrity and facilitate the long-term hypertrophic response.7,8 Fibroblasts and fibro-adipogenic progenitors contribute heavily to ECM deposition and fibrotic protein production, but numerous ECM-related genes are epigenetically regulated specifically in myonuclei during MOV, suggesting muscle fiber participation in these processes.100 Recent work using muscle-specific genetic mice reinforce the important role muscle fibers play in ECM regulation during adaptation.101,102 MPC EVs also increased gene expression of some genes in myotubes, although the precise explanation for this deserves further study. Interestingly, Nr4a1 was elevated by EVs, and nuclear receptors were recently identified via large-scale meta-analyses as the most highly responsive genes to any type of exercise in skeletal muscle, including resistance exercise.64,103 Classically associated with substrate utilization in skeletal muscle,104–106Nr4a1 is epigenetically modified with resistance retraining after detraining in muscle,107 controls adult muscle mass,108,109 and is a potent inhibitor of fibrosis.110 Satellite cell control of ECM dynamics via intercellular communication is consistent with the broader influence of satellite cell secretory products reported by us and others,7,8,20,22,111–114 and the effects of satellite cell EVs directly on muscle fibers should not be overlooked.
The rigidity of the myonuclear domain has recently been disputed,5,115 but numerous investigations across species show at least modest flexibility in the myonuclear domain of adult skeletal muscle during hypertrophy.18 Satellite cell fusion early during hypertrophy was posited to ameliorate muscle fiber membrane damage to support late-stage adaptation13; however, muscle fiber membrane repair is efficiently carried out by resident intracellular machinery,116–120 and MOV-induced muscle fiber hypertrophy in our hands proceeds even when satellite cell fusion is delayed during loading. In context with our present findings, we speculate that when satellite cell fusion is effectively prevented by genetic means,13,76,79 that other secretory functions of the cell may also be affected which could negatively affect hypertrophy.121 Along these lines, mTOR activation in skeletal muscle is preserved during MOV in the complete absence of satellite cells10 but is abolished in the presence of myomaker-null fusion-incompetent satellite cells,76 which could point to altered satellite cell secretory function. In our hands, N-WASp-deficient MPCs eventually fuse to form large myotubes, which is congruent with our 8-week N-WASp MOV data. N-WASp depletion in our model involves excision of exons six to nine and results in a truncated protein generated by exons one through five,122 which may explain why we did not observe the complete prevention of fusion that was reported using a floxed exon two construct36 and may also explain intact secretory function. We speculate that impaired fusion with N-WASp disruption could be due to compromised MPC migration123 since we observed that differences between conditions were less apparent at higher seeding densities in vitro. It is also conceivable that compensatory mechanisms were activated that circumvented the loss of N-WASp during prolonged MOV, or that the path of satellite cell myogenic progression and fusion differs during hypertrophy versus developmental growth and regeneration.75 Nevertheless, delayed satellite cell fusion during MOV did not inhibit long-term skeletal muscle hypertrophy or result in excess ECM accumulation.
Numerous investigations have linked the level of satellite cell proliferation and subsequent myonuclear accretion to the magnitude of skeletal muscle hypertrophy.14,15,65,124,125 Our data suggest that robust satellite cell proliferation in response to loading may promote long-term muscle fiber growth through fusion-dependent as well as -independent mechanisms. Our results also indicate that Cre-induced cytoplasmic-localized fluorescent reporters should be used with caution for accurately determining the contribution of satellite cell fusion to muscle fibers.29–32,126 Future experiments involving satellite cell-specific inhibition of EV release, as well as genetic manipulation of specific miRNAs in satellite cells in vivo will help provide more detailed insight into satellite cell regulation of muscle fiber gene expression by fusion-independent mechanisms. Satellite cells coordinate adaptation to support maximal long-term muscle hypertrophy,19 which may in part be due to EV delivery of miRNA to muscle fibers that affect ECM-related gene expression.
Materials and Methods
Animals
Pax7CreERT/+ tdTomatofl/+ mice were generated by crossing male Pax7CreERT/CreERT mice to female ai9 Rosa26 tdTomatofl/fl mice,127 purchased from the Jackson Laboratory; these were called Pax7-tdT. Pax7CreERT/+ N-WASpfl/fl tdTomatofl/+ mice were generated by crossing male Pax7CreERT/CreERT mice with female N-WASpfl/fl mice,122 that through several rounds of breeding, produced male Pax7CreER/+ N-WASPfl/fl mice (N-WASp). Male double-mutant mice were crossed to female N-WASpfl/fl tdTomatofl/fl mice that were bred to homozygosity by crossing tdTomatofl/fl mice with N-WASpfl/fl mice; the triple-mutant mouse was termed N-WASp/tdT. Pax7CreERT/+ Rosa26DTA/+ were generated by crossing male Pax7CreERT/CreERT mice with female Rosa26DTA/DTA mice,10,128,129 both purchased from the Jackson Laboratory (Bar Harbor, ME, USA); this mouse was termed Pax7-DTA. All Pax7-tdT, N-WASp/tdT, and N-WASp experiments involved a balanced mix of males and females, and Pax7-DTA experiments involved male mice. In all mice, tamoxifen treatment induces Cre activity, which recombines out stop cassettes in front of DTA or tdT constructs that were knocked into the Rosa26 locus, and recombines out exons six through nine flanked by loxP sites in the N-WASp mice. Administration of tamoxifen to Pax7-tdT and N-WASp/tdT mice results in robust expression of tdT (shown here and elsewhere29–32) and depletion of N-WASp (in N-WASp/tdT), and to Pax7-DTA mice results in specific and significant depletion of satellite cells across various skeletal muscles regardless of age.10,130,131 Mice were genotyped via tail snip and NaOH digestion, then polymerase chain reaction for 40 cycles using GoTaq G2 chemistry (Promega, Madison, WI, USA), followed by electroporation on 1.5%–2.0% agarose gel to visualize DNA bands (see Supplemental Table S5 for genotyping primer sequences). C57BL/6J mice (obtained from the Jackson Laboratory) were used to obtain primary MPCs for in vitro experiments. Experiments and animal care were performed in accordance with the University of Kentucky Institutional Animal Care and Use Committee. All mice were housed in a temperature- and humidity-controlled room and maintained on a 14:10 light:dark cycle, with standard chow and water ad libitum. All mice were >4 months old at the time of experimentation, consistent with our previous work.6
Mouse Treatment, Synergist Ablation MOV Surgery, and Tissue Collection
Mice were injected intraperitoneally with vehicle (15% ethanol in sunflower seed oil) or tamoxifen (2 mg/day, suspended in ethanol and sunflower seed oil) for 5 days, then given a 2-week washout period before any experimentation ensued.10 Mice underwent bilateral synergist ablation surgery to induce hypertrophy of the plantaris muscle, as previously described by our laboratory.7,66 Briefly, mice were anesthetized using 95% oxygen and 5% isoflurane gas, then approximately one-third of the gastrocnemius/soleus complex was removed, careful not to disturb neural or vascular supply. Sham surgery involved all procedures except for excision of muscle. Following recovery from surgery, mice were euthanized via lethal dosage of sodium pentobarbital and cervical dislocation at the appropriate time point. Hind limb muscles for immunohistochemistry analysis were weighed, covered in Tissue-Tek Optimal Cutting Temperature compound (Sakura Finetek, Torrance, CA, USA), frozen in liquid nitrogen-cooled isopentane, and stored at −80°C until the time of sectioning. For single muscle fiber myonuclear count analysis, whole hind limbs were excised and the ankle was fixed at 90° (to control for sarcomere spacing) in 4% paraformaldehyde (PFA) for 48 h. For N-WASp/tdT mice, after 48 h in PFA, the top one-fourth of the plantaris was removed and incubated in 30% sucrose overnight at 4°C, then frozen for visualization of tdT, as described elsewhere.31 The remainder of the muscle was stored in phosphate-buffered saline (PBS) until the time of single fiber analysis. The entire plantaris muscle was flash-frozen and used for molecular analysis in Pax7-DTA experiments.
Muscle Histology and Quantification
All histological analyses are described in detail in previous publications from our laboratory.6–8,10 In brief, frozen tissue was sectioned (7 µm) and air-dried for at least 1 h. For Pax7, sections were fixed in 4% PFA then subjected to epitope retrieval using sodium citrate (10 mM, pH 6.5) at 92°C for 20 min. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide in PBS for 7 min, followed by a mouse-on-mouse block (Vector Laboratories, Burlingame, VT, USA). Sections were then incubated overnight with a Pax7 antibody and a laminin antibody, followed by incubation with a biotin-conjugated secondary antibody for Pax7, and a fluorophore-conjugated antibody for laminin; sections were then incubated with 4′6-diamidino-2-phenylindole (DAPI, 1:10 000) for 10 min. The Pax7 signal was amplified using streptavidin horseradish peroxidase (SA-HRP) followed by a tyramide signal amplification (TSA) fluorophore. Pax7 staining was conducted only in conjunction with laminin, accepting the limitation that a very small percentage of myonuclei may costain with Pax7 protein for some unknown reason.132 For fiber cross-sectional area, sections were incubated with an antibody against dystrophin, followed by a fluorescent secondary antibody. To analyze ECM, sections were incubated with directly-conjugated α-wheat germ agglutinin (WGA, 1 mg/mL, Invitrogen) for 2 h. All sections with fluorescence were mounted using VectaShield (Vector) and stored in the dark until the time of imaging. To visualize endogenous tdT expression, PFA-fixed sections were cut then dried for 1 h in the dark, fixed again in 4% PFA for 10 min, washed in PBS, then mounted with VectaShield.31 See Supplemental 5 for antibody information. Muscle sections were imaged using a Zeiss upright microscope (AxioImager M1, Oberkochen, Germany) and analyzed with Zen software or MyoVision software.133 A size correction factor previously published by our laboratory was used to normalize fiber cross-sectional area for experiments involving both male and female mice.9 For Pax7, entire muscle cross sections were imaged at ×20 magnification using the mosaic image stitching function in Zen. Pax7+ satellite cells were identified as Pax7+/DAPI+ and residing underneath the basement membrane and were quantified manually by a blinded using tools in the Zen software (normalized to muscle fiber number). Similarly, entire muscle cross sections were captured at ×20 magnification, and muscle fiber cross-sectional area was determined using semiautomated analysis software.133 WGA was analyzed by capturing 2–3 artifact-free ×20 images that were then analyzed using a threshold tool in Zen.
Single Muscle Fiber Isolation and Quantification
Single fiber myonuclear counts were obtained according to previously published methods.6,10,134 In brief, 48-h PFA-fixed muscles were manually dissected into bundles then incubated in 40% NaOH for 2 h at room temperature while rotating. Fibers were then washed liberally with PBS atop a 40 µm strainer, gently moved into tubes using tweezers, triturated lightly until completely separated using low retention pipette tips, then dispersed on glass slides. Fibers were mounted with a glass coverslip using VectaShield with DAPI (Vector). For analysis, linear sections (∼300–500 µm) from 30 randomly selected individual muscle fibers per muscle were imaged at ×20 magnification using the z-stack function in Zen. Myonuclei were manually counted by a blinded trained technician, and fiber length was determined using a tool in Zen. An aggregate mean value from the 30 fibers analyzed from each mouse was used for statistics. To visualize tdT-expressing satellite cells on longitudinal muscle fibers (see Figure 1A), PFA-fixed muscle fibers were manually dissected in PBS using a dissecting microscope. Isolated muscle fibers were plated on glass slides and mounted in a mixture of phalloidin in PBS (1:100) and VectaShield with DAPI. Satellite cells were visualized at ×40 magnification using a Zeiss LSM 880 with Airyscan confocal microscope.
Primary MPC Isolation and Maintenance
To obtain MPCs for in vitro experiments, the detailed FACS protocol from the Rando Laboratory was employed.33 In brief, minced muscle was sequentially digested with collagenase, then collagenase and dispase at 37°C while rocking. Suspensions were aspirated with a 20 gauge needle, strained through 40 µm filters, pelleted at 500 × g for 5 min, then resuspended. The single-cell suspensions were incubated with antibodies against Vcam, Cd31, Cd45, and Sca1 at 4°C; a biotinylated secondary antibody was used for Vcam. Cells were pelleted at 500 × g, resuspended, and sorted using an iCyt FACS machine (Sony Biotechnology, Champaign, IL, USA). Appropriate isotype-specific controls were used to validate antibody specificity, and dead cells were gated out using forward/side scatter profiles and/or propidium iodide (non-tdT experiments). Satellite cells were identified as Vcam+/Cd31−/Cd45−/Sca1−. MPCs were maintained in 5% O2 and expanded in growth media made of Ham’s F10 + 15% normal horse serum + 1% penicillin/streptomycin supplemented with basic fibroblast growth factor (5 ng/mL, Millipore) on plates coated with ECM gel diluted 1:100 (Sigma-Aldrich). Cells were differentiated on ECM-coated plates (35 mm or 8-well plastic chamber slides) in differentiation media (DM) made of high-glucose DMEM + 10% normal horse serum + 1% penicillin/streptomycin. Cells from in vivo vehicle or tamoxifen-treated mice had a minimum 2-week washout before being isolated. See Supplemental 5 for antibody information.
EV Isolation and Characterization
To obtain EVs from MPCs, 10 cm plates were expanded to high confluence. MPC conditioned growth media was collected after 24–48 h and stored at −80°C (note: EVs from fresh media were used for imaging analysis, described below). At the time of experimentation, EVs were isolated using ExoQuick-TC according to System Bioscience’s protocol and as previously described by our laboratory.7 For further validation, EVs were isolated via UC and DGUC (see Figure S1). Briefly, for ExoQuick-TC isolation of EVs, MPC-conditioned media was cleared of cellular debris via centrifugation for 15 min at 3000 × g, moved to a new sterile vessel, incubated with ExoQuick-TC for a minimum of 12 h at 4°C, then centrifuged for 30 min at 1500 × g. The EV pellet was then resuspended in 300 µL of PBS or DM (depending on downstream application). Particle size and concentration were carried out on a ZetaView nanoparticle tracking analyzer (NTA, Particle Metrix, Meerbusch, Germany) after filtration through a 0.22 µm filter (1:3000 in PBS, sensitivity 76, shutter 350). For UC isolation of EVs, conditioned media was cleared of debris as described above, diluted in PBS, then spun at 118 000 × g for 6 h. The resulting EV pellet was reconstituted in 100 µL of PBS and used for NTA, RNA, protein, and high-resolution imaging analysis. For additional DGUC EV analyses, we used an OptiPrep gradient as previously described by Jimenez et al.135 Briefly, ExoQuick-TC-collected EVs were layered on top of a discontinuous density gradient of 5%, 10%, 20%, and 40% iodixanol. The iodixanol dilutions were prepared by diluting OptiPrep (60% aqueous iodixanol) with 0.25 M sucrose/10 mM Tris, pH 7.5. After an 18-h centrifugation at 100 000 × g, 12 density gradient fractions were collected, diluted in PBS, and centrifuged at 100 000 × g for 6 h. NTA analysis and protein quantification validated the fraction enriched with EVs, and this fraction was then used for protein and RNA analysis.
FCS and High-Magnification Fluorescent Microscopy of Individual Vesicles
To fluorescently label vesicles, 5 µM DiI was added to the vesicle suspension and incubated at 37°C for 30 min. Vesicles were then purified using a 0.45 µm sterile syringe filter. For FCS measurements, 40 µL of freshly prepared fluorescently-labeled sample was placed onto a coverslip mounted on an Olympus IX83 microscope (Olympus, Shinjuku, Tokyo, Japan) equipped with a PicoQuant PicoHarp 300 Time-Correlated Single Photon Counting system (PicoQuant, Berlin, Germany). A ×60 water immersion objective was used to focus a 532 nm CW laser into the sample solution. Two avalanche photodiodes were used for photon detection. All FCS results are an average of at least three measurements. To diminish the background signal from immobilized molecules, we performed all measurements 30 µm above the glass surface in the sample solution. Data analysis was performed using SymPhoTime 64 software (PicoQuant). An autocorrelation of the fluorescence time trace was then calculated and fit to extract the diffusion time.136,137 Measurements of vesicles labeled with DiI and of free recombinant tdT protein were used to establish the expected diffusion time of tdT in vesicles as compared to freely diffusing protein. To visualize tdT-containing vesicles in solution, a 40 µL sample of the vesicle suspension was placed on a glass coverslip. The fluorescence was imaged on an inverted microscope (Olympus IX81) using a 60X 1.49 NA oil immersion objective, an electron-multiplying charge couple device, and 561 nm laser excitation. Sequential frames (10 ms) were collected for a duration of 1 min. Images were processed and analyzed using ImageJ.
Western Blot Analysis of EVs
Protein was isolated from EVs using RIPA buffer supplemented with protease and phosphatase inhibitors (Thermo Fisher). Protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Bio-Rad, Hercules, CA, USA). For quantification of protein abundance, 27 µg of protein was loaded, separated on a discontinuous acrylamide gel (4%–15%, Bio-Rad), and transferred to a nitrocellulose membrane. Total protein transfer was visualized using Revert 700 total protein stain (LI-COR, Lincoln, NE, USA). The membrane was blocked using 5% nonfat dry milk in TBS + 0.1% Tween (TBS-T) for 1 h at room temperature, followed by overnight incubation at 4°C with primary antibodies (1:1000 dilution in 5% nonfat dry milk). After primary antibody incubation, membranes were washed with TBS-T and incubated with appropriate goat anti-rabbit secondary antibodies at 1:10 000 dilution for 1 h at room temperature in 5% nonfat dry milk. Blots were developed using enhanced chemiluminescence (Clarity Western ECL substrate, Bio-Rad), and imaged on a ChemiDoc Imaging System (Bio-Rad).
Immunocytochemistry
To determine N-WASp+/tdT− and N-WASp−/tdT+ MPC proliferation rate, 1 × 104 cells per well were plated on eight-well chamber slides. MPC growth media was spiked with five µM of 5-ethynyl-2′-deoxyuridine (EdU) for 24 h. Cells were fixed in warmed 4% PFA for 15 min, then EdU was visualized using a detection cocktail described by Kirby et al.9 Briefly, fixed cells were permeabilized in 0.5% Triton X-100 (Sigma-Aldrich), washed with 3% bovine serum albumin (BSA) in PBS, then incubated in the EdU detection cocktail containing tris base (100 mM), biotin azide (100 µM), copper sulfate (4 mM), ascorbic acid (100 mM), and deionized H2O. Following PBS washes, cells were incubated in SA-HRP followed by fluorescent TSA. Slides were mounted using VectaShield with DAPI, images were captured at ×20 magnification, and EdU+/− cells were quantified relative to DAPI. To determine fusion dynamics, MyHC area and fusion index were quantified. On eight-well chamber slides, 5 × 104 cells were differentiated for 30 h, then fixed in warm 4% PFA. To visualize MyHC, myotubes were washed with 0.2% Triton X-100, blocked with 1% BSA for an hour, and incubated with a pan-myosin primary antibody in PBS (1:100). Myotubes were then incubated with a biotinylated secondary antibody, followed by a fluorophore-conjugated streptavidin. After being mounted with DAPI, 2–3 images were captured at ×20 magnification, and MyHC area normalized to DAPI content was quantified using a quantification tool in Zen. Fusion index (myotubes containing ≥5 myonuclei) was quantified by a trained technician. To determine whether tdT message could be transferred to myotubes, 2 × 104 C57BL/6J MPCs were differentiated in eight-well chamber slides until large multinucleated myotubes were observed (30 h), then myotubes were incubated with EVs from N-WASp+/tdT− or N-WASp−/tdT+ MPCs for 20 h. Myotubes were then fixed with warm 4% PFA, incubated with phalloidin in PBS (1:100) for 2 h, then mounted with DAPI to visualize tdT fluorescence. Cell culture experiments were conducted at or before passage six. Antibody information is found in Supplemental 5.
Gene Expression Experiments Involving MPCs, Myotubes, EVs, and Muscle Tissue
For cellular and tissue RNA extraction, cells were lysed in TRIzol reagent, and RNA was extracted using the Zymo MiniPrep kit (Zymo Research, Irvine, CA, USA) with slight modification; the aqueous phase from TRIzol combined with 3-bromo-chloropropane following phase separation was added directly to the spin column, and not the TRIzol itself (as recommended by the manufacturer). RNA concentration and quality were assessed via NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and Bioanalyzer using Nano chips (Agilent, Santa Clara, CA, USA). For qPCR on cells, 2 ng of complementary DNA (cDNA) was used per reaction, and reactions were performed using SYBR chemistry on a QuantStudio 3 (Thermo Fisher Scientific) in duplicate with Gapdh as the housekeeping gene. Reaction efficiency was evaluated using LinReg software,138 and data are presented as relative gene expression.
To model in vitro what satellite cells may communicate to muscle fibers, 3 × 105 C57BL/6J MPCs were differentiated on 35 mm plates until large multi-nucleated myotubes were observed (32 h), and ExoQuick-isolated EVs from C57BL/6J MPCs were then incubated with myotubes in DM for 12 and 24 h; ExoQuick-isolated particles from non-MPC conditioned GM served as the control treatment. For RNA sequencing, myotube RNA was prepared as a 250–300 bp nonstrand-specific library using the NEBNext Ultra RNA Library Prep Kit (NEB) followed by paired-end sequencing using an Illumina NovaSeq 6000 s4 platform. Mapping to the mouse mm10 transcriptome was performed using HISAT2, data were normalized according to Novogene’s standard practices,139 quality was assessed and reads were filtered by removing reads that contained adapters, reads that contained N > 0.1% (N represents base that could not be determined), and low-quality reads (quality value of over 50% of bases of the read ≤20), and differential analysis was performed using DESeq2 v2 with a model based on the negative binomial distribution. To evaluate gene expression in EVs via qPCR, total RNA was extracted using the ExoRNeasy Serum/Plasma kit (Qiagen). RNA concentration was evaluated using a bioanalyzer with small or pico chips (Agilent), cDNA was generated using the Vilo IV reverse transcription kit (Thermo Fisher Scientific), and qPCR was used to detect tdT message (normalized to 18s, which was present in EVs according to Agilent analysis as well as a previous investigation of myogenic cell EVs37). Reactions were performed using SYBR chemistry on a QuantStudio 3 (Thermo Fisher Scientific) in duplicate with 0.2 ng of cDNA, and presented as relative gene expression (2−Δ00394;Ct). Primer sequences for qPCR are found in Supplemental 5. From homogenized Pax7-DTA muscle tissue, microarray analysis was carried out as previously described by our laboratory140; the microarray results from the Pax7-DTA experiments are part of a larger cohort of mice with additional time points that has not been published yet, but we present the sham and 7-day time points here. Briefly, RNA quality, concentration, and integrity were determined by spectrophotometer and bioanalyzer, and 250 ng of RNA per sample was submitted to the University of Kentucky Genomics core for analysis using an Affymetrix GCS3000 7G scanner and mouse gene 2.0 ST array chips (Thermo Fisher Scientific). Similarly, for microarray analysis of MPC EV miRNA (isolated from multiple 10 cm cell culture plates of proliferating C57BL/6J primary MPCs), 70 ng of pooled EV RNA was submitted to the University of Kentucky microarray core. Using the FlashTag Biotin HSR kit (Thermo Fisher Scientific) and following a prolonged ∼40-h hybridization step, miRNA was analyzed using an Affymetrix GeneChip miRNA 4.0 array. Microarray data were obtained using GeneChip Command Console software, analyzed with Affymetrix Expression Console software (Thermo Fisher Scientific), and normalized using the robust multichip analysis method.
Pathway and in silico Analyses
Gene expression pathway analysis was carried out using ConsensusPathDB with the overrepresentation feature for mouse genes in KEGG, Reactome, and WikiPathway databases, and a minimum input list overlap of “2” and P-value cutoff of P = 0.01.141 DIANA miRPath was used to determine which pathways were targeted by MPC EV miRNAs,142 and specific in silico miRNA-mRNA 3′-UTR interactions were predicted using miRWalk.143
Statistical Analyses
For statistical comparison of two groups, unpaired two-tailed Student’s t-tests were employed. For comparisons of two groups with two conditions, two-way analysis of variances were employed and, when an interaction effect was identified, a Tukey’s HSD post hoc test was employed. All data were normal according to the Kolmogorov–Smirnov test. Differences among groups were considered significant when the probability value, P < 0.05. Outliers greater than 2 SD from the mean were excluded from analysis. For RNA sequencing, false discovery rate was controlled for using the Benjamini and Hochberg’s approach, with adjusted P < 0.05 and a log2 fold change >1.
Supplementary Material
Acknowledgments
The authors wish to thank Jennifer Strange of the University of Kentucky Flow Cytometry Core and Dr. Yuan Wen for his technical assistance. The graphical abstract was created using BioRender.
Conflict of interest statement
The authors have no conflicts to declare.
Funding
This work was supported by funding from the NIH National Institutes of Arthritis and Musculoskeletal and Skin Diseases (AR060701 to C.A.P. and J.J.M. and AR071753 to K.A.M.), National Institute on Aging (AG049086 to C.A.P. and J.J.M. and AG063994 to K.A.M.), and the National Institute of Diabetes and Digestive and Kidney Diseases (DK119619 to C.A.P. and J.J.M.).
Authors’ contributions
K.A.M., C.A.P., and J.J.M. designed experiments. K.A.M., I.J.V., D.W.V.P., S.E.C., C.M.D., V.C.F., X.F., K.K., C.I.R., and C.S.F. performed experiments. K.A.M., I.J.V., D.W.V.P., S.E.C., X.F., C.S.F., and C.I.R. analyzed data. K.A.M. wrote the manuscript and prepared the figures. C.A.P. and J.J.M provided funding support and supervised the study. C.A.P. and J.J.M. assisted with data interpretation and manuscript writing. All authors edited and approved the final manuscript.
References
- 1. Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA.. Pax7 is required for the specification of myogenic satellite cells. Cell 2000;102(6):777–786. [DOI] [PubMed] [Google Scholar]
- 2. White RB, Biérinx A-S, Gnocchi VF, Zammit PS.. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol 2010;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bachman JF, Klose A, Liu W. et al. Prepubertal skeletal muscle growth requires Pax7-expressing satellite cell-derived myonuclear contribution. Development 2018;145(20):dev167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egner IM, Bruusgaard JC, Gundersen K.. Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development 2016;143:2898–2906. [DOI] [PubMed] [Google Scholar]
- 5. McCarthy JJ, Dupont-Versteegden EE, Fry CS, Murach KA, Peterson CA.. Methodological issues limit interpretation of negative effects of satellite cell depletion on adult muscle hypertrophy. Development 2017;144:1363–1365. [DOI] [PubMed] [Google Scholar]
- 6. Murach KA, White SH, Wen Y. et al. Differential requirement for satellite cells during overload-induced muscle hypertrophy in growing versus mature mice. Skelet Muscle 2017;7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA.. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 2017;20:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fry CS, Lee JD, Jackson JR. et al. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J 2014;28:1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirby TJ, Patel RM, McClintock TS, Dupont-Versteegden EE, Peterson CA, McCarthy JJ.. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol Biol Cell 2016;27:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCarthy JJ, Mula J, Miyazaki M. et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 2011;138:3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Englund DA, Murach KA, Dungan CM. et al. Depletion of resident muscle stem cells negatively impacts running volume, physical function and muscle hypertrophy in response to lifelong physical activity. Am J Physiol Cell Physiol 2020;318(6):C1178–C1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruusgaard JC, Johansen I, Egner I, Rana Z, Gundersen K.. Myonuclei acquired by overload exercise precede hypertrophy and are not lost on detraining. Proc Natl Acad Sci USA 2010;107:15111–15116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goh Q, Song T, Petrany MJ. et al. Myonuclear accretion is a determinant of exercise-induced remodeling in skeletal muscle. eLife 2019;8:e44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conceicao MS, Vechin FC, Lixandrao M. et al. Muscle fiber hypertrophy and myonuclei addition: a systematic review and meta-analysis. Med Sci Sports Exerc 2018;50(7):1385–1392. [DOI] [PubMed] [Google Scholar]
- 15. Petrella JK, Kim JS, Cross JM, Kosek DJ, Bamman MM.. Efficacy of myonuclear addition may explain differential myofiber growth among resistance-trained young and older men and women. Am J Physiol Endocrinol Metab 2006;291:E937–E946. [DOI] [PubMed] [Google Scholar]
- 16. Petrella JK, Kim JS, Mayhew DL, Cross JM, Bamman MM.. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: a cluster analysis. J Appl Physiol 2008;104:1736–1742. [DOI] [PubMed] [Google Scholar]
- 17. Fry CS, Noehren B, Mula J. et al. Fibre type-specific satellite cell response to aerobic training in sedentary adults. J Physiol 2014;592:2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murach KA, Englund DA, Dupont-Versteegden EE, McCarthy JJ, Peterson CA.. Myonuclear domain flexibility challenges rigid assumptions on satellite cell contribution to skeletal muscle fiber hypertrophy. Front Physiol 2018;9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murach KA, Fry CS, Kirby TJ. et al. Starring or supporting role? Satellite cells and skeletal muscle fiber size regulation. Physiology 2018;33:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G.. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 2011;138:3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patsalos A, Pap A, Varga T. et al. In situ macrophage phenotypic transition is affected by altered cellular composition prior to acute sterile muscle injury. J Physiol 2017;595(17):5815–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verma M, Asakura Y, Murakonda BSR. et al. Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling. Cell Stem Cell 2018;23:530–543.e539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cossetti C, Lugini L, Astrologo L, Saggio I, Fais S, Spadafora C.. Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes. PLoS One 2014;9:e101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ortin-Martinez A, Tsai ELS, Nickerson PE. et al. A reinterpretation of cell transplantation: GFP transfer from donor to host photoreceptors. Stem Cells 2017;35:932–939. [DOI] [PubMed] [Google Scholar]
- 25. Pearson R, Gonzalez-Cordero A, West E. et al. Donor and host photoreceptors engage in material transfer following transplantation of post-mitotic photoreceptor precursors. Nat Commun 2016;7:13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santos-Ferreira T, Llonch S, Borsch O, Postel K, Haas J, Ader M.. Retinal transplantation of photoreceptors results in donor–host cytoplasmic exchange. Nat Commun 2016;7:13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H-X, Qiu Y-R, Zhong X-P.. Intercellular protein transfer from thymocytes to thymic epithelial cells. PLoS One 2016;11(3): e0152641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flaherty SE 3rd, Grijalva A, Xu X, Ables E, Nomani A, Ferrante AW Jr.. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 2019;363:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keefe AC, Lawson JA, Flygare SD. et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat Commun 2015;6:7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matthews BG, Torreggiani E, Roeder E, Matic I, Grcevic D, Kalajzic I.. Osteogenic potential of alpha smooth muscle actin expressing muscle resident progenitor cells. Bone 2016;84:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pawlikowski B, Pulliam C, Betta ND, Kardon G, Olwin BB.. Pervasive satellite cell contribution to uninjured adult muscle fibers. Skelet Muscle 2015;5:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Summers MA, Mikulec K, Peacock L, Little DG, Schindeler A.. Limitations of the Pax7-creER(T2) transgene for driving deletion of Nf1 in adult mouse muscle. Int J Dev Biol 2017;61:531–536. [DOI] [PubMed] [Google Scholar]
- 33. Liu L, Cheung TH, Charville GW, Rando TA.. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc 2015;10:1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forterre A, Jalabert A, Berger E. et al. Proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: a new paradigm for myoblast-myotube cross talk? PLoS One 2014;9:e84153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanada M, Bachmann MH, Hardy JW. et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc Natl Acad Sci USA 2015;112:E1433–E1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gruenbaum-Cohen Y, Harel I, Umansky KB. et al. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci USA 2012;109:11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Bihan M-C, Bigot A, Jensen SS. et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J Proteomics 2012;77:344–356. [DOI] [PubMed] [Google Scholar]
- 38. Vechetti IJ Jr, Valentino T, Mobley CB, McCarthy JJ.. The role of extracellular vesicles in skeletal muscle and systematic adaption to exercise. J Physiol 2020. doi: 10.1113/JP278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO.. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654. [DOI] [PubMed] [Google Scholar]
- 40. Deshmukh AS, Cox J, Jensen LJ, Meissner F, Mann M.. Secretome analysis of lipid-induced insulin resistance in skeletal muscle cells by a combined experimental and bioinformatics workflow. J Proteome Res 2015;14:4885–4895. [DOI] [PubMed] [Google Scholar]
- 41. Carmeli E, Moas M, Reznick AZ, Coleman R.. Matrix metalloproteinases and skeletal muscle: a brief review. Muscle Nerve 2004;29:191–197. [DOI] [PubMed] [Google Scholar]
- 42. Player D, Martin N, Passey S, Sharples A, Mudera V, Lewis M.. Acute mechanical overload increases IGF-I and MMP-9 mRNA in 3D tissue-engineered skeletal muscle. Biotechnol Lett 2014;36:1113–1124. [DOI] [PubMed] [Google Scholar]
- 43. Li H, Mittal A, Paul PK. et al. Tumor necrosis factor-related weak inducer of apoptosis augments matrix metalloproteinase 9 (MMP-9) production in skeletal muscle through the activation of nuclear factor-κB-inducing kinase and p38 mitogen-activated protein kinase. J Biol Chem 2009;284:4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chenette DM, Cadwallader AB, Antwine TL. et al. Targeted mRNA decay by RNA binding protein AUF1 regulates adult muscle stem cell fate, promoting skeletal muscle integrity. Cell Rep 2016;16:1379–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yeghiazaryan M, Zybura-Broda K, Cabaj A. et al. Fine-structural distribution of MMP-2 and MMP-9 activities in the rat skeletal muscle upon training: a study by high-resolution in situ zymography. Histochem Cell Biol 2012;138:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimowska M, Brzoska E, Swierczynska M, Streminska W, Moraczewski J.. Distinct patterns of MMP-9 and MMP-2 activity in slow and fast twitch skeletal muscle regeneration in vivo. Int J Dev Biol 2008;52:307–314. [DOI] [PubMed] [Google Scholar]
- 47. Cai W, Zhang J, Yang J. et al. MicroRNA-24 attenuates vascular remodeling in diabetic rats through PI3K/Akt signaling pathway. Nutr Metab Cardiovas Dis 2019;29:621–632. [DOI] [PubMed] [Google Scholar]
- 48. Liu Z, Liu Z, Zhang Y, Li Y, Liu B, Zhang K.. miR-24 represses metastasis of human osteosarcoma cells by targeting Ack1 via AKT/MMPs pathway. Biochem Biophys Res Commun 2017;486:211–217. [DOI] [PubMed] [Google Scholar]
- 49. Zhang S, Zhang C, Liu W. et al. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol 2015;47:1351–1360. [DOI] [PubMed] [Google Scholar]
- 50. Feng Q, Zhang H, Nie X, Li Y, Chen W-D, Wang Y-D.. miR-149* Suppresses liver cancer progression by down-regulating tumor necrosis factor receptor 1–associated death domain protein expression. Am J Pathol 2020;190:469–483. [DOI] [PubMed] [Google Scholar]
- 51. Hentati-Kallel M, Le Jan S, Bernard P, Antonicelli F, Trussardi-Régnier A.. Histone deacetylases meet microRNA-associated MMP-9 expression regulation in glucocorticoid-sensitive and-resistant cell lines. Int J Oncol 2017;50:717–726. [DOI] [PubMed] [Google Scholar]
- 52. Liu G, Yin L, Ouyang X, Zeng K, Xiao Y, Li Y.. M2 Macrophages promote HCC cells invasion and migration via miR-149-5p/MMP9 signaling. J Cancer 2020;11:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Palmieri D, Capponi S, Geroldi A, Mura M, Mandich P, Palombo D.. TNFα induces the expression of genes associated with endothelial dysfunction through p38MAPK-mediated down-regulation of miR-149. Biochem Biophys Res Commun 2014;443:246–251. [DOI] [PubMed] [Google Scholar]
- 54. Xu K, Liu X, Mao X. et al. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem 2015;35:499–515. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Q, Su J, Wang Z. et al. MicroRNA-149* suppresses hepatic inflammatory response through antagonizing STAT3 signaling pathway. Oncotarget 2017;8:65397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hu H, Xu H, Lu F. et al. Exosome-derived miR-486-5p regulates cell cycle, proliferation and metastasis in lung adenocarcinoma via targeting NEK2. Front Bioeng Biotechnol 2020;8:259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Zhang Y, Fu J, Zhang Z, Qin H.. miR-486-5p regulates the migration and invasion of colorectal cancer cells through targeting PIK3R1. Oncol Lett 2018;15:7243–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ventayol M, Viñas JL, Sola A. et al. miRNA let-7e targeting MMP9 is involved in adipose-derived stem cell differentiation toward epithelia. Cell Death Dis 2014;5:e1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Y-Q, Zhang Z-H, Zheng Y-F, Feng S-Q.. Dysregulated miR-133a mediates loss of type II collagen by directly targeting matrix metalloproteinase 9 (MMP9) in human intervertebral disc degeneration. Spine 2016;41:E717–E724. [DOI] [PubMed] [Google Scholar]
- 60. Zhen Y, Liu J, Huang Y, Wang Y, Li W, Wu J.. miR-133b inhibits cell growth, migration, and invasion by targeting MMP9 in non-small cell lung cancer. Oncol Res 2017;25:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bronisz A, Godlewski J, Wallace J. et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 2012;14:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vlachos IS, Kostoulas N, Vergoulis T. et al. DIANA miRPath v. 2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 2012;40:W498–W504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Herwig R, Hardt C, Lienhard M, Kamburov A.. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc 2016;11:1889. [DOI] [PubMed] [Google Scholar]
- 64. Reibe S, Hjorth M, Febbraio MA, Whitham M.. GeneXX: an online tool for the exploration of transcript changes in skeletal muscle associated with exercise. Physiol Genomics 2018;50:376–384. [DOI] [PubMed] [Google Scholar]
- 65. Lundberg TR, Martinez-Aranda LM, Sanz G. et al. Early accentuated muscle hypertrophy is strongly associated with myonuclear accretion. J Appl Physiol 2020;319(1):R50–R58. [DOI] [PubMed] [Google Scholar]
- 66. Kirby TJ, McCarthy JJ, Peterson CA, Fry CS.. Synergist ablation as a rodent model to study satellite cell dynamics in adult skeletal muscle. Methods Mol Biol 2016;1460:43–52. [DOI] [PubMed] [Google Scholar]
- 67. Englund D, Peck B, Murach K. et al. Resident muscle stem cells are not required for testosterone-induced skeletal muscle hypertrophy. Am J Physiol 2019;317(4):C719–C724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee S-J, Huynh TV, Lee Y-S. et al. Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc Natl Acad Sci USA 2012;109:E2353–E2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang Q, McPherron AC.. Myostatin inhibition induces muscle fibre hypertrophy prior to satellite cell activation. J Physiol 2012;590:2151–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Amthor H, Otto A, Vulin A. et al. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc Natl Acad Sci USA 2009;106:7479–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blaauw B, Canato M, Agatea L. et al. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J 2009;23:3896–3905. [DOI] [PubMed] [Google Scholar]
- 72. Omairi S, Matsakas A, Degens H. et al. Enhanced exercise and regenerative capacity in a mouse model that violates size constraints of oxidative muscle fibres. eLife 2016;5:e16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Raffaello A, Milan G, Masiero E. et al. JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J Cell Biol 2010;191:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Welle S, Bhatt K, Pinkert CA, Tawil R, Thornton CA.. Muscle growth after postdevelopmental myostatin gene knockout. Am J Physiol Endocrinol Metab 2007;292:E985–E991. [DOI] [PubMed] [Google Scholar]
- 75. Fukuda S, Kaneshige A, Kaji T. et al. Sustained expression of HeyL is critical for the proliferation of muscle stem cells in overloaded muscle. eLife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goh Q, Millay DP.. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. eLife 2017;6:e20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hindi SM, Shin J, Gallot YS. et al. MyD88 promotes myoblast fusion in a cell-autonomous manner. Nat Commun 2017;8:1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moriya N, Miyazaki M.. Akt1 deficiency diminishes skeletal muscle hypertrophy by reducing satellite cell proliferation. Am J Physiol Regul Integr Comp Physiol 2018;314(5):R741–R751. [DOI] [PubMed] [Google Scholar]
- 79. Randrianarison-Huetz V, Papaefthymiou A, Herledan G. et al. Srf controls satellite cell fusion through the maintenance of actin architecture. J Cell Biol 2018;217:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Farup J, Madaro L, Puri P, Mikkelsen UR.. Interactions between muscle stem cells, mesenchymal-derived cells and immune cells in muscle homeostasis, regeneration and disease. Cell Death Dis 2015;6:e1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mitchell R, Mellows B, Sheard J. et al. Secretome of adipose-derived mesenchymal stem cells promotes skeletal muscle regeneration through synergistic action of extracellular vesicle cargo and soluble proteins. Stem Cell Res Ther 2019;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakamura Y, Miyaki S, Ishitobi H. et al. Mesenchymal‐stem‐cell‐derived exosomes accelerate skeletal muscle regeneration. FEBS Lett 2015;589:1257–1265. [DOI] [PubMed] [Google Scholar]
- 83. Sicco CL, Reverberi D, Balbi C. et al. Mesenchymal stem cell‐derived extracellular vesicles as mediators of anti‐inflammatory effects: endorsement of macrophage polarization. Stem Cells Trans Med 2017;6:1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wosczyna MN, Rando TA.. A muscle stem cell support group: coordinated cellular responses in muscle regeneration. Dev Cell 2018;46:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Altanerova U, Jakubechova J, Benejova K. et al. Prodrug suicide gene therapy for cancer targeted intracellular by mesenchymal stem cell exosomes. Int J Cancer 2019;144:897–908. [DOI] [PubMed] [Google Scholar]
- 86. Álvarez-Aznar A, Martinez-Corral I, Daubel N, Betsholtz C, Mäkinen T, Gängel K.. Tamoxifen-independent recombination of reporter genes limits lineage tracing and mosaic analysis using CreER T2 lines. Transgenic Res 2020;29:53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chappell‐Maor L, Kolesnikov M, Kim JS. et al. Comparative analysis of CreER transgenic mice for the study of brain macrophages: a case study. Eur J Immunol 2019;50(3):353–362. [DOI] [PubMed] [Google Scholar]
- 88. Stifter SA, Greter M.. STOP floxing around: specificity and leakiness of inducible Cre/loxP systems. Eur J Immunol 2020;50:338–341. [DOI] [PubMed] [Google Scholar]
- 89. Van Hove H, Antunes ARP, De Vlaminck K, Scheyltjens I, Van Ginderachter JA, Movahedi K.. Identifying the variables that drive tamoxifen‐independent CreERT2 recombination: implications for microglial fate mapping and gene deletions. Eur J Immunol 2020;50:459–463. [DOI] [PubMed] [Google Scholar]
- 90. Suhail Y, Lee J, Walker M. et al. Modeling intercellular transfer of biomolecules through tunneling nanotubes. Bull Math Biol 2013;75:1400–1416. [DOI] [PubMed] [Google Scholar]
- 91. Tavi P, Korhonen T, Hänninen SL. et al. Myogenic skeletal muscle satellite cells communicate by tunnelling nanotubes. J Cell Physiol 2010;223:376–383. [DOI] [PubMed] [Google Scholar]
- 92. Choi JS, Yoon HI, Lee KS. et al. Exosomes from differentiating human skeletal muscle cells trigger myogenesis of stem cells and provide biochemical cues for skeletal muscle regeneration. J Control Release 2016;222:107–115. [DOI] [PubMed] [Google Scholar]
- 93. Zimowska M, Olszynski KH, Swierczynska M, Streminska W, Ciemerych MA.. Decrease of MMP-9 activity improves soleus muscle regeneration. Tissue Eng Part A 2012;18:1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li H, Mittal A, Makonchuk DY, Bhatnagar S, Kumar A.. Matrix metalloproteinase-9 inhibition ameliorates pathogenesis and improves skeletal muscle regeneration in muscular dystrophy. Hum Mol Genet 2009;18:2584–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shiba N, Miyazaki D, Yoshizawa T. et al. Differential roles of MMP-9 in early and late stages of dystrophic muscles in a mouse model of Duchenne muscular dystrophy. Biochim Biophys Acta 2015;1852:2170–2182. [DOI] [PubMed] [Google Scholar]
- 96. Mehan RS, Greybeck BJ, Emmons K, Byrnes WC, Allen DL.. Matrix metalloproteinase-9 deficiency results in decreased fiber cross-sectional area and alters fiber type distribution in mouse hindlimb skeletal muscle. Cells Tissues Organs 2011;194:510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dahiya S, Bhatnagar S, Hindi SM. et al. Elevated levels of active matrix metalloproteinase-9 cause hypertrophy in skeletal muscle of normal and dystrophin-deficient mdx mice. Hum Mol Genet 2011;20:4345–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Deryugina EI, Quigley JP.. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: contrasting, overlapping and compensatory functions. Biochim Biophys Acta 2010;1803:103–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Weng C-H, Chung F-P, Chen Y-C. et al. Pleiotropic effects of myocardial MMP-9 inhibition to prevent ventricular arrhythmia. Sci Rep 2016;6:38894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. von Walden F, Rea M, Mobley CB. et al. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics 2020; doi:10.1080/15592294.2020.1755581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Accornero F, Kanisicak O, Tjondrokoesoemo A, Attia AC, McNally EM, Molkentin JD.. Myofiber-specific inhibition of TGFβ signaling protects skeletal muscle from injury and dystrophic disease in mice. Hum Mol Genet 2014;23:6903–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Petrosino JM, Leask A, Accornero F.. Genetic manipulation of CCN2/CTGF unveils cell-specific ECM-remodeling effects in injured skeletal muscle. FASEB J 2019;33:2047–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pillon NJ, Gabriel BM, Dollet L. et al. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat Commun 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF.. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol 2007;21:2152–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kanzleiter T, Preston E, Wilks D. et al. Overexpression of the orphan receptor Nur77 alters glucose metabolism in rat muscle cells and rat muscle in vivo. Diabetologia 2010;53:1174–1183. [DOI] [PubMed] [Google Scholar]
- 106. Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GE.. Nur77 regulates lipolysis in skeletal muscle cells J Biol Chem 2005;280:12573–12584. [DOI] [PubMed] [Google Scholar]
- 107. Seaborne RA, Strauss J, Cocks M. et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep 2018;8:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Cortez-Toledo O, Schnair C, Sangngern P, Metzger D, Chao LC.. Nur77 deletion impairs muscle growth during developmental myogenesis and muscle regeneration in mice. PLoS One 2017;12(2):e0171268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tontonoz P, Cortez-Toledo O, Wroblewski K. et al. The orphan nuclear receptor Nur77 is a determinant of myofiber size and muscle mass in mice. Mol Cell Biol 2015;35:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Palumbo-Zerr K, Zerr P, Distler A. et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med 2015;21:150. [DOI] [PubMed] [Google Scholar]
- 111. Alexakis C, Partridge T, Bou-Gharios G.. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am J Physiol Cell Physiol 2007;293:C661–C669. [DOI] [PubMed] [Google Scholar]
- 112. El Fahime E, Torrente Y, Caron N, Bresolin M, Tremblay J.. In vivo migration of transplanted myoblasts requires matrix metalloproteinase activity. Exp Cell Res 2000;258:279–287. [DOI] [PubMed] [Google Scholar]
- 113. Guérin CW, Holland PC.. Synthesis and secretion of matrix‐degrading metalloproteases by human skeletal muscle satellite cells. Dev Dynam 1995;202:91–99. [DOI] [PubMed] [Google Scholar]
- 114. Tierney MT, Gromova A, Sesillo FB.. et al. Autonomous extracellular matrix remodeling controls a progressive adaptation in muscle stem cell regenerative capacity during development. Cell Rep 2016;14:1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Egner IM, Bruusgaard JC, Gundersen K.. An apparent lack of effect of satellite cell depletion on hypertrophy could be due to methodological limitations. Response to ‘Methodological issues limit interpretation of negative effects of satellite cell depletion on adult muscle hypertrophy’. Development 2017;144:1365–1367. [DOI] [PubMed] [Google Scholar]
- 116. Cai C, Masumiya H, Weisleder N. et al. MG53 nucleates assembly of cell membrane repair machinery. Nat Cell Biol 2009;11:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Demonbreun AR, Quattrocelli M, Barefield DY. et al. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J Cell Biol 2016;213:705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Han R, Campbell KP.. Dysferlin and muscle membrane repair. Curr Opin Cell Biol 2007;19:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lek A, Evesson FJ, Lemckert FA. et al. Calpains, cleaved mini-dysferlinC72, and L-type channels underpin calcium-dependent muscle membrane repair. J Neurosci 2013;33:5085–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Scheffer LL, Sreetama SC, Sharma N. et al. Mechanism of Ca 2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat Commun 2014;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Blaauw B. Platelet‐derived growth factor signaling and the role of cellular crosstalk in functional muscle growth. FEBS Lett 2017;591:690–692. [DOI] [PubMed] [Google Scholar]
- 122. Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R.. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep 2001;2:850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kawamura K, Takano K, Suetsugu S. et al. N-WASP and WAVE2 acting downstream of phosphatidylinositol 3-kinase are required for myogenic cell migration induced by hepatocyte growth factor. J Biol Chem 2004;279:54862–54871. [DOI] [PubMed] [Google Scholar]
- 124. Bellamy LM, Joanisse S, Grubb A. et al. The acute satellite cell response and skeletal muscle hypertrophy following resistance training. PLoS One 2014;9:e109739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kadi F, Thornell L-E.. Concomitant increases in myonuclear and satellite cell content in female trapezius muscle following strength training. Histochem Cell Biol 2000;113:99–103. [DOI] [PubMed] [Google Scholar]
- 126. Collins BC,, Arpke RW, Larson AA. et al. Estrogen regulates the satellite cell compartment in females. Cell Rep 2019;28:368–381.e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Madisen L, Zwingman TA, Sunkin SM. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nishijo K, Hosoyama T, Bjornson CR. et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J 2009;23:2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wu S, Wu Y, Capecchi MR.. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development 2006;133:581–590. [DOI] [PubMed] [Google Scholar]
- 130. Fry CS, Lee JD, Mula J. et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat Med 2015;21:76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Murach KA, Confides AL, Ho A. et al. Depletion of Pax7+ satellite cells does not affect diaphragm adaptations to running in young or aged mice. J Physiol 2017;595:6299–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schultz E, Chamberlain C, McCormick KM, Mozdziak PE.. Satellite cells express distinct patterns of myogenic proteins in immature skeletal muscle. Dev Dynam 2006;235:3230–3239. [DOI] [PubMed] [Google Scholar]
- 133. Wen Y, Murach KA, Vechetti IJ Jr. et al. MyoVision: software for automated high-content analysis of skeletal muscle immunohistochemistry. J Appl Physiol 2017;124(1):40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Brack AS, Bildsoe H, Hughes SM.. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age-related muscle atrophy. J Cell Sci 2005;118:4813–4821. [DOI] [PubMed] [Google Scholar]
- 135. Jimenez L, Yu H, McKenzie AJ. et al. Quantitative proteomic analysis of small and large extracellular vesicles (EVs) reveals enrichment of adhesion proteins in small EVs. J Proteome Res 2019;18:947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Moonschi FH, Effinger AK, Zhang X. et al. Cell-derived vesicles for single-molecule imaging of membrane proteins. Angew Chem 2015;54:481–484. [DOI] [PubMed] [Google Scholar]
- 137. Wyss R, Grasso L, Wolf C, Grosse W, Demurtas D, Vogel H.. Molecular and dimensional profiling of highly purified extracellular vesicles by fluorescence fluctuation spectroscopy. Anal Chem 2014;86:7229–7233. [DOI] [PubMed] [Google Scholar]
- 138. Ruijter J, Ramakers C, Hoogaars W. et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009;37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Dillies M-A, Rau A, Aubert J. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 2013;14:671–683. [DOI] [PubMed] [Google Scholar]
- 140. Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ.. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol 2013;115:1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kamburov A, Stelzl U, Lehrach H, Herwig R.. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2012;41:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Vlachos IS, Zagganas K, Paraskevopoulou MD. et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sticht C, De La Torre C, Parveen A, Gretz N.. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One 2018;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.