Abstract
Tumor protein p53 is a key regulator of several cellular pathways, including DNA repair, cell cycle and angiogenesis. Kevetrin exhibits p53-dependent as well as-independent activity in solid tumors, while its effects on leukemic cells remain unknown. The aim of the present study was to analyze the response of acute myeloid leukemia (AML) cell lines (TP53 wild-type: OCI-AML3 and MOLM-13; and TP53-mutant: KASUMI-1 and NOMO-1) to kevetrin at a concentration range of 85–340 µM. The cellular and molecular effects of the treatment were analyzed in terms of cell growth, viability [Annexin V-propidium iodide (PI) staining] and cell cycle alterations (PI staining). Gene expression profiling, western blotting and immunofluorescence were performed to elucidate the pathways underlying kevetrin activity. Pulsed exposure exerted no effect on the wild-type cells, but was effective on mutant cells. After continuous treatment, significant cell growth arrest and apoptosis were observed in all cell lines, with TP53-mutant models displaying a higher sensitivity and p53 induction. Kevetrin also displayed efficacy against TP53 wild-type and mutant primary AML, with a preferential cytotoxic activity against blast cells. Gene expression profiling revealed a common core transcriptional program altered by drug exposure and the downregulation of glycolysis, DNA repair and unfolded protein response signatures. These findings suggest that kevetrin may be a promising therapeutic option for patients with both wild-type and TP53-mutant AML.
Keywords: acute myeloid leukemia, gene expression profiling, kevetrin, apoptosis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease defined by clonal expansion of immature myeloid cells that infiltrate the bone marrow and other tissues (1). The therapeutic strategies AML have remained largely unchanged over the past 30 years, and the disease is currently curable in 35–40% of patients aged ≤60 years, and in 5–15% of those aged >60 years (2). The development of next-generation sequencing (NGS) technologies have led to the identification of eight functional categories of significantly mutated genes, including tumor-suppressor genes, such as tumor protein p53 (TP53) (3). TP53 mutations occur in 8–14% of the cases (4) and they are associated with complex karyotype AML (5), in particular typical complex karyotype (6) and chromothripsis (7,8), and confer a very poor prognosis (4,9). Moreover, the p53-transcriptional program is generally silenced in aneuploid AML (10), and TP53 mutations and aneuploidy define a specific molecular subgroup in the recent AML genomic classification and prognostic stratification (9).
TP53 is the most frequently mutated gene in cancer (11,12) and is a critical regulator of several genes involved in DNA repair [e.g., growth arrest and DNA damage (GADD45)], cell cycle [e.g., cyclin-dependent kinase inhibitor 1A (CDKN1A)], apoptosis [e.g., BCL2-associated X protein, BCL2-binding component 3] and angiogenesis (13). Under normal conditions, p53 is maintained at a low level and is mainly regulated by E3-ubiquitin-ligase mouse double minute 2 (MDM2), which promotes its proteasomal degradation (14). Moreover, its rapid turnover is associated with correct folding by chaperone proteins, such as heat shock protein 90 (Hsp90), which has been proven to be fundamental for the stability and DNA-binding capacity of the wild-type (wt) protein (15). Under stress conditions, p53 is phosphorylated by various sensor kinases, promoting the dissociation of the p53-MDM2 complex and p53 stabilization and activation (16,17).
Kevetrin (thioureidobutyronitrile or 3-cyanopropyl carbamimidothioate hydrochloride, C5H10ClN3S) is a small-molecule compound that exhibits p53-dependent and -independent activity in solid tumors, including lung, breast, colon and ovarian cancer cell and xenograft models (18–20). In TP53-wt models, kevetrin induces cell cycle arrest and apoptosis through the alteration of the E3 ligase processivity of MDM2 and activation and stabilization of p53, with increased expression of its targets, including protein 21 (p21) and p53-upregulated modulator of apoptosis (18,19). A p53-independent upregulation of p21 expression was observed in TP53-mutant ovarian cancer cell lines (20). Accordingly, in a phase I trial evaluating the effect of kevetrin on solid cancers, 48% of patients exhibited a ≥10% increase in p21 expression in the peripheral blood 7–24 h after the treatment initiation (NCT01664000). Moreover, it was hypothesized that kevetrin induced downregulation of histone deacetylase 6 (HDAC6), negatively affected the HDAC6-Hsp90 chaperone axis, resulting in degradation of p53 in the mutated models (19). Kevetrin treatment led to the forced expression of the pro-apoptotic protein BID and decreased levels of the anti-apoptotic protein MCL1. It also had an effect on the Rb-E2F tumor suppressor pathway by downregulating E2F1 and its target genes in TP53-mutant and wt models (19).
To the best of our knowledge, the present study is the first to investigate the effects of kevetrin exposure on AML cell lines and primary cells characterized by different TP53 mutational status.
Materials and methods
Cell lines and culture
Four AML cell lines, MOLM-13 (AML M5), KASUMI-1 (AML M2), OCI-AML3 (AML M4) and NOMO-1 (AML M5) were obtained from the American Type Culture Collection, and were mycoplasma-tested and authenticated using the LGC Standards Cell Line Authentication service. The cell lines were cultured at 37°C in a 5% CO2 atmosphere at a density of 0.3×106 cells/ml in complete medium, in T75 flasks. MOLM-13 and KASUMI-1 cells were cultured in RPMI-1640 (Euroclone) supplemented with 20% heat-inactivated FBS (GE Healthcare), 2 mM L-glutamine (GE Healthcare), 100 U/ml penicillin, 100 µg/ml streptomycin (GE Healthcare) and 0.2% Mycozap (Lonza, Inc.). OCI-AML3 cells were cultured in α-MEM (Lonza, Inc.) with 20% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. NOMO-1 cells were grown in RPMI-1640 with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
Drug
Kevetrin powder was kindly provided by Innovation Pharmaceuticals, dissolved in sterile water in a 3.4 mM stock solution, stored at 4°C and used within 1 month. Cells were seeded in 96-well or 6-well plates at 0.5×106/ml in 100 and 3,000 µl of medium, respectively, and treated with increasing drug concentrations (85–340 µM), according to peak plasma concentrations measured in the phase I clinical trial (NCT01664000). For pulsed experiments, cells were exposed to the drug for 6 h and then washed and replated in complete medium [wash-out (wo)]. After 66 h, cells were reseeded in fresh medium containing the drug for 6 h, followed by a 66-h wo. The pulsed treatment was repeated 2–3 times.
Primary cell cultures
Samples were collected at Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS from 4 AML patients at diagnosis (inclusion criteria: Age ≥18 years, confirmed AML diagnosis, available clinical data for review and obtained written informed consent) between December 2018 and October 2019 (Table SI). Bone marrow mononuclear cells (BMMCs) and peripheral blood mononuclear cells (PBMCs) were collected by density gradient centrifugation using Lymphosep (BioWest SAS), then lysed in RLT buffer (Qiagen, Ltd.) supplemented with 1% β-mercaptoethanol, and/or cryopreserved in 90% FBS and 10% DMSO (Sigma-Aldrich; Merck KGaA). After thawing, BMMCs were primed for 24 h with a cytokine cocktail [20 ng/ml Fms-related tyrosine kinase 3 ligand (FLT3-L), interleukin (IL)-3, IL-6, stem cell factor and granulocyte colony-stimulating factor (Miltenyi Biotec GmbH)] and live cells [collected using the Dead Cell Removal Kit (Miltenyi Biotec GmbH)] were then treated with increasing doses of kevetrin (85–340 µM) for 48 h.
Cell viability assay
Cell viability was determined using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation), according to the manufacturer's instructions. The optical density was determined after 3 h at a wavelength of 490 nm by the Thermo Multiskan EX microplate reader (Thermo Fisher Scientific, Inc.). Cell viability in primary samples was evaluated by the trypan blue exclusion assay.
Annexin V staining
Phosphatidylserine externalization was evaluated using the fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection kit (eBioscence; Thermo Fisher Scientific, Inc.). After treatment, cells were incubated with 25 µl/ml of Annexin V-FITC for 15 min at 37°C in a humidified atmosphere in the dark. Prior to flow cytometric analysis, propidium iodide (PI) was added to a final concentration of 5 µg/ml. Flow cytometric analysis was performed using a FACSCanto flow cytometer (Becton, Dickinson and Co.) equipped with 488 nm (blue) and 633 nm (red) lasers, and 10,000 events were recorded for each sample. Data acquisition and analysis were performed using FACSDiva software v.6.1.3. (Becton, Dickinson and Co.). In primary samples, Annexin V staining was combined with surface markers using the following antibodies: CD45-APC Vio770 (cat. no. 130-110-635), CD33-APC (cat. no. 130-111-020), CD14-PerCP Vio 700 (cat. no. 130-110-523), CD3-PE (cat. no. 130-113-139) (all from Miltenyi Biotec GmbH, dilution 1:50) and CD19-PE Cy7 (cat. no. 302216, 1:10, BioLegend, Inc.).
Mitochondrial membrane potential (ΔΨm) depolarization analysis
Mitochondrial membrane polarization was evaluated using the Mito-PT JC-1 Assay Kit (ImmunoChemistry Technologies, LLC). Following kevetrin exposure, cells were incubated according to the manufacturer's instructions in JC-1 working solution for 15 min at 37°C and 5% CO2 in a humidified atmosphere in the dark, suspended in 1X assay buffer and analyzed by flow cytometry. A positive control treated with 50 µM of carbonyl cyanide 3-chlorophenylhydrazone was used for each experiment.
TUNEL assay
Fragmented DNA was detected by the TUNEL assay (Roche Diagnostics GmbH). After each treatment, samples were fixed in 1% formaldehyde on ice for 15 min, suspended in 70% ice-cold ethanol and stored overnight at −20°C. Cells were incubated for 5 min at 4°C in PBS containing 0.1% Triton X-100 (Bio-Rad Laboratories, Inc.). Thereafter, samples were suspended in 50 µl of solution containing TdT and FITC-conjugated dUTP deoxynucleotides 1:1 (Roche Diagnostics GmbH) and incubated for 90 min at 37°C in a humidified atmosphere in the dark. Samples were counterstained with 2.5 µg/ml of PI (MP Biomedicals, LLC) and 10 kU/ml of RNAse (Sigma-Aldrich; Merck KGaA) for 30 min at 4°C in the dark, then analyzed by flow cytometry. A positive control treated with 80 kU/ml of DNase (Sigma-Aldrich; Merck KGaA) was included for each experiment.
Active caspase-3 assay
The percentage of active caspase-3 was measured using the FITC Active Caspase-3 Apoptosis Kit (BD Biosciences). After treatment, cells were collected, washed with ice-cold PBS 1X and incubated for 20 min at 4°C in Cytofix/Cytoperm buffer (BD Biosciences). The samples were then incubated with 20 µl of anti-active caspase-3 antibody (cat. no. 550480, 1:5, BD Biosciences) for 30 min at room temperature in the dark, washed in Perm/Wash buffer and analyzed by flow cytometry. A positive control treated with 5 µM camptothecin was used for each experiment.
Cell cycle analysis
After treatment, cells were washed in 1X PBS, fixed in 70% ice-cold ethanol, stained with 10 µg/ml PI (MP Biomedicals), 10 kU/ml RNAse (Sigma-Aldrich; Merck KGaA) and 0.01% Nonidet™ P40 (NP40; Sigma-Aldrich; Merck KGaA) overnight at 4°C in the dark and analyzed by flow cytometry. Data analysis was performed using ModFit 4.1 (DNA Modelling System, Verity Software House, Inc.).
NGS and variant calling
DNA was extracted from primary mononuclear cells (BMMCs or PBMCs) using the Maxwell® RSC Blood DNA Kit (Promega Corporation) according to the manufacturer's instructions. The mutational profile of patients was determined using SOPHiA Myeloid Solution™ (SOPHiA GENETICS), a CE-IVD marked molecular diagnostic application. Libraries were prepared according to the manufacturer's instructions and quantified using the Qubit® dsDNA High Sensitivity Assay on a Qubit® Fluorometer (Thermo Fisher Scientific, Inc.). The median average of amplicons was determined by capillary electrophoresis using Agilent High Sensitivity DNA kit (Agilent Technologies, Inc.). The pooled libraries were paired-end (2×301) and sequenced with v3 chemistry on a MiSeq™ instrument (Illumina, Inc.), as described in the manufacturer's protocol. FASTQ sequencing files were uploaded onto the SOPHiA DDM® platform (version 4), which uses patented advanced technologies for variant calling and annotation. Human Genome Build 19 (Hg19) was used as the reference for sequence alignment. A minimum coverage depth of 1,000× was recommended. A filtering tool was set to exclude known single-nucleotide polymorphisms, variants localized in intronic and UTR regions and synonymous variants. Only exonic and splice site variants with a variant allele frequency (VAF) ≥2.5% were evaluated.
Gene expression profiling
RNA was isolated using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) from cells untreated or treated for 6 and 48 h with kevetrin at 340 µM. Labeled single-stranded complementary DNA was prepared from 100 ng of RNA and hybridized to Human Transcriptome Array 2.0 (Thermo Fisher Scientific, Inc.), according to the manufacturer's recommendations. Three independent replicates of each condition per cell line were analyzed. Data quality control and normalization were carried out by Expression Console software v1.4.1.46 (Thermo Fisher Scientific, Inc.), while supervised analysis was performed with Transcriptome Analysis Console software v.3.0 (Thermo Fisher Scientific, Inc.). Functional annotation clustering was performed using David Bioinformatics Resources 6.8 (National Institute of Allergy and Infectious Diseases, National Institutes of Health) (21). Gene set enrichment analysis (GSEA) was performed with GSEA software v.3.0 (Broad Institute) (22,23).
Western blot analysis
After a 48-h treatment, total protein extracts were prepared in ice-cold lysis buffer (0.5% NP40, 250 mM NaCl, 50 mM HEPES, 5 mM EDTA and 0.5 mM EGTA) containing phosphatase inhibitor cocktail 2 (Sigma-Aldrich; Merck KGaA), protease inhibitor (Clontech Laboratories, Inc.), and DTT (Invitrogen; Thermo Fisher Scientific, Inc.). Before use, extract concentrations were normalized using a BCA protein assay kit (Bio-Rad Laboratories, Inc.). Proteins (50–100 µg) were separated by SDS-PAGE using 4–20% polyacrylamide precast gel (Bio-Rad Laboratories, Inc.) and were transferred to PVDF membranes using a TransBlot Turbo system (Bio-Rad Laboratories, Inc.). Blocking and antibody incubations were performed in Tris-buffered saline with 0.1% Tween-20 (TBST) plus 5% dry milk or in TBST plus 5% BSA (Sigma-Aldrich; Merck KGaA). The following antibodies were used: Anti-phosphorylated-p53 (Ser15) (cat. no. 9286, clone 16G8, 1:1,000), anti-p53 (cat. no. 2527, clone 7F5, 1:1,000) and anti-p21 (cat. no. 2946, clone DCS60, 1:2,000), all from Cell Signaling Technologies, Inc. Detection was performed using horseradish peroxidase-conjugated anti-rabbit (cat. no. NA934, 1:5,000) and anti-mouse (cat. no. NA931, 1:10,000) secondary antibodies (GE Healthcare) with WesternBright Sirius (Advansta, Inc.) or SuperSignal West Femto (Thermo Fisher Scientific, Inc.) substrates. Membranes were imaged on a ChemiDoc MP system (Bio-Rad Laboratories, Inc.). Membranes were stripped and reprobed with antibody against β-actin (cat. no. ab49900, clone AC-15, 1:25,000, Abcam) as normalizer. QuantityOne 4.6.8 software (Bio-Rad Laboratories, Inc.) was used for analysis.
Immunofluorescence
Cells were harvested after a 48-h treatment, washed in 1X PBS and fixed in 10% non-buffered formalin in a 37°C water bath for 1 h. Samples were then washed in 1X PBS and resuspended in 1 ml of 70% EtOH. Slides were prepared by cytospin centrifugation at 133 × g for 5 min at room temperature and 5 min incubation at room temperature in ethanol solutions with increasing concentration (50–70–100%). Blocking was performed with 1% BSA and 0.3% Triton X-100 in 1X PBS for 1 h. Slides were incubated overnight at 4°C with anti-p53 antibody (clone 7F5, 1:1,600 Cell Signaling Technologies, Inc.), washed and stained with goat anti-rabbit Alexa Fluor 594 secondary antibody (1:1,000, Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The samples were washed three times in 1X PBS and mounted using ProLong Antifade DAPI (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were imaged with a N-SIM E laser confocal microscope (Nikon Corporation) at a magnification of ×60 and analyzed with NIS Elements software 5.11 (Nikon Corporation) and ImageJ software 1.52a (National Institutes of Health).
Statistical analysis
Statistical analysis was carried out with GraphPad Prism 8.0.1 software (GraphPad Software, Inc.). Comparisons between two groups were performed using Student's t-test, whereas multiple comparisons were performed using one-way analysis of variance with Dunnett's post hoc test. Values represent the mean ± standard deviation of three independent experiments. P<0.05 was considered to indicate statistically significant differences.
Results
Kevetrin-pulsed treatment decreases the viability of only the KASUMI-1 cell line
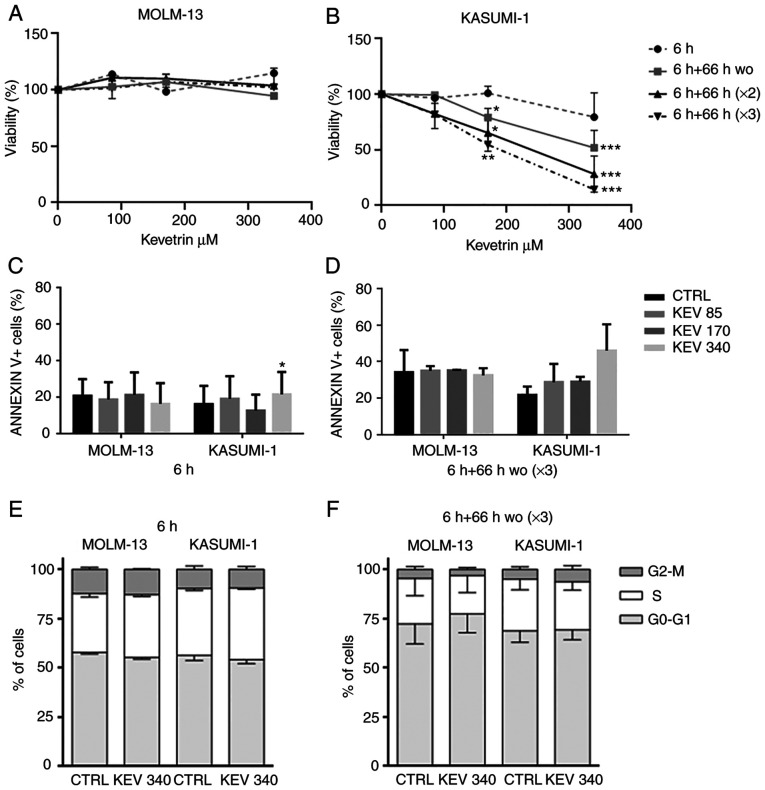
The response of MOLM-13 (TP53-wt) and KASUMI-1 [mutated (homozygous single-nucleotide variant, p.R248Q)] cells to kevetrin exposure was analyzed, initially using a pulsed treatment to simulate the dosing schedules used in clinical practice. Cell viability was evaluated after exposure to kevetrin doses of 85, 170 and 340 µM for 6 h, or for 6 h followed by a 66-h wo. The latter schedule was repeated consecutively two and three times, respectively. No effect on the viability of MOLM-13 cells was observed (Fig. 1A), whereas a dose- and time-dependent decrease was evident in the KASUMI-1 cell line (Fig. 1B). The role of kevetrin treatment in apoptosis induction was then evaluated by examining phosphatidylserine externalization following exposure to the highest concentration in KASUMI-1 cells, detecting a trend towards increased percentage of Annexin V+ cells (Fig. 1C and D and Fig. S1A and B). Kevetrin treatment did not induce cell cycle alterations (Fig. 1E and F and Fig. S1C and D).
Figure 1.
Effect of pulsed kevetrin on AML cell line viability, cell cycle progression and apoptosis induction. Viability of (A) MOLM-13 and (B) KASUMI-1 cell lines treated with different concentrations of kevetrin (85, 170 and 340 µM) for up to 3 pulsations. (C and D) Quantification of Annexin V+ cells in MOLM-13 and KASUMI-1 models before and after treatment with 85, 170 and 340 µM kevetrin for 6 and 6+66 h wo (×3). Cell cycle analysis of MOLM-13 and KASUMI-1 cells after (E) 6 h and (F) 6+66 h wo (×3) kevetrin treatment. Values represent the mean ± standard deviation of 3 biological replicates (*P<0.05, **P<0.01, ***P<0.001). AML, acute myeloid leukemia; wo, wash-out; CTRL, control.
Short-term kevetrin treatment induces metallothionein (MT) expression in AML cells
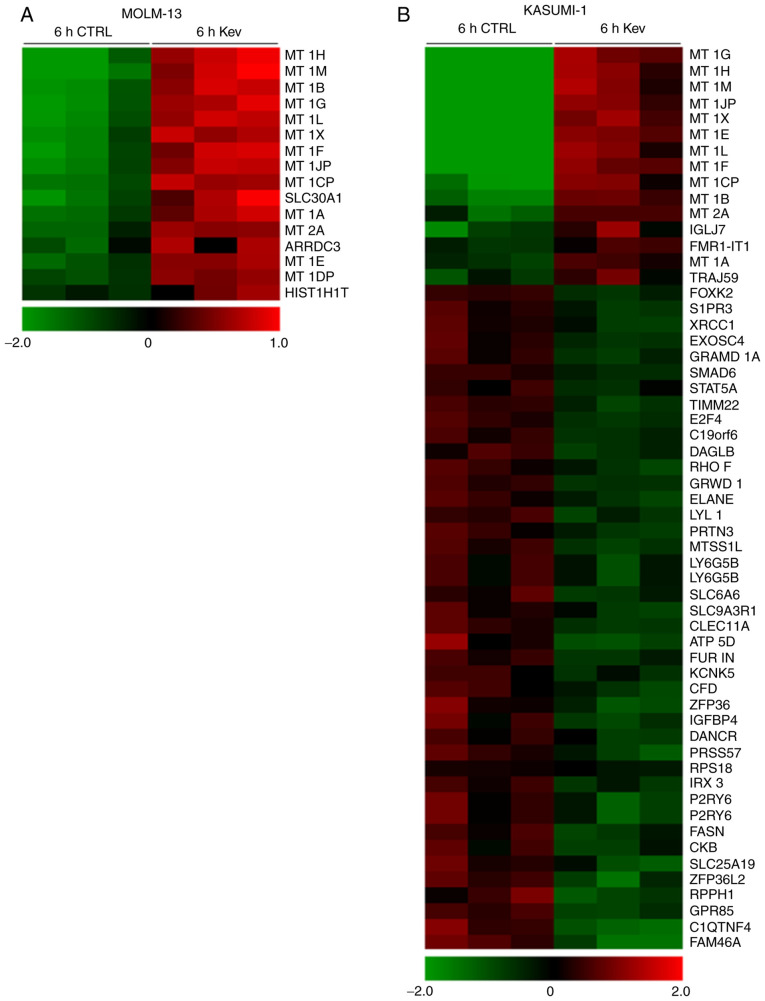
Gene expression profile analysis of MOLM-13 and KASUMI-1 cells was performed following treatment with kevetrin or the vehicle for 6 h to identify the transcriptional program regulated by short-term kevetrin exposure. This strategy enabled us to select early kevetrin-responsive genes. The overall transcriptional program was barely affected and we observed a differential expression of a restricted subset of genes (Fig. 2A and B), including the regulator of WNT/β-catenin signaling forkhead box K2 and the transcription factor signal transducer and activator of transcription 5A (STAT5A), which were downregulated 2-fold in KASUMI-1 cells after treatment (Fig. 2B). Moreover, MT 1 and 2, which are involved in the scavenging of oxygen-free radicals, were upregulated by kevetrin exposure in both cell lines (Fig. 2A and B). In addition, KASUMI-1 cells exhibited a significant downregulation of genes in relation to p53 activity, including E2F transcription factor 4 (E2F4), glutamate-rich WD repeat containing 1 (GRWD1), solute carrier family 6 member and elastase, neutrophil expressed (ELANE) (Fig. 2B).
Figure 2.
Short-term gene expression profile of kevetrin-treated AML cell lines. Genes differentially expressed in (A) MOLM-13 and (B) KASUMI-1 cells untreated (6 h CTRL) or treated (6 h Kev) for 6 h with kevetrin 340 µM. Color changes within a row indicate gene expression levels relative to the mean (red and green indicate upregulation and downregulation, respectively). AML, acute myeloid leukemia; Kev, kevetrin.
Continuous kevetrin treatment induces apoptosis in AML cell lines
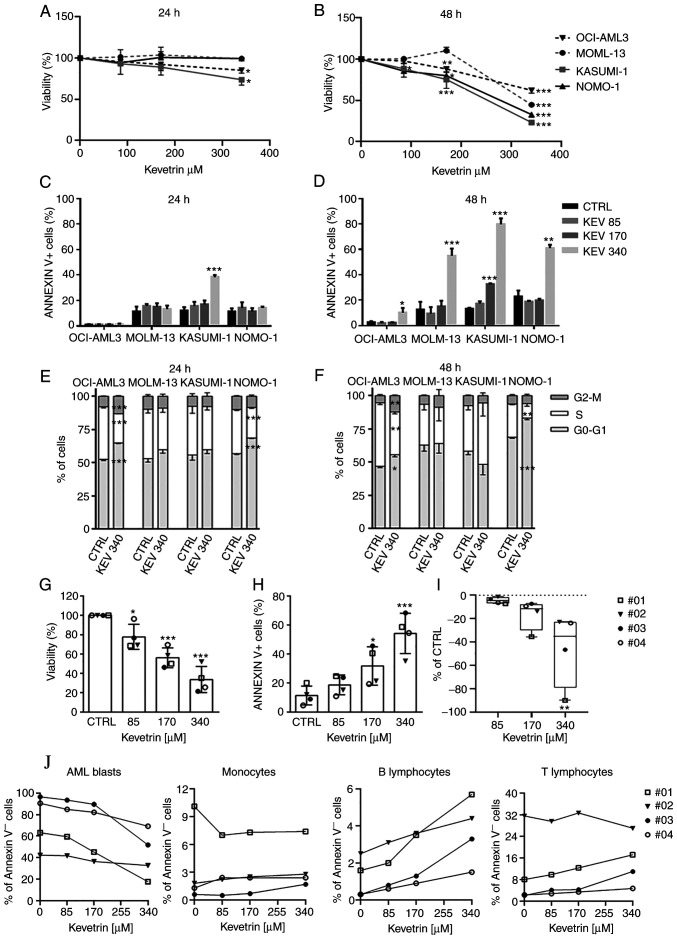
The effects of prolonged kevetrin exposure were investigated by evaluating cell viability after treatment for 24, 48 and 72 h (Fig. S2) using the same concentration range tested in the pulsed experiments. In addition to being tested on MOLM-13 and KASUMI-1 cells, continuous kevetrin treatment was also tested on TP53-wt OCI-AML3 cells and TP53-mutant (heterozygous frameshift deletion, p.C242fs) NOMO-1 models. Cell viability was barely altered after 24 h of treatment (Fig. 3A). At 48 h, a significant decrease in cell viability was observed in MOLM-13 cells at the highest concentration and in OCI-AML3 cells at 170 and 340 µM (Fig. 3B). A dose-dependent inhibition was also detected in KASUMI-1 and NOMO-1 cells (Fig. 3B). To better define the mechanism of action of kevetrin, apoptosis and cell cycle progression were analyzed. In KASUMI-1 cells, a significant apoptosis induction was observed after 24 h of treatment at the highest concentration (Fig. 3C and Fig. S3). After a 48-h exposure to the highest kevetrin concentration (340 µM), a significant increase was observed in Annexin V+ cells among MOLM-13 (54.95±5.63% in kevetrin-treated cells vs. 12.53±6.15% in the control) and NOMO-1 (60.93±2.63% in kevetrin-treated cells vs. 22.90±4.63% in the control) cells, and a mild but significant effect in OCI-AML3 cells (10.03±3.79% in kevetrin-treated cells vs. 2.60±0.70% in the control; Fig. 3D and Fig. S4). KASUMI-1 cells exhibited a dose-dependent response at 48 h, with 79.70±4.57% of apoptotic cells at 340 µM (compared with 13.18±0.80% in the control; Fig. 3D and Fig. S4). Apoptotic data in MOLM-13 and KASUMI-1 cells were confirmed in terms of mitochondrial depolarization, DNA fragmentation and caspase-3 activation (Fig. S5A-F). No cell cycle alterations were observed in MOLM-13 and KASUMI-1 cells, while NOMO-1 and OCI-AML3 cells displayed an accumulation of cells in the G0/G1 phase and a decrease of S phase cells after 24 and 48 h of kevetrin treatment at any concentration (Fig. 3E and F and Fig. S6A-D).
Figure 3.
Effect of continuous kevetrin treatment on viability and apoptosis induction in AML cell lines and primary samples. (A and B) Viability of TP53-wt OCI-AML3 and MOLM-13 and TP53-mut KASUMI-1 and NOMO-1 cell lines treated with different concentrations of kevetrin (85, 170 and 340 µM) for 24 and 48 h. (C and D) Quantification of Annexin V+ cells in AML cell lines treated with kevetrin at different concentrations (85, 170 and 340 µM) for 24 and 48 h. (E and F) Cell cycle analysis of cell lines treated with 340 µM kevetrin for 24 and 48 h. Values represent the mean ± standard deviation of three biological replicates. (G) Cell viability and (H) percentage of Annexin V+ cells in bone marrow mononuclear cells from AML patients exposed to increasing concentrations of kevetrin. (I) Percentage of AML blasts relative to control. Values represent the mean ± standard deviation. Symbols indicate samples from AML patients (no. 01: TP53-mut AML; nos. 02, 03 and 04: TP53-wt AML). (J) Percentage of Annexin V− cells in AML blasts, monocytes and lymphocytes. Blasts were defined as CD33+CD14− cells in the CD45/side scatter ‘blast gate’, monocytes as CD33+CD14+, B lymphocytes as CD19+ and T lymphocytes as CD3+. *P<0.05, **P<0.01, ***P<0.001. AML, acute myeloid leukemia; KEV, kevetrin; CTRL, control; wt, wild-type.
Kevetrin induces apoptosis in primary AML blast cells
BMMCs from 4 patients were treated with increasing kevetrin concentrations to evaluate its effect on primary AML cells. The mutational status of patients was assessed by targeted NGS (Table SII). One of the cases carried a TP53 mutation [no. 01: NM_000546, c.764T>A, p.(Ile255Asn), VAF 92.3%]. A dose-dependent decrease in cell viability (85 µM: 77.8±12.9%, 170 µM: 56.1±10.3% and 340 µM: 33.6.1±13.5%; Fig. 3G) and a significant increase in the proportion of apoptotic cells (170 µM: 31.8±13.3% and 340 µM: 54.3±13.9% vs. control: 11.4±6.5%; Fig. 3H and Fig. S6E) were observed. Kevetrin cytotoxicity was then assessed in BM cell populations and a preferential activity against AML blast cells was noted, indicating a significant dose-dependent decrease in the percentage of alive cells with respect to control, with the TP53-mutant sample exhibiting high sensitivity (Fig. 3I and J). Conversely, the percentage of AnnexinV− monocytes and B and T lymphocytes was only marginally affected by kevetrin treatment (Fig. 3J), suggesting a selective cytotoxic activity of this drug.
Prolonged kevetrin treatment results in the alteration of a shared core transcriptional program in AML cell lines
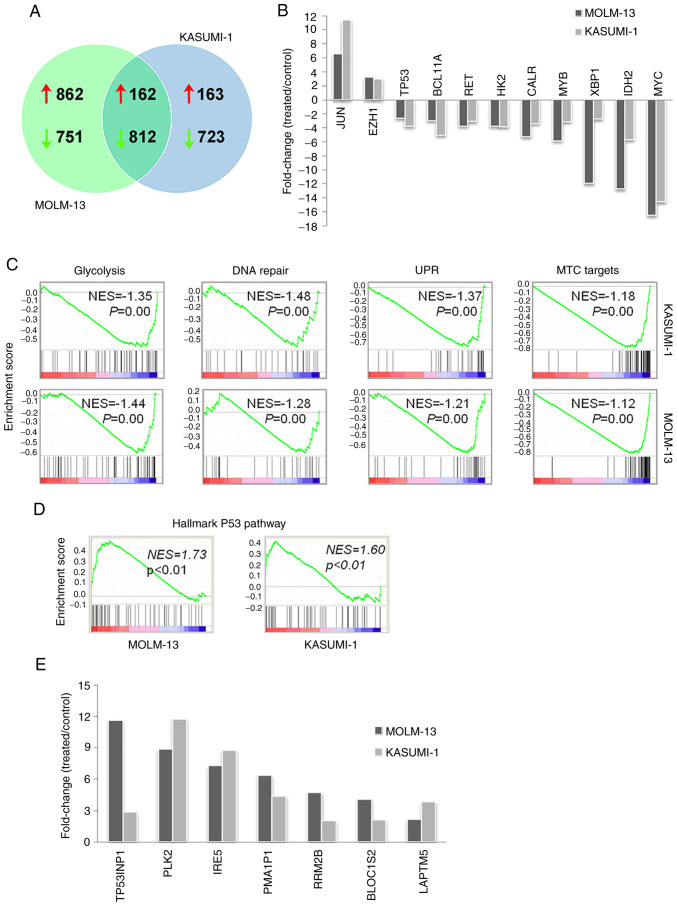
To better understand the molecular and biological effects of kevetrin exposure, gene expression profiling of MOLM-13 and KASUMI-1 cells after a 48-h treatment was performed. A total of 1,024 upregulated and 1,563 downregulated genes were identified in MOLM-13 cells, whereas KASUMI-1 cells exhibited increased expression of 325 genes and decreased levels of 1,535 genes. Of note, although a number of genes were uniquely altered, there was also a high degree of overlap in the transcriptional changes between MOLM-13 and KASUMI-1 cells, with a core transcriptional program of 162 upregulated and 812 downregulated genes in both cell lines (Fig. 4A). Enrichment analysis of the core transcripts revealed that upregulated genes were mainly involved in transcription, nucleosome assembly and telomere organization (HIST4H4, HIST1H3H, HIST2H4B and HIST1H4H), apoptosis (CASP10 and CASP8), autophagy (GABARAPL1, ATG14 and WIPI1), nuclear factor-κB pathway regulation (TRIM38, RIPK1, NFKBIA, IL1B, HSPA1A, HSPA1B, S100A4, CASP10, PLK2, CASP8, TLR6, PTGS2 and IL1B) and mitogen-activated protein kinase activity (DUSP1, JUN, MAP3K8, DUSP10, IL1B, HSPA1A, HSPA1B, GADD45B and DUSP22) (Table SIII). Downregulated genes were mostly involved in cell cycle (e.g., CDK4 and CDC7), DNA repair (e.g., EXO1 and FANCI), biosynthetic processes (e.g., PPAT involved in purine nucleotide biosynthesis and FASN in fatty acid synthesis), bioenergetics [e.g., glycolytic enzyme hexokinase 2 (HK2) and mitochondrial electron transport member COX5A), translation (e.g., RPL34 and RPSA), telomere maintenance (e.g., DKC1) and splicing (e.g., HNRNPR and PTBP1) (Table SIV). The common core transcriptional program included critical regulators of myelopoiesis and leukemogenesis, such as MYC, MYB and BCL11A, leukemia-related genes involved in unfolded protein response (UPR; XBP1 and CALR), signaling (RET), glycolysis (HK2) and DNA methylation [isocitrate dehydrogenase 2 (IDH2)], all of which were downregulated by kevetrin treatment (Fig. 4B). Conversely, increased levels of EZH1 and JUN were detected after exposure to kevetrin. Notably, GSEA of microarray data revealed that kevetrin-treated MOLM-13 and KASUMI-1 cells shared the downregulation of glycolysis, DNA repair, UPR and MYC target gene sets (Fig. 4C), in line with the results obtained by pathway analysis.
Figure 4.
Transcriptional changes induced by a 48-h exposure to 340 µM kevetrin. (A) Number of genes upregulated and downregulated in MOLM-13 and KASUMI-1 cells. (B) Fold-change of expression for leukemia-related genes from the common core transcriptional program in the two lines (ratio between kevetrin-treated and control cells). (C) Downregulation of glycolysis, DNA repair, UPR and MYC target signatures identified by gene set enrichment analysis. (D) Enrichment of a p53-related transcriptional signature in treated MOLM-13 and KASUMI-1 cells. (E) p53 target genes were upregulated in both cell lines following exposure to kevetrin (340 µM for 48 h). NES, normalized enrichment score; UPR, unfolded protein response.
Kevetrin affects the expression of p53 and its related proteins
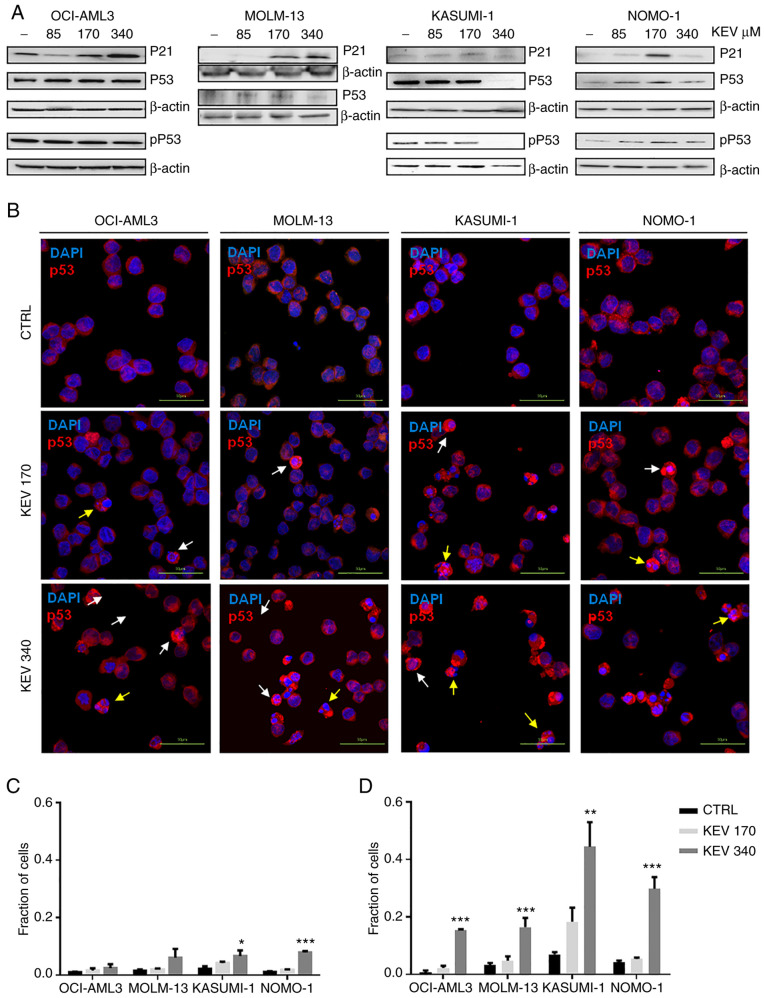
The effects of kevetrin treatment on the p53 pathway were then analyzed. High kevetrin doses induced a mild reduction in p53 mRNA levels. However, treated cells were enriched with a transcriptional gene signature involved in the p53 pathway (Fig. 4D). A total of 24 bona fide p53 targets (24) exhibited increased expression in MOLM-13 cells, including p21 (CDKN1A), MDM2 and NOXA (PMAIP1) (Table I). The latter gene was also upregulated in KASUMI-1 cells, along with 8 additional p53 targets (Table I). Among these, BLOC1S2, PLK2, RRM2B, TP53INP1, IER5 and LAPTM5 also exhibited increased expression in MOLM-13 cells following kevetrin treatment (Fig. 4E). To better understand the dynamics of p53 expression and its related proteins under kevetrin treatment, western blot and immunofluorescence analyses were performed after a 48-h exposure to increasing kevetrin doses. Western blot analysis did not reveal relevant alterations in the level of total p53 or its active form (phosphorylated on Serine 15), with the exception of KASUMI-1 cells that exhibited reduced p53 expression at the highest kevetrin dose. This may be associated with the high numbers of apoptotic cells lost during the washing steps required for pellet preparation. Immunofluorescence analysis at the highest kevetrin dose revealed an increased p53 expression compared with control cells, which resulted from a fraction of intact cells displaying nuclear p53 staining and apoptotic cells (identified by nuclear fragmentation) with very high p53 levels (Fig. 5B-D). This phenomenon was observed across all cell lines, particularly TP53-mutant ones (Fig. 5C and D). MOLM-13 cells generally displayed lower p53 expression levels across all conditions, thus hampering the detection of its active form by western blotting. Its target protein, p21, exhibited a dose-dependent upregulation following kevetrin treatment in TP53-wt models, in line with transcriptomic data obtained on MOLM-13 cells (Fig. 5A and Table I).
Table I.
p53 targets modulated by kevetrin treatment.
| Genes | MOLM-13 cells fold change (treated/control) | KASUMI-1 cells fold change (treated/control) |
|---|---|---|
| CDKN1A | 2.82 | |
| MDM2 | 11.00 | |
| BTG2 | 5.67 | |
| TNFRSF10B | 2.38 | |
| ZMAT3 | 5.20 | |
| PPM1D | 3.41 | |
| FAS | 2.48 | |
| NINJ1 | 2.48 | |
| SERTAD1 | 3.66 | |
| SESN1 | 10.67 | |
| SLC30A1 | 2.28 | |
| ARHGEF3 | 4.11 | |
| FUCA1 | 2.33 | |
| PRDM1 | 11.78 | |
| RNF19B | 2.86 | |
| TNFRSF10D | 2.52 | |
| ASCC3 | 2.21 | |
| BLOC1S2 | 4.09 | 2.06 |
| PLK2 | 8.92 | 11.76 |
| RRM2B | 4.73 | 2.04 |
| TP53INP1 | 11.64 | 2.85 |
| IER5 | 7.28 | 8.78 |
| LAPTM5 | 2.11 | 3.83 |
| PMAIP1 | 6.37 | 4.35 |
| ANKRA2 | 2.24 | |
| CD82 | 2.74 |
Figure 5.
Effect of kevetrin on the expression of p53 in AML cell lines. (A) Representative western blots of pSer15 p53 (pP53), P53 and P21 expression levels in MOLM-13, KASUMI-1, OCI-AML3 and NOMO-1 cell lines treated with kevetrin at 85, 170 and 340 µM for 48 h. β-actin was used as loading control. The figures show one representative of three independent experiments. (B) Immunofluorescence analysis of p53 in AML cell lines treated with 170 and 340 µM kevetrin for 48 h. The nuclei were stained with DAPI. White and yellow arrows indicate intact cells with nuclear p53 and nuclear-fragmented cells expressing high p53, respectively. (C) Fraction of intact cells with nuclear p53 localization or (D) p53-high apoptotic cells in kevetrin-treated cells. Values represent the mean ± standard deviation of 3 biological replicates (*P<0.05, **P<0.01, ***P<0.001). AML, acute myeloid leukemia; KEV, kevetrin; CTRL, control.
Discussion
Tumor suppressor p53 is a fundamental regulator of several responses to stress signals, such as DNA damage and hypoxia, also modulating various genes that play a key role in cell cycle arrest and DNA repair (13). Recently, p53 was found to regulate metabolism, stem cell maintenance, invasion and communication within the tumor microenvironment (16), suggesting a more comprehensive role in orchestrating cellular responses. TP53 is one of the most frequently mutated genes in human tumors (12) and mutant p53 exerts a negative effect on the wt protein or acquires gain-of-function properties (25,26). The aim of the present study was to evaluate the cellular and molecular effects of kevetrin on AML models with different TP53 mutational status, as well as on primary AML cells.
After both pulsed and continuous treatments, a higher sensitivity of the mutated cell lines compared with wt cells was observed. Kevetrin exhibited a dose-dependent efficacy after a 48-h treatment, with a stronger effect on mutated models. We did not observe cell cycle alterations in KASUMI-1 or MOLM-13 models, whereas a significant accumulation of cells in the G0/G1 phase was observed in OCI-AML3 and NOMO-1 cells. Only TP53-wt OCI-AML3 cells displayed an increase in the percentage of G2/M cells after treatment, as previously reported in the TP53-wt A549 lung carcinoma cell line (18). The difference observed between KASUMI-1 and NOMO-1 mutated models may be attributable to the predominant apoptotic effect induced by kevetrin on KASUMI-1 cells and to the different TP53 mutational status. KASUMI-1 cells are characterized by a homozygous mutation resulting in an inactive protein, whereas NOMO-1 cells harbor a frameshift deletion (p.C242fs) leading to the synthesis of a truncated protein. Of note, kevetrin was effective against primary AML cells. Both TP53-wt and TP53-mutant cells responded to ex vivo treatment, with mutant cells exhibiting promising sensitivity. Kevetrin induced apoptosis in leukemic blasts, whilst largely sparing the immune microenvironment, which points to a less toxic drug profile.
Early response to kevetrin treatment, which was assessed after a 6-h drug exposure, was characterized by upregulation of the low-molecular weight cysteine-rich proteins MT1 and 2 in both cell lines. Several studies have reported that MTs promote cell growth and protect cells from drug-induced oxidative stress in cancer models (27). Thus, increased MT expression shortly after kevetrin treatment may represent an attempt to resist chemotherapeutic drugs (28). Although little is known on the role of MTs in hematological malignancies, it was inferred that MT upregulation exerted a tumor-suppressive function in our models, as was recently suggested for MT3 overexpression in pediatric AML, which inhibits proliferation and induces apoptosis (29). Of note, MTs are known to interact with both wt and inactive p53 protein (30), and to modulate p53 transcriptional activity through modification of its conformation (31), as reported in breast cancer and osteosarcoma cell lines (31,32). Downregulation of STAT5, a mediator of KIT-driven leukemogenesis, only occurred in the mutated model and may be correlated with the downregulation of the signaling pathway activated by KIT mutation in KASUMI-1 cells (33).
With regard to TP53 interactors, the mutated model exhibited downregulation of E2F4, which is known to be involved in p53-dependent gene repression (34), and GRWD1, which appears to interact with p53, negatively regulating its transcriptional activity (35). The downregulation of another member of the E2F transcription factor family, E2F1, was documented by Kumar et al in MDA-MB-231 breast cancer cells, MIA PaCa-2 pancreatic cancer cells and K-562 TP53-mutant leukemic cells (19,36). A reduction in ELANE levels, a target highly expressed in leukemia patients and negatively correlated with TP53, was also observed (37).
The p53 pathway was modulated by a 48-h kevetrin treatment, with transcriptional upregulation of several p53 targets in TP53-wt and -mutant models. In the wt cell lines, kevetrin induced a dose-dependent upregulation of the p21 protein, in line with the trend observed in TP53-wt solid tumor cells (18,20). In the TP53-mutant models, p21 upregulation was observed at an intermediate kevetrin dose, with increased p53 nuclear localization (also detected at the highest dose), together with a high fraction of p53-positive cells with nuclear fragmentation. The differences between western blot analysis and immunofluorescence may be partly attributed to the strong effects of the treatment on cell viability (with loss of apoptotic cells in the washing steps required for pellet preparation). Moreover, immunofluorescence analysis is characterized by higher sensitivity. Overall, the data of the present study revealed that kevetrin exerted a therapeutic effect against both TP53-wt and -mutant AML cells, the latter being more sensitive in terms of apoptosis and p53 induction. The results obtained from wt lines, i.e., no significant p53 nuclear translocation and few p53-related transcriptional changes at early timepoints, suggest an indirect kevetrin effect on p53-wt AML cells.
Several cellular processes, including glycolysis, DNA repair and UPR, were deregulated in response to high-dose kevetrin treatment. Kevetrin exposure targeted a common core transcriptional program including genes involved in myelopoiesis and leukemogenesis. Among these, JUN and enhancer of zeste homolog 1 (EZH1) were upregulated. JUN, which plays a key role in cellular proliferation, differentiation and apoptosis, is highly expressed in AML (38). Kevetrin-induced upregulation of JUN is in line with previous findings indicating that c-JUN represses p53 transcription and downregulates its protein levels, thereby controlling cell cycle progression (39). EZH1 is involved in the methylation of the histone H3 and may be mutated in AML (3). Its increased expression in our models may have been caused by deregulated chromatin modification following drug exposure. Moreover, JUN or EZH1/2 inhibition exerts a therapeutic effect against leukemic cells, suggesting that these genes may represent potential targets for kevetrin-based combination strategies (40,41).
Kevetrin modulates several oncogenic pathways, including those involving c-MYC, MYB and B-cell lymphoma/leukemia 11A (BCL11A). c-MYC, a gene playing a key role in hematopoiesis, is frequently overexpressed and activated in AML and is often associated with leukemogenesis (42,43). Hoffman et al documented the importance of c-MYC in apoptosis induction, highlighting its crosstalk with p53 (44). c-MYB plays an important role in cellular proliferation, while BCL11A is involved in normal hematopoiesis and lymphoid malignancies (45) and inhibits p53 activity (46). We also observed downregulation of two additional key leukemia-related genes: Mitochondrial HK2, frequently upregulated by the presence of internal tandem duplications of the FLT3 gene, which, in turn, promotes glycolysis, and the isocitrate dehydrogenase 2 (IDH2), which is commonly mutated in AML, with a consequent impact on DNA methylation and 2-hydroxiglutarate metabolism (47). This evidence, along with the repression of a glycolysis-related gene set in drug-treated cells, suggests that kevetrin altered cellular metabolism in our models.
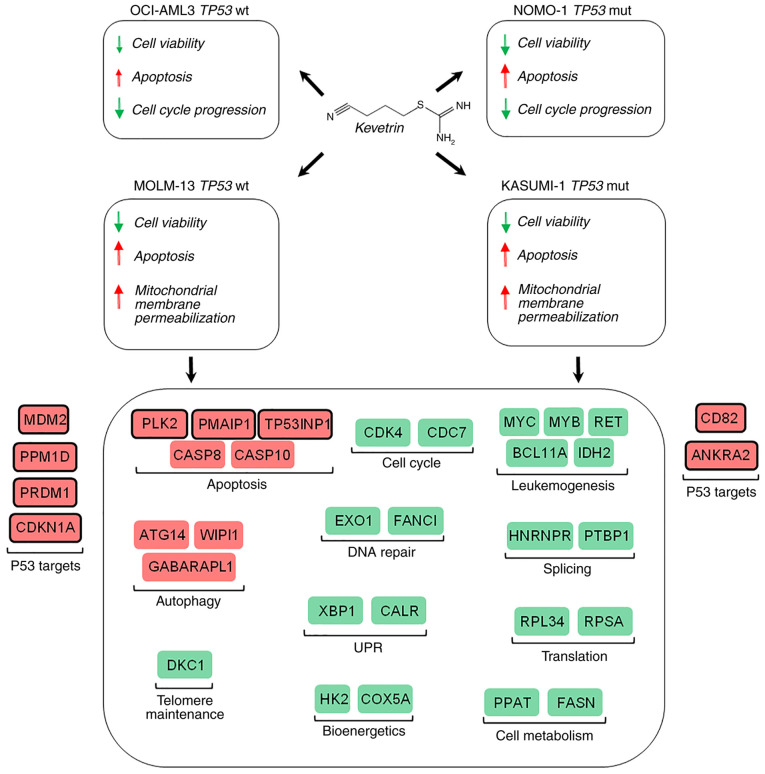
Given the importance of the TP53 gene in cancer and the frequency of its mutation, several compounds have been tested with the aim of restoring wt p53 function or of degrading the mutated protein to promote apoptosis. Despite the lack of in vivo experiments, which represents a limitation to the present study, the results suggest that kevetrin may be a promising novel drug for the treatment of AML patients carrying either wt or mutant TP53, with the latter representing an imperative medical need due to its associated dismal prognosis (Fig. 6) (9). A phase I clinical trial evaluating kevetrin activity in advanced solid tumors has been successfully completed, and its results indicate good tolerability and the potential for therapeutic response (NCT01664000) (36,48). The data presented herein provide a rationale for an experimental trial in AML patients, particularly those carrying TP53 mutation, for whom the therapeutic options are currently limited.
Figure 6.
Schematic representation of the effects of kevetrin on AML cell lines. Red indicates ‘increase’, while green indicates ‘decrease’. Representative genes for the most relevant affected process are shown. UPR, unfolded protein response.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated during the current study are available in the GEO repository, under the accession number GSE137574.
Authors' contributions
Conceptualization, RN, SC, GS, AC, GMu and GMa; Data curation, SDM, SC, DC and GS; formal analysis, RN, SB, GA, LC, MG, MTB, CL, LM and GS; investigation, RN, SDM and KM; supervision, AC and GMa; writing of the original draft, RN, SDM and GS; review and editing, SC, KM, GMu and GMa. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Primary samples were obtained after informed consent, as approved by the Institutional Ethics Committees in accordance with the Declaration of Helsinki (protocol 112/2014/U/Tess of Policlinico Sant'Orsola-Malpighi).
Patient consent for publication
Not applicable.
Competing interests
KM is a co-founder of Innovation Pharmaceuticals and he is the inventor of the patent (US 8338454 B2) for the kevetrin molecule used in this study. GM has received honoraria from Novartis, BNS, Roche, Pfizer, Ariad and EMSB. All other authors declare that they have no competing interests.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson AG, Hoadley K, Triche TJ, Jr, Laird PW, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 5.Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 6.Leung GMK, Zhang C, Ng NKL, Yang N, Lam SSY, Au CH, Chan TL, Ma ESK, Tsui SP, Ip HW, et al. Distinct mutation spectrum, clinical outcome and therapeutic responses of typical complex/monosomy karyotype acute myeloid leukemia carrying TP53 mutations. Am J Hematol. 2019;94:650–657. doi: 10.1002/ajh.25469. [DOI] [PubMed] [Google Scholar]
- 7.Fontana MC, Marconi G, Feenstra JDM, Fonzi E, Papayannidis C, Ghelli Luserna di Rorá A, Padella A, Solli V, Franchini E, Ottaviani E, et al. Chromothripsis in acute myeloid leukemia: Biological features and impact on survival. Leukemia. 2018;32:1609–1620. doi: 10.1038/s41375-018-0035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rücker FG, Dolnik A, Blätte TJ, Teleanu V, Ernst A, Thol F, Heuser M, Ganser A, Döhner H, Döhner K, Bullinger L. Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica. 2018;103:e17–e20. doi: 10.3324/haematol.2017.180497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonetti G, Padella A, do Valle IF, Fontana MC, Fonzi E, Bruno S, Baldazzi C, Guadagnuolo V, Manfrini M, Ferrari A, et al. Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer. 2019;125:712–725. doi: 10.1002/cncr.31837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 12.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scoumanne A, Chen X. Protein methylation: A new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23:1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 15.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 16.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, Hiran T, Holden SA, Chafai-Fadela K, Rogers S, Ram S, Menon K. Kevetrin™, a novel small molecule, activates p53, enhances expression of p21, induces cell cycle arrest and apoptosis in a human cancer cell line. Cancer Res. 2011;71:4470. [Google Scholar]
- 19.Kumar A, Holden SA, Chafai-Fadela K, Ram S, Menon KE. Kevetrin targets both MDM2-p53 and Rb-E2F pathways in tumor suppression. Cancer Res. 2012;72:2874. [Google Scholar]
- 20.Kumar A, Brennan DP, Chafai-Fadela K, Holden SA, Ram S, Shapiro GI, Menon GI. Kevetrin induces p53-dependent and independent cell cycle arrest and apoptosis in ovarian cancer cell lines representing heterogeneous histologies. Cancer Res. 2017;77:322. [Google Scholar]
- 21.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller PA, Vousden KH. P53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 26.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood. 2017;130:699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi S. Molecular functions of metallothionein and its role in hematological malignancies. J Hematol Oncol. 2012;5:41. doi: 10.1186/1756-8722-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu A, Krishnamurthy S. Cellular responses to cisplatin-induced DNA damage. J Nucleic Acids. 2010;2010:201367. doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao YF, Xu LX, Lu J, Cao L, Li ZH, Hu SY, Wang NN, Du XJ, Sun LC, Zhao WL, et al. Metallothionein III (MT3) is a putative tumor suppressor gene that is frequently inactivated in pediatric acute myeloid leukemia by promoter hypermethylation. J Transl Med. 2014;12:182. doi: 10.1186/1479-5876-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrakhovitch EA, Olsson PE, Jiang S, Cherian MG. Interaction of metallothionein with tumor suppressor p53 protein. FEBS Lett. 2006;580:1235–1238. doi: 10.1016/j.febslet.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 31.Méplan C, Richard MJ, Hainaut P. Metalloregulation of the tumor suppressor protein p53: Zinc mediates the renaturation of p53 after exposure to metal chelators in vitro and in intact cells. Oncogene. 2000;19:5227–5236. doi: 10.1038/sj.onc.1203907. [DOI] [PubMed] [Google Scholar]
- 32.Habel N, Hamidouche Z, Girault I, Patiño-García A, Lecanda F, Marie PJ, Fromigué O. Zinc chelation: A metallothionein 2A's mechanism of action involved in osteosarcoma cell death and chemotherapy resistance. Cell Death Dis. 2013;4:e874. doi: 10.1038/cddis.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A, Ghosh J, Ramdas B, Mali RS, Martin H, Kobayashi M, Vemula S, Canela VH, Waskow ER, Visconte V, et al. Regulation of Stat5 by FAK and PAK1 in oncogenic FLT3- and KIT-driven leukemogenesis. Cell Rep. 2014;9:1333–1348. doi: 10.1016/j.celrep.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson EK, Mungamuri SK, Attie O, Kracikova M, Sachidanandam R, Manfredi JJ, Aaronson SA. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene. 2014;33:3959–3969. doi: 10.1038/onc.2013.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayama K, Watanabe S, Takafuji T, Tsuji T, Hironaka K, Matsumoto M, Nakayama KI, Enari M, Kohno T, Shiraishi K, et al. GRWD1 negatively regulates p53 via the RPL11-MDM2 pathway and promotes tumorigenesis. EMBO Rep. 2017;18:123–137. doi: 10.15252/embr.201642444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro G, Mier JW, Hilton JF, Gandhi L, Chau NG, Bullock AJ, Supko JG, Verselis SJ, Murgo K, Sze C, et al. A phase 1, dose-escalation, safety, pharmacokinetic, pharmacodynamic study of thioureidobutyronitrile, a novel p53 targeted therapy, in patients with advanced solid tumors. J Clin Oncol. 2015;33(Suppl 15):TPS2613. doi: 10.1200/jco.2015.33.15_suppl.tps2603. [DOI] [Google Scholar]
- 37.Zhao Y, Si L, Zhang W, Huang W, Wang R. ELANE is highly expressed in leukemia patients and predicts poor survival. Int J Clin Exp Med. 2019;12:3153–3160. [Google Scholar]
- 38.Rangatia J, Vangala RK, Singh SM, Zada AAP, Elsässer A, Kohlmann A, Haferlach T, Tenen DG, Hiddemann W, Behre G. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPalpha DNA binding via leucine zipper domain interaction. Oncogene. 2003;22:4760–4764. doi: 10.1038/sj.onc.1206664. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita S, Honma D, Adachi N, Araki K, Takamatsu E, Katsumoto T, Yamagata K, Akashi K, Aoyama K, Iwama A, Kitabayashi I. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia. 2018;32:855–864. doi: 10.1038/leu.2017.300. [DOI] [PubMed] [Google Scholar]
- 41.Zhou C, Martinez E, Di Marcantonio D, Solanki-Patel N, Aghayev T, Peri S, Ferraro F, Skorski T, Scholl C, Fröhling S, et al. JUN is a key transcriptional regulator of the unfolded protein response in acute myeloid leukemia. Leukemia. 2017;31:1196–1205. doi: 10.1038/leu.2016.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolores Delgado M, León J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–616. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 45.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 46.Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, Boyiadzis M. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: Associations with prognosis and potential treatment strategies. Leukemia. 2014;28:1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro G, Supko JG, Cho DC, Hilton JF, Hadfield M, Pruitt-Thompson S, Bordoli-Trachsela E, Zvereva N, Wolanski A, Sato-DiLorenzo A, et al. A phase I, dose-escalation, safety, pharmacokinetic, pharmacodynamic study of thioureidobutyronitrile, a novel p53 targeted therapy, in patients with advanced solid tumors. J Clin Oncol. 2013;31(Suppl 15):TPS2627. doi: 10.1200/jco.2013.31.15_suppl.tps2627. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the GEO repository, under the accession number GSE137574.