Summary
The mechanisms underlying ribonucleoprotein (RNP) granule assembly, including the basis for establishing and maintaining RNP granules with distinct composition, are unknown. One prominent type of RNP granule is the stress granule (SG), a dynamic and reversible cytoplasmic assembly formed in eukaryotic cells in response to stress. Here we show that SGs assemble through liquid-liquid phase separation (LLPS) arising from interactions distributed unevenly across a core protein-RNA interaction network. The central node of this network is G3BP1, which functions as a molecular switch that triggers RNA-dependent LLPS in response to a rise in intracellular free RNA concentrations. Moreover, we show that interplay between three distinct intrinsically disordered regions (IDRs) in G3BP1 regulates its intrinsic propensity for LLPS, and this is fine-tuned by phosphorylation within the IDRs. Further regulation of SG assembly arises through positive or negative cooperativity by extrinsic G3BP1-binding factors that strengthen or weaken, respectively, the core SG network.
Introduction
Biomolecular condensation is increasingly recognized as a vital strategy of cellular organization governing a variety of biological functions (Banani et al., 2017). One such function is the regulation of RNA metabolism, which occurs in condensates known as ribonucleoprotein (RNP) granules (Buchan, 2014; Gomes and Shorter, 2019). RNP granules are dynamic assemblies of RNA and protein that are abundant in both the nucleus (e.g., nucleoli, Cajal bodies, speckles, paraspeckles, promyelocytic leukemia bodies (Dundr, 2012)) and cytoplasm (e.g., P bodies, stress granules (SGs), RNA transport granules (Kiebler and Bassell, 2006)). Each type of RNP granule has a distinct identity defined by its constituents, material properties (e.g., surface tension, elasticity, viscosity), morphology, subcellular localization, and functions.
RNP granules assemble by liquid-liquid phase separation (LLPS), which occurs when protein-laden RNAs that are dispersed in the cytoplasm or nucleoplasm (soluble phase) coalesce into a concentrated state (condensed phase). In this condensed phase, the highly concentrated RNAs and RNA-binding proteins (RBPs) behave as a single organelle with liquid-like properties. The constituents of membraneless organelles remain in dynamic equilibrium with the surrounding nucleoplasm or cytoplasm and may form transiently or persist indefinitely. Some RBPs, particularly those harboring low complexity domains (LCDs), undergo concentration-dependent LLPS in vitro (Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015). However, how the LLPS of an individual RBP relates to assembly of an RNP granule consisting of hundreds of similar RBPs has remained unclear. Indeed, depletion or knockout of many individual RBPs that are capable of LLPS in vitro has no apparent impact on assembly of the RNP granule to which they belong (Buchan et al., 2013).
One prominent type of RNP granule is the SG, a cytoplasmic assembly formed in eukaryotic cells in response to a variety of stressors (Ivanov et al., 2019). SG assembly correlates with arrest of translation initiation, which is accompanied by polysome disassembly and a rise in the cytoplasmic concentration of uncoated mRNAs (Panas et al., 2016). The mechanism underlying SG assembly has remained ill-defined, although insights have emerged from proteomic analysis of SG cores (Jain et al., 2016) and spatial proteomics (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018), which have illuminated the protein content of SGs, along with similar studies that have characterized RNA content (Khong et al., 2017). These studies implicate hundreds of different proteins and thousands of different RNAs as constituents of SGs, but do not reveal the mechanism of SG assembly or the relative contribution of individual constituents. Whereas RNA self-assembly contributes to SG assembly (Van Treeck et al., 2018), evidence indicates an important role for RBPs as well. Several studies have implicated various RBPs as “essential” to SG assembly, including TIA1 (Gilks et al., 2004), HDAC6 (Kwon et al., 2007), G3BP1 and G3BP2 (Kedersha et al., 2016), PRRC2C (Youn et al., 2018), CSDE1 (Youn et al., 2018), and UBAP2L (Markmiller et al., 2018). Yet, insight into the relative importance of these proteins and how they relate to one another to govern assembly has been lacking. Indeed, a key gap in knowledge permitting us to understand the molecular basis of SG assembly and dynamics is lack of a conceptual framework built from fundamental biophysical principles.
Here we took an unbiased approach to obtain a holistic view of the determinants of SG formation and how these factors work together to assemble this RNP granule. This approach revealed that SG formation is encoded by a core protein-RNA interaction network. When the collective interactions of this network breach a threshold saturation concentration, LLPS ensues and seeds SG assembly. The driving forces underlying LLPS are distributed unevenly across this network, with some nodes more important than others. G3BP1 and 2 are the nodes of highest centrality within this network and by virtue of this position are essential for SG assembly under certain conditions. Biophysical studies revealed details of how G3BP1 functions as a molecular switch that triggers RNA-dependent LLPS. Importantly, interplay between three distinct intrinsically disordered regions (IDRs) in G3BP1 regulates its intrinsic propensity for LLPS, and this is fine-tuned by phosphorylation within the IDRs. Further regulation of SG assembly arises through positive or negative cooperativity by extrinsic G3BP1-binding factors that strengthen or weaken, respectively, the core SG network.
Results
G3BP Proteins Have the Highest Centrality within the Core SG Network
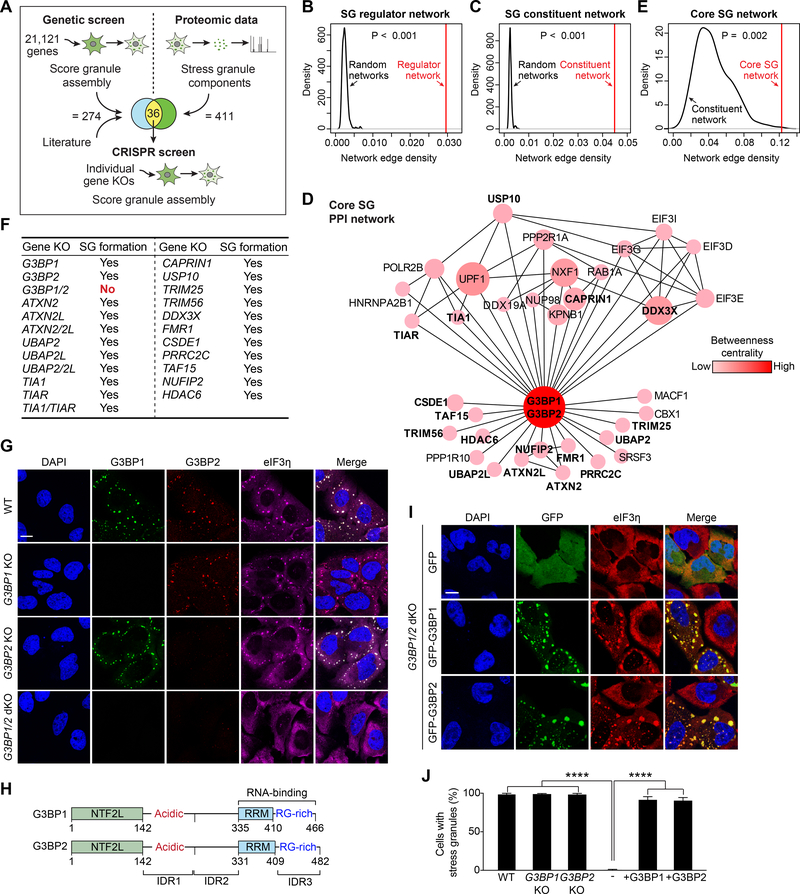
To identify genes essential for SG assembly, we conducted a genome-wide RNAi screen in which we targeted 21,121 genes with pools of four different siRNAs per gene (84,484 total siRNAs) in a U2OS cell line expressing a G3BP1-GFP reporter (Figures 1A, S1A–E). After confirmation in quadruplicate, we identified 210 genes whose depletion limited SG assembly in response to an exogenous stressor (Table S1). Gene ontological analysis revealed that this group was enriched in genes that regulate RNA metabolism and nucleocytoplasmic transport (Figure S1F). Of the 210 genes identified, 81 had been implicated in SG assembly in response to arsenite by a smaller-scale RNAi screen targeting 7,317 genes (Ohn et al., 2008). This prior screen also identified 51 genes that regulate arsenite-dependent SG assembly that were not identified in our screen (Figure S1G, Table S1). Thirteen genes implicated in SG assembly in previous studies were not identified in either screen; thus, at least 274 genes are implicated in regulating the assembly of SGs in response to a variety of stressors (Figure 1A). We used these 274 genes to construct an interaction network of genes that regulate SG assembly. This “SG regulator network” was more densely connected (i.e., with higher edge density) than random networks (Figure 1B), suggesting that this network of genes operates within a common functional module (Jeong et al., 2001).
Figure 1. G3BP Is the Node of Highest Centrality within the Core Stress Granule (SG) Network.
(A) Approach used to identify the core SG network. A genome-wide siRNA screen was performed in heat-shocked U2OS cells expressing G3BP1-GFP. Results from this screen were combined with published results and integrated with proteomic datasets. Genes that overlapped between the genetic screen and the proteomic datasets were assessed individually by CRISPR-Cas9-based knockout (KO).
(B-C) Network analyses showing connectivity (network edge density) of the SG regulator network (B) and SG constituent network (C). P values were estimated by empirically calculating the probability of observing a denser network by randomly sampling 1,000 subnetworks of similar size from the entire interactome.
(D) The core SG network of 36 core genes identified using the approach illustrated in (A). The color density and size of each node is proportional to its betweenness centrality in the network. Genes tested by CRISPR-Cas9 in (F) are in bold.
(E) Network analysis of 36 core SG genes comparing connectivity with the SG constituent network. The P value was estimated similarly as in (B-C) but using the network of SG constituents as background.
(F) U2OS cell lines with CRISPR-Cas9-based single or double KOs. Cells were exposed to sodium arsenite (500 μM, 1 h) and SG assembly was assessed by staining with a panel of 17 SG markers.
(G) U2OS cells with KO of G3BP1, G3BP2, or both G3BP1 and G3BP2 were exposed to sodium arsenite (500 μM, 1 h) and stained for the SG marker eIF3η. Scale bar, 20 μm.
(H) Comparison of human G3BP1 and G3BP2 proteins. NTF2L, NTF2-like; IDR, intrinsically disordered region; RRM, RNA-recognition motif; RG-rich: arginine-glycine rich.
(I-J) G3BP1/2 dKO cells were transfected with GFP-G3BP1 or GFP-G3BP2, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Cells were imaged (I) and the percentage of cells positive for SGs was quantified (J). Error bars indicate SD. ****P < 0.0001 by one-way ANOVA, Tukey’s multiple comparisons test. Scale bar, 20 μm.
We next integrated this regulatory network with the 411 proteins previously defined as the SG proteome (Jain et al., 2016). This latter group formed a “SG constituent network” with higher edge density than random networks (Figure 1C), suggesting that this network of proteins also operates within a common functional module. The SG regulator network and SG constituent network had 36 proteins in common, which themselves constitute a network of very high edge density and topological centrality that we have termed the “core SG network” (Figure 1D–E). Interestingly, this network included all seven proteins that have been described as “essential” to SG assembly: TIA1 (Gilks et al., 2004), HDAC6 (Kwon et al., 2007), G3BP1 and G3BP2 (Kedersha et al., 2016), PRRC2C (Youn et al., 2018), CSDE1 (Youn et al., 2018), and UBAP2L (Markmiller et al., 2018). Also worth noting, this integration of genetic screening and SG proteomics revealed that individual depletion of most SG proteins had no appreciable impact on SG assembly, indicating that although they are constituents of the the SG they do not contribute significantly to its assembly.
Proteins within the Core SG Network Contribute to SG Assembly to Varying Degrees
To examine the importance of the 36 proteins in the core SG network, we used CRISPR/Cas9 to create 23 U2OS cell lines harboring knockout of one or both isoforms of these genes and assessed SG assembly in response to arsenite using 17 SG markers (16 protein markers and polyadenylated (polyA) RNA, Table S2, and Figure 1F). To our surprise, nearly all of these genes were not essential for SG assembly (Figure 1F). Indeed, even cells with double knockout of paralogous isoforms of supposed “essential” genes (e.g., UBAP2 and UBAP2L, TIA1 and TIAR) still formed SGs (Figures 1F, S1H). Nevertheless, knockout of genes in the core SG network were consistently associated with reduced and/or delayed SG assembly. In contrast, cells with double knockout of G3BP1 and G3BP2 (hereafter referred to as G3BP1/2 dKO cells) were devoid of SGs (Figure 1G, Table S2), as previously reported (Kedersha et al., 2016). Notably, G3BP1 and G3BP2 have the highest betweenness centrality within the core SG network (Figure 1D). Indeed, the impact of each gene deletion on SG assembly correlated with betweenness centrality within the core SG network (data not shown). Nevertheless, whereas G3BP1/2 dKO cells formed no SGs in response to arsenite, these cells did assemble SGs in response to heat shock or osmotic stress (Figure S1I). Taken together, these observations indicate that the factors driving SG assembly differ depending on the initiating stimulus, and suggest that the relative importance of these factors in the assembly process correlates with the degree of their centrality within a core SG network. These results also highlight G3BP as an important node within the network of interactions that underlie SG assembly and as an essential factor in the context of arsenite stress. However, the mechanism whereby G3BP1 and G3BP2 contribute to SG assembly is unknown, and elucidating this became the focus of our study.
G3BP1 and G3BP2 are paralogous proteins that share a common domain architecture, with 65% identity and 80% similarity in primary amino acid sequence (Figure 1H). Expression of GFP-G3BP1 or GFP-G3BP2 were each sufficient to restore SG assembly in response to arsenite in G3BP1/2 dKO cells based on 17 markers, indicating that these two proteins are redundant with respect to supporting SG assembly (Figure 1I–J, Table S2). These data illustrate the utility of G3BP1/2 dKO cells in assaying the ability of exogenous G3BP to restore SG assembly. Given the homology and functional redundancy between G3BP1 and G3BP2, we focused most of our subsequent studies on G3BP1.
G3BP1 Undergoes RNA-dependent Phase Separation
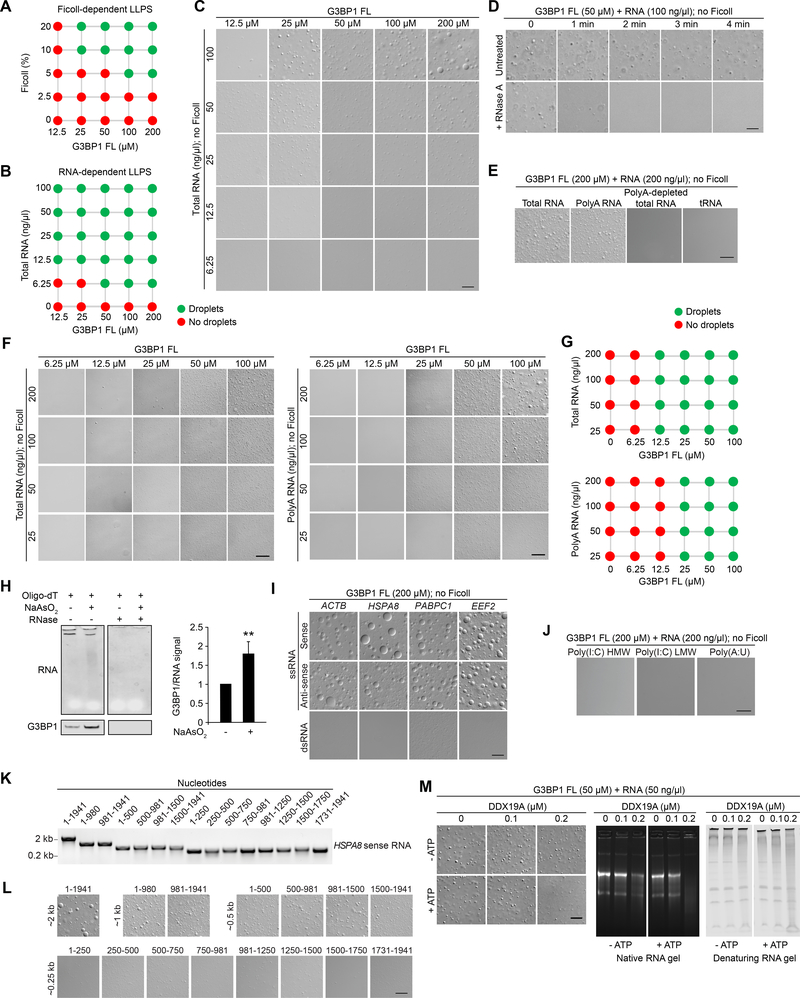
Many protein constituents of SGs harbor IDRs that facilitate LLPS in vitro, and these findings have led to the proposal that IDRs are important drivers of SG assembly in cells (Lin et al., 2015; Molliex et al., 2015; Nott et al., 2015; Patel et al., 2015). Given that G3BP1 has multiple IDRs, we examined whether this protein undergoes LLPS in vitro. To our surprise, at physiological salt concentration (150 mM NaCl) we found no evidence of LLPS across a range of G3BP1 concentrations (Figures 2A, S2A). G3BP1 did exhibit concentration-dependent LLPS upon addition of the molecular crowding agent Ficoll (Figures 2A, S2A), although the physiological relevance of this mono-component, crowder-dependent LLPS is questionable based on subsequent observations below.
Figure 2. G3BP1 Undergoes LLPS with RNAs that Have Specific Features.
(A) Summary of phase separation behaviors of G3BP1 FL with increasing concentration of Ficoll (crowding agent). Corresponding images are shown in Figure S2A.
(B-C) LLPS of purified recombinant G3BP1 in the absence of a crowding agent and increasing concentrations of total RNA purified from human cells. Scale bar, 20 μm.
(D) LLPS of purified recombinant G3BP1 with 100 ng/μl total RNA, with or without the addition of 1 μg/ml RNase A. Scale bar, 50 μm.
(E) LLPS of purified recombinant G3BP1 with the addition of RNA species as indicated. Scale bar, 20 μm.
(F) LLPS of purified recombinant G3BP1 with increasing concentrations of total (left panels) or polyA (right panels) RNA. Scale bars, 50 μm.
(G) Summary of phase separation behaviors of G3BP1 in (F).
(H) PolyA pulldown from U2OS cells exposed to sodium arsenite (500 μM, 1 h) revealing RNA (SYBR Gold staining) and G3BP1 pulldown (immunoblot). Representative image from 4 experiments is shown. The level of G3BP1 pulled down by polyA RNA was normalized to the total RNA level. Quantification was performed using 4 replicates. Error bar indicates SD. **P = 0.0038 by unpaired t-test.
(I) LLPS of purified recombinant G3BP1 in the absence of a crowding agent and the addition of mRNAs as indicated. Scale bar, 20 μm.
(J) LLPS of purified recombinant G3BP1 in the presence of long double strands of poly(I:C) or poly(A:U). Scale bar, 20 μm.
(K) In vitro-transcribed HSPA8 sense-strand RNA of different lengths was assessed by agarose gel and ethidium bromide staining.
(L) In vitro-transcribed HSPA8 sense-strand RNA was mixed with 200 μM recombinant G3BP1 to assess its ability to trigger LLPS in the absence of a crowding agent. Scale bar, 20 μm.
(M) DIC images and RNA gels showing effect of helicase pre-treatment of RNA on G3BP1-RNA LLPS. Scale bar, 10 μm.
All in vitro LLPS experiments were performed in 150 mM NaCl.
Since SG assembly correlates with a rise in the cytoplasmic concentration of uncoated mRNAs upon arrest of translation (Panas et al., 2016), we next characterized the influence of RNA on LLPS of G3BP1. We purified total RNA from U2OS cells and added increasing concentrations of this RNA to G3BP1 protein in vitro in the absence of Ficoll, and found that this RNA strongly promoted LLPS of G3BP1 (Figure 2B–C). The addition of RNase A rapidly reversed RNA-triggered LLPS in vitro (Figure 2D) and disrupted SGs in cells (Figure S2B, Video S1), confirming the role of RNA in enhancing LLPS of G3BP1.
Notably, whereas neither polyA-depleted cellular RNA nor tRNA triggered LLPS of G3BP1 in the absence of Ficoll (Figure 2E), the polyA fraction of total cellular RNA triggered LLPS of G3BP1 nearly as potently as total RNA (Figure 2F–G). To pursue this finding in cells, we performed polyA RNA pulldown from U2OS cells. This pulldown recovered low levels of endogenous G3BP1, which increased upon arsenite treatment, indicating increased G3BP1-mRNA interaction in response to stress (Figure 2H). Consistent results were obtained with G3BP2 (data not shown). The binding affinity of G3BP1 for mRNA was not significantly altered after arsenite treatment (Figure S2C), suggesting that the observed increase in G3BP1-mRNA interaction reflects increased availability of mRNA under stressed conditions. To further investigate the G3BP1-mRNA interaction, we used in vitro transcription to generate transcripts of four mRNAs found in SGs (ACTB, HSPA8, PABPC1, EEF2) (Khong et al., 2017). Both sense and antisense transcripts of these four mRNAs strongly triggered LLPS of G3BP1 (Figure 2I). However, annealing the sense and antisense transcripts to generate double-stranded RNA abolished the effect on LLPS with G3BP1 (Figure 2I). We also found that long (~1–10 kB) homopolymeric RNAs (polyA, polyC, polyU, and polyG) promoted LLPS by G3BP1 (Figure S2D–E). However, long double strands of poly(I:C) and poly(A:U) failed to undergo LLPS with G3BP1 (Figure 2J). From these results, we conclude that regions of single-strandedness in RNA, but not specific sequence motifs, are required to trigger LLPS with G3BP1, consistent with previous reports that G3BP1 binds RNA in cells without clear preference for a consensus sequence (Edupuganti et al., 2017; Martin et al., 2016b).
To determine whether mRNA length influences LLPS with G3BP1, we in vitro-transcribed full-length HSPA8 (~2 kb) or smaller fragments thereof, covering the length of this transcript. We found a strong correlation between mRNA length and the ability to drive LLPS of G3BP1, with a lower limit of ~250 nucleotides (Figure 2K–L). The lower limit mRNA length is about 10 times longer than the minimum RNA length necessary for binding G3BP1 (data not shown). These results may explain why RNA enrichment in SGs appears to be correlated with RNA length rather than sequence motifs (Khong et al., 2017; Van Treeck et al., 2018). Furthermore, pre-treatment of total RNA with the RNA helicase DDX19A (a constituent of the core SG network) and ATP strongly mitigated the ability of RNA to promote LLPS of G3BP1, presumably by limiting intermolecular RNA-RNA interactions (Figure 2M). In summary, these results indicate that beyond binding to G3BP1, the features of RNA that favor LLPS with G3BP1 include long length, single-strandedness, and the availability of RNA-RNA interactions. This latter observation is consistent with a recent report implicating RNA-RNA interactions in contributing to SG assembly in cells (Van Treeck and Parker, 2018).
In Vitro Phase Separation of G3BP1 with RNA Predicts SG Reconstitution in Cells
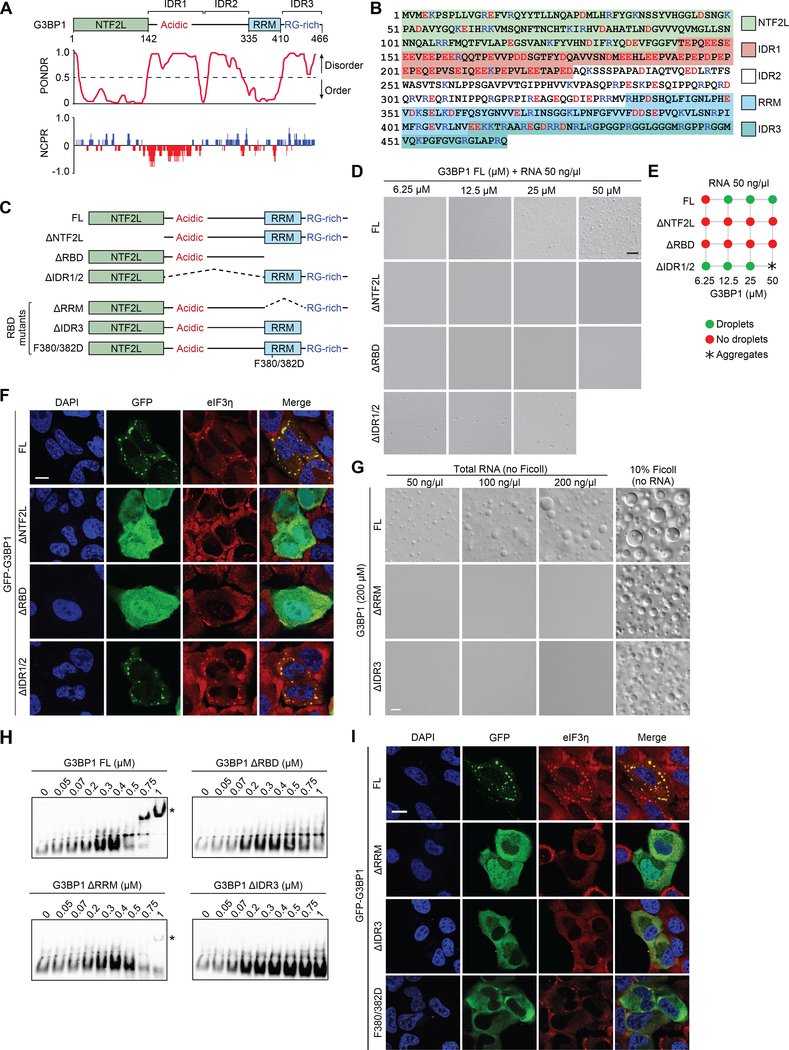
G3BP1 and G3BP2 have three major domains: an N-terminal NTF2-like (NTF2L) domain, a central segment predicted to be largely disordered, and a C-terminal RNA-binding domain (RBD) (Figures 1H, 3A, and S3A–B). The long, central IDR of G3BP1 has a non-uniform charge distribution separated by a region of predicted order and may be considered as two segments (Figure 3A–B): IDR1 (aa 143–225), which is highly negatively charged and highly acidic, and IDR2 (aa 226–334), which is slightly positively charged. The RBD is composed of a folded RNA-recognition motif (RRM) in tandem with an RG-rich IDR that is net positively charged and predicted to be disordered, which we designate IDR3 (Figure 3A–B).
Figure 3. G3BP1 Phase Separation Correlates with SG Reconstitution.
(A) G3BP1 domains aligned with results of PONDR (Predictor of Natural Disordered Regions) and NCPR (net charge per residue; 5-amino acid window) analyses.
(B) Human G3BP1 protein is shown with major domains marked by highlighted colors. Charged residues are shown in blue (basic; Arg and Lys) and red (acidic; Asp and Glu).
(C) Constructs used to investigate the function of individual domains of G3BP1.
(D) LLPS of purified recombinant G3BP1 in 150 mM NaCl and 50 ng/μl total RNA and the absence of a crowding agent. Scale bar, 10 μm.
(E) Summary of phase separation behaviors of G3BP1 shown in (D).
(F) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(G) LLPS of recombinant G3BP1 proteins in 150 mM NaCl, with or without crowding agent or total RNA from human cells. Scale bar, 10 μm.
(H) Electrophoretic mobility shift assay of purified G3BP1 proteins showing biotinylated RNA as detected by HRP-conjugated streptavidin. Asterisk indicates RNA-G3BP1 complex.
(I) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
See also Figure S3.
To understand the contribution of individual domains of G3BP1 to LLPS, we generated constructs in which NTF2L, the long, central IDR (IDR1/2), or the RBD were deleted (Figure 3C). Deletion of NTF2L or the RBD abolished LLPS of G3BP1 in the presence of RNA (Figure 3C–E). Interestingly, IDR1/2 was dispensable for LLPS. Indeed, deletion of IDR1/2 promoted LLPS at lower threshold concentrations, whereas at higher concentrations, G3BP1 ΔIDR1/2 had poor solubility and was prone to aggregation (Figure 3C–E). Similar results were obtained when RNA was excluded and LLPS was driven by the addition of Ficoll (Figure S3C–D). We next assessed the ability of G3BP1 deletion mutants to reconstitute SG assembly in G3BP1/2 dKO cells in response to arsenite. Consistent with their in vitro behavior, neither GFP-G3BP1 ΔNTF2L nor GFP-G3BP1 ΔRBD restored SG formation, whereas expression of G3BP1 ΔIDR1/2 did restore SG assembly (Figure 3F). Indeed, despite the frequent observation that IDRs in many RBPs mediate LLPS in vitro, we found that the long central IDR of G3BP1 is dispensable for both LLPS and SG assembly in cells.
Two RNA-binding Entities in G3BP1 Contribute Independently to SG Assembly
The RBD of G3BP1 comprises two distinct RNA-binding entities: an RRM, a stable, folded structure that NMR spectroscopy shows to be composed of amino acids 335–410 (Mathieu et al., in preparation), and IDR3 (amino acids 411–466). To investigate the contributions of these domains to LLPS of G3BP1, we generated G3BP1 constructs in which these domains were individually deleted (Figure 3C). RNA-dependent LLPS was severely impaired by either of these deletions, whereas Ficoll-dependent LLPS was not appreciably affected (Figure 3G). Importantly, this observation indicates mechanistic differences in LLPS in the presence of RNA vs. Ficoll.
We next assessed the RNA-binding capacity of these mutants using electrophoretic mobility shift assay (EMSA). Removal of both the RRM and IDR3 domains (ΔRBD) abolished RNA binding (Figure 3H). Removal of the RRM significantly reduced RNA binding, whereas removal of IDR3 abolished RNA binding (Figure 3H), consistent with impaired RNA-dependent LLPS of these mutants (Figure 3G). Interestingly, when introduced into G3BP1/2 dKO cells, G3BP1 ΔRRM and ΔIDR3 mutants exhibited severely impaired ability to support SG formation (Figures 3I, S3E–F). We next further examined the RRM domain by introducing two point mutations (F380/382D) that impair RNA binding by this domain. This mutant also failed to support SG assembly, confirming the importance of RNA binding by the RRM domain (Figure 3C, I). A prior study reported that the RRM domain is dispensable for SG assembly (Kedersha et al., 2016), although that observation was based on a different deletion (residues 340–415). To address this apparent discrepancy, we recreated the G3BP1 Δ340–415 mutant and quantified rescue of SG assembly in G3BP1/2 dKO cells. Despite good expression, we found no assembly of SGs in response to arsenite in ~70% of cells and few, small SGs in ~30% of cells (Figure S3G–H). Thus, using three different RRM mutants, we observed strong impairment of SG formation. Based on these observations we conclude that both the RRM domain and IDR3 contribute to RNA-dependent LLPS and SG assembly.
Multivalent RNA Interaction Promotes SG Assembly
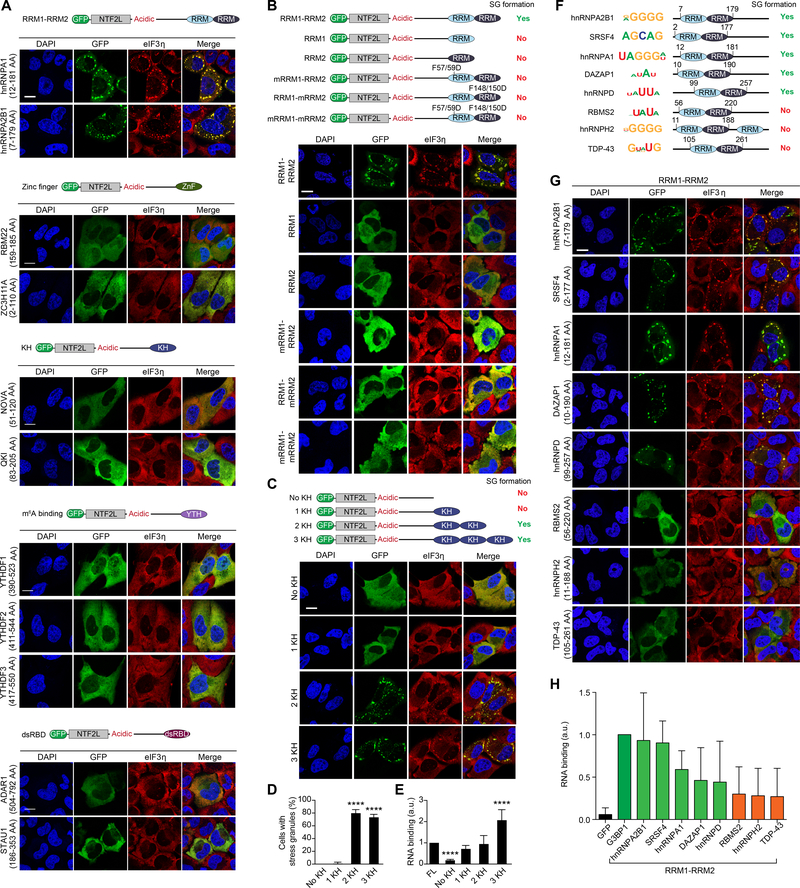
We next conducted domain swapping experiments to investigate the mechanism whereby RNA binding by G3BP1 contributes to SG assembly. We exchanged the RBD of G3BP1 with five different classes of RBDs: the tandem RRMs from hnRNPA1 and hnRNPA2B1, the zinc finger motifs from RBM22 and ZC3H11A, the KH domains from NOVA and QKI, the YTH domains from YTHDF1, YTHDF2, and YTHDF3 (three proteins that specifically bind m6A-RNA (Zhao et al., 2017)), and the dsRBDs from ADAR1 and STAU1 (Figure 4A). Despite similar levels of expression (Figure S4A), only the tandem RRMs from hnRNPA1 and hnRNPA2B1 reconstituted SG assembly in G3BP1/2 dKO cells (Figure 4A). Importantly, the ability of these chimeric proteins to reconstitute SG assembly did not correlate with RNA-binding capacity (Figure S4B).
Figure 4. Multivalency in RNA Binding Mediates SG Formation.
(A-C) G3BP1/2 dKO cells were transfected with G3BP1 constructs in which the RBD was substituted with different types of RBDs as indicated. Cells were exposed to sodium arsenite (500 μM, 1 h) and stained for eIF3η. Scale bars, 20 μm.
(D) Cells from (C) were imaged and the percentage of cells positive for SGs was quantified. Error bars indicate SD. ****P < 0.0001 vs. no KH by one-way ANOVA, Dunnett’s multiple comparisons test.
(E) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs. Immunoprecipitated G3BP1 cross-linked to RNA was assessed by immunoblotting for biotin (RNA) and GFP (G3BP1). Binding of RNA was quantified and normalized to protein signal. Results show quantification of triplicate experiments. Error bars indicate SD. ****P < 0.0001 vs. FL by one-way ANOVA, Dunnett’s multiple comparisons test.
(F) RNA-binding proteins with tandem RRM motifs are shown together with their top binding motif and domain structure.
(G) G3BP1/2 dKO cells were transfected with G3BP1 constructs in which the RBD was substituted with indicated tandem RRM domains. Cells were exposed to sodium arsenite and stained as in (A). Scale bar, 20 μm.
(H) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs and analyzed as in (E). Green bars correspond to functional swap mutants for SG formation, orange bars correspond to non-functional swap mutants. Results show quantification from triplicate experiments. Error bars indicate SD.
See also Figure S4.
Since two distinct RNA-binding entities (i.e., RRM and IDR3) are important for SG assembly (Figure 3I), and the only domain swaps that successfully replaced the native RBD were also multivalent (i.e., tandem RRM1-RRM2 of hnRNPA1 and hnRNPA2B1) (Figure 4A), we hypothesized that the RNA-binding valency of G3BP1 may be an important feature in SG assembly. To test this idea, we reduced the RNA-binding valency of the G3BP1 chimera with tandem RRM domains. Deletion of either RRM1 or RRM2, or introduction of missense mutations that interrupt RNA binding by either or both RRM1 and RRM2, all abolished the ability of the G3BP1 chimera to support SG formation (Figures 4B, S4C). As a further test, we increased RNA-binding valency by swapping the native RBD of G3BP1 with one, two, or three KH domains from QKI (Figure 4C–D). Whereas a single KH domain substitution restored the RNA-binding capacity to that of G3BP1 WT (Figure 4E), this was not sufficient to support SG assembly. In contrast, substitution with two or three KH domains reconstituted SG formation (Figure 4C–D) despite lower level expression of these proteins (Figure S4C). We found identical results upon increasing the valency of ZnF domains (Figure S4D). Based on these observations we conclude that multivalency for RNA binding contributes to RNA-dependent LLPS and SG assembly.
Interestingly, whereas the efficiency of SG reconstitution was equivalent with the introduction of two vs. three KH or ZnF domains, and the SGs were morphologically indistinguishable, the material properties of these SGs were different. Specifically, SGs assembled with three KH or ZnF domains were significantly less dynamic than those assembled with two KH or ZnF domains (Figure S4E), suggesting that the material properties of SGs are influenced by the degree of association of G3BP1 with RNA. We also examined the consequence of duplicating the G3BP1 RBD domain (2xRBD). G3BP1 2xRBD, which has double valency and RNA-binding capacity, efficiently reconstituted SGs in G3BP1/2 dKO cells, but these were also less dynamic (Figure S4F–J). This inverse correlation between fluidity of a biomolecular condensate and interaction valency of its constituents is consistent with observations in vitro (Li et al., 2012) and may reflect a general principle controlling material properties of biomolecular condensates.
SG Assembly Does Not Require G3BP1 Interaction with Specific RNA Sequence Motifs
The observation that different RNA-binding entities can substitute for the native RBD of G3BP1 suggests that no specific RNA motif interactions or RNA targets are essential for SG assembly. To investigate this further, we generated G3BP1 chimeras using six additional multivalent RBDs (Figure 4F). When expressed at equivalent levels (Figure S4K), the ability of these RBDs to support SG assembly did not correlate with any motif interaction preferences, but rather with relative RNA binding capacity (Figure 4G–H). Thus, the ability of G3BP1 to support SG assembly requires not only multivalent interactions with RNA, but also sufficient RNA-binding capacity above a threshold concentration. We note that IDR3 has been reported to interact with two ribosomal proteins (RPS6 and RPS23), leading to the suggestion that G3BP1 interaction with the 40s ribosome may be important for SG assembly (Kedersha et al., 2016). The ability of multiple heterologous RNA-binding entities to replace IDR3 and reconstitute SG assembly suggests that IDR3 interaction with ribosomal proteins may not be essential for supporting SG assembly.
The NTF2L Domain Mediates Dimerization Without Significant Higher-Order Oligomerization
Previous studies have reported conflicting results regarding higher-order oligomerization (i.e., trimers and tetramers) of G3BP1 via the NTF2L domain (Tourriere et al., 2003; Vognsen et al., 2013). Given the frequency of oligomerization as a strategy to promote LLPS, including precedent for higher-order oligomerization of dimers to promote LLPS (Marzahn et al., 2016), we sought to clarify the ability of the NTF2L domain to mediate the assembly of multimers greater than dimers. Size exclusion chromatography coupled to multi-angle light scattering revealed exclusively dimer formation by G3BP1 across a range of concentrations (Figure S5A). Analytical ultracentrifugation analysis of purified G3BP1 at 50 μM confirmed the dimeric state of the protein, with less than 2% found in higher-order species (Figure S5B–C). Finally, using in vitro cross-linking of purified protein with bissulfosuccinimidyl suberate (BS3), we confirmed the role of NTF2L in mediating this dimerization and saw no significant higher-order oligomerization (Figure S5D). Consistent with these results, BS3-cross-linked G3BP1 from cells with or without arsenite treatment confirmed the NTF2L domain-dependent dimeric state of the purified protein and no significant higher-order oligomerization (Figure S5E–F).
G3BP1 Dimerization Is Essential for LLPS In Vitro and SG Assembly in Cells
The NTF2L domain mediates interaction with several other SG proteins and these interactions influence the extent of SG assembly. For example, interaction of NTF2L with caprin-1 and UBAP2L promotes SG assembly, whereas interaction with USP10 limits assembly (Kedersha et al., 2016; Markmiller et al., 2018; Panas et al., 2015; Youn et al., 2018). Here we showed that G3BP1 lacking the NTF2L domain failed to reconstitute SG assembly in G3BP1/2 dKO cells, illustrating that this domain is essential for SG assembly (Figure 3F). Theoretically, this requirement in cells could be due to essential interactions of the NTF2L domain with binding partners; yet we found that NTF2L is also essential for LLPS of G3BP1 with RNA in vitro where these partners are not present (Figure 3D). Therefore, an alternative possibility is that the critical role of NTF2L is dimerization, which increases the valency of RBDs to promote LLPS of G3BP with RNA. We examined each of these possibilities in turn.
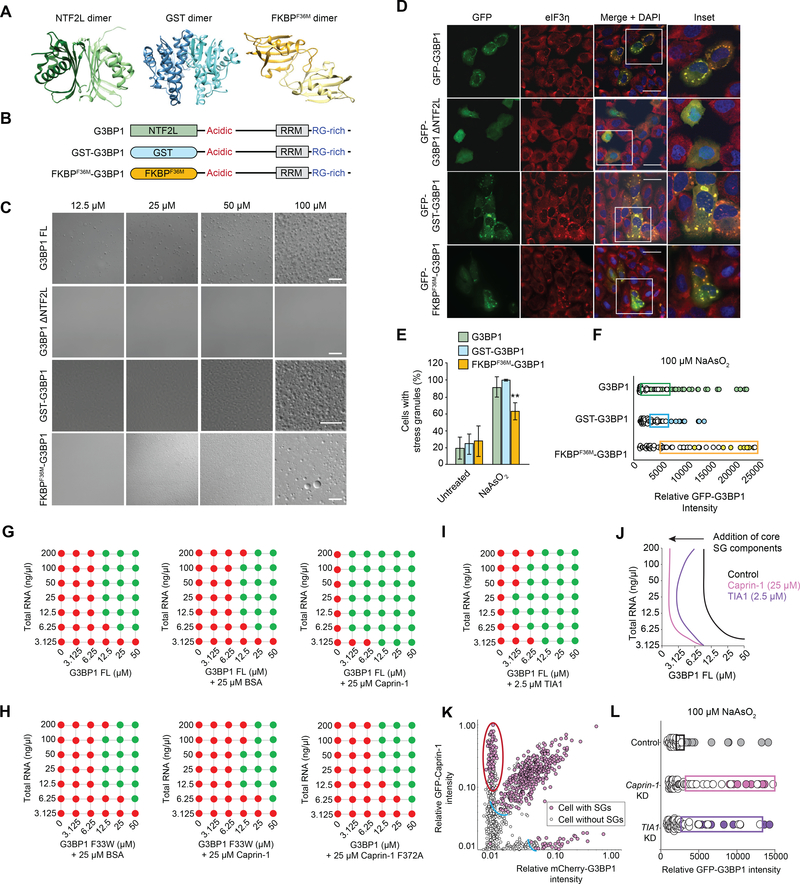
First, we fully replaced the NTF2L domain of G3BP1 with alternative dimerization domains that form constitutive dimers. Substituting NTF2L with glutathione S-transferase (GST, 211 amino acids) (Fabrini et al., 2009) or a FK506-binding protein mutant (FKBPF36M, 108 amino acids) (Rollins et al., 2000) each restored in vitro LLPS behavior to G3BP1 (Figure 5A–C). When expressed in G3BP1/2 dKO cells, both chimeric proteins successfully reconstituted SGs, although the efficiency of the FKBPF36M chimera was reduced (Figure 5D–E, Table S2). Thus, dimerization via the N-terminal domain per se, independent of the nature of the dimer domain, is necessary and sufficient to support both LLPS in vitro and SG assembly in cells. The observed difference in reconstitution efficiency of the two chimeras tested may reflect their respective dimer dissociation constants (Kd for GST = 0.34 μM, Kd for FKBPF36M = 40 μM). Notably, using U2OS cells expressing tdTomato-tagged endogenous G3BP1, we confirmed that these chimeric versions of G3BP1 co-mingle in SGs with endogenous G3BP1 (Figure S5G).
To fully assess the ability of these chimeric proteins with different dimer domains to reconstitute SG assembly, we mapped their intracellular phase separation thresholds by taking advantage of the natural variation in expression of GFP-G3BP1 when transiently transfected into G3BP1/2 dKO cells. Simultaneous cell-by-cell analysis of GFP-G3BP1 expression level and SG formation permitted a determination of the G3BP1 concentration threshold that initiates SG assembly. The concentration threshold for SG assembly was similar for G3BP1 WT and GST-G3BP1, whereas this threshold concentration was markedly higher for FKBPF36M-G3BP1 (Figure 5F). These data illustrate that an essential function of the NTFL2 domain is to support LLPS through dimerization, independent of any NTFL2-specific interactions.
Figure 5. NTF2L Domain-Mediated Dimerization and Interaction with Core SG Components Regulate SG Assembly.
(A) Structures of the G3BP1 NTF2L (PDBID: 4FCJ), GST (PDBID: 1UA5), and FKBPF36M (PDBID: 1YEM) dimers.
(B) Constructs used to investigate the function of the NTF2L domain.
(C) LLPS of purified recombinant G3BP1 in the presence of a crowding agent and 150 mM NaCl. Scale bar, 50 μm.
(D-E) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Cells were imaged (D) and the percentage of cells positive for SGs was quantified (E). Scale bars, 50 μm. Error bars indicate SD. **P = 0.0098 vs. G3BP1 by two-way ANOVA, Sidak’s multiple comparisons test.
(F) Intracellular phase diagrams of indicated G3BP1 constructs transfected into G3BP1/2 dKO cells. Cells were exposed to sodium arsenite (30 min), fixed, and stained for PABP. SG formation and GFP intensities were assessed cell-by-cell. Cells with SGs are plotted as filled circles; cells without SGs are plotted as empty circles. Boxes highlight the 25% highest levels of expression among SG-negative cells.
(G) Co-phase separation of purified recombinant G3BP1 with BSA or caprin-1 in the absence of a crowding reagent, in 150 mM NaCl, and with increasing concentrations of total RNA from human cells.
(H) Co-phase separation of purified recombinant G3BP1 and caprin-1 variants as in (G).
(I) Co-phase separation of purified recombinant G3BP1 and TIA1 as in (G).
(J) Phase diagram of G3BP1 with caprin-1 and TIA1 as shown in (G-I).
(K) Intracellular phase diagrams of G3BP1 with addition of caprin-1. G3BP1/2 dKO cells were co-transfected with mCherry-G3BP1 and GFP-caprin-1 and intracellular phase diagrams were measured as in (F). Expression of caprin-1 reduces the G3BP1 threshold for SG formation (blue arc). Cells with high caprin-1 levels assembled SGs at very low levels of G3BP1 (red oval).
(L) Intracellular phase diagrams of G3BP1 after knockdown of caprin-1 or TIA1. G3BP1/2 dKO cells were co-transfected with GFP-G3BP1 WT and a pool of siRNA targeting expression of caprin-1 or TIA1 and intracellular phase diagrams were measured as in (F).
G3BP1 Dimerization Is Essential to Maintain SG Assembly in Cells
FKBPF36M homodimers can be dissociated by a small molecule ligand such as the rapamycin derivative AP21998 or “D/D solubilizer” (Rollins et al., 2000). Addition of this ligand in sub-stoichiometric amounts abolished LLPS by FKBPF36M-G3BP1 but had no effect on LLPS of G3BP1 WT, further illustrating that dimerization per se is required for LLPS (Figure S5H). To examine the effect of disrupting G3BP1 dimer formation on SG assembly in cells, we pre-incubated G3BP1/2 dKO cells expressing FKBPF36M-G3BP1 with sub-stoichiometric amounts of this ligand and found that arsenite-induced SG assembly was abolished, confirming the importance of G3BP1 dimerization for this assembly (Figure S5I–J). Remarkably, fully assembled SGs rapidly disassembled within 30 seconds by the addition of ligand to the culture medium (Figure S5K–L, Video S2). These results indicate that dimerization of G3BP1 is not only essential for SG assembly, but also for maintenance of SGs, an observation that may have implications for the mechanism of SG disassembly upon removal of stress.
Cooperative Interactions between Members of the Core SG Network Regulate Assembly
Given that NTF2L-mediated interactions are not essential for SG formation, the mechanism whereby interactors of G3BP1 exert influence over assembly was unclear. We hypothesized that interactors of G3BP1 contribute to and regulate SG assembly by contributing to the overall sum of interactions within the SG network (Figure 1D). In support of this hypothesis, we found that the presence of an additional SG constituent (i.e., caprin-1) that binds the NTF2L domain of G3BP1 significantly reduced the threshold concentration of G3BP1 and RNA necessary for LLPS, thereby exhibiting positive cooperativity (Figure 5G). This cooperativity reflects a specific interaction between caprin-1 and G3BP1, as evidenced by point mutations that disrupt binding between caprin-1 and G3BP1 (G3BP1 F33W and caprin-1 F372A), each of which abolished the ability of caprin-1 to promote LLPS of G3BP1 with RNA (Figure 5H). Similar to caprin-1, we found that TIA1 also exhibited positive cooperativity with G3BP1 and RNA, significantly reducing the threshold concentration necessary for LLPS (Figure 5I–J).
To examine the significance of this biophysical behavior in cells, we measured the intracellular phase separation threshold for SG assembly as a function of intracellular concentrations of these binding partners. Consistent with in vitro results, over-expression of caprin-1 with G3BP1 in G3BP1/2 dKO cells significantly reduced the concentration threshold of G3BP1 necessary to initiate SG assembly even in the absence of arsenite (Figure 5K). Interestingly, in cells with very high caprin-1 levels, even cells with very low levels of G3BP1 were able to assemble SGs. Consistent with these results, we found that depletion of either caprin-1 or TIA1 by siRNA-mediated knockdown significantly raised the critical concentration threshold of G3BP1 necessary to initiate SG assembly (Figures 5L, S5M). Thus, both in vitro and cell-based results suggest that components of the core SG network influence SG assembly through positive (e.g., caprin-1, UBAP2L, TIA1) or negative (e.g., USP10) cooperativity, collectively setting the critical threshold concentration of constituents necessary for LLPS of G3BP1 with RNA (Figure 5J). However, no specific interaction appears to be essential, since SGs still assemble in cells with knockout of individual binding partners (e.g., caprin-1, UBAP2L, TIA1) or paralogous pairs (e.g., TIA1/TIAR, UBAP2/UBAP2L) (Figure 1F).
G3BP1 IDR1/2 Regulates SG Dynamics and Composition
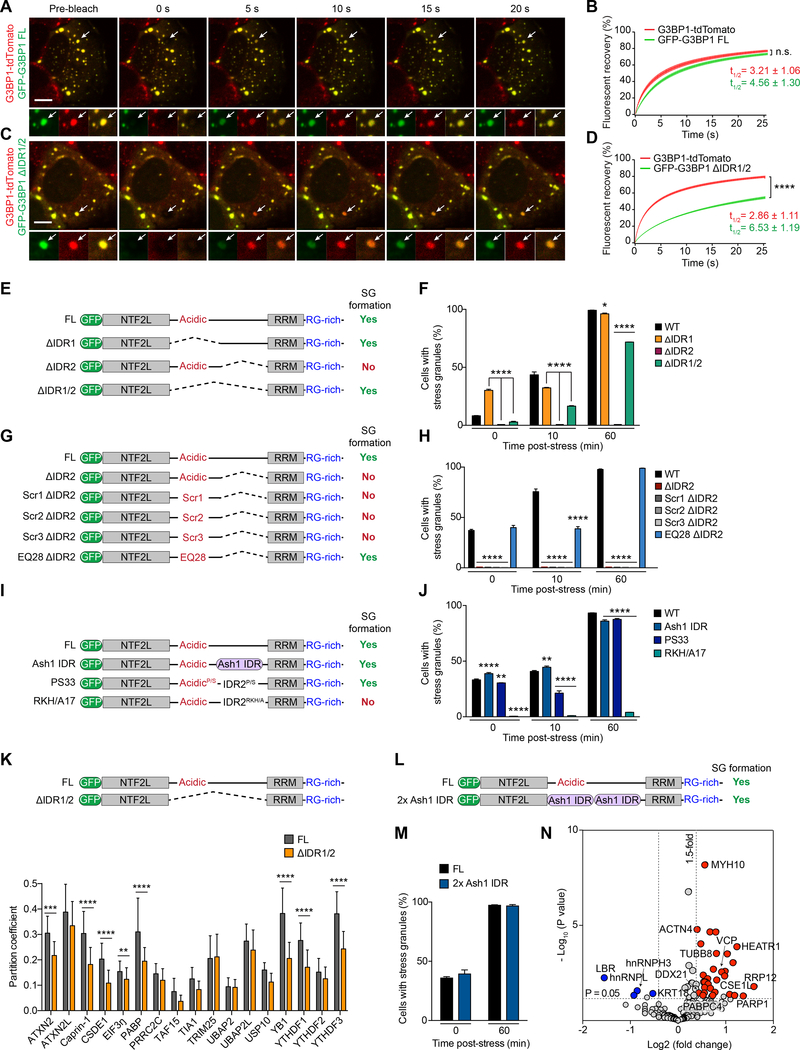
The precise role of IDRs in regulating the assembly or material properties of SGs remains poorly understood. To gain insight into this role, we pursued our surprising finding that the long central IDR (IDR1/2) of G3BP1 is not essential for LLPS of G3BP1 in vitro nor for SG assembly in cells (Figure 3C–F). The IDR1/2 segment did not exhibit LLPS in vitro even at high concentration (400 μM) and the presence of a crowding agent (Figure S6A), and did not reconstitute SG assembly when expressed in G3BP1/2 dKO cells (Figure S6B). Indeed, even when expressed in WT cells, the IDR1/2 segment was not recruited into arsenite-induced SGs arising from endogenous G3BP (Figure S6C). These data indicate that IDR1/2 does not encode interactions that are essential for the assembly and/or maintenance of SGs.
However, deletion of IDR1/2 did alter the biophysical properties of G3BP1. Specifically, G3BP1 ΔIDR1/2 showed reduced solubility and underwent LLPS at lower concentrations in vitro (Figure 3D–E), and had relatively poor solubility in cells (Figure S6D). Additionally, although G3BP1 ΔIDR1/2 reconstituted SG assembly in G3BP1/2 dKO cells, efficiency was reduced (Figure S6E).
To explore the role of IDR1/2 in SG dynamics, we used U2OS cells with endogenous G3BP1 tagged with tdTomato. We introduced exogenous GFP-tagged G3BP1 (WT or ΔIDR1/2) to permit simultaneous monitoring of their mobility within SGs by FRAP. As expected, when exogenous G3BP1 WT was introduced into these cells, endogenous and exogenous WT proteins showed identical rates of FRAP (Figure 6A–B, Video S3). However, exogenous G3BP1 ΔIDR1/2 showed significantly slower FRAP and a greater immobile fraction compared with endogenous WT protein, revealing a role for IDR1/2 in defining G3BP1 dynamics (Figure 6C–D, Video S4). By co-immunoprecipitation assay, we confirmed that GFP-G3BP1 (both WT and ΔIDR1/2) formed homodimers as well as heterodimers with HA-G3BP1 WT (Figure S6F), suggesting that these FRAP experiments likely underestimate the extent to which the exchange rate of G3BP1 is impaired by deletion of IDR1/2.
Figure 6. The Long Central IDR of G3BP1 Regulates SG Assembly, Dynamics, and Composition; G3BP1 IDR1 Is an Autoinhibitory Element.
(A-D) U2OS cells expressing tdTomato-tagged endogenous G3BP1 were transfected with indicated GFP-tagged G3BP1 constructs, exposed to sodium arsenite (500 μM, 1 h), and the relative mobility of endogenous tdTomato-G3BP1 was compared to exogenous GFP-G3BP1 by FRAP. Error bars indicate SEM. n.s., not significant, ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparison test. Mobile fractions were 76% (tdTomato-G3BP1 FL), 73% (GFP-G3BP1 FL) in (A-B); 79% (tdTomato-G3BP1 FL) and 54% (GFP-G3BP1 ΔIDR1/2) in (C-D). Scale bar, 10 μm.
(E-F) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs and exposed to sodium arsenite (500 μM). Cells were imaged and the percentage of cells positive for SGs was quantified prior to arsenite exposure (0 min) or 10 min or 60 min after arsenite exposure. Error bars indicate SEM. *P = 0.0224, ****P < 0.0001 by two-way ANOVA, Dunnett’s multiple comparisons test.
(G-H) G3BP1/2 dKO cells were transfected, stressed, imaged, and quantified as in (E-F). G3BP1 mutant constructs had deletion of IDR2 and either a scrambled IDR1 sequence (Scr) or mutation of all 28 glutamates in IDR1 to glutamine (EQ28). Error bars indicate SEM. ****P < 0.0001 by two-way ANOVA, Dunnett’s multiple comparisons test.
(I-J) G3BP1/2 dKO cells were transfected, stressed, imaged, and quantified as in (E-F). G3BP1 mutant constructs had substitution of IDR2 with Ash1 IDR, substitution of all 33 proline residues in IDR1/2 to serine (PS33), or substitution of 17 positively charged residues to alanines (RKH/A17). Error bars indicate SEM. **P = 0.0077 (0 min) and 0.0014 (10 min), ****P < 0.0001 by two-way ANOVA, Dunnett’s multiple comparisons test.
(K) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, exposed to sodium arsenite (500 μM, 1 h), fixed, and stained with SG markers as indicated. Confocal images were taken for partition coefficient analysis. Error bars indicate SD. **P =0.0011, ***P =0.0001, ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparisons test.
(L-M) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs (L) and exposed to sodium arsenite (500 μM, 1 h). Cells were imaged and the percentage of cells positive for SGs was quantified (M). Error bars indicate SD.
(N) Fold change of spectral counts of proteins identified by APEX2 proximity labeling and P values are plotted. Colored circles indicate proteins more enriched in SGs formed with G3BP1–2xAsh1 IDR (red; 10 representative proteins are labeled) or with G3BP1 FL (blue).
The Acidic IDR1 Domain of G3BP1 Is an Autoinhibitory Element
To further address the role of IDR1 and IDR2 in SG assembly, we directly compared SG assembly in G3BP1/2 dKO cells reconstituted with G3BP1 mutants with alterations in IDR1 or IDR2. We examined assembly prior to treatment with arsenite (“spontaneous SGs”), as well as time points after arsenite treatment. Expression of G3BP1 ΔIDR1 not only fully reconstituted SG assembly following stress, but resulted in strongly increased formation of spontaneous SGs (Figures 6E–F, S6G–H). In contrast, expression of G3BP1 ΔIDR2 failed to support SG assembly (Figures 6E–F, S6G–H), which contrasts sharply with the ability of G3BP1 ΔIDR1/2 to reconstitute SG assembly. Taken together, these results reveal that the acidic region comprising IDR1 is an autoinhibitory element, and further that this inhibition is mitigated by the presence of IDR2.
We reasoned that the negative charge density present within IDR1, rather than motifs encoded in the primary amino acid sequence, might mediate its inhibitory effect on SG assembly. To test this notion, we generated three different G3BP1 mutants in which the primary IDR1 sequence was scrambled and the adjacent IDR2 was deleted (Figures 6G, S6I). All three of these mutants failed to reconstitute SG assembly, indicating that the inhibitory activity of IDR1 was preserved (Figures 6H, S6J). We also generated a mutant in which all 28 glutamates in IDR1 were substituted with charge-neutral glutamines (EQ28 ΔIDR2, Figure S6I). By contrast with the scrambled mutants, this mutant supported robust SG assembly in G3BP1/2 dKO cells (Figures 6H, S6K). Together, these results indicate that the inhibitory activity of IDR1 is encoded by the negative charge density in this region rather than the primary sequence.
We next turned to IDR2, which has a net positive charge density and is enriched in proline residues that may confer functionally relevant rigidity to this segment. To investigate the importance of spacing, composition, and primary sequence of IDR2, we engineered a series of substitutions or mutations. First, we substituted IDR2 with an IDR of similar length and composition derived from the S. cerevisiae transcription factor Ash1 (G3BP1-Ash1 IDR). This 79-residue proline-rich IDR adopts expanded coil-like conformations (Martin et al., 2016a). When expressed in G3BP1/2 dKO cells, G3BP1-Ash1 IDR supported WT levels of SG assembly (Figures 6I–J, S6L), suggesting that the spacing and/or composition within this segment, rather than its primary sequence, is important for suppressing the inhibitory activity of IDR1.
To assess the importance of spacing, we introduced of a second copy of IDR2 (G3BP1–2xIDR2) to double the putative spacing afforded by IDR2. This second copy of IDR2 significantly enhanced SG assembly relative to G3BP1 WT, perhaps by separating the negatively charged acidic IDR1 from the positively charged IDR3 within the RBD, or by acting as a decoy interrupting this interaction (Figure S6M–N). We also assessed the importance of the proline residues in IDR2 by substituting all 33 proline residues in IDR1/2 with serine (PS33). When expressed in G3BP1/2 dKO cells, this mutant supported WT levels of SG assembly, indicating that these proline residues are not critically important for the mitigating effect of IDR2 on IDR1 inhibition (Figures 6I–J, S6L). On the other hand, a mutant with all 17 positively charged residues in IDR2 substituted with alanine (RKH/A17) was unable to support SG formation when expressed in G3BP1/2 dKO cells, suggesting that beyond the spacing afforded by IDR2, this domain might engage in charge-dependent interaction with IDR1, mitigating the interaction between IDR1 and IDR3 (Figures 6I–J, S6L).
Notably, when we reconstituted SGs with G3BP1 ΔIDR1/2, 8 proteins from a panel of 17 SG markers showed reduced recruitment to SGs (Figure 6K). This observation suggested that in addition to regulating SG assembly and dynamics, IDR1/2 might play a role in determining the composition of SGs. To comprehensively assess the role of IDR1/2 in SG composition, we performed proximity proteomics using APEX2-labeled G3BP1. For this analysis we compared G3BP1 WT to G3BP1–2xAsh1 IDR, in which both IDR1 and IDR2 were replaced by Ash1 IDR sequences. We selected this mutant for head-to-head comparison because G3BP1–2xAsh1 affords highly efficient SG assembly despite harboring a completely different IDR primary sequence (Figure 6L–M). Despite indistinguishable kinetics and frequency of SG assembly in response to arsenite, altering this IDR sequence resulted in significant differences in SG composition. Specifically, nine proteins that are consistently reported in SGs (Jain et al., 2016; Markmiller et al., 2018) were absent from SGs seeded by G3BP1–2xAsh1 IDR, and recruitment of an additional four SG proteins was significantly reduced (Figure 6N, Table S3). Moreover, SGs seeded with G3BP1–2xAsh1 IDR recruited 43 proteins not previously reported in SGs (Jain et al., 2016; Markmiller et al., 2018), and 43 other proteins showed significantly increased recruitment to these mutant SGs (Figure 6N, Table S3). Thus, the long central IDR plays a role in defining SG composition.
IDR1 Phosphorylation Tunes Interplay between 3 IDRs and Regulates LLPS
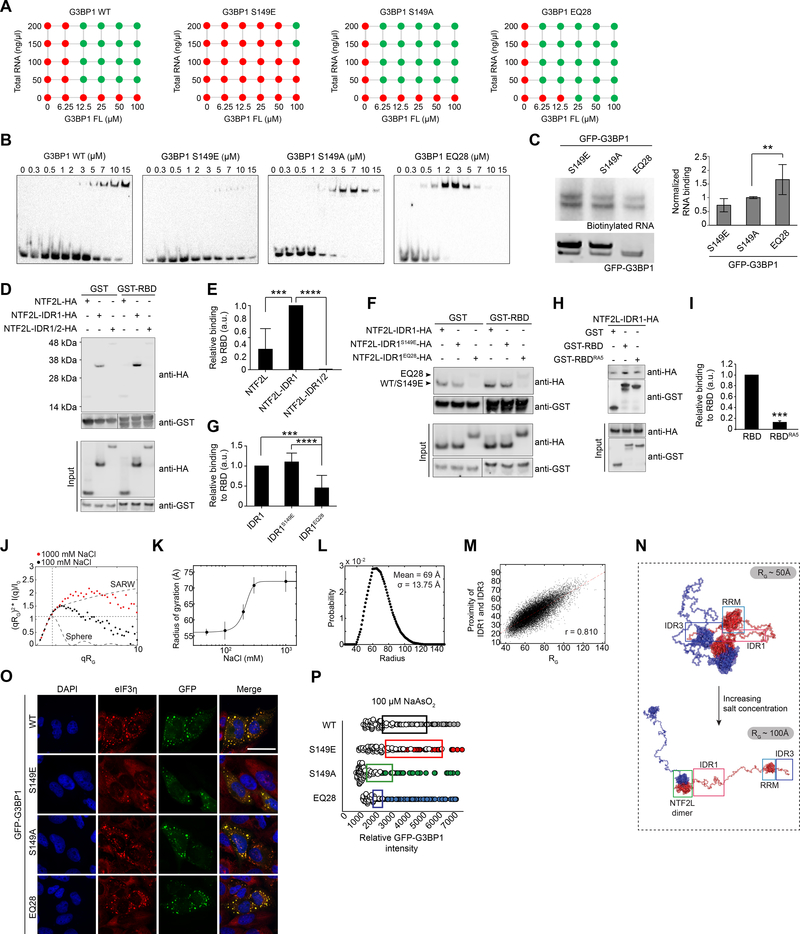
Several studies have suggested that post-translational modifications regulate G3BP1 (Sahoo et al., 2018; Tourriere et al., 2003; Tsai et al., 2016). Among these, phosphorylation of S149 in IDR1 has been implicated as a regulator of SG assembly. Whereas the fact that S149 can be phosphorylated is clear, the precise role of this post-translational modification in SG assembly has been uncertain (Panas et al., 2019; Sahoo et al., 2018; Tourriere et al., 2003). Nevertheless, recent compelling evidence has been reported that G3BP1-S149 phosphorylation is functionally relevant during physiological stress in neurons (Sahoo et al., 2018).
To illuminate the biophysical mechanism of G3BP1 regulation by S149 phosphorylation, we generated G3BP1 variants in which the charge content of IDR1 was altered: a phosphomimetic form (S149E), in which the negative charge density is modestly increased, a non-phosphorylatable form (S149A) that does not alter charge, and EQ28, in which the negative charge density of IDR1 is substantially reduced. Consistent with a recent report (Panas et al., 2019), the phosphorylation state of S149 did not change the ability of G3BP1 to dimerize as assessed by in vitro cross-linking analysis (Figure S7A–C). We then examined the impact of altered IDR1 charge content on LLPS in vitro. Compared to G3BP1 WT, the S149E, S149A, and EQ28 variants had no significant influence on Ficoll-dependent phase transition (Figure S7D). In contrast, RNA-dependent LLPS of G3BP1 was strongly influenced by the charge content of IDR1: whereas G3BP1 S149A behaved similarly to WT, the S149E variant strongly impaired LLPS of G3BP1 with RNA, shifting the phase separation threshold to significantly higher concentrations of G3BP1 and RNA (Figures 7A, S7E). The charge-neutralizing EQ28 variant had the opposite effect and strongly promoted LLPS of G3BP1 with RNA (Figures 7A, S7E). Since only RNA-dependent LLPS of G3BP1 was influenced by the charge content of IDR1, we next tested the influence of IDR1 charge content on RNA binding by G3BP1. As predicted by the LLPS results, EMSA using purified protein and RNA revealed similar RNA-binding capacity by G3BP1 WT and S149A, but decreased RNA binding by G3BP1 S149E and increased RNA binding by G3BP1 EQ28 (Figure 7B). Consistent results were obtained in cells, where S149E showed reduced RNA binding and EQ28 showed increased RNA binding (Figure 7C). We hypothesized that the autoinhibitory activity of IDR1 reflects electrostatic intramolecular interaction with the RBD, thereby competing with RNA for binding. Further, we hypothesized that increased negative charge in IDR1 caused by phosphorylation enhances its autoinhibitory activity by strengthening the intramolecular IDR1-RBD interaction. To test these ideas, we performed GST pulldown experiments to investigate a putative IDR1-RBD interaction. We tested the ability of G3BP1 protein fragments (NTF2L domain alone, NTF2L-IDR1, and NTF2L-IDR1/2) to interact with a fragment consisting of the RBD (which contains RRM-IDR3). No significant interaction was detected between the RBD and the NTF2L domain alone, as expected, but strong interaction was detected when IDR1 was included (NTF2L-IDR1) (Figures 7D–E). The presence of IDR2 (NTF2L-IDR1/2) mitigated this interaction (Figures 7D–E), consistent with the ability of IDR2 to mitigate the autoinhibitory activity of IDR1 in SG assembly (Figures 6I–J). This interaction was enhanced by the S149E mutation and diminished by the charge-neutralizing EQ28 mutations (Figures 7F–G). We reasoned that electrostatic interaction between the negatively charged IDR1 and RBD was mediated by the positive charge density in IDR3. To test this notion, we reduced the positive charge in IDR3 by introducing five arginine-to-alanine substitutions (RBDRA5). RBDRA5 had greatly reduced interaction with NTF2L-IDR1, confirming electrostatic interaction between IDR1 and the RBD, mediated by IDR3 (Figures 7H–I). The positive charges in IDR3 are also essential for RNA binding, as illustrated by lack of RNA-dependent LLPS of G3BP1-RBDRA5 and the inability of this mutant to reconstitute SG assembly in G3BP1/2 dKO cells (Figures S7F–G). Of note, G3BP1-RBDRA5 provides a further example of differences in RNA-dependent and Ficoll-dependent LLPS (Figure S7F). Substitution of these same arginines in IDR3 with lysines (RBDRK5) did permit RNA-dependent LLPS in vitro and reconstituted SG assembly in G3BP1/2 dKO cells, illustrating that charge rather than other properties specific to arginine residues underlie these activities (Figures S7F–G).
Figure 7. IDR1 Phosphorylation Tunes Interplay between 3 IDRs and Regulates LLPS.
(A) Summary of RNA-triggered LLPS of G3BP1 under indicated conditions, in the presence of 150 mM NaCl. Corresponding images are shown in Figure S7E.
(B) Electrophoretic mobility shift assay of G3BP1 WT and IDR1 mutants.
(C) CLIP analysis of G3BP1 IDR1 mutants. Indicated G3BP1 constructs were transiently expressed in G3BP1/2 dKO cells and analyzed as in Figure 4E. Error bars indicate SD. **P = 0.005 vs. S149A by one-way ANOVA, Dunnett’s multiple comparisons test.
(D-E) GST pulldown of purified GST-RBD with HA-G3BP1 N-terminal mutants indicates an intramolecular interaction between G3BP1 RBD and IDR1, and IDR2 mitigates this interaction. Results are quantified in (H). Error bars indicate SD. ***P = 0.0002 and ****P < 0.0001 vs. NTF2L-IDR1-HA by one-way ANOVA, Dunnett’s multiple comparisons test.
(F-G) GST pulldown of GST-RBD with NTF2L-IDR1-HA. RBD interactions with WT or mutant IDR1 (S149E, EQ28) were assessed as in (G) and quantified in (J). Error bars indicate SD. ***P = 0.0006 and ****P < 0.0001 by one-way ANOVA, Tukey’s multiple comparisons test.
(H-I) GST pulldown of GST-RBD mutants with NTF2L-IDR1-HA. IDR1 interactions with WT or RBDRA5 were assessed as in (G) and quantified in (L). Error bar indicates SD. ***P = 0.0002 by unpaired t-test.
(J) Small-angle X-ray scattering (SAXS) data for G3BP1 WT at high and low NaCl concentration. Data are presented as normalized Kratky plots where the intensity is normalized by the zero-angle scattering and the momentum transfer (q) is normalized by the radius of gyration (Rg). The intersection of dashed lines indicates the theoretical maximum of a solid sphere. Experimental data with a higher maximum coupled to a shift to the right is caused by conformational flexibility. Comparing the experimental data to synthetic data for a sphere and a self-avoiding random walk (SARW) indicates that G3BP1 is flexible in both conditions and expands toward maximal random dimensions at high NaCl concentration.
(K) Rgs were extracted from SAXS data by Guinier analysis at 50–1000 mM NaCl. Data were fit to a logistic function suggesting a minimum dimension of ~55Å at low NaCl concentration to a maximum extension of ~72 Å at high NaCl concentration with a transition with a midpoint at ~215 mM NaCl, indicating an interaction between oppositely charged regions of the protein. Error bars indicate SD.
(L) Distribution of G3BP1 radii in randomly generated conformations.
(M) In random synthetic conformations, the distance between IDR1 and IDR3 is positively correlated with the total RG.
(N) Representative conformations of G3BP1. Conformations with representative radii were sampled from the randomly generated pool shown in (E) to show compact conformations enriched in buffers with ionic strength below 200 mM (top) and extended conformations that would be enriched at high ionic strength (bottom).
(O) G3BP1/2 dKO cells transiently expressing indicated G3BP1 constructs were exposed to sodium arsenite (500 μM, 1 h), fixed, and stained for eIF3η. Scale bar, 50 μm.
(P) Intracellular phase diagram of indicated G3BP1 constructs transfected into G3BP1/2 dKO cells. Cells were exposed to 100 μM sodium arsenite (30 min), fixed, and stained for PABP. SG formation and GFP intensities were assessed cell-by-cell. Cells with SGs are plotted as filled circles; cells without SGs are plotted as empty circles. Boxes highlight the 25% highest levels of expression among SG-negative cells.
Vertical lines in blots in (I) and (K) indicate noncontiguous lanes from the same gel.
See also Figure S7.
We further explored the influence of IDR1-IDR3 interaction on G3BP1 conformation by using small angle X-ray scattering (SAXS) to analyze purified G3BP1 across a range of salt concentrations. Increasing NaCl concentration shifted the protein from an ensemble of compact conformations with a mean radius of gyration (⟨RG⟩) of 55 Å to extended conformations with ⟨RG⟩ of 72 Å. The sharp transition from compact to extended conformations occurred within the range of physiological ionic strength, indicating that G3BP1 size and shape is determined by electrostatic interactions. Moreover, at physiological ionic strength, this conformational change in G3BP1 is likely sensitive to the small changes in charge that occur with phosphorylation (Figures 7J–K). To further characterize the conformational landscape of G3BP1, we compared experimental SAXS data with an ensemble of in silico-generated conformations where the IDRs were modeled as self-avoiding polymers. The in silico ensemble had a mean radius of 69 Å, consistent with the ⟨RG⟩ measured at high NaCl concentration by SAXS (Figures 7K–L). Thus, at lower, physiological ionic strength, the IDRs are compacted by attractive electrostatic interactions relative to an unrestrained self-avoiding polymer. We then measured the distances within the G3BP1 dimer between the two negatively charged IDR1s and the positively charged IDR3s and observed a linear correlation between the overall radius of the protein and the distance separating IDR1 and IDR3, indicating that overall compaction likely reports on close association between these domains (Figures 7M–N). These observations strongly implicate intramolecular interaction between IDR1 and IDR3 that controls RNA interaction and RNA-dependent LLPS.
Finally, we investigated the role of the G3BP1 conformational switch on the assembly of SGs. G3BP1 WT, S149E, S149A, and EQ28 all rescued SG formation in G3BP1/2 dKO cells (Figure 7O). By measuring the intracellular G3BP1 concentration threshold for phase separation by these mutants, we found that G3BP1 S149A required significantly lower concentrations to initiate SG assembly relative to G3BP1 WT (Figure 7P), as expected since G3BP1 WT is found as a mixture of phosphorylated and unphosphorylated forms in cultured cells (Gallouzi et al., 1998). Consistent with this observation, G3BP1 EQ28 initiated SG assembly at very low concentrations (Figure 7P). In contrast, G3BP1 S149E required higher concentrations to initiate SG assembly relative to WT (Figure 7P). Together, these data indicate that IDR1 regulates G3BP1-RNA LLPS through a conformational switch that controls RNA binding and the subsequent assembly of SGs in cells and, further, that this switch can be tuned by phosphorylation status.
We were intrigued by the observation that each of the three major domains of G3BP1 were either non-essential or could be substituted by a heterologous domain with similar properties. This raised the question of whether any precise sequence or structure encoded by G3BP1 was essential to its function with respect to SG assembly. To address this question, we created a “synthetic G3BP1” molecule in which GST served as an N-terminal dimerization domain, tandem Ash-1 IDR sequences served as a disordered spacer segment, and tandem RRMs from hnRNPA1 served as a multivalent RNA-binding domain at the C terminus (Figure S7H). Remarkably, synthetic G3BP1 restored SG formation in G3BP1/2 dKO cells (Figure S7H), although the fine-tuning ability through IDR1-IDR3 interaction and phosphorylation of IDR1 was lost.
Discussion
Cells partition a large fraction of RNAs and associated proteins into RNP granules, an organizational strategy that not only governs RNA metabolism but also generates cellular compartments that do double duty by regulating other biological pathways. SGs, for example, compartmentalize a fraction of mRNAs stalled in translation, which may limit the accumulation of irreversible RNA aggregates, but also serve as a compartment that promotes other stress-related functions (Onomoto et al., 2012; Samir et al., 2019; Thedieck et al., 2013; Tsai and Lloyd, 2014; Wippich et al., 2013). The dynamic nature of RNP granules, including SGs, is essential to their function; indeed, impairment in the dynamics and material properties of RNP granules is strongly implicated as a factor in neurodegenerative diseases (Nedelsky and Taylor, 2019).
Here we introduce and functionally validate a conceptual framework for SG assembly in which assembly is driven by LLPS that occurs when the collective interactions of a core protein-RNA interaction network breach a threshold saturation concentration. This core network consists of ~36 proteins and associated mRNAs. Importantly, the interactions driving this LLPS are distributed unevenly across the core network, with some nodes more important than other. The central node of this network is G3BP1, which functions as a molecular switch that triggers RNA-dependent LLPS in response to a rise in intracellular free RNA concentrations. The threshold for phase separation – and corresponding SG assembly – is set predominantly by intrinsic properties of G3BP1 and RNA, and further regulated through positive or negative cooperativity by extrinsic factors that act upon core SG network.
With regard to the intrinsic properties of RNA that influence LLPS with G3BP, we found that length (> 250 nucleotides) and single-strandedness form the basis for this interaction. Furthermore, although intermolecular RNA-RNA interactions are not essential, they can promote assembly and lower the threshold of constituents required for LLPS. These conclusions are consistent with the observation that G3BP1 association with polyA RNA significantly increases after arsenite stress, as well as the previous report that the SG transcriptome is enriched predominantly in mRNAs and to a lesser extent non-coding RNAs (Khong et al., 2017). We found that no specific RNA sequence is required for LLPS with G3BP1, which is consistent with cross-linking experiments conducted in cells showing that G3BP1 binds a variety of coding and non-coding RNAs without a specific consensus sequence (Edupuganti et al., 2017; Martin et al., 2016b).
The intrinsic properties of G3BP1 that underlie LLPS with RNA are dimerization via the NTF2L domain and multivalency for RNA binding. Importantly, RNA binding by the RBDs (and therefore, LLPS) is controlled by a molecular switch. This switch in G3BP1 arises from competition between intra- and inter-molecular interactions (Figure S7I). When RNA concentrations are low, intramolecular interaction between the acidic IDR1 and the basic IDR3 is favored, creating a compact, “closed” conformation. Above a threshold concentration, RNA displaces IDR1 to bind IDR3, which permits the G3BP1 homodimer to adopt an expanded, “open” conformation, initiating LLPS. Thus, IDR1 functions as an autoinhibitory element that sets the threshold concentration of RNA necessary for LLPS. The autoinhibitory activity of IDR1 is regulated by IDR2, which is weakly basic and can impede IDR1 interaction with IDR3, and also by phosphorylation of IDR1, which strengthens the interaction with IDR3. Consistent findings regarding the molecular switch encoded by G3BP1 were found in the companion manuscript by Guillen-Boixet et al. (personal communication). The domain architecture of dimeric G3BP1 is another intrinsic property that influences LLPS. Dimerization via NTF2L not only doubles the valency for RNA binding, but also determines spacing between the two RBDs, with NTF2L dimerization establishing the center of the volume occupied by dimeric G3BP1 and the RBDs able to explore the periphery of this volume, which likely influence the ability of a G3BP1 homodimer to cross-link different RNA molecules and promote LLPS. In sum, our findings regarding the IDRs of G3BP demonstrate that, in contrast to the conventional view that the role of IDRs is to “drive” LLPS, IDRs in G3BP1 may have evolved to fine-tune the saturation concentration for LLPS. Beyond this important role, the IDRs in G3BP1 influence both the material properties and the composition of SGs.
Whereas intrinsic properties of G3BP1 and RNA are the most important determinants of SG formation, other proteins influence this assembly. For example, overexpression of caprin-1 and TIA1 can lead to the formation of “spontaneous” SGs in the absence of an exogenous stress (Gilks et al., 2004). This phenomenon can now be fully explained within the conceptual framework for SG assembly that we advance here. The existence of a saturation concentration for LLPS is indicative of a cooperative process (Crick et al., 2013; Posey et al., 2018). A change in the network of interactions that lowers the saturation concentration for LLPS, thus impacting the cooperative transition positively, is termed “positive cooperativity.” In contrast, a change in the network of interactions that increases the saturation concentration represents “negative cooperativity.” The impact of a variety of extrinsic factors that regulate SG assembly can be understood to act through positive or negative cooperativity. For example, our data illustrate that introduction of either caprin-1 or TIA1 to a mixture of G3BP1 and RNA strengthens the collective network of interactions and lowers the saturation concentration for LLPS, exhibiting positive cooperativity. We document this same phenomenon in cells, where the concentration of endogenous caprin-1 or TIA1 influences the G3BP1 saturation concentration necessary to trigger SG assembly. We speculate that many components of the core SG network function in this manner, lowering the saturation concentration of G3BP1 and RNA necessary for SG assembly. On the other hand, it has also been observed that overexpression of USP10, another component of the core SG network, inhibits SG assembly (Kedersha et al., 2016). How might negative cooperativity be introduced to this system? USP10 and caprin-1 compete for overlapping binding sites in the NTF2L domain and exhibit mutually exclusive interaction with G3BP1 (Kedersha et al., 2016). It is highly likely that USP10, upon being substituted for caprin-1, weakens the collective network of interactions. Notably, affinity capture proteomics using the NTF2L domain as bait reveals that a large number of G3BP1 interactors in the core SG network can be traced to NTF2L-mediated interactions (Peiguo Yang, unpublished observation), highlighting this domain as a central hub controlling network connectivity. Consistent findings regarding a central hub of interaction encoded by the NTF2L domain of G3BP1 were found in the companion manuscript by Sanders et al. (personal communication).
In conclusion, these studies illuminate the molecular basis of SG assembly and provide a conceptual framework for understanding regulation of this process that builds upon fundamental biophysical principles. Importantly, this conceptual framework accommodates all known biophysical principles and regulators of SG assembly, including the role of other proteins, post-translational modifications of proteins, duplexing or unwinding of RNA, or chemical modification of RNAs, all of which may be viewed as impinging upon the collective interactions of a core protein-RNA interaction network. Furthermore, the principles established here are likely generalizable to many biomolecular condensates. A “scaffold/client” model was recently advanced based on observations in simple, reconstituted systems where the assembly of condensates was regulated by the valency, concentration, and molar ratio of a small number of scaffold molecules (Banani et al., 2016). In light of the work presented here we can add network centrality as an important feature driving biomolecular condensation and providing a conceptual framework for how this condensation is regulated.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, J. Paul Taylor (jpaul.taylor@stjude.org). All reagents generated in this study will be made available on request but may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
HEK293T and U2OS cells (both of female origin) were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum and 1% antibiotics, and maintained at 37°C in a humified incubator with 95% air and 5% CO2. Cells were authenticated by short tandem repeat (STR) profiling. For bacterial cell culture, TOP10 and Rosetta 2(DE3) chemically competent bacteria strains were cultured in LB media containing appropriate antibiotic as described below, at 37°C in a shaker incubator (200 rpm).
METHODS DETAILS
Cell Culture and Transfection
U2OS cells were purchased from ATCC (HTB-96). U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum (HyClone SH30071.03 and SH30396.03) and maintained at 37°C in a humidified incubator with 5% CO2. FuGENE 6 (Promega, E2691) or Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) were used for transient transfections per the manufacturer’s instructions. G3BP1/2 dKO cells have been previously described (Zhang et al., 2018). U2OS cells stably expressing G3BP1-GFP have been previously described (Figley et al., 2014).
Constructs
DNA fragments encoding human G3BP1 and G3BP2 were PCR-amplified from a U2OS cDNA library. The cDNA library was created by SuperScript III First-Strand Synthesis System (Thermo, 18080051). The FKBPF36M fragment was PCR-amplified from pLVX-rHom-1 (Takara, 635062). DNA fragments encoding G3BP1 or G3BP2 were inserted into the pEGFP-C3 backbone using NEBuilder HiFi DNA Assembly Master Mix kit (NEB, E2621). Deletion mutants of EGFP-G3BP1 and EGFP-G3BP1 S149E or S149A were created by the Q5 Site-Directed Mutagenesis kit (NEB, E0554S). To create EGFP-G3BP1 swap mutants with different RNA-binding domains, RNA-binding domain fragments encoding hnRNPA1, hnRNPA2B1, hnRNPD, DAZAP1, SRSF4, TDP-43, RBMS2, hnRNPH2, RBM22, ZC3H11A, NOVA, QKI, YTHDF1, YTHDF2, YTHDF3, ADAR1, or STAU1 were amplified from a cDNA library and assembled into pEGFP-G3BP using NEBuilder HiFi DNA Assembly Master Mix. EGFP-G3BP1 swap mutants with different dimerization domains (GST or FKBPF36M) were obtained by overlapping extension-PCR methodology. Briefly, overlapping fragments encoding GST or FKBPF36M and G3BP1 ΔNTF2L were amplified by PCR, mixed in an equimolar ratio, and used as a template to generate the fusion proteins. The resulting PCR product was then cloned into pGEX-2T or pEGFP-C3 plasmids using BamHI/EcoRI and BglII/ApaI restriction enzymes, respectively. The EQ28 coding sequence, IDR1 scrambled sequences, and RKH-17A IDR2 sequence were synthesized as dsDNA gene blocks through IDT and were further assembled into a pEGFP-G3BP1 construct. The Ash1 coding sequence was amplified from a His-Ash1 plasmid reported previously (Martin et al., 2016a) and further assembled into a pEGFP-G3BP1 plasmid. For protein expression, G3BP1-coding DNA was cut by BamHI and EcoRI and ligated into the pGEX-2T plasmid in which the thrombin site had been replaced with a TEV site using the Q5 Site-Directed Mutagenesis kit (NEB, E0554S). G3BP1 deletion mutants and G3BP1 S149E or S149A were further created through the Q5 Site-Directed Mutagenesis kit (NEB, E0554S). To create tandem KH, ZnF, and tandem G3BP1-RBD constructs, XbaI-SpeI mixed sites and BioBrick Assembly method was used. All constructs were confirmed by sequencing.
Genome-Wide RNAi Screen
The human kinome collection (Dharmacon) was reverse-transfected into U2OS cells stably expressing G3BP1-GFP using Lipofectamine RNAiMAX (Life Technologies) at a final concentration of 25 nM. Briefly, reverse transfection is a process by which cells are overlaid onto a lipid/siRNA mix. Reverse transfection was employed since this process is more amenable to high-throughput assays. Cells were incubated at 43°C for 2 h at 48 h post-transfection. The cells were then fixed in 4% paraformaldehyde (EMS), stained for DAPI, and imaged with an IN Cell 6000 imaging platform (GE Healthcare). Intensity, area, count of nuclei, and GFP-positive SGs were measured using the IN Cell Developer program. A robust SSMD (strictly standardized mean difference)-based algorithm on GUItars (a GUI tool for analysis of high-throughput RNA interference screening) was used for hit selection (Goktug et al., 2012). Briefly, SSMD is defined as the ratio of the mean to the SD of the random variable representing the difference between two independent populations. The bigger the magnitude of the SSMD between two populations, the more the two populations are separate from each other. An siRNA with an SSMD ≤ −1.5 is considered a fairly moderate hit. In high-throughput assays, the data analysis methods should be robust to outliers. The robust version of SSMD (robust SSMD or SSMD*) can be obtained by replacing the mean with the median and SD with median absolute deviation (Zhang, 2011). The higher the SSMD score, the higher possibility that the siRNA is a hit (Zhang, 2007; Zhang et al., 2006). Putative positive regulators of SG assembly from the arrayed siRNA screen were identified as having a robust SSMD value of ≤ −1.5. The primary screen was performed in a single replicate, with each well containing a pool of 4 siRNAs that target the same gene. The hits with SSMD ≤ −1.5 were assayed in a confirmatory screen where the same putative hits were screened in quadruplicate. The hits from the confirmatory screen (SSMD ≤ −1.5) were further tested in a deconvolution screen where we tested the individual siRNAs for the targeted gene in quadruplicate.
Gene Ontology
Gene-annotation enrichment and functional annotation clustering of the hits acquired from the genome-wide RNAi screen were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources (Huang da et al., 2009).
Protein-Protein Interaction Network
A composite protein-protein interaction (PPI) network was built by combining STRING (v10) (Szklarczyk et al., 2015), BioPlex (Huttlin et al., 2015), and InWeb (Hopkins et al., 2017), as well as three unbiased SG interactomes (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018). Only physical protein-protein interactions were considered, and the interactome was filtered by edge confidence scores, with thresholds determined by best fitting the scale-free network property (Barabasi and Albert, 1999; Zhang and Horvath, 2005).
Network Enrichment Analysis
The significance of the generated subnetworks (e.g., RNAi and SG subnetworks) was evaluated by empirically estimating the probability of observing a denser network by random sampling 1,000 subnetworks from the entire interactome (or any background network to be tested). Network density (Wasserman and Faust, 1994) was calculated using R–igraph version 1.2.4.1.
Network Centrality Analysis
The network of 36 genes that was co-identified by the genetic screen and proteomics data was extracted from our curated PPI network. We used betweenness as the metric of node centrality and calculated the betweenness for each node in the 36-gene network using R–igraph version 1.2.4.1. Cytoscape version 3.7.1 (Shannon et al., 2003) was used for the visualization of this network in Figure 1D.
CRISPR-Cas9-Mediated Knockout Cells
gRNA for each gene was designed through http://crispr.mit.edu/ and synthesized DNA oligos were ligated into BbsI- (NEB) digested px459 vectors (Addgene, 62988) (Ran et al., 2013). Targeting vectors were verified by Sanger sequencing. WT U2OS cells were transiently transfected with gRNA vector for 1–3 days, followed by the addition of 2 μg/ml puromycin for 3 days. Knockout clones were obtained by single-cell dilution in 96-well plates and successful knockout was verified by antibody staining. For certain gene knockouts, a mixture of puromycin-selected cells were stained directly without single-cell isolation. We note that TIA1/TIAR KO cells still contain one WT allele of TIAR. TIA1/TIAR dKO cells were lethal, similar to the situation in HEK293 cells (Meyer et al., 2018).
CRISPR-Cas9-Mediated Knockin Cells
Genetically modified U2OS cells were generated using CRISPR-Cas9. Briefly, 400,000 U2OS cells were transiently co-transfected with 200 ng gRNA expression plasmid (cloned into Addgene, 43860), 500 ng Cas9 expression plasmid (Addgene, 43945), and, if required, 500 ng donor plasmid via nucleofection (Lonza, 4D-Nucleofector X-unit) using solution P3, program CM-104 in small cuvettes according to the manufacturer’s protocol. For the G3BP1-tdTomato tagged cell line, cells were single-cell sorted by FACS for tdTomato-positive cells into 96-well plates. After sorting, cells were clonally expanded and screened for the desired modification using targeted next generation sequencing and/or PCR-based assays.
Immunofluorescence and Microscopy
Cells were grown in 4-well or 8-well chamber slides (Millipore). Transfected cells were fixed with 4% paraformaldehyde (Electron Microscopy Science) in PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and then blocked with 1% BSA for 1 h, with all steps performed at room temperature. Samples were further incubated with primary antibodies in blocking buffer overnight at 4°C, then washed 3 times with PBST (0.1% Tween) and incubated with secondary antibody for 1 h at room temperature. Primary antibodies used in this study are listed in the Key Resources Table. Host-specific Alexa Fluor 488/555/647 (Thermo Fisher) secondary antibodies were used for visualization. For microscopic imaging, slides were mounted with ProLong Gold Antifade reagent with DAPI (Invitrogen). Images were captured using a Leica TCS SP8 STED 3X confocal microscope with a 63x oil objective.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ATXN2 (22/Ataxin-2) antibody | BD Biosciences | 611378; RRID: AB_398900 |
| Mouse monoclonal anti-G3BP (23/G3BP) antibody | BD Biosciences | 611126; RRID: AB_398437 |
| Rabbit polyclonal anti-ATXN2L antibody | Bethyl Laboratories | A301–370A; RRID: AB_937724 |
| Rabbit polyclonal anti-CAPRIN1 antibody | Proteintech | 15112–1-AP; RRID: AB_2070016 |
| Rabbit polyclonal anti-CSDE1 antibody | Bethyl Laboratories | A303–158A; RRID: AB_10895727 |
| Mouse monoclonal anti-DDX3 antibody | Santa Cruz | sc-81247; RRID: AB_2092867 |
| Rabbit polyclonal anti-FMR1 antibody | Bethyl Laboratories | A305–200A; RRID: AB_2631593 |
| Rabbit polyclonal anti-G3BP2 antibody | Bethyl Laboratories | A302–041A; RRID: AB_1576543 |
| Rabbit polyclonal anti-G3BP2 antibody | Proteintech | 16276–1-AP |
| Mouse monoclonal anti-GAPDH | Santa Cruz | sc47724; RRID: AB_627678 |
| Mouse monoclonal anti-GFP antibody | Santa Cruz | sc-9996; RRID: AB_627695 |
| Rabbit polyclonal anti-GST antibody | Abcam | ab19256, RRID: AB_444809 |
| Rabbit polyclonal anti-HA-tag antibody | Abcam | ab9110, RRID: AB_307019 |
| Rabbit monoclonal anti-HDAC6 antibody | Abcam | ab133493 |
| Rabbit polyclonal anti-NUFIP2 antibody | Bethyl Laboratories | A301–600A; RRID: AB_1078870 |
| Rabbit polyclonal anti-PABP antibody | Abcam | ab21060; RRID: AB_777008 |
| Rabbit polyclonal anti-PRRC2C antibody | Abcam | ab117790; RRID: AB_10903523 |
| Rabbit monoclonal anti-TAF15 antibody | Abcam | ab134916; RRID: AB_2614922 |
| Goat polyclonal anti-TIA1 antibody | Santa Cruz | sc-1751; RRID: AB_2201433 |
| Mouse monoclonal anti-TIAR (6/TIAR) antibody | BD Biosciences | 610352; RRID: AB_397742 |
| Rabbit monoclonal anti-TRIM25 antibody | Abcam | ab167154; RRID: AB_2721902 |
| Rabbit monoclonal anti-TRIM56 antibody | Abcam | ab154862; |
| Rabbit polyclonal anti-UBAP2 antibody | Bethyl Laboratories | A304–627A; RRID: AB_2620822 |
| Rabbit polyclonal anti-UBAP2L antibody | Abcam | ab138309; |
| Rabbit polyclonal anti-USP10 antibody | Proteintech | 19374–1-AP; RRID: AB_10858617 |
| Rabbit polyclonal anti-YB1 antibody | Cell Signaling Technology | 4202S; |
| Rabbit polyclonal anti-YTHDF1 antibody | Proteintech | 17479–1-AP; RRID: AB_2217473 |
| Rabbit polyclonal anti-YTHDF2 antibody | Proteintech | 24744–1-AP; AB_2687435 |
| Mouse monoclonal anti-YTHDF3 antibody | Santa Cruz | sc-377119; RRID: AB_2687436 |
| Goat polyclonal anti-eIF3η (N-20) | Santa Cruz | sc-16377; RRID: AB_671941 |
| IRDye 680LT Streptavidin | LI-COR | P/N 926–68031 |
| IRDye 800CW Goat anti-Rabbit IgG (H + L) | LI-COR | P/N 926–32211 |
| IRDye 680RD Goat anti-Rabbit IgG (H + L) | LI-COR | P/N 926–68071 |
| IRDye 800CW Goat anti-Mouse IgG (H + L) | LI-COR | P/N 926–32210 |
| IRDye 680RD Goat anti-Mouse IgG (H + L) | LI-COR | P/N 926–68070 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-31572; RRID: AB_162543 |
| Donkey anti-Goat IgG (H+L) Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-21432; RRID: AB_2535853 |
| Donkey anti-Goat IgG (H+L) Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | A-21447; RRID: AB_141844 |
| Donkey anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-21202; RRID: AB_141607 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-11008; RRID: AB_143165 |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-11029; RRID: AB_138404 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-21428; RRID: AB_141784 |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-21422; RRID: AB_141822 |
| Bacterial and Virus Strains | ||
| Rosetta 2(DE3) Competent Cells | Millipore | 71400–4 |
| HI-Control BL21(DE3) Chemically Competent Cells (SOLOs) | Lucigen | 60435–1 |
| HI-Control 10G Chemically Competent Cells (SOLOs) | Lucigen | 60110–1 |
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | C404003 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine 2000 | Invitrogen | 1168019 |
| Fugene | Promega | E2691 |
| Dharmafect Duo | Dharmacon | T-2010 |
| Protease inhibitor | Roche | 11697498001 |
| OligodT25 dynabeads | Thermo Fisher Scientific | 61005 |
| GFP-Trap_MA beads | ChromoTek | gtma-20 |
| NuPAGE LDS sample buffer (4X) | Thermo Fisher Scientific | NP0007 |
| 20% paraformaldehyde | Electron Microscopy Science | 15713-S |
| ProLong Gold Antifade Mountant with DAPI | Thermo Fisher Scientific | P36931 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | 89900 |
| SimplyBlue™ SafeStain | Thermo Fisher Scientific | LC6065 |
| SYBR™ Gold Nucleic Acid Gel Stain | Thermo Fisher Scientific | S11494 |
| TRIzol Reagent | Thermo Fisher Scientific | 15596018 |
| RNaseA | Thermo Fisher Scientific | EN0531 |
| RNase I | Life Technologies | AM2294 |
| RNase IN | Promega | N2111 |
| Turbo DNase | Thermo Fisher Scientific | AM2238 |
| Alkaline Phosphatase, Calf Intestinal | Promega | M1821 |
| Sodium Arsenite Solution | Sigma | 35000–1L-R |
| Ficoll400 | Sigma | F2637 |
| Ni-NTA agarose | GE | 17–5318-02 |
| Glutathione Sepharose 4B | GE | 17075601 |
| EZ view Red Protein A affinity gel | Millipore-Sigma | P6486 |
| EZ view Red Protein G affinity gel | Millipore Sigma | E3403 |
| EZ view Red Anti-Flag M2 affinity gel | Millipore Sigma | F2426 |
| FLAG peptide | Sigma | F3290 |
| Glutathione reduced | Sigma | G4251 |
| Imidazole | Sigma | I2399 |
| (Isopropyl β-D-1-thiogalactopyranoside) IPTG | Goldbio | 12481C100 |
| Dithiothreitol (DTT) | Sigma | 43815 |
| D/D compound | Takara | 635054 |
| SMARTpool: ON-TARGETplus CAPRIN1 siRNA | Dharmacon | L-016057–00–0005 |
| SMARTpool: ON-TARGETplus TIA1 siRNA | Dharmacon | L-013042–02–0005 |
| tRNA | Sigma | 10109495001 |
| polyA | Sigma | 10108626001 |
| polyG | Sigma | P4404–5MG |
| polyC | Sigma | P4903–10MG |
| polyU | Sigma | P9528–10MG |
| Poly(I:C)HMW | Invivogen | tlrl-pic |
| Poly(I:C)LMW | Invivogen | tlrl-picw |
| Poly(A:U) | Invivogen | tlrl-pau |
| bis(sulfosuccinimidyl)suberate (BS3) | Thermo Fisher Scientific | 21580 |
| Recombinant DDX19A | MyBioSource | MBS1371734 |
| Biotin tyramide | Sigma | SML2135 |
| H2O2 | Sigma | H1009 |
| Sodium Ascorbate | Sigma | A7631 |
| Trolox | Sigma | 238813 |
| Dynabeads™ MyOne™ Streptavidin C1 | Thermo Fisher Scientific | 65001 |
| Biotin | Sigma | B4501 |
| BSA | Sigma | A3294S |
| ATP | Sigma | A2383 |
| Critical Commercial Assays | ||
| HiScribe high yield RNA synthesis kit | New England Biolab | E2040S |
| RNA 3′ end biotinylation kit | Thermo Fisher Scientific | 20160 |
| Chemiluminescent Nucleic Acid Detection Module | Thermo Fisher Scientific | 89880 |
| polyA Spin mRNA Isolation Kit | New England Biolabs | S1560S |
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | E2621S |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0554S |
| Monarch RNA Cleanup Kit | New England Biolabs | T2040S |
| NEBNext Magnesium RNA Fragmentation Module | New England Biolabs | E6150S |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | 18080051 |
| Magnetic mRNA Isolation Kit | New England Biolabs | S1550S |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human: U-2 OS | ATCC | HTB-96 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| gRNA sequence for SG gene KO | ||
| ATXN2 | AAAGTACAGAATCCAGTTCG | |
| ATXN2L | TTTCCGGTGCACAGCATCCA | |
| CAPRIN1 | CGACAAGAAACTTCGGAACC | |
| CSDE1 | TATCATCGGACCGACGGACT | |
| DDX3 | AGTGGAAAATGCGCTCGGGC | |
| FMR1 | ATCCTTATGTGCCGCCTCTT | |
| G3BP1 | TAGTCCCCTGCTGGTCGGGC | |
| G3BP2 | CGCCCTACAAGCAGCGGACT | |
| HDAC6 | GGTGGAATCCTGGCCGGTTG | |
| NUFIP2 | TGCTCATGTTTCAGCGGCTT | |
| PRRC2C | GGTGGACGTAAACTGGGTCC | |
| TAF15 | GGAAGTTACGGTCAGTCTGG | |
| TIA1 | TATGTACTCTTTGGAGCGGG | |
| TIAR | CATATGGGCGGTCAGTGGTT | |
| TRIM25 | GTCGCGCCTGGTAGACGGCG | |
| TRIM56 | GCACTGTCTCGCGGCACTCG | |
| UBAP2 | TTTCCCGAGCACCTCGACAA | |
| UBAP2L | AGAGACTATAGTCGGCGACG | |
| USP10 | GCCTGGGTACTGGCAGTCGA | |
| Recombinant DNA | ||
| Plasmid: EGFP-C3-G3BP1 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP2b | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔNTF2L[1–142] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1ΔIDR1/2[143–334] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1ΔRBD[335–466] | This paper | N/A |
| Plasmid: EGFP-C3-GST-G3BP1[143–466] | This paper | N/A |
| Plasmid: EGFP-C3-FKBPF36M-G3BP1[143–466] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 F380/382D | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1–2xIDR2 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 S149A | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 S149D | This paper | N/A |
| Plasmid: CMV-HA-G3BP1 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPA1RRM[12–181] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPA2B1RRM[7–179] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPDRRM[99–257] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-DAZAP1RRM[10–190] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-SRSF4-RRM[2–177] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-TDP43RRM[105–261] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-RBMS2RRM[56–220] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPH2RRM[11–188] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-RBM22-ZF[159–185] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-ZC3H11A-ZF[2–110] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-NOVA-KH[51–120] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-QKI-KH[83–205] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-QKI-2KH[83–205] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-QKI-3KH[83–205] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-RBM222ZF[159–185] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-RBM223ZF[159–185] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-YTHDF1YTH[390–523] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-YTHDF2YTH[411–544] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-YTHDF3YTH[417–550] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPA1RRM[12–181] F57/59D | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPA1RRM[12–181] F148/150D | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1[1–334]-HnRNPA1RRM[12–181] F57/59/148/150D | This paper | N/A |
| Plasmid: EGFP-C3-GST-2xAsh1-IDR-HnRNPA1RRM[12–181] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 EQ28 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1–2xRBD | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1ΔIDR1[143–226] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1ΔIDR2[227–334] | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR2[227–334]-Acidic-scrambled-1 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR2[227–334]-Acidic-scrambled-2 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR2[227–334]-Acidic-scrambled-3 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1ΔIDR2[227–334]-EQ28 | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 Δ143–334]+Ash1-IDR | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR1[143–226]+Ash1-IDR | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR2[227–334]+Ash1-IDR | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1 ΔIDR [143–334]+2xAsh1-IDR | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1-IDR1/2(P-S) | This paper | N/A |
| Plasmid: EGFP-C3-G3BP1-IDR2(RKH-A17) | This paper | N/A |
| Plasmid: mCherry-G3BP1 | This paper | N/A |
| Plasmid: EGFP-Caprin-1 | This paper | N/A |
| Plasmid: CMV-Flag-G3BP1 | This paper | N/A |
| Plasmid: CMV-HA-G3BP1 | This paper | N/A |
| Plasmid: APEX2-NES-G3BP1 | This paper | N/A |
| Plasmid: APEX2-NES-G3BP1Δ IDR[143–334]+2xAsh1-IDR | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 F33W | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1Δ NTF2L[1–142] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1-NTF2L[1–142] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1-NTF2L[1–142] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1Δ IDR1/2[143–334] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1-IDR1/2[143–334] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1Δ RBD[335–466] | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1-Δ RRM[335–410] | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1-Δ RGG[411–466] | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1-RBD[335–466] | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-FKBPF36MG3BP1[143–466] | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1-TEV-HIS S149A | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1-TEV-HIS S149E | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 EQ28 | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1 NTF2L-HA | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1 NTF2L-IDR1HA | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1 NTF2LIDR1/2-HA | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 RBD | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 RBD-RA5 | This paper | N/A |
| Plasmid: pGEX-2T-thrombin-G3BP1 IDR1-S149EHA | This paper | N/A |
| Plasmid: pGEX-2T-TEV-G3BP1 IDR1-EQ28-HA | This paper | N/A |
| Plasmid: HIS-SUMO-Caprin1 | This paper | N/A |
| Plasmid: HIS-SUMO-Caprin1 F372A | This paper | N/A |
| Plasmid: HIS-SUMO-TIA1 | Mackenzie et al., 2017 | N/A |
| Plasmid: pSpCas9(BB)-2A-Puro (PX459) V2.0 | Ran et al., 2013 | Addgene 62988 |
| Plasmid: pRK793 | Kapust et al., 2001 | Addgene 8827 |
| pLVX-rHom-1 | Takara | 635062 |
| APEX2-NES | Lam et al., 2015 | Addgene 49386 |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Image Studio | LI-COR | https://www.licor.com/bio/products/software/image_studio_lite/ |
| GraphPad Prism Software | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Igor Pro 6 | Wavemetrics | https://www.wavemetrics.com/ |
| SlideBook 6 software | Intelligent Imaging Innovations | https://www.intelligent-imaging.com/slidebook.php |
| CellProfiler | Broad Institute | https://cellprofiler.org/ |
| R–igraph version 1.2.4.1 | igraph | https://rdrr.io/cran/igraph/ |
| Cytoscape version 3.7.1 | Cytoscape Consortium | https://cytoscape.org/release_notes_3_7_1.html |
| Other | ||
G3BP1 Intracellular Phase Diagrams
U2OS G3BP1/2 dKO cells were transfected with GFP-G3BP1 WT along with an siRNA pool targeting caprin-1 or TIA1, or were transfected with either GFP-G3BP1 WT or mutants (GST-G3BP1, FKBPF36M-G3BP1, S149A, S149E, or EQ28) or were co-transfected with GFP-Caprin-1 and mCherry-G3BP1. After 48 h of expression, cells were placed in fresh medium for 1 h and stressed with 100 μM sodium arsenite for 30 min at 37°C. After treatment, cells were fixed with 4% PFA and stained for PABP. Images were captured using Leica DMi8 microscope with a 20x objective. Phase diagrams were constructed by measuring GFP fluorescence intensity in each cell and assessing the presence of SGs using PABP as a marker, using Fiji software. Boxes highlight the 25% highest levels of expression of GFP-G3BP1 that did not support SG formation.
Bacterial Cell Culture
cDNA clones were transformed into One Shot TOP10 chemically competent E. coli (Thermo Fisher Scientific). Single colonies were grown overnight at 37°C in LB media containing selection antibiotic (50 μg/ml kanamycin for pEGFP vectors, 100 μg/ml ampicillin for pGEX-2T vectors). Cells were pelleted by centrifugation and lysed for harvesting DNA. All competent bacterial cells were stored at −80°C until transformation for cloning and recombinant protein expression.
Protein Purification
G3BP1 full-length and mutants were expressed and purified from E. coli Rosetta 2(DE3) cells (Millipore) and purified under native conditions unless otherwise noted. G3BP1-expressing constructs contain a TEV cleavage site between the N-terminal GST and fusion protein. The ΔNTF2L mutant as well as IDR fragments contain 2x GGS linkers between the TEV site and fusion proteins for efficient TEV cleavage. E. coli were grown to OD600 of 0.8 and induced with 0.6 mM IPTG at 16°C overnight. Pelleted cells were resuspended in lysis buffer (250 mM NaCl, 50 mM HEPES 7.5, 1 mM DTT, protease inhibitor). After sonication, lysates were pelleted at 30,000 x g at 4°C for 30 min. Supernatants were applied to packed GST columns with 10 ml GST beads (GE) prewashed with lysis buffer at room temperature. Proteins were eluted with 10 mM glutathione (Sigma) in lysis buffer. The eluted proteins were incubated with TEV protease at 4°C overnight (Kapust et al., 2001). The proteins were further treated with 0.1 mg/ml RNaseA (Thermo Fisher) to remove RNA (Molliex et al., 2015). Cleaved proteins were further purified by ion exchange with a HiTrap SP or Q column. The fractions were analyzed by SDS-PAGE, pooled, and concentrated. The proteins were then purified by Superdex 200 16/200 column (GE) equilibrated in SEC buffer (400 mM NaCl, 50 mM HEPES 7.5, 1 mM DTT). The fractions were analyzed by SDS-PAGE, pooled, concentrated, filtered, flash frozen in liquid nitrogen, and stored at −80°C.
Liquid-Liquid Phase Separation
In vitro LLPS experiments were performed at room temperature unless otherwise indicated. LLPS of G3BP1 was induced by addition of indicated concentrations of Ficoll 400 or RNA. The samples were mixed in low binding tubes (COSTAR 3206) and transferred to a sandwiched chamber created by cover glass and a glass slide with a double-sided spacer (Sigma GBL654002). Samples were observed under a DIC microscope using a Leica DMi8 microscope with a 20x objective. All imaged were captured within 5 min after LLPS induction. For LLPS experiments in which RNA was added, total RNA was isolated from U2OS cells using TRIzol (Thermo Fisher Scientific) and the concentration of RNA was measured by Nanodrop (Thermo Fisher Scientific). For LLPS experiments in which we tested mRNA length-dependent G3BP1 LLPS, we maintained a constant mass of mRNA; thus shorter mRNAs have an increased molar concentration.
D/D Solubilizer Treatment
GFP-tagged FKBPF36M-G3BP1 was transfected into U2OS G3BP1/2 dKO cells for 48 h, allowing the cells to reach 80% confluence. GFP-G3BP1 WT was used as a control. U2OS G3BP1/2 dKO cells were pre-incubated with 250 nM D/D solubilizer (Takara) for 30 min before addition of 500 μM sodium arsenite and further treated for 1 h at 37°C, 5% CO2. After treatment, the cells were fixed and stained for SG markers and DAPI.
FKBP Live-Cell Dissolution
Live cell experiments were performed with the Opterra II Swept Field Confocal Microscope (Bruker) using Prairie View 5.5 Software. G3BP1/2 dKO cells were seeded into 35-mm poly-D-lysine-coated glass bottom dishes No. 1.5 (MatTek) 48 h prior to imaging. Cells were transfected with GFP-G3BP1 WT or GFP-FKBPF36M-G3BP1 24 h prior to imaging using FuGENE 6 (Promega). Immediately before imaging, the medium was changed to 1 mL complete phenol red-free DMEM medium (HyClone). During imaging, cells were maintained at 37°C and supplied with 5% CO2 using a Bold Line Cage Incubator (Okolabs) and an objective heater (Bioptechs). Imaging was performed using a 60x Plan Apo 1.40NA oil objective and Perfect Focus (Nikon) was engaged for the duration of the capture. A multipoint time lapse was taken, imaging each field every 30 sec with the 488-nm laser set at 80 power with 75-ms exposure using a 35-μm slit. Two min into imaging, 500 μM sodium arsenite (Sigma) was added to the sample. Thirty-two min into imaging, 250 nM D/D solubilizer was added while maintaining the 500 μM sodium arsenite concentration. Images were imported using the Bio-Formats plugin and analyzed in ImageJ 1.51J (NIH). The intensity of the SGs was quantified on the frame immediately before and after D/D solubilizer addition, and set relative to the cytoplasmic intensity of the cell for the frame. The data was normalized to the maximum intensity of GFP-G3BP1 WT or GFP-FKBPF36M-G3BP1 on the frame prior to addition of D/D solubilizer.
Protein Solubility Assay
G3BP1/2 dKO U2OS cells were transfected with GFP-tagged G3BP1 FL or IDR1/2 deletion constructs. ~24 h after transfection, cells were washed with ice-cold 1x PBS and lysed in ice-cold RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; Thermo Fisher, 89900). Cell lysates were then passed through a 21-gauge needle 10 times before being cleared by centrifugation at 21,000 x g for 30 min at 4°C to generate the RIPA-soluble fraction. The pellets were washed twice with ice-cold RIPA buffer and re-centrifuged for 10 min. RIPA-insoluble pellets and soluble fractions were mixed with 1x LDS sample buffer containing 0.1 M DTT and further boiled at 75°C for 5 min. Proteins were resolved by NuPAGE Novex 4%–12% Bis-Tris Gel (Thermo Fisher) and followed by Western blotting for GFP.
Western Blotting
Cells were washed with PBS and lysed with RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; Pierce, 89901) supplemented with proteinase inhibitor cocktail (Roche, 1186153001). Lysates were centrifuged for 10 min at 4°C at 21,000 x g. 4X NuPAGE LDS sample buffer (Thermo Fisher Scientific, NP0008) with 25 mM DTT was added to the supernatant and samples were boiled at 70°C for 5 min. Samples were separated in 4–12% NuPAGE Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes using an iBlot 2 transfer device (Thermo Fisher Scientific). Membranes were blocked with Odyssey blocking buffer (LI-COR) and then incubated with primary antibodies at 4°C overnight. Membranes were washed 3 times with PBST (0.1% Tween) and further incubated with dye-labeled secondary antibodies. Membranes were visualized with an Odyssey Fc imaging system (LI-COR).
Fluorescence Recovery After Photobleaching
FRAP experiments were performed with the Opterra II Swept Field Confocal Microscope (Bruker) using Prairie View 5.5 Software. G3BP1/2 dKO cells or G3BP1-tdTomato knock-in cells were seeded into a 4-well lab-Tek chambered coverglass (Nunc) 48 h prior to imaging. Cells were transfected with GFP-G3BP1 WT or mutants 24 h prior to imaging using FuGENE 6 (Promega). Immediately before imaging, the medium was changed to 1 mL complete phenol red-free DMEM medium (HyClone). During imaging, cells were maintained at 37°C and supplied with 5% CO2 using a Bold Line Cage Incubator (Okolabs) and an objective heater (Bioptechs). Imaging was performed using a 60x Plan Apo 1.40NA oil objective and Perfect Focus (Nikon) was engaged for the duration of the capture.
For imaging, cells without spontaneous SGs and of similar intensity were selected and the fields were stored as stage locations. The cells were then incubated with 500 μM sodium arsenite (Sigma) for 30 min. Following incubation, time lapses were taken using both the 488-nm and 561-nm imaging lasers set at 100 power and 100-ms exposure each set at max speed (~0.27 ms period) for 100 frames. Photobleaching of SGs occurred 2 sec into capture, using the 488-nm FRAP laser to bleach both the green and red channels simultaneously. A 3 μm-diameter circle was used for all FRAP experiments. Data was repeated in triplicate for each condition, with each replicate having at least n = 15 cells.
Data was opened in ImageJ 1.51J (NIH) using the Prairie Reader plugin. ROIs were generated in the photobleach region, a non-photobleached cell, and the background for each time lapse, and the mean intensity of each was extracted. These values were exported into Igor Pro 7.0 (WaveMetrics), where photobleach and background correction were performed, and fit FRAP curves were generated.
PolyA RNA Co-Immunoprecipitation
U2OS cells were exposed to 500 μM sodium arsenite for 1 h at 37°C. After treatment, cells were washed 3 times with PBS and harvested in 200 μL lysis buffer containing 20 mM Tris HCl, pH 7.5, 2.5 mM MgCl2, 100 mM NaCl, 0.5% Triton, protease inhibitors, 0.2 U/mL RNase inhibitors (Promega). Cells were incubated 15 min on ice and centrifuged for 5 min at 1,000 x g at 4°C. 100 μL oligo-dT Dynabeads (Invitrogen) pre-equilibrated with lysis buffer were added and binding was performed for 30 min at 4°C on a rotating wheel. Dynabeads were washed five times with 20 mM Tris HCl, pH 8, 150 mM NaCl, 0.02% NP-40, 0.1 U/mL RNase inhibitors, for 5 min on ice. Following this step, 50 μL of 20 mM Tris HCl, pH 7.5, was added to the beads and heated for 45 sec at 80°C. Supernatant was then quickly separated from beads. Half of the sample was analyzed by Western blot using anti-G3BP1 antibody and the other half was run on TBE-urea 6% gels and stained with SYBR Gold (Invitrogen) to detect RNA. The same experiment was performed in the absence of RNaseIn and in the presence of 0.1 U/mL RNase A (Invitrogen) as a negative control.
Depletion of PolyA RNA
Total cellular RNA was isolated from U2OS cells using TRIzol (Thermo Fisher, 15596018) and dissolved in H2O. The concentration of RNA was measured by Nanodrop (Thermo Fisher Scientific). For polyA depletion, 100 μg total RNA was mixed with 500 μl lysis/binding buffer (NEB) and 100 μl washed magnetic oligo d(T)25 beads (NEB, S1550S) was added to the RNA sample. After incubating the sample at room temperature for 5 min, another 100 μl fresh beads were added. After 2 rounds of polyA RNA extraction, the leftover RNA was precipitated by isopropanol and washed by 75% ethanol and further resuspended in H2O.
In Vitro Transcription
DNA templates encoding full length ACTB, HSPA8, PABPC1, EEF2 and fragments of HSPA8 were amplified from a U2OS cDNA library. T7 promoter sequence: TAATACGACTCACTATAGGGAG was added in the 5′ end of the DNA template. Transcription was carried out overnight at 37°C in a final volume of 20 μL and using 0.5 μg DNA template using HiScribe RNA synthesis kit (NEB, E2040S). The reaction was stopped by the addition of DNase I, transcription and quality of RNA was checked by agarose gel, and RNA was purified using Monarch RNA purification kit (NEB).
RNA Treatment with DDX19A Helicase
1 μg/μl total cellular RNA was incubated with 0, 0.1, or 0.2 μM purified DDX19A (MyBioSource) overnight at 37°C in 30 mM HEPES, pH 7.5, 75 mM NaCl, 5 mM MgCl2, 0.2 mM DTT, in presence or absence of 2 mM ATP, to allow the disruption of RNA-RNA interactions. Pre-treated RNA was then diluted 10 times in 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT. 50 ng/μL helicase-treated RNA was then mixed with 50 μM purified G3BP1 and imaged by DIC using a Leica DMi8 microscope with a 20x objective. In parallel, integrity and unwinding of RNA was analyzed on denaturing TBE-urea and native TBE RNA gels respectively. RNA was detected using SYBR Gold.
RNA Biotinylation
Purified RNA was biotinylated using a Thermo Fisher RNA 3′ end biotinylation kit as recommended by the manufacturer. Briefly, a final concentration of 500 μM RNA was incubated with 2 mM biotinylated cytosine, T4 RNA ligase, PEG, and RNase inhibitors overnight at 16°C. RNA was then purified using a Monarch Total RNA Miniprep Kit and quantified by absorbance at 260 nm.
Electrophoretic Mobility Shift Assay
RNA binding of purified G3BP1 WT and mutants was assessed by incubating increasing concentrations of G3BP1 with 2 nM biotinylated RNA (20 bases –AGAUUCCACCACAAAGACCC - for purified G3BP1 and 100 bases – AGCCUGAAUCUCAGAUUCCACCACAAAGACCUCAGCGGGAUCAAAGAGUGCGAGA ACAACGAAUAAAUAUUCCUCCCCAAAGGGGACCCAGACCAAUCCG - for FLAG-tagged G3BP1 and purified G3BP1 RBD mutants) for 30 min at RT, in HEPES 50 mM, pH 7.5, 150 mM NaCl, 1 mM DTT, 10 mM MgCl2. Complexes were then separated on TBE 4–12% gels at 100 V in TBE buffer 0.5X and transferred on a nylon membrane. RNA was revealed using Thermo Fisher Chemiluminescent Nucleic Acid Detection Module Kit according to manufacturer instructions.
GST Pulldown
100 nM GST, GST-RBD, or GST-RBDRA5 were incubated with 100 nM NTF2L-HA, NTF2-IDR1-HA WT or mutants, or NTF2L-IDR1/2-HA for 1 h at room temperature, in 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT. Complexes were pulled down using Glutathione-Sepharose 4B resin for 2 h at 4°C, washed 3 times with 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and analyzed by Western blot using anti-GST and anti-HA antibodies.
Size Exclusion Chromatography Multi-Angle Light Scattering
Size exclusion chromatography multi-angle light scattering (SEC-MALS) experiments were carried out using a WTC-0150S5 (MW range 5000–1,250,000 Da) size-exclusion column (Wyatt Technologies) with 3 detectors connected in series: an Agilent 1200 ultraviolet (UV) detector (Agilent Technologies), a Wyatt DAWN-HELEOS-II multi-angle light-scattering (MALS) detector, and a Wyatt Optilab T-rEX differential refractive index (RI) detector (Wyatt Technologies). The column was equilibrated with PBS. All data were collected at 25°C. A 100-μL sample was injected into the column using an auto-sample injection method with a flow rate of 0.5 ml/min. Protein in the eluent was detected via UV absorbance at 280 nm, light scattering, and refractive index detectors. The data were recorded and analyzed with Wyatt Astra software (version 7.0). The refractive index increment, dn/dc, was assumed to be 0.185 ml/g for measuring the concentration of the protein samples. EASI Graphs (Astra software) were exported and plotted as a molar mass distribution superimposed on light scattering and UV traces versus elution volume.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) following standard protocols unless mentioned otherwise (Zhao et al., 2013). Samples (50 μM G3BP1) in buffer containing 20 μM PBS, 50 mM HEPES, pH 7, 400 mM NaCl, 5 mM DTT were loaded into cell assemblies composed of double sector charcoal-filled centerpieces with a 12-mm path length and sapphire windows. Buffer density and viscosity were determined in a DMA 5000 M density meter (Anton Paar) and an AMVn automated micro-viscometer (Anton Paar), respectively. The partial specific volumes and the molecular masses of the proteins were calculated based on their amino acid compositions in SEDFIT (https://sedfitsedphat.nibib.nih.gov/software/default.aspx) (Cohn and Edsall, 1943). The cell assemblies, containing identical sample and reference buffer volumes of 390 μL, were placed in a rotor and temperature equilibrated at rest at 20°C for 2 h before it was accelerated from 0 to 50,000 rpm. Rayleigh interference optical data were collected at 1-min intervals for 12 h. The velocity data were modeled with diffusion-deconvoluted sedimentation coefficient distributions c(s) in SEDFIT (https://sedfitsedphat.nibib.nih.gov/software/default.aspx) using algebraic noise decomposition and with signal-average frictional ratio and meniscus position refined with non-linear regression (Schuck, 2000). The s-value was corrected for time and finite acceleration of the rotor was accounted for in the evaluation of Lamm equation solutions (Zhao et al., 2015). Maximum entropy regularization was applied at a confidence level of P −0.68. The partial specific volumes of the proteins, based on their amino acid composition, were calculated in SEDFIT. All plots were created in GUSSI (http://biophysics.swmed.edu/MBR/software.html) (provided by Dr. Chad Brautigam). A two-dimensional size-shape distribution, c(s, f/f0) (with the one dimension the s-distribution and the other the f/f0-distribution) was calculated with an equidistant f/f0-grid of 0.25 steps that varied from 1 to 3.5, a linear s-grid from 0.2 to 8 S with 100 s-values. Tikhonov-Phillips regularization was at one standard deviation. The velocity data were transformed to c(M,f/f0), c(s,f/f0), and c(s,M) distributions, with M the molecular weight, f/f0 the frictional ratio, s the sedimentation coefficient, and plotted as contour plots. This analysis was with regularization. The color temperature of the contour lines indicates the population of species. As in one-dimensional c(s), the peak width in c(s,M) contains contributions both from regularization (reflecting limited resolution given the signal-to-noise ratio of the data) and from true heterogeneity. The dotted lines of c(M, f/f0) indicate constant s and for the c(s,M) plot constant f/f0. The distributions were not normalized (Brown and Schuck, 2006; Zhao et al., 2013).
Cross-Linking of Recombinant Proteins
10 μM recombinant proteins were incubated with 100 μM amine-specific cross-linker BS3 in a final volume of 20 μl for 30 min at room temperature. The cross-linking reaction was quenched by the addition of 50 mM Tris HCl, pH 7.5, and incubated for at least 15 min at room temperature. Samples were then analyzed by SDS-PAGE.
Cross-Linking of Cell Extracts
U2OS cells or U2OS G3BP1/2 dKO cells expressing indicating constructs were harvested in PBS, pelleted at 800 x g for 5 min and resuspended in 200 μl PBS buffer, pH 7, supplemented with 0.1% Triton X-100 and protease inhibitors. Cells were then incubated on ice for 15 min and centrifuged for 5 min at 1000 x g at 4°C. Supernatants were collected and protein concentration was assessed using Bradford reagent and BSA. 1 mg/ml protein lysate was then incubated with 2.5 mM BS3 for 30 min at room temperature in a final volume of 200 μl. The reaction was quenched by the addition of 50 mM Tris HCl, pH 7.5, and incubated for 15 min at room temperature. Samples were then separated by Tris-acetate 3–8% SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked with 5% BSA prepared in TBS + 0.1% Tween-20 and analyzed using mouse anti-G3BP1 antibody.
RNA Binding Assay of Recombinant G3BP1 with Isolated RNA
5 μM recombinant GST-tagged-G3BP1 was mixed with 50 ng/μl indicated RNA in 50 mM HEPES buffer, 150 mM NaCl, 1 mM DTT. Samples were incubated for 10 min at room temperature to allow RNA binding, followed by exposure to UV (254 nm, 400 mJ/cm2 followed by 200 mJ/cm2). Four volumes of binding buffer containing 20 mM Tris HCl, pH 7.5, 137 mM NaCl, 1% Triton X-100, 2 mM EDTA, and 1x protease inhibitor (Roche) were added to one volume of G3BP1/RNA mix. Samples were treated with 4 U/mL of RNase I for 3 min at 37°C and the reaction was stopped by the addition of RNaseIn (0.4 U/μL). GST-G3BP1/RNA complexes were then isolated with Glutathione-Sepharose 4B beads for 2 h at 4°C on a rotating wheel. For RNA detection, RNA-G3BP1 complexes were treated with alkaline phosphatase (Promega, M1821) and RNA was biotinylated using an RNA 3’ End Biotinylation Kit (Pierce) as described below.
RNA Binding Assay of Cellular G3BP1 WT and Mutants
The RNA binding assay was adapted from a previous report (Valentin-Vega et al., 2016). U2OS G3BP1/2 dKO cells expressing GFP-G3BP1 WT or indicated mutants were washed with PBS and exposed to UV (254 nm, 400 mJ/cm2 followed by 200 mJ/cm2). Cells were then harvested in lysis buffer containing 20 mM Tris HCl, pH 7.5, 137 mM NaCl, 1% Triton X-100, 2 mM EDTA, 1x protease inhibitor (Roche) and incubated for 10 min on ice. Lysates were then successively treated with 8 U/mL DNase I for 5 min at 37°C and 4 U/mL RNase I for 3 min at 37°C. The reaction was stopped by the addition of RNaseIn (0.4 U/μL). Lysates were then centrifuged at 21,000 x g for 10 min at 4°C and 10 μl GFP-Trap beads (Chromotek, gtma-20) were incubated with the supernatant fraction at 4°C overnight. The beads were washed twice with lysis buffer, twice with 1 M NaCl, and twice with lysis buffer again. Beads were further suspended in 100 μl 10 mM Tris-HCl, pH 7.5, and treated with 2 units of calf intestinal alkaline phosphatase (Promega, M182A) at 37°C for 10 min at 1000 rpm in a Thermomixer. Beads were washed with lysis buffer twice and RNA labeling was performed with an RNA 3′ End Biotinylation Kit (Pierce, 20160). After washing with lysis buffer twice, beads were boiled in 1x LDS sample buffer (Life Technologies, NP007) with 100 mM DTT. Protein-RNA complexes were separated by 4–12% NuPAGE Bis-Tris gels, transferred to nitrocellulose membranes, and blotted with IRDye 680LT streptavidin (LI-COR, 926–68031).
Measurement of G3BP1 RNA-Binding Affinity Before and After Stress
FLAG-G3BP1 was transiently expressed in U2OS G3BP1/2 dKO cells for 24 h. Cells were then exposed to 500 μM sodium arsenite for 1 h, quickly washed with PBS, and harvested in lysis buffer containing PBS, pH 7, 5 mM EDTA, 0.2% NP-40, 10% glycerol, 10 μg/mL RNase A, 0.01 u/mL DNase I and protease inhibitors. Cells were further lysed by passing through a 21-gauge needle attached to a 1 mL syringe 5 times. Samples were centrifuged for 5 min at 1000 x g at 4°C, and the supernatant was collected. The lysate was pre-cleared for 1 h at 4°C using protein A/G beads and then incubated with anti-FLAG M2 beads for 90 min at 4°C on a wheel. Beads were successively washed with lysis buffer (twice), lysis buffer + 250 mM NaCl (400 mM NaCl final concentration; twice) and lysis buffer (twice). Finally, FLAG-G3BP1 was eluted by incubating the beads in lysis buffer + 200 μg/mL FLAG peptide for 30 min at room temperature while shaking. Supernatant containing FLAG-G3BP1 was collected and dialyzed against 50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT overnight at 4°C. Relative FLAG-G3BP1 concentrations were assessed by Western blot and equal concentrations were used for EMSA. Free and bound RNA fractions were quantified using FIJI software and data were plotted against the Hill-Langmuir equation. Midpoint values for G3BP1 before and after stress were 0.211 ± 0.028 and 0.173 ± 0.095 respectively.
APEX2-Mediated Proximity Labeling of Stress Granules
APEX2-G3BP1 and APEX2-G3BP1–2xAsh1 plasmids were constructed using pcDNA3 APEX2-NES, a gift from Alice Ting (Addgene plasmid 49386; http://n2t.net/addgene:49386; RRID:Addgene_49386). 2 μg APEX2-G3BP1 and APEX2-G3BP1–2xAsh1 plasmids were transfected into 10-cm dishes of G3BP1/2 dKO U2OS cells with Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) overnight. Cells were then exposed to 0.5 mM sodium arsenite for 1 h. After 30 min arsenite treatment, 0.5 mM biotin phenol (Sigma, SML2135) was added into the medium for a total of 30 min of incubation. Cells were further treated with 1 mM H2O2 for 1 min followed by quenching buffer (10 mM sodium ascorbate and 5 mM Trolox in 1x PBS). Cells were lysed with 1 ml RIPA buffer (25 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail, 10 mM sodium ascorbate, 5 mM Trolox). The lysates were cleared by centrifugation at 21,000 x g for 10 min at 4°C and then dialyzed in 500 ml RIPA buffer (25 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) using Slide-A-Lyzer Dialysis Cassettes (10K MWCO) (Thermo Fisher Scientific, 66380) at 4°C overnight to remove free biotin phenol. Dialyzed lysates were incubated with 100 μl streptavidin magnetic beads (Thermo Fisher Scientific, 65001) at 4°C overnight with gentle rotation. Beads were then washed with 2× 1 mL RIPA lysis buffer, 1× 1 ml of 1 M NaCl, 1× 1 mL of 2 M urea in 10 mM Tris HCl, pH 8.0, and again with 2× 1 mL RIPA lysis buffer. To elute the biotinylated proteins, the beads were boiled for 10 min in 50 μl 1x NuPAGE LDS sample buffer (Thermo Fisher Scientific, NP0007) supplemented with 20 mM dithiothreitol DTT and 2 mM biotin. The streptavidin eluate was collected and run on an 10% SDS-PAGE gel for mass spectrometry. Proteins identified were plotted based on fold change of G3BP1_FL/2xAsh1 Swap as well as P value.
Small Angle X-ray Scattering
Small angle X-ray scattering (SAXS) experiments were performed at BioCAT (beamline 18ID at the Advanced Photon Source, Chicago) with in-line size exclusion chromatography (SEC-SAXS) to separate dimeric protein from aggregates and ensure the best possible buffer subtraction. Samples were injected onto a Superdex 200 Increase 5/150 GL column (GE Lifesciences) preequilibrated in the desired buffer and ionic strength using an FPLC running at 0.45 mL/min. The output of the column passed through UV and conductance monitors prior to injection into a coflow capillary system (Kirby et al., 2016) in which the sample is sheathed in a matched buffer allowing for slower flow without risk of radiation damage. Scattering intensity was recorded using a Pilatus3 1M (Dectris) detector placed 3.5 m from the sample providing access to a q-range from 0.004–0.4 Å−1. 0.5-second exposures were acquired every second during the elution. Data were reduced using BioXTAS RAW 1.4.0 (Hopkins et al., 2017). The contribution of the buffer to the X-ray scattering curve was determined by averaging frames from the SEC eluent, which gave rise to baseline levels of integrated X-ray scattering, UV absorbance and conductance. Frames were selected as close to the protein elution as possible and, ideally, frames pre- and post-elution were averaged. Final q versus I(q) data sets were generated by subtracting the average buffer trace from all elution frames and averaging curves from elution volumes close to the maximum integrated scattering intensity; these frames were statistically similar in both small and large angles. Buffer subtraction, subsequent Guinier fits, and Kratky transformations were done using custom MATLAB (Mathworks) scripts.
In Silico Modeling
The size distribution of G3BP1 was determined by generating a large pool of conformations where the structures of the folded domains were held fixed and the IDRs were modeled as random self-avoiding chains. The NTF2L dimer structure was fixed to those in the crystal structure 4FCM (Vognsen et al., 2013). Presently there exists no set of coordinates for the G3BP1 RRM; however, substantial sequence conservation with known RRMs allowed for reliable homology modeling implemented in Swiss-Prot (Waterhouse et al., 2018). The IDRs linking the folded regions and C-terminal IDR3 were modeled using the “Random” algorithm in Ranch (Bernado et al., 2007). This algorithm builds random chains via addition of residues using a pool of accepted peptide backbone angles. The only restriction imposed by the algorithm on the conformations is that there are no steric clashes. Sidechains are ignored, and the resulting IDRs are simply an alpha carbon trace of the peptide backbone. The Ranch algorithm implemented in this way generates a pool of conformations in which all IDRs are close to the self-avoiding random walk limit, i.e., there are no attractive interactions between IDRs and/or folded domains. For this study, 20,000 independent conformations were generated to ensure proper sampling of the conformational space.
Radii of gyration for the random pool of conformations were calculated by using the ‘measure radius’ function in VMD (Humphrey et al., 1996) on a selection containing all alpha carbons. This method does not account for the contribution of hydration shell water to the radius; thus the numerical values are a slight underestimate with respect to values measured by SAXS. Analysis of the distribution of radii was performed using MATLAB (Mathworks). The distances between different regions of G3BP1 were calculated by first measuring the location of the centers of mass of the relevant atoms, then calculating the vector distance between the two centers of mass. In order to produce a relevant metric to correlate the IDR distances to the global radius, we wanted to create a value that informed on whether IDR3 of each monomer was close in space to IDR1 of either monomer. This value was computed using the equation:
where rxx represent the relevant distances between IDR3 on monomer “a” and “b”.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Stress Granule Rescue in U2OS Cells by G3BP1 Mutants
U2OS G3BP1/2 dKO cells transiently expressing indicated G3BP1 mutants were exposed to 500 μM sodium arsenite for indicated times (0, 10 min, or 60 min) to induce SG formation, fixed, and stained for eIF3η. All images were taken with the same settings to allow the comparison of expression level between different conditions. For each cell, average GFP intensity was measured using Fiji software to control for expression level of the GFP-fused proteins, and cells were scored for the presence of SGs using eIF3η as a SG marker. When possible, only equivalent levels of expression were used to quantify SG rescue. Otherwise, all cells were considered for the quantification.
Enrichment of GFP-G3BP1 WT and Mutants in Stress Granules
The SG enrichment percentage of G3BP1 FL protein and G3BP1 2xRBD protein were quantified by CellProfiler as previously described (Mackenzie et al., 2017). Briefly, the cell boundary and the boundary of SGs were identified using CellProfiler. The ratios of GFP fluorescent signal intensity in the SGs to that in the entire cell were used as indicators for the SG enrichment percentages of these two proteins. At least 100 cells were included in each condition. ****P < 0.0001 by unpaired t-test.
Statistical Analysis
P > 0.05 was considered not significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by two-tailed Student’s t test, one-way ANOVA, or Log-rank (Mantel-Cox) test. Statistical analysis was performed in GraphPad Prism or Excel.
DATA AND CODE AVAILABILITY
Data Resources
The published article includes all datasets generated or analyzed during this study.
Supplementary Material
Figure S1. Genome-Wide Screen Followed by CRISPR-Cas9 Knockout Identifies G3BP1/2 as Factors Uniquely Required for SG Assembly, Related to Figure 1
(A) Experimental workflow. U2OS cell lines were transfected with siRNA. Forty-eight hours post-transfection, cells were incubated at 43°C for 2 h, then fixed in 4% paraformaldehyde and stained for DAPI.
(B) Workflow for assay development of the high-throughput screen.
(C) Primary screen hits plotted in a waterfall diagram. Hits with SSMD ≤ 1.5 were selected for a further confirmation screen.
(D) Confirmatory screen hits plotted in a waterfall diagram. Hits with SSMD ≤ −1.5 were selected for further deconvolution.
(E) Results from the deconvolution screen showing the number of individual siRNAs (of a total of four siRNAs per gene) that yielded a significant effect on SG assembly.
(F) Gene ontology analysis of the hits of the deconvolution screen. For each functional category, the number of genes in that category that were identified in our screen is shown in blue; these numbers are graphed in proportion to the total number of genes implicated in that functional category in the dataset used for our analysis.
(G) Comparison of genes found in our screen to regulate assembly of SGs during heat shock compared to those previously identified as regulators of SG assembly during arsenite stress (Ohn et al., 2008).
(H) U2OS cell lines were generated using CRISPR-Cas9 to knock out genes identified as potential regulators of SG assembly. Cells were exposed to sodium arsenite (500 μM, 1 h), stained for G3BP1 to identify SGs, and imaged. All knockout lines shown retained the ability to form SGs in response to arsenite. 70% of UBAP2/UBAP2L dKO cells showed normal SG assembly, but 30% failed to assemble SGs. Scale bar, 20 μm.
(I) WT and G3BP1/2 dKO cells were incubated at 43°C for 1 h or exposed to 0.4 M sorbitol for 1 h, stained for G3BP1 and eIF3η, and imaged. G3BP1/2 dKO cells still formed eIF3η-positive foci under both heat shock and osmotic stresses. Scale bar, 10 μm.
Figure S2 Undergoes RNA-dependent Phase Separation, Related to Figure 2
(A) LLPS of purified recombinant G3BP1 full-length (FL) protein was assessed in the context of increasing concentrations of the crowding reagent Ficoll and increasing concentrations of protein. LLPS of G3BP1 was dependent on G3BP1 concentration and the concentration of Ficoll, Scale bar, 20 μm.
(B) U2OS cells stably expressing G3BP1-GFP were exposed to sodium arsenite (500 μM) and further treated with 0.2% Tween 20 in PBS for 10 min. Cells were imaged in PBS alone or with RNase I (1 U/μl) + RNase A (100 μg/ml) in PBS and imaged for indicated time. Scale bar, 50 μm.
(C) G3BP1/2 dKO cells were transiently transfected with FLAG-G3BP1 and exposed to sodium arsenite (500 μM, 1 h). FLAG-G3BP1 was isolated and the RNA-binding affinity of FLAG-G3BP1 was assessed by EMSA. Free and bound RNA fractions were quantified and plotted as a function of relative concentrations of FLAG-G3BP1. Error bars indicate SD.
(D) 5 μM GST-G3BP1 WT was incubated with 50 ng/μl of indicated RNA at room temperature and exposed to UV. Complexes were treated with 4 U/ml RNase I for 3 min and isolated by GST pulldown. RNA was then biotinylated for further detection. Complexes were analyzed by anti-GST immunoblot to detect G3BP1and streptavidin to detect RNA.v(E) LLPS of purified recombinant G3BP1 was assessed in the absence of a crowding reagent, 150 mM NaCl, and the addition of long single-stranded PolyA, PolyG, PolyC, or PolyU.
See also Video S1.
Figure S3. In Vitro Phase Separation of G3BP1 with RNA Strongly Predicts SG Reconstitution in Cells, Related to Figure 3
(A) G3BP2 domains are shown aligned with results of PONDR (Predictor of Natural Disordered Regions) and NCPR (net charge per residue; 5-amino acid window) analyses.
(B) The amino acid sequence of human G3BP2 is shown with major domains marked by highlighted colors. Charged residues are shown in blue (basic; Arg and Lys) and red (acidic; Asp and Glu).
(C) LLPS of G3BP1 deletion constructs in which the NTF2L domain, IDR1/2, or RBD have been removed. LLPS of purified recombinant protein was assessed in the presence of a crowding agent. G3BP1 lacking the NTF2L or RBD domains failed to phase separate, whereas G3BP1 lacking IDR1/2 did phase separate. Scale bars, 50 μm.
(D) Representation of the phase separation results obtained in (C).
(E) G3BP1 ΔIDR3 fails to assemble SGs. We tested two IDR3 deletion mutants, G3BP1 ΔIDR3 (Δ410–466) and G3BP1 ΔIDR3 (Δ428–466). Although G3BP1 ΔIDR3 (Δ410–466) accumulated in cytoplasmic puncta, these puntca formed in the absence of stress, did not contain SG markers, and did not resolve after the removal of stress, suggesting that these were insoluble aggregates, not SGs. Modifying these boundaries to residue 428 to 466 (G3BP1 ΔIDR3 (Δ428–466)) alleviated this aggregation, but G3BP1/2 dKO cells expressing this construct failed to assemble SGs. Scale bar, 20 μm.
(F) Puncta formed by G3BP1 ΔIDR3 (410–466) were analyzed by FRAP and showed poor fluorescence recovery. Error bars indicate SD. ***P < 0.001, ****P < 0.0001, ns; not significant by two-way ANOVA, Sidak’s multiple comparisons test.
(G) G3BP1/2 dKO cells were transfected with GFP-tagged G3BP1 Δ340–415, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. The majority of cells transfected with G3BP1 Δ340–415 did not form SGs, whereas few cells showed weak rescue of SG formation. Scale bar, 20 μm.
(H) Quantification of result shown in (G). Error bars indicate SD. **P = 0.0021 by unpaired t-test.
Figure S4. Multivalency in RNA Binding Mediates SG Formation, Related to Figure 4
(A) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. Twenty-four hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
(B) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. Cellular protein-RNA complexes were cross-linked with UV and cells were lysed. Immunoprecipitated RNA cross-linked to G3BP1 was then treated with alkaline phosphatase and biotinylated. Complexes were subjected to SDS-PAGE and immunoblotting for biotin (detection of RNA) and GFP (detection of G3BP1). RNA binding was quantified and normalized to protein signal. Results show quantification from duplicate experiments. Error bars indicate SD. *P = 0.0136 vs. FL by one-way ANOVA, Dunnett’s multiple comparisons test.
(C) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants and assessed by immunoblot as in (A).
(D) G3BP1/2 dKO cells were transfected with G3BP1 constructs in which the RBD was substituted with indicated combinations of ZnF domains. Cells were exposed to sodium arsenite (500 μM, 1 h) and stained for eIF3η. G3BP1 with a single ZnF domain did not support SG formation, but G3BP1 with two or more ZnF domains did support SG formation. Scale bar, 20 μm.
(E) SGs formed by indicated constructs were analyzed by FRAP. SGs assembled with 3 KH or ZnF domains were significantly less dynamic than those assembled with 2 KH or ZnF domains. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparisons test.
(F) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or G3BP1 2xRBD and exposed to sodium arsenite (500 μM). SG formation was monitored using live cell imaging beginning when arsenite was added to the medium. Duplication of the G3BP1 RBD dramatically enhanced SG assembly. Scale bar, 20 μm.
(G) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or G3BP1 2xRBD for 24 h. SGs were counted as spontaneous (no arsenite) or arsenite-induced (500 μM, 1 h). Cells were immediately fixed with 4% PFA and representative images were shown for each condition. Scale bar, 50 μm.
(H) Quantification of (G). The percentage of cells with spontaneous or arsenite-induced SGs are shown. Cells with at least 3 granules were counted as a SG-positive cell. *P = 0.0431 by two-way ANOVA, Sidak’s multiple comparisons test. Error bars indicate SD.
(I) The ratio of GFP signal in SGs to the entire cell (GFP enrichment in SGs) from results obtained in (G). The SG enrichment percentage of G3BP1 FL protein and G3BP1 2xRBD protein were quantified by CellProfiler as previously described (Mackenzie et al., 2017). Briefly, the cell boundary and the boundary of SGs were identified by CellProfiler. The ratios of GFP fluorescent signal intensity in the SGs to that in the entire cell were used as indicators for the SG enrichment percentages of these two proteins. At least 100 cells were included in each condition. ****P < 0.0001 by unpaired t-test. Error bars indicate SD.
(J) SGs formed by G3BP1 2xRBD were analyzed by FRAP and showed poor fluorescence recovery. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparisons test.
(K) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. 16 to 24 hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
Figure S5. NTF2L Domain-Mediated Dimerization and Interaction with Core SG Components Regulate Assembly of SGs, Related to Figure 5
(A) Size exclusion chromatography (SEC) multi-angle light scattering (MALS) on purified G3BP1 at concentrations of 20, 50, or 100 μM protein. Inset shows a distribution of average molecular weights superimposed on the chromatogram of UV absorbance at 280 nm.
(B) Sedimentation velocity analytical ultracentrifugation (AUC) of G3BP1. The sedimentation velocity profiles (fringe displacement) were fitted to a continuous sedimentation coefficient distribution model. Data were acquired at 1–20 min for high molecular mass species and 30–130 min for low molecular mass species. Table at right shows (1) total concentration of sample (mg/ml); (2) weight-average sedimentation coefficient s20 calculated from the 2D c(s, f/f0) model with percentage protein amount of total protein in parentheses; (3) standard sedimentation coefficient (s20,w -value) in water at 20°C; (4) estimated molar mass calculated from (sw, f/f0,w) from the 2D c(s, f/f0) model, with the theoretical dimer molar mass shown below table; and (5) weight-average frictional ratio (f/f0) calculated from the 2D c(s, f/f0) model.
(C) Two-dimensional size-and-shape distribution analyses of the sedimentation velocity data of G3BP1. The transformed velocity data are shown as contour plots (heat maps) of c(M, f/f0), c(s, f/f0), and c(s,M) with 0 fringes/S (white) to maximum value fringes/S (red), with increasing color temperature indicating higher values. Table below shows (1) total concentration of sample (mg/ml); (2) weight-average sedimentation coefficient sw calculated from the 2D c(s,f/f0) model with percentage protein amount of total protein in parentheses; (3) standard sedimentation coefficient (s20,w -value) in water at 20°C; (4) estimated molar mass calculated from (sw,f/f0,w) from the 2D c(s,f/f0) model, with the theoretical dimer molar mass shown below table; (5) weight-average frictional ratio (f/f0w ) calculated from the 2D c(s,f/f0) model; and (6) Stokes radius (nm).
(D) Indicated recombinant proteins were incubated with the cross-linker bissulfosuccinimidyl suberate (BS3) and then analyzed by SDS-PAGE and Coomassie Blue staining. M, monomer; D, dimer.
(E) Extracts from G3BP1/2 dKO cells transfected with GFP-tagged G3BP1 (G3BP1 FL, G3BP1 ΔNTF2L, or G3BP1 with the NTF2L domain replaced with GST) were cross-linked using BS3 and analyzed by immunoblotting with GFP antibody. M, monomer; D, dimer. Asterisk indicates expected location of dimer formed by G3BP1 ΔNTF2L.
(F) U2OS cells were exposed to sodium arsenite (500 μM, 1 h), cross-linked using BS3, and endogenous G3BP1 was analyzed by immunoblot with G3BP1 antibody. M, monomer; D, dimer.
(G) U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with GFP-tagged G3BP1 constructs as indicated, exposed to sodium arsenite (500 μM, 1 h), fixed, and imaged. Scale bar, 50 μm.
(H) LLPS of purified recombinant G3BP1 was assessed in the presence of a crowding agent, 150 mM NaCl, and indicated concentrations of D/D solubilizer. Increasing concentrations of the D/D solubilizer disrupted LLPS of FKBPF36M-G3BP1, but not of G3BP1 WT. Scale bar, 50 μm.
(I-J) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, pre-incubated with 250 nM D/D solubilizer for 30 min, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Cells were imaged and the percentage of cells positive for SGs was quantified. Addition of D/D solubilizer disrupted SG formation in cells expressing FKBPF36M-G3BP1, but not in cells expressing G3BP1 WT. Scale bars, 50 μm. Error bars indicate SD; ****P < 0.0001 vs. G3BP1 by two-way ANOVA, Sidak’s multiple comparisons test.
(K-L) G3BP1/2 dKO cells were transfected with G3BP1-WT or FKBPF36M-G3BP1 and exposed to sodium arsenite (500 μM). After 30 min of exposure, 250 nM of D/D solubilizer was added and cells were further incubated for 30 min. Images show live-cell imaging prior to and within 2 min after addition of D/D solubilizer. SG dissolution following treatment with D/D solubilizer was assessed by measuring the intensity of the SGs on the frame immediately before and after D/D solubilizer addition. Error bars indicate SD; ***P < 0.001 by one-way ANOVA, Tukey’s multiple comparisons test. Scale bar, 50 μm.
(M) Knockdown of caprin-1 and TIA1 in G3BP1/2 dKO cells. G3BP1/2 dKO cells were co-transfected with GFP-G3BP1 and a pool of siRNA targeting caprin-1 or TIA1. After 48 h, cells were harvested, lysed in RIPA buffer, and expression levels of caprin-1 and TIA1 were analyzed by western blot. Error bars indicate SD; **P = 0.0023 for caprin-1 and **P = 0.0039 for TIA1 by unpaired t-test.
Figure S6. The Long Central IDR of G3BP1 Regulates SG Assembly, Dynamics, and Composition; G3BP1 IDR1 Is an Autoinhibitory Element, Related to Figure 6
(A) LLPS of purified recombinant full-length G3BP1 (FL) and IDR1/2 (a 192-amino acid fragment of G3BP1 representing the IDR1/2 segment) was assessed in the presence of a crowding agent and 150 mM NaCl. Purified IDR1/2 failed to phase separate. Scale bar, 50 μm.
(B-C) G3BP1/2 dKO cells (B) or WT U2OS cells (C) were transfected with GFP-tagged IDR1/2, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Expression of IDR1/2 protein segment did not reconstitute SG assembly in G3BP1/2 dKO cells (B), nor was the IDR1/2 protein segment recruited to SGs in WT cells (C). Scale bar, 20 μm.
(D) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or ΔIDR1/2. Lysates were extracted with RIPA buffer, separated by SDS-PAGE, and immunoblotted for GFP and GAPDH. G3BP1 FL was largely found in the soluble fraction (S), whereas G3BP1 ΔIDR1/2 was largely recovered in the RIPA-insoluble pellet (I). Representative images from 3 replicates are shown. Band intensities were quantified based on 3 replicates. Error bars indicate SEM. ***P = 0.0022 by unpaired t-test.
(E) G3BP1/2 dKO cells were transfected with GFP-G3BP1 or GFP-G3BP1 ΔIDR1/2. Cells were exposed to sodium arsenite (500 μM, 1 h), and SG assembly was assessed by formation of G3BP1-positive structures. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparison test.
(F) G3BP1/2 dKO cells were co-transfected with GFP-G3BP1 FL or GFP-G3BP1 ΔIDR1/2 together with HA-G3BP1 FL. Lysates were extracted with RIPA buffer and incubated with GFP magnetic beads for IP. Washed beads were subjected to SDS-PAGE and blotted with GFP as well as HA antibodies. Both G3BP1 FL and ΔIDR1/2 could form dimers with HA-tagged G3BP1 FL.
(G) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. 16 to 24 hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
(H) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 (WT, ΔIDR1, or ΔIDR2), exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(I) Amino acid sequence of constructs shown in Figure 6G.
(J-L) G3BP1/2 dKO cells were transfected with indicated G3BP1 mutants, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bars, 20 μm.
(M-N) G3BP1/2 dKO cells were transfected with either WT or a mutant G3BP1 construct in which the IDR2 was duplicated. Cells were exposed to sodium arsenite (500 μM, 1 h) and stained for eIF3η. Cells were imaged and the percentage of cells positive for SGs was quantified. G3BP1–2xIDR2 supported SG formation more efficiently than G3BP1 WT. Scale bar, 20 μm. Error bars indicate SD. ***P =0.0004 by unpaired t-test.
Figure S7. IDR1 Phosphorylation Tunes Interplay between 3 IDRs and Regulates LLPS, Related to Figure 7
(A) Phosphorylation-dependent charge distribution of G3BP1. Phosphorylation on S149 increases negative charges in IDR1.
(B) Indicated recombinant proteins were incubated with the cross-linker bissulfosuccinimidyl suberate (BS3) and then analyzed by SDS-PAGE and Coomassie Blue staining. M, monomer; D, dimer.
(C) Extracts obtained from G3BP1/2 dKO cells transfected with indicated GFP-tagged G3BP1 constructs were cross-linked using BS3 and analyzed by immunoblotting with GFP antibody. M, monomer; D, dimer.
(D) Ficoll-dependent LLPS of G3BP1 WT and IDR1 mutants (S149E, S149A, or EQ28). Indicated concentrations of purified G3BP1 were mixed with 10% Ficoll in HEPES 50 mM, pH 7.5, 150 mM NaCl, 1 mM DTT, and imaged by DIC. Scale bar, 50 μm.
(E) RNA-dependent LLPS of G3BP1 WT and IDR1 mutants. Indicated concentrations of purified G3BP1 were mixed with increasing concentration of total RNA purified from cells in HEPES 50 mM, pH 7.5, 150 mM NaCl, 1 mM DTT, and imaged by DIC. Scale bars, 50 μm.
(F) RNA-dependent (left) and Ficoll-dependent (right) LLPS of G3BP1RA5 and G3BP1RK5. 200 μM of purified G3BP1 was mixed with either 200 ng/μL total RNA or 10% Ficoll and imaged by DIC. Substitution of five arginines in IDR3 with lysines (RBDRK5) permitted RNA-dependent LLPS in vitro, whereas substitution of the same arginines with alanines did not, illustrating that charge rather than other properties specific to arginine residues is important for LLPS. Scale bars, 10 μm.
(G) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 (RA5 or RK5), exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(H) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 or GFP-tagged synthetic G3BP1, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(I) The molecular switch in G3BP1 arises from competition between intra- and inter-molecular interactions of three IDRs. Phosphorylation of IDR1 favors the closed conformation by enhancing intramolecular interaction between IDR1 (which is highly acidic) and IDR3 (the basic component of the RBD). Upon increased RNA concentration, RNA displaces IDR1 to bind IDR3, which permits the G3BP1 homodimer to adopt an expanded, open conformation, initiating LLPS.
Table S1, Related to Figures 1 and S1. List of 210 genes whose depletion limited SG assembly in response to heat shock
Table S2, Related to Figures 1, 5, S1, and S5. Validation of SG markers in different cell lines used
Table S3, Related to Figure 6. Summary of protein-protein interaction data for APEX-G3BP1 FL and APEX-G3BP1 2xAsh1 IDR
Video S1. RNase Treatment Leads to Dissolution of Stress Granules, Related to Figures 2 and S2.
U2OS cells stably expressing G3BP1-GFP were exposed to sodium arsenite (500 μM) and further treated with 0.2% Tween 20 in PBS for 10 min. Cells were imaged in PBS alone or with RNase I (1 U/μl) + RNase A (100 μg/ml) in PBS as indicated.
Video S2. Disrupting Dimerization of FKBPF36M-G3BP1 by D/D Solubilizer Causes Disassembly of Stress Granules in Cells, Related to Figures 5 and S5.
G3BP1/2 dKO cells were transfected with G3BP1-WT or FKBPF36M-G3BP1 and exposed to sodium arsenite (500 μM). After 30 min of exposure, 250 nM of D/D solubilizer was added and cells were further incubated for 30 min. Images show live-cell imaging prior to and within 2 min after addition of D/D solubilizer.
Video S3. FRAP Analysis of GFP-G3BP1 WT, Related to Figure 6.
U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with exogenous full-length GFP-G3BP1. The relative mobility of endogenous (tdTomato-tagged) G3BP1 was compared to exogenous (GFP-tagged) G3BP1 by FRAP.
Video S4. FRAP Analysis of GFP-G3BP1 ΔIDR1/2, Related to Figure 6.
U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with exogenous GFP-G3BP1 ΔIDR1/2. The relative mobility of endogenous (tdTomato-tagged) G3BP1 was compared to exogenous (GFP-tagged) G3BP1 by FRAP.
ACKNOWLEDGMENTS
We thank Natalia Nedelsky for editorial assistance; the CAGE at St. Jude Children’s Research Hospital for assistance with Cas9/CRISPR-modified cell lines; Taosheng Chen for assistance with high-throughput RNAi screening; Amanda Nourse and Sivaraja Vaithiyalingam for assistance with AUC and MALS; and Alex Holehouse and Rohit Pappu for valuable discussions. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. Beamline 18ID (BioCAT) is supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health (NIH). This work was supported by grants from HHMI and NIH (R35NS097974) to J.P.T., and from the St. Jude Research Collaborative on Membraneless Organelles to T.M. and J.P.T.
Footnotes
DECLARATION OF INTERESTS
J.P.T. is a consultant for 5AM and Third Rock Ventures.
References
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, and Albert R (1999). Emergence of scaling in random networks. Science 286, 509–512. [DOI] [PubMed] [Google Scholar]
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, and Svergun DI (2007). Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc 129, 5656–5664. [DOI] [PubMed] [Google Scholar]
- Brown PH, and Schuck P (2006). Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J 90, 4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR (2014). mRNP granules. Assembly, function, and connections with disease. RNA Biol 11, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, and Parker R (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn EJ, and Edsall JT (1943). Density and apparent specific volume of proteins In Proteins, amino acids, and peptides, Cohn EJ, and Edsall JT, eds. (Princeton, NJ: Van Nostrand-Reinhold; ), pp. 370–381. [Google Scholar]
- Crick SL, Ruff KM, Garai K, Frieden C, and Pappu RV (2013). Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci U S A 110, 20075–20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M (2012). Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 24, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen P, Rossa M, et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrini R, De Luca A, Stella L, Mei G, Orioni B, Ciccone S, Federici G, Lo Bello M, and Ricci G (2009). Monomer-dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry 48, 10473–10482. [DOI] [PubMed] [Google Scholar]
- Figley MD, Bieri G, Kolaitis RM, Taylor JP, and Gitler AD (2014). Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci 34, 8083–8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, and Tazi J (1998). A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol 18, 3956–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, and Anderson P (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15, 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goktug AN, Ong SS, and Chen T (2012). GUItars: a GUI tool for analysis of high-throughput RNA interference screening data. PLoS One 7, e49386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E, and Shorter J (2019). The molecular language of membraneless organelles. J Biol Chem 294, 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, and Skou S (2017). BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr 50, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, and Anderson P (2019). Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, and Oltvai ZN (2001). Lethality and centrality in protein networks. Nature 411, 41–42. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, and Waugh DS (2001). Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14, 993–1000. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820 e805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, and Bassell GJ (2006). Neuronal RNA granules: movers and makers. Neuron 51, 685–690. [DOI] [PubMed] [Google Scholar]
- Kirby N, Cowieson N, Hawley AM, Mudie ST, McGillivray DJ, Kusel M, Samardzic-Boban V, and Ryan TM (2016). Improved radiation dose efficiency in solution SAXS using a sheath flow sample environment. Acta Crystallogr D Struct Biol 72, 1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, and Matthias P (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21, 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, and Parker R (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. (2017). TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95, 808–816 e809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Grace CR, Hughes A, Pappu RV, and Mittag T (2016a). Sequence Determinants of the Conformational Properties of an Intrinsically Disordered Protein Prior to and upon Multisite Phosphorylation. J Am Chem Soc 138, 15323–15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Bellora N, Gonzalez-Vallinas J, Irimia M, Chebli K, de Toledo M, Raabe M, Eyras E, Urlaub H, Blencowe BJ, et al. (2016b). Preferential binding of a stable G3BP ribonucleoprotein complex to intron-retaining transcripts in mouse brain and modulation of their expression in the cerebellum. J Neurochem 139, 349–368. [DOI] [PubMed] [Google Scholar]
- Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben-Nissan G, Kolaitis RM, Peters JL, Pounds S, et al. (2016). Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J 35, 1254–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Mazzola M, Gerstberger S, Molina H, and Tuschl T (2018). The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol Cell 69, 622–635 e626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelsky NB, and Taylor JP (2019). Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol 15, 272–286. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57, 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, and Anderson P (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, et al. (2012). Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7, e43031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Ivanov P, and Anderson P (2016). Mechanistic insights into mammalian stress granule dynamics. J Cell Biol 215, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Kedersha N, Schulte T, Branca RM, Ivanov P, and Anderson P (2019). Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol 218, 2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, and McInerney GM (2015). Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11, e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, Diamond MI, and Pappu RV (2018). Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J Biol Chem 293, 3734–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins CT, Rivera VM, Woolfson DN, Keenan T, Hatada M, Adams SE, Andrade LJ, Yaeger D, van Schravendijk MR, Holt DA, et al. (2000). A ligand-reversible dimerization system for controlling protein-protein interactions. Proc Natl Acad Sci U S A 97, 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo PK, Lee SJ, Jaiswal PB, Alber S, Kar AN, Miller-Randolph S, Taylor EE, Smith T, Singh B, Ho TS, et al. (2018). Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat Commun 9, 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samir P, Kesavardhana S, Patmore DM, Gingras S, Malireddi RKS, Karki R, Guy CS, Briard B, Place DE, Bhattacharya A, et al. (2019). DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Klasener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, et al. (2013). Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, and Lloyd RE (2016). Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J Biol Chem 291, 22671–22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, and Lloyd RE (2014). Cytoplasmic RNA Granules and Viral Infection. Annu Rev Virol 1, 147–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep 6, 25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, and Parker R (2018). Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, and Parker R (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A 115, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vognsen T, Moller IR, and Kristensen O (2013). Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS One 8, e80947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S, and Faust K (1994). Social Network Analysis: Methods and Applications (Cambridge University Press; ). [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, and Pelkmans L (2013). Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805. [DOI] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. (2018). High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69, 517–532 e511. [DOI] [PubMed] [Google Scholar]
- Zhang B, and Horvath S (2005). A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4, Article17. [DOI] [PubMed] [Google Scholar]
- Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, et al. (2018). Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 173, 958–971 e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD (2007). A new method with flexible and balanced control of false negatives and false positives for hit selection in RNA interference high-throughput screening assays. J Biomol Screen 12, 645–655. [DOI] [PubMed] [Google Scholar]
- Zhang XD (2011). Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screen 16, 775–785. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, Holder DJ, Ferrer M, and Espeseth AS (2006). Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics 7, 299–309. [DOI] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, and He C (2017). Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Brautigam CA, Ghirlando R, and Schuck P (2013). Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci Chapter 20, Unit20 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ghirlando R, Alfonso C, Arisaka F, Attali I, Bain DL, Bakhtina MM, Becker DF, Bedwell GJ, Bekdemir A, et al. (2015). A multilaboratory comparison of calibration accuracy and the performance of external references in analytical ultracentrifugation. PLoS One 10, e0126420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genome-Wide Screen Followed by CRISPR-Cas9 Knockout Identifies G3BP1/2 as Factors Uniquely Required for SG Assembly, Related to Figure 1
(A) Experimental workflow. U2OS cell lines were transfected with siRNA. Forty-eight hours post-transfection, cells were incubated at 43°C for 2 h, then fixed in 4% paraformaldehyde and stained for DAPI.
(B) Workflow for assay development of the high-throughput screen.
(C) Primary screen hits plotted in a waterfall diagram. Hits with SSMD ≤ 1.5 were selected for a further confirmation screen.
(D) Confirmatory screen hits plotted in a waterfall diagram. Hits with SSMD ≤ −1.5 were selected for further deconvolution.
(E) Results from the deconvolution screen showing the number of individual siRNAs (of a total of four siRNAs per gene) that yielded a significant effect on SG assembly.
(F) Gene ontology analysis of the hits of the deconvolution screen. For each functional category, the number of genes in that category that were identified in our screen is shown in blue; these numbers are graphed in proportion to the total number of genes implicated in that functional category in the dataset used for our analysis.
(G) Comparison of genes found in our screen to regulate assembly of SGs during heat shock compared to those previously identified as regulators of SG assembly during arsenite stress (Ohn et al., 2008).
(H) U2OS cell lines were generated using CRISPR-Cas9 to knock out genes identified as potential regulators of SG assembly. Cells were exposed to sodium arsenite (500 μM, 1 h), stained for G3BP1 to identify SGs, and imaged. All knockout lines shown retained the ability to form SGs in response to arsenite. 70% of UBAP2/UBAP2L dKO cells showed normal SG assembly, but 30% failed to assemble SGs. Scale bar, 20 μm.
(I) WT and G3BP1/2 dKO cells were incubated at 43°C for 1 h or exposed to 0.4 M sorbitol for 1 h, stained for G3BP1 and eIF3η, and imaged. G3BP1/2 dKO cells still formed eIF3η-positive foci under both heat shock and osmotic stresses. Scale bar, 10 μm.
Figure S2 Undergoes RNA-dependent Phase Separation, Related to Figure 2
(A) LLPS of purified recombinant G3BP1 full-length (FL) protein was assessed in the context of increasing concentrations of the crowding reagent Ficoll and increasing concentrations of protein. LLPS of G3BP1 was dependent on G3BP1 concentration and the concentration of Ficoll, Scale bar, 20 μm.
(B) U2OS cells stably expressing G3BP1-GFP were exposed to sodium arsenite (500 μM) and further treated with 0.2% Tween 20 in PBS for 10 min. Cells were imaged in PBS alone or with RNase I (1 U/μl) + RNase A (100 μg/ml) in PBS and imaged for indicated time. Scale bar, 50 μm.
(C) G3BP1/2 dKO cells were transiently transfected with FLAG-G3BP1 and exposed to sodium arsenite (500 μM, 1 h). FLAG-G3BP1 was isolated and the RNA-binding affinity of FLAG-G3BP1 was assessed by EMSA. Free and bound RNA fractions were quantified and plotted as a function of relative concentrations of FLAG-G3BP1. Error bars indicate SD.
(D) 5 μM GST-G3BP1 WT was incubated with 50 ng/μl of indicated RNA at room temperature and exposed to UV. Complexes were treated with 4 U/ml RNase I for 3 min and isolated by GST pulldown. RNA was then biotinylated for further detection. Complexes were analyzed by anti-GST immunoblot to detect G3BP1and streptavidin to detect RNA.v(E) LLPS of purified recombinant G3BP1 was assessed in the absence of a crowding reagent, 150 mM NaCl, and the addition of long single-stranded PolyA, PolyG, PolyC, or PolyU.
See also Video S1.
Figure S3. In Vitro Phase Separation of G3BP1 with RNA Strongly Predicts SG Reconstitution in Cells, Related to Figure 3
(A) G3BP2 domains are shown aligned with results of PONDR (Predictor of Natural Disordered Regions) and NCPR (net charge per residue; 5-amino acid window) analyses.
(B) The amino acid sequence of human G3BP2 is shown with major domains marked by highlighted colors. Charged residues are shown in blue (basic; Arg and Lys) and red (acidic; Asp and Glu).
(C) LLPS of G3BP1 deletion constructs in which the NTF2L domain, IDR1/2, or RBD have been removed. LLPS of purified recombinant protein was assessed in the presence of a crowding agent. G3BP1 lacking the NTF2L or RBD domains failed to phase separate, whereas G3BP1 lacking IDR1/2 did phase separate. Scale bars, 50 μm.
(D) Representation of the phase separation results obtained in (C).
(E) G3BP1 ΔIDR3 fails to assemble SGs. We tested two IDR3 deletion mutants, G3BP1 ΔIDR3 (Δ410–466) and G3BP1 ΔIDR3 (Δ428–466). Although G3BP1 ΔIDR3 (Δ410–466) accumulated in cytoplasmic puncta, these puntca formed in the absence of stress, did not contain SG markers, and did not resolve after the removal of stress, suggesting that these were insoluble aggregates, not SGs. Modifying these boundaries to residue 428 to 466 (G3BP1 ΔIDR3 (Δ428–466)) alleviated this aggregation, but G3BP1/2 dKO cells expressing this construct failed to assemble SGs. Scale bar, 20 μm.
(F) Puncta formed by G3BP1 ΔIDR3 (410–466) were analyzed by FRAP and showed poor fluorescence recovery. Error bars indicate SD. ***P < 0.001, ****P < 0.0001, ns; not significant by two-way ANOVA, Sidak’s multiple comparisons test.
(G) G3BP1/2 dKO cells were transfected with GFP-tagged G3BP1 Δ340–415, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. The majority of cells transfected with G3BP1 Δ340–415 did not form SGs, whereas few cells showed weak rescue of SG formation. Scale bar, 20 μm.
(H) Quantification of result shown in (G). Error bars indicate SD. **P = 0.0021 by unpaired t-test.
Figure S4. Multivalency in RNA Binding Mediates SG Formation, Related to Figure 4
(A) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. Twenty-four hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
(B) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. Cellular protein-RNA complexes were cross-linked with UV and cells were lysed. Immunoprecipitated RNA cross-linked to G3BP1 was then treated with alkaline phosphatase and biotinylated. Complexes were subjected to SDS-PAGE and immunoblotting for biotin (detection of RNA) and GFP (detection of G3BP1). RNA binding was quantified and normalized to protein signal. Results show quantification from duplicate experiments. Error bars indicate SD. *P = 0.0136 vs. FL by one-way ANOVA, Dunnett’s multiple comparisons test.
(C) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants and assessed by immunoblot as in (A).
(D) G3BP1/2 dKO cells were transfected with G3BP1 constructs in which the RBD was substituted with indicated combinations of ZnF domains. Cells were exposed to sodium arsenite (500 μM, 1 h) and stained for eIF3η. G3BP1 with a single ZnF domain did not support SG formation, but G3BP1 with two or more ZnF domains did support SG formation. Scale bar, 20 μm.
(E) SGs formed by indicated constructs were analyzed by FRAP. SGs assembled with 3 KH or ZnF domains were significantly less dynamic than those assembled with 2 KH or ZnF domains. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparisons test.
(F) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or G3BP1 2xRBD and exposed to sodium arsenite (500 μM). SG formation was monitored using live cell imaging beginning when arsenite was added to the medium. Duplication of the G3BP1 RBD dramatically enhanced SG assembly. Scale bar, 20 μm.
(G) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or G3BP1 2xRBD for 24 h. SGs were counted as spontaneous (no arsenite) or arsenite-induced (500 μM, 1 h). Cells were immediately fixed with 4% PFA and representative images were shown for each condition. Scale bar, 50 μm.
(H) Quantification of (G). The percentage of cells with spontaneous or arsenite-induced SGs are shown. Cells with at least 3 granules were counted as a SG-positive cell. *P = 0.0431 by two-way ANOVA, Sidak’s multiple comparisons test. Error bars indicate SD.
(I) The ratio of GFP signal in SGs to the entire cell (GFP enrichment in SGs) from results obtained in (G). The SG enrichment percentage of G3BP1 FL protein and G3BP1 2xRBD protein were quantified by CellProfiler as previously described (Mackenzie et al., 2017). Briefly, the cell boundary and the boundary of SGs were identified by CellProfiler. The ratios of GFP fluorescent signal intensity in the SGs to that in the entire cell were used as indicators for the SG enrichment percentages of these two proteins. At least 100 cells were included in each condition. ****P < 0.0001 by unpaired t-test. Error bars indicate SD.
(J) SGs formed by G3BP1 2xRBD were analyzed by FRAP and showed poor fluorescence recovery. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparisons test.
(K) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. 16 to 24 hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
Figure S5. NTF2L Domain-Mediated Dimerization and Interaction with Core SG Components Regulate Assembly of SGs, Related to Figure 5
(A) Size exclusion chromatography (SEC) multi-angle light scattering (MALS) on purified G3BP1 at concentrations of 20, 50, or 100 μM protein. Inset shows a distribution of average molecular weights superimposed on the chromatogram of UV absorbance at 280 nm.
(B) Sedimentation velocity analytical ultracentrifugation (AUC) of G3BP1. The sedimentation velocity profiles (fringe displacement) were fitted to a continuous sedimentation coefficient distribution model. Data were acquired at 1–20 min for high molecular mass species and 30–130 min for low molecular mass species. Table at right shows (1) total concentration of sample (mg/ml); (2) weight-average sedimentation coefficient s20 calculated from the 2D c(s, f/f0) model with percentage protein amount of total protein in parentheses; (3) standard sedimentation coefficient (s20,w -value) in water at 20°C; (4) estimated molar mass calculated from (sw, f/f0,w) from the 2D c(s, f/f0) model, with the theoretical dimer molar mass shown below table; and (5) weight-average frictional ratio (f/f0) calculated from the 2D c(s, f/f0) model.
(C) Two-dimensional size-and-shape distribution analyses of the sedimentation velocity data of G3BP1. The transformed velocity data are shown as contour plots (heat maps) of c(M, f/f0), c(s, f/f0), and c(s,M) with 0 fringes/S (white) to maximum value fringes/S (red), with increasing color temperature indicating higher values. Table below shows (1) total concentration of sample (mg/ml); (2) weight-average sedimentation coefficient sw calculated from the 2D c(s,f/f0) model with percentage protein amount of total protein in parentheses; (3) standard sedimentation coefficient (s20,w -value) in water at 20°C; (4) estimated molar mass calculated from (sw,f/f0,w) from the 2D c(s,f/f0) model, with the theoretical dimer molar mass shown below table; (5) weight-average frictional ratio (f/f0w ) calculated from the 2D c(s,f/f0) model; and (6) Stokes radius (nm).
(D) Indicated recombinant proteins were incubated with the cross-linker bissulfosuccinimidyl suberate (BS3) and then analyzed by SDS-PAGE and Coomassie Blue staining. M, monomer; D, dimer.
(E) Extracts from G3BP1/2 dKO cells transfected with GFP-tagged G3BP1 (G3BP1 FL, G3BP1 ΔNTF2L, or G3BP1 with the NTF2L domain replaced with GST) were cross-linked using BS3 and analyzed by immunoblotting with GFP antibody. M, monomer; D, dimer. Asterisk indicates expected location of dimer formed by G3BP1 ΔNTF2L.
(F) U2OS cells were exposed to sodium arsenite (500 μM, 1 h), cross-linked using BS3, and endogenous G3BP1 was analyzed by immunoblot with G3BP1 antibody. M, monomer; D, dimer.
(G) U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with GFP-tagged G3BP1 constructs as indicated, exposed to sodium arsenite (500 μM, 1 h), fixed, and imaged. Scale bar, 50 μm.
(H) LLPS of purified recombinant G3BP1 was assessed in the presence of a crowding agent, 150 mM NaCl, and indicated concentrations of D/D solubilizer. Increasing concentrations of the D/D solubilizer disrupted LLPS of FKBPF36M-G3BP1, but not of G3BP1 WT. Scale bar, 50 μm.
(I-J) G3BP1/2 dKO cells were transfected with indicated G3BP1 constructs, pre-incubated with 250 nM D/D solubilizer for 30 min, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Cells were imaged and the percentage of cells positive for SGs was quantified. Addition of D/D solubilizer disrupted SG formation in cells expressing FKBPF36M-G3BP1, but not in cells expressing G3BP1 WT. Scale bars, 50 μm. Error bars indicate SD; ****P < 0.0001 vs. G3BP1 by two-way ANOVA, Sidak’s multiple comparisons test.
(K-L) G3BP1/2 dKO cells were transfected with G3BP1-WT or FKBPF36M-G3BP1 and exposed to sodium arsenite (500 μM). After 30 min of exposure, 250 nM of D/D solubilizer was added and cells were further incubated for 30 min. Images show live-cell imaging prior to and within 2 min after addition of D/D solubilizer. SG dissolution following treatment with D/D solubilizer was assessed by measuring the intensity of the SGs on the frame immediately before and after D/D solubilizer addition. Error bars indicate SD; ***P < 0.001 by one-way ANOVA, Tukey’s multiple comparisons test. Scale bar, 50 μm.
(M) Knockdown of caprin-1 and TIA1 in G3BP1/2 dKO cells. G3BP1/2 dKO cells were co-transfected with GFP-G3BP1 and a pool of siRNA targeting caprin-1 or TIA1. After 48 h, cells were harvested, lysed in RIPA buffer, and expression levels of caprin-1 and TIA1 were analyzed by western blot. Error bars indicate SD; **P = 0.0023 for caprin-1 and **P = 0.0039 for TIA1 by unpaired t-test.
Figure S6. The Long Central IDR of G3BP1 Regulates SG Assembly, Dynamics, and Composition; G3BP1 IDR1 Is an Autoinhibitory Element, Related to Figure 6
(A) LLPS of purified recombinant full-length G3BP1 (FL) and IDR1/2 (a 192-amino acid fragment of G3BP1 representing the IDR1/2 segment) was assessed in the presence of a crowding agent and 150 mM NaCl. Purified IDR1/2 failed to phase separate. Scale bar, 50 μm.
(B-C) G3BP1/2 dKO cells (B) or WT U2OS cells (C) were transfected with GFP-tagged IDR1/2, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Expression of IDR1/2 protein segment did not reconstitute SG assembly in G3BP1/2 dKO cells (B), nor was the IDR1/2 protein segment recruited to SGs in WT cells (C). Scale bar, 20 μm.
(D) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or ΔIDR1/2. Lysates were extracted with RIPA buffer, separated by SDS-PAGE, and immunoblotted for GFP and GAPDH. G3BP1 FL was largely found in the soluble fraction (S), whereas G3BP1 ΔIDR1/2 was largely recovered in the RIPA-insoluble pellet (I). Representative images from 3 replicates are shown. Band intensities were quantified based on 3 replicates. Error bars indicate SEM. ***P = 0.0022 by unpaired t-test.
(E) G3BP1/2 dKO cells were transfected with GFP-G3BP1 or GFP-G3BP1 ΔIDR1/2. Cells were exposed to sodium arsenite (500 μM, 1 h), and SG assembly was assessed by formation of G3BP1-positive structures. Error bars indicate SD. ****P < 0.0001 by two-way ANOVA, Sidak’s multiple comparison test.
(F) G3BP1/2 dKO cells were co-transfected with GFP-G3BP1 FL or GFP-G3BP1 ΔIDR1/2 together with HA-G3BP1 FL. Lysates were extracted with RIPA buffer and incubated with GFP magnetic beads for IP. Washed beads were subjected to SDS-PAGE and blotted with GFP as well as HA antibodies. Both G3BP1 FL and ΔIDR1/2 could form dimers with HA-tagged G3BP1 FL.
(G) G3BP1/2 dKO cells were transfected with GFP-G3BP1 FL or indicated GFP-G3BP1 mutants. 16 to 24 hours post-transfection, cells were lysed in 1x NuPAGE sample buffer and subjected to SDS-PAGE and immunoblotting for GFP (detection of G3BP1) and GAPDH (loading control).
(H) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 (WT, ΔIDR1, or ΔIDR2), exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(I) Amino acid sequence of constructs shown in Figure 6G.
(J-L) G3BP1/2 dKO cells were transfected with indicated G3BP1 mutants, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bars, 20 μm.
(M-N) G3BP1/2 dKO cells were transfected with either WT or a mutant G3BP1 construct in which the IDR2 was duplicated. Cells were exposed to sodium arsenite (500 μM, 1 h) and stained for eIF3η. Cells were imaged and the percentage of cells positive for SGs was quantified. G3BP1–2xIDR2 supported SG formation more efficiently than G3BP1 WT. Scale bar, 20 μm. Error bars indicate SD. ***P =0.0004 by unpaired t-test.
Figure S7. IDR1 Phosphorylation Tunes Interplay between 3 IDRs and Regulates LLPS, Related to Figure 7
(A) Phosphorylation-dependent charge distribution of G3BP1. Phosphorylation on S149 increases negative charges in IDR1.
(B) Indicated recombinant proteins were incubated with the cross-linker bissulfosuccinimidyl suberate (BS3) and then analyzed by SDS-PAGE and Coomassie Blue staining. M, monomer; D, dimer.
(C) Extracts obtained from G3BP1/2 dKO cells transfected with indicated GFP-tagged G3BP1 constructs were cross-linked using BS3 and analyzed by immunoblotting with GFP antibody. M, monomer; D, dimer.
(D) Ficoll-dependent LLPS of G3BP1 WT and IDR1 mutants (S149E, S149A, or EQ28). Indicated concentrations of purified G3BP1 were mixed with 10% Ficoll in HEPES 50 mM, pH 7.5, 150 mM NaCl, 1 mM DTT, and imaged by DIC. Scale bar, 50 μm.
(E) RNA-dependent LLPS of G3BP1 WT and IDR1 mutants. Indicated concentrations of purified G3BP1 were mixed with increasing concentration of total RNA purified from cells in HEPES 50 mM, pH 7.5, 150 mM NaCl, 1 mM DTT, and imaged by DIC. Scale bars, 50 μm.
(F) RNA-dependent (left) and Ficoll-dependent (right) LLPS of G3BP1RA5 and G3BP1RK5. 200 μM of purified G3BP1 was mixed with either 200 ng/μL total RNA or 10% Ficoll and imaged by DIC. Substitution of five arginines in IDR3 with lysines (RBDRK5) permitted RNA-dependent LLPS in vitro, whereas substitution of the same arginines with alanines did not, illustrating that charge rather than other properties specific to arginine residues is important for LLPS. Scale bars, 10 μm.
(G) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 (RA5 or RK5), exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(H) G3BP1/2 dKO cells were reconstituted with GFP-tagged G3BP1 or GFP-tagged synthetic G3BP1, exposed to sodium arsenite (500 μM, 1 h), and stained for eIF3η. Scale bar, 20 μm.
(I) The molecular switch in G3BP1 arises from competition between intra- and inter-molecular interactions of three IDRs. Phosphorylation of IDR1 favors the closed conformation by enhancing intramolecular interaction between IDR1 (which is highly acidic) and IDR3 (the basic component of the RBD). Upon increased RNA concentration, RNA displaces IDR1 to bind IDR3, which permits the G3BP1 homodimer to adopt an expanded, open conformation, initiating LLPS.
Table S1, Related to Figures 1 and S1. List of 210 genes whose depletion limited SG assembly in response to heat shock
Table S2, Related to Figures 1, 5, S1, and S5. Validation of SG markers in different cell lines used
Table S3, Related to Figure 6. Summary of protein-protein interaction data for APEX-G3BP1 FL and APEX-G3BP1 2xAsh1 IDR
Video S1. RNase Treatment Leads to Dissolution of Stress Granules, Related to Figures 2 and S2.
U2OS cells stably expressing G3BP1-GFP were exposed to sodium arsenite (500 μM) and further treated with 0.2% Tween 20 in PBS for 10 min. Cells were imaged in PBS alone or with RNase I (1 U/μl) + RNase A (100 μg/ml) in PBS as indicated.
Video S2. Disrupting Dimerization of FKBPF36M-G3BP1 by D/D Solubilizer Causes Disassembly of Stress Granules in Cells, Related to Figures 5 and S5.
G3BP1/2 dKO cells were transfected with G3BP1-WT or FKBPF36M-G3BP1 and exposed to sodium arsenite (500 μM). After 30 min of exposure, 250 nM of D/D solubilizer was added and cells were further incubated for 30 min. Images show live-cell imaging prior to and within 2 min after addition of D/D solubilizer.
Video S3. FRAP Analysis of GFP-G3BP1 WT, Related to Figure 6.
U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with exogenous full-length GFP-G3BP1. The relative mobility of endogenous (tdTomato-tagged) G3BP1 was compared to exogenous (GFP-tagged) G3BP1 by FRAP.
Video S4. FRAP Analysis of GFP-G3BP1 ΔIDR1/2, Related to Figure 6.
U2OS cells were constructed using CRISPR-Cas9 to express tdTomato-tagged endogenous G3BP1. Cells were then transfected with exogenous GFP-G3BP1 ΔIDR1/2. The relative mobility of endogenous (tdTomato-tagged) G3BP1 was compared to exogenous (GFP-tagged) G3BP1 by FRAP.
Data Availability Statement
Data Resources
The published article includes all datasets generated or analyzed during this study.