Abstract
The aberrant expression of long non-coding RNAs (lncRNAs), including LINC00958, has been demonstrated in several types cancers. The present study aimed to investigate the role of LINC00958 in colorectal cancer (CRC) and identify the possible underlying mechanisms. The expression of LINC00958 and microRNA (miR)-3619-5p was detected in several human CRC cell lines using reverse transcription-quantitative PCR. Then, short hairpin RNA (shRNA)-LINC00958 was transfected into the cells. The results revealed that the expression of LINC00958 was notably upregulated, whereas miR-3619-5p was downregulated in CRC cells. Transfection with shRNA-LINC00958 inhibited the proliferation, invasion and migration of CRC cells. Moreover, the rate of apoptosis was enhanced, accompanied by a decrease in the expression of Bcl-2 and an increase in the expression of Bax and caspase-3. A luciferase reporter assay was conducted to verify the target binding site between LINC00958 and miR-3619-5p. The luciferase reporter assay confirmed that miR-3619-5p could be directly targeted by LINC00958. Furthermore, the miR-3619-5p inhibitor reversed the effects of LINC00958 silencing on proliferation, invasion, migration and apoptosis. Taken together, the findings suggest that the downregulation of LINC00958 suppresses the proliferation, invasion and migration, and promotes the apoptosis of CRC cells by targeting miR-3619-5p in vitro, which provides a theoretical basis and therapeutic strategy for the treatment of CRC.
Keywords: colorectal cancer, proliferation, invasion, migration, apoptosis, LINC00958
Introduction
Colorectal cancer (CRC) is a common digestive system malignancy and a major cause of cancer-related mortality worldwide (1). CRC is a fatal disease that is responsible for more than 600,000 deaths each year (2). Lifestyle and dietary factors contribute to the increasing incidence of CRC each year (3). In the past decades, combined therapies for CRC have improved treatment efficacy. These therapies primarily includes surgical resection, chemotherapy and radiotherapy, but none have proven to be optimal due to the fact that the majority of patients with CRC are often diagnosed at an advanced stage (4,5). Notably, the primary reason for poor prognosis in patients with advanced CRC is tumor metastasis and recurrence (6). Thus, elucidating the molecular mechanisms underlying the progression of CRC may reveal potential prognostic biomarkers and novel treatment strategies for CRC.
Long non-coding RNAs (lncRNAs) are a class of non-protein coding transcripts that are >200 nucleotides in length. It has been well reported that lncRNAs play significant roles in cancer biology, including proliferation, invasion, apoptosis and metastasis (7,8). Aberrant expression of lncRNAs has been observed in the occurrence and development of a variety of cancers, suggesting that they may serve as powerful mediators in carcinogenesis (9–11). Over the past few decades, researchers have focused on investigating the function of lncRNAs in the pathogenesis of CRC (12,13). LINC00958 is a newly discovered lncRNA, which was identified in recent years (14). The abnormally high expression of LINC00958 has been demonstrated in patients with gastric cancer, and was found to be associated with metastasis and unfavorable prognosis (15). Additionally, evidence indicates that LINC00958 accelerates the progression of oral squamous cell carcinoma and pancreatic cancer by acting as a microRNA (miR/miRNA) sponge (16,17). However, to the best of our knowledge, the function of LINC00958 in CRC has not yet been studied, which has peaked our interest.
In the present study, the expression of LINC00958 was detected in several CRC cell lines. Subsequently, the effects of LINC00958 on proliferation, invasion, migration and apoptosis of CRC cells, as well as its underlying mechanisms were investigated. It is hoped that this study provides the theoretical basis for a novel therapeutic strategy for the treatment of CRC.
Materials and methods
Cell culture
The colon cancer cell lines (GEO, SNU-C1, COLO205, HCT-116 and SW480), colorectal cancer cell line (HT-29) and normal colorectal epithelial cell line (NCM460) used in this study were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and the cell lines were authenticated using STR profiles. Cell lines were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in incubators containing 5% CO2 at 37°C.
Cell transfection
SW480 cells were seeded in a 24-well plate. When cell confluence reached 60%, 20 nM of short hairpin RNA (shRNA) targeted against LINC00958 (shRNA-LINC00958-1 or shRNA-LINC00958-2) or scramble shRNA (negative control for LINC00958 shRNA, shRNA-NC), 40 nM of miR-3619-5p mimic, miR-3619-5p inhibitor or miRNA negative control (miR-NC) were transfected into SW480 cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. The above oligonucleotides were purchased from Shanghai GenePharma Co., Ltd. SW480 cells were seeded in a 24-well plate. SW480 cells without transfection served as the control group. At 48 h post-transfection, cells were harvested and successful transfection was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Cell Counting Kit-8 (CCK-8) assay
Cells were grown in 96-well plates and viability was evaluated using a CCK-8 reagent (Sigma-Aldrich; Merck KGaA), according to the manufacturer's protocol when 80% confluency was reached. After transfection for 24, 48 and 72 h, 10 µl CCK-8 solution was added to each well, then the optical density was measured at 450 nm using a microplate reader.
EdU (5-ethynyl-20-deoxyuridine) incorporation assay
The proliferation of SW480 cells was detected using an EdU assay kit (Ribobio, China) in accordance with the manufacturer's guidelines. Briefly, cells were incubated with 10 µM EdU and subsequently fixed in 4% paraformaldehyde. After EdU staining, cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The images of EDU-positive cells were observed and captured randomly in five fields under a confocal fluorescence microscope (magnification, ×100; Nikon, Tokyo, Japan).
Transwell assay
A Transwell assay was used to determine the invasive activity of SW480 cells. Briefly, 200 µl serum-free DMEM containing 5×104 cells were plated into the apical chamber, which consisted of 8-µm pore inserts coated with Matrigel (BD Biosciences) in culture plates. DMEM supplemented with 10% FBS as a chemotactic factor was added in the basolateral chambers. Then, 4% polyformaldehyde was used to fix cells and 0.1% crystal violet was employed to stain cells. Finally, the number of invasive cells were counted and images were captured by a light microscope (Olympus Corporation) at ×200 magnification.
Scratch wound healing assay
SW480 cells were seeded into 6-well plates (6×105 cells per well) for adherent culture. When cells reached 80% confluence, a thin wound was created on the cell monolayer using a 10-µl sterile pipette tip. The detached cells were washed off twice, and the medium was replaced with DMEM containing 1% FBS. Subsequently, the migration of cells was observed and images were acquired at 0 and 24 h after wounding using a phase contrast microscope (magnification, ×100; IX711; Olympus Corporation). The relative wound healing closure was calculated.
Cell apoptosis assay
SW480 cells were seeded into 6-well plates (6×105 per well). Then, each group of cells was treated as needed. Following this, cells were double stained with Annexin V and propidium iodide (PI) using the Annexin V-FITC Apoptosis Detection kit (Vazyme Biotech Co., Ltd.) in accordance with the manufacturer's instructions. The activity of Annexin V/PI was then examined using flow cytometry (BD Biosciences) and quantified by Flow Jo software (version 7.6.1; FlowJo LLC).
RT-qPCR
Cells were subjected to total RNA extraction using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The total RNA (2 µg) was then reverse transcribed into cDNA using a Reverse Transcription kit (Beijing Transgen Biotech Co., Ltd.), according to the manufacturer's protocols. qPCR was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, Inc.) on an ABI 7500 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). Relative expression of the target gene was normalized against U6 and GAPDH and calculated based on the 2−ΔΔCq method (18).
Western blot analysis
Total protein was isolated from SW480 cells using a protein lysis buffer (RIPA; Beyotime Institute of Biotechnology), followed by quantification of protein concentration using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Protein samples (40 µg/lane) were loaded on a 10% gel, resolved via SDS-PAGE, and then subsequently transferred to a PVDF membrane (EMD Millipore). After blocking with skimmed milk, these membranes were incubated with primary antibodies overnight at 4°C. Subsequently, these blots were probed with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.). Finally, the immunoreactive bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences). Anti-Bcl-2 (1:1,000 dilution; cat. no. 3498T), anti-Bax (1:1,000 dilution; cat. no. 5023T), anti-caspase-3 (1:1,000 dilution; cat. no. 14220T) anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:1,000; cat. no. 5174T) antibodies were the products of Cell Signaling Technology, Inc. GAPDH was used as an internal control. The intensity of the bands was quantified using ImageJ software (version 1.52r; National Institutes of Health).
Luciferase reporter assay
The potential miRNAs which may have binding sites with LINC00958 were predicted using the bioinformatics tool StarBase version 3.0 (http://starbase.sysu.edu.cn/) (19). Wild-type and mutant reporter plasmids of LINC00958 (WT-LINC00958 and MUT-LINC00958), which contained miR-3619-5p mimic or miR-NC binding sites, were synthesized by Shanghai GenePharma Co., Ltd. (cat. no. H00001). 293T cells were seeded in 24-well plates and cultured at 37°C until they reached 70% confluence. To perform the luciferase reporter assay, cells were first co-transfected with LINC00958 3′-UTR-WT or LINC00958 3′-UTR-MUT and miR-3619-5p mimic or miR-NC using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), following which luciferase activity was detected using a Dual-Luciferase® Reporter Assay System (Promega Corp.) 48 h after transfection. Renilla luciferase activity was used as control.
Statistical analysis
Each experiment was repeated in triplicate and data are presented as the mean ± standard deviation. Data were analyzed using GraphPad Prism version 6.0 (GraphPad Software, Inc.). Comparisons between groups were performed using a two-tailed Student's t-test, whereas multiple comparisons were analyzed with one-way ANOVA followed by a post hoc Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
LINC00958 is highly expressed in CRC cell lines
To investigate the role of LINC00958 in CRC, the expression of LINC00958 was detected using RT-qPCR in several human CRC cell lines (GEO, SNU-C1, COLO205, HCT-116, HT-29 and SW480) and one normal colorectal epithelial cell line (NCM460). As presented in Fig. 1A, LINC00958 expression was notably upregulated in CRC cell lines compared with that noted in the NCM460 cells, and the expression of LINC00958 was the highest in the SW480 cells. Therefore, SW480 cells were used to perform the following experiments.
Figure 1.
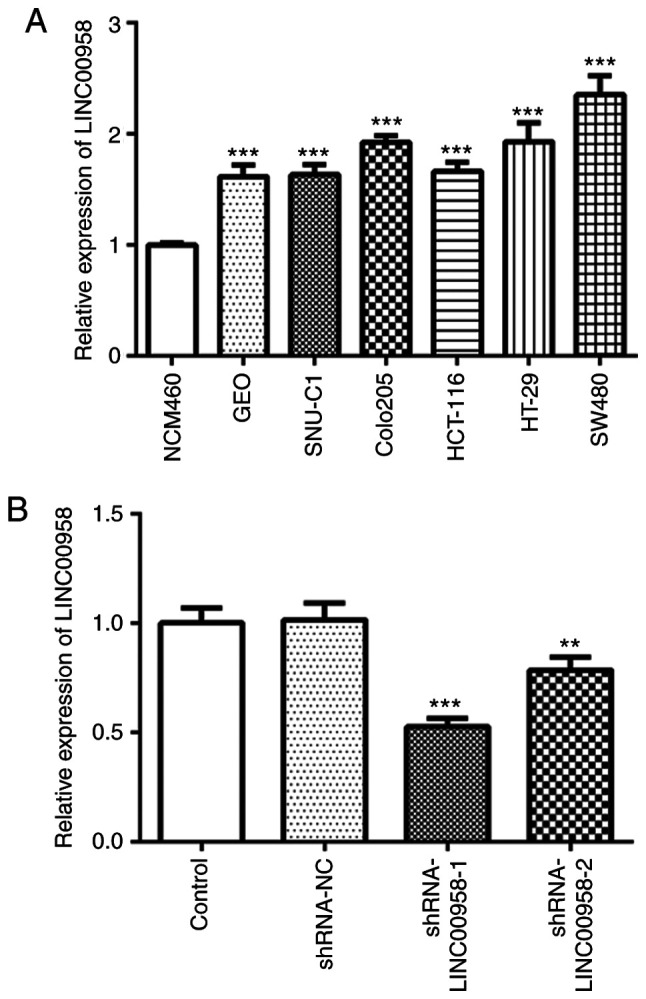
Expression of LINC00958 is upregulated in CRC cell lines. (A) The expression of LINC00958 was examined in several CRC cell lines (GEO, SNU-C1, COLO205, HCT-116, HT-29 and SW480) using reverse transcription-quantitative PCR. The results were generated from three independent repeats (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a two-tailed Student's t test. ***P<0.001 vs. NCM460. (B) The expression of LINC00958 was determined using western blotting after transfection with shRNA-LINC00958-1 or shRNA-LINC00958-2. All experiments were repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a two-tailed Student's t test. **P<0.01, ***P<0.001 vs. shRNA-NC. shRNA, short hairpin RNA; NC, negative control.
LINC00958 silencing inhibits the proliferation, invasion and migration of CRC cells
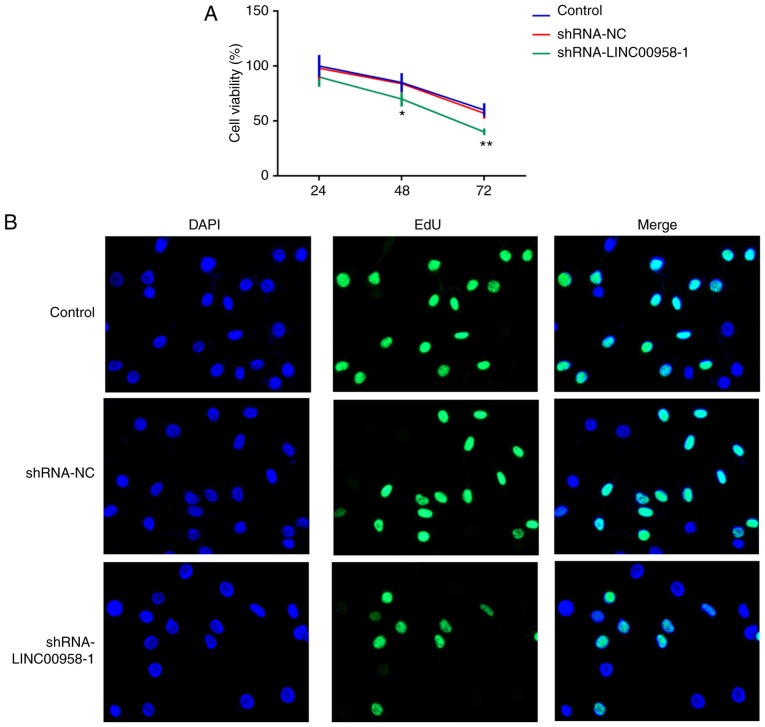
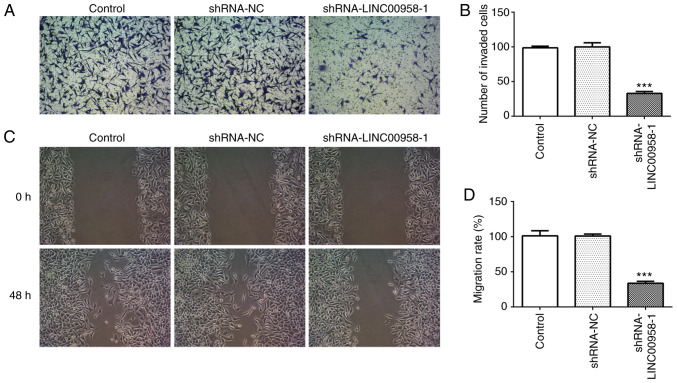
To explore the effects of LINC00958 in regards to the functions of SW480 cells, shRNA-LINC00958-1 or shRNA-LINC00958-2 was transfected into the cells. As shown in Fig. 1B, the expression of LINC00958 was notably decreased after transfection compared with the shRNA-NC group. shRNA-LINC00958-1 was used to perform the following experiments. A CCK-8 assay was employed to measure the proliferation of SW-480 cells. As exhibited in Fig. 2A and B, LINC00958 silencing significantly suppressed the proliferative ability of SW-480 cells compared with the shRNA-NC group. In addition, Transwell and wound healing assays were performed to evaluate the effects of LINC00958 on the invasion and migration of CRC cells, respectively. As demonstrated in Fig. 3A and B, the invasive ability of SW480 cells was significantly decreased relative to the shRNA-NC group following transfection with shRNA-LINC00958-1. Concurrently, notable inhibition of SW480 cell migration was found in the shRNA-LINC00958-1 group compared with the shRNA-NC group (Fig. 3C and D). These findings indicated that LINC00958 silencing suppressed the proliferation, invasion and migration of CRC cells.
Figure 2.
Downregulation of LINC00958 inhibits the proliferation of SW480 cells. Cell proliferation was detected using (A) Cell Counting Kit-8 assay and (B) EdU incorporation assay, respectively. All experiments were repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a two-tailed Student's t test. *P<0.05, **P<0.01, vs. shRNA-NC. EdU, 5-ethynyl-20-deoxyuridine; shRNA, short hairpin RNA; NC, negative control.
Figure 3.
Downregulation of LINC00958 inhibits the invasion and migration of SW480 cells. (A) The invasive ability of SW480 cells was evaluated using a Transwell assay. (B) Relative number of invaded cells. (C) Migratory activity of SW480 cells was determined using a scratch wound healing assay. (D) Relative migratory rate of SW480 cells. The experiments were generated from three independent repeats (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a two-tailed Student's t test. ***P<0.001, vs. shRNA-NC. shRNA, short hairpin RNA; NC, negative control.
LINC00958 silencing promotes the apoptosis of CRC cells
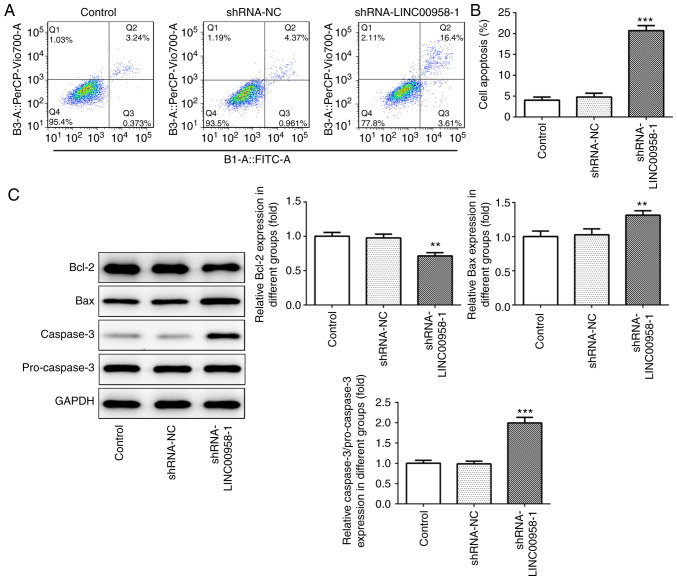
Flow cytometry was performed to examine the effects of LINC00958-1 transfection on the apoptosis of SW480 cells. As presented in Fig. 4A and B, silencing of LINC00958 notably elevated the rate of cell apoptosis in the SW480 cells compared with the shRNA-NC group. Subsequently, the expression levels of apoptosis-related proteins were measured using western blotting. As shown in Fig. 4C, the expression of Bcl-2 was significantly reduced after transfection with shRNA-LINC00958-1, which was accompanied by a significant increase in Bax and caspase-3 expression. These results indicate that downregulation of LINC00958 promoted the apoptosis of CRC cells.
Figure 4.
LINC00958 silencing promotes the apoptosis of SW480 cells. (A) Flow cytometry was used to measure the apoptotic rate of SW480 cells. (B) Cell apoptosis was quantified. (C) The expression of apoptosis-related proteins was detected using western blotting. All experiments were repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a two-tailed Student's t test. **P<0.01, ***P<0.001 vs. shRNA-NC. shRNA, short hairpin RNA; NC, negative control.
LINC00958 is identified as a potential target of miR-3619-5p
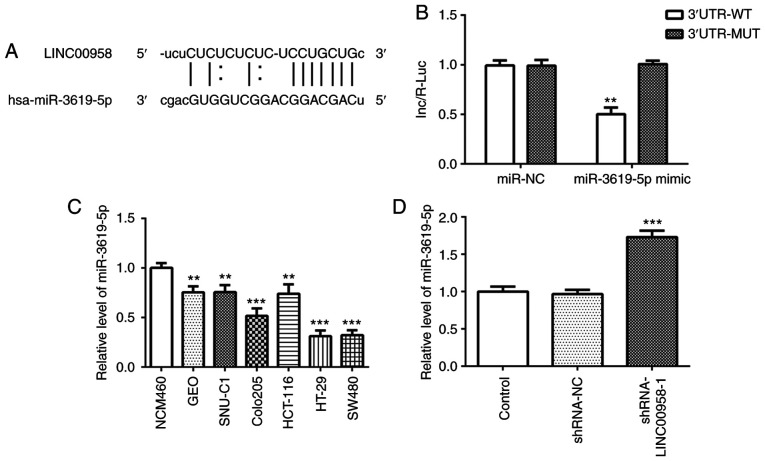
To explore the underlying regulatory mechanisms of LINC00958 in CRC, StarBase bioinformatics database (http://starbase.sysu.edu.cn/) was used to predict the potential miRNAs which may have binding sites with LINC00958. The binding site of miR-3619-5p on LINC00958 was predicted by StarBase (Fig. 5A). The location of binding site on LINC00958 is chr11:13011075-13011081[-]. Then, a dual luciferase assay was adopted to verify the target relationship between LINC00958 and miR-3619-5p. As presented in Fig. 5B, the luciferase intensity was significantly decreased after co-transfection with a miR-3619-5p mimic and LINC00958-WT. These observations revealed that LINC00958 is a potential direct target of miR-3619-5p.
Figure 5.
miR-3619-5p is directly targeted by LINC00958. (A) Binding region between miR-3619-5p and LINC00958. (B) A luciferase reporter assay was performed to detect the relative luciferase activity. Each experiment was repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were analyzed by a two-tailed Student's t test. **P<0.01 vs. 3′untranslated region-MUT. (C) The expression of miR-3619-5p was measured in several CRC cell lines (GEO, SNU-C1, COLO205, HCT-116, HT-29 and SW480) using RT-qPCR. The experiments were generated from three independent repeats (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were analyzed by a two-tailed Student's t test. **P<0.01, ***P<0.001 vs. NCM460. (D) The expression of miR-3619-5p was detected using RT-qPCR after transfection with shRNA-LINC00958-1. Each experiment was repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were analyzed by a two-tailed Student's t-test. ***P<0.001 vs. shRNA-NC. shRNA, short hairpin RNA; NC, negative control; miR, microRNA; MUT, mutant; WT, wild-type; RT-qPCR, reverse transcription-quantitative PCR.
miR-3619-5p inhibitor reverses the inhibitory effects of LINC00958 silencing on proliferation, invasion and migration of CRC cells
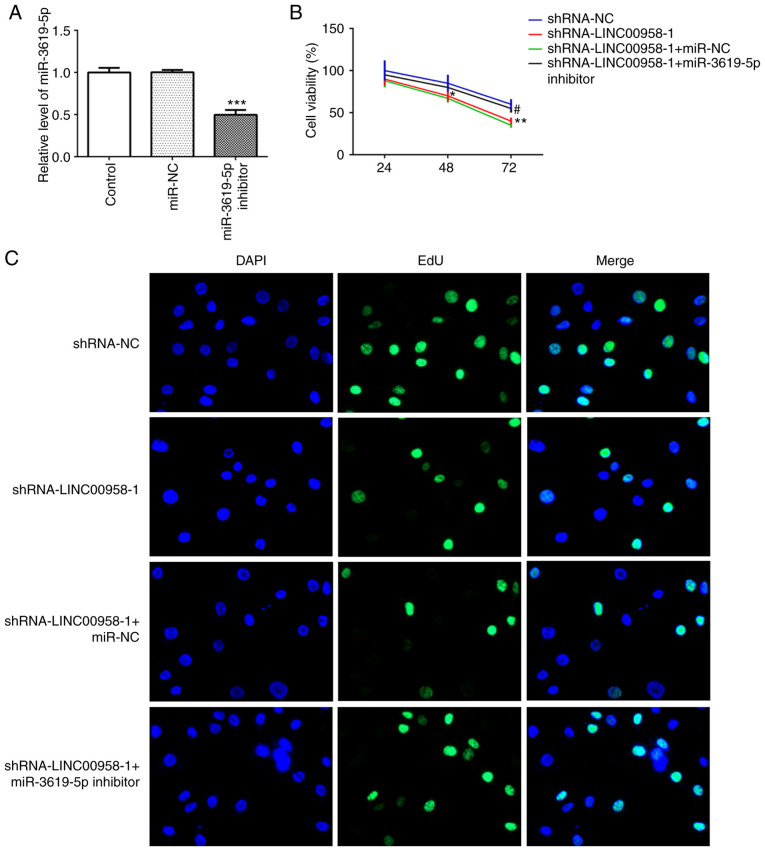
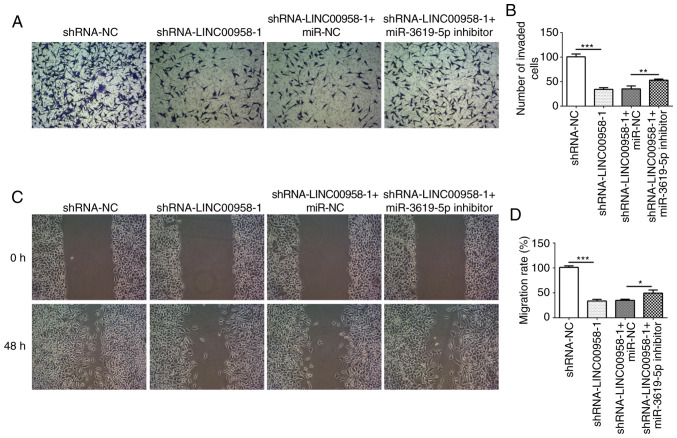
The expression of miR-3619-5p was assessed in several cell lines using RT-qPCR. As shown in Fig. 5C, significantly reduced expression of miR-3619-5p was observed in the CRC cell lines, especially in SW480 cells. Subsequently, the level of this miRNA was detected in SW480 cells following transfection with shRNA-LINC00958-1. It was found that the expression of miR-3619-5p was notably upregulated following LINC00958 silencing (Fig. 5D). Then, a miR-3619-5p inhibitor was transfected into SW480 cells and successful transfection is presented in Fig. 6A. In addition, the proliferation of SW480 cells was measured using a CCK-8 assay. As exhibited in Fig. 6B and C, miR-3619-5p inhibitor reversed the effects of LINC00958 silencing on cell viability in SW480 cells. Moreover, the changes in invasive (Fig. 7A and B) and migratory activity (Fig. 7C and D) were consistent with the results of the proliferation assay. These data indicated that the miR-3619-5p inhibitor attenuated the inhibitory effects of LINC00958 silencing on proliferation, invasion and migration of CRC cells.
Figure 6.
miR-3619-5p inhibitor reverses the inhibitory effects of LINC00958 silencing on proliferation of CRC cells. (A) The expression of miR-3619-5p was evaluated using reverse transcription-quantitative PCR after transfection with a miR-3619-5p inhibitor. The experiments were generated from three independent repeats (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were analyzed by a two-tailed Student's t test. ***P<0.001 vs. miR-NC. Cell proliferation was detected using (B) Cell Counting Kit-8 assay and (C) EdU incorporation assay, respectively. Each experiment was repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were analyzed by a one-way ANOVA followed by a post hoc Tukey's test. *P<0.05, **P<0.01 vs. shRNA-NC; #P<0.05 vs. shRNA-LINC00958-1+miR-NC. EdU, 5-ethynyl-20-deoxyuridine; shRNA, short hairpin RNA; NC, negative control; miR, microRNA.
Figure 7.
miR-3619-5p inhibitor reverses the inhibitory effects of LINC00958 silencing on invasion and migration of CRC cells. (A) The invasive ability of SW480 cells was detected using a Transwell assay. (B) Relative number of invaded cells. (C) Migratory activity of SW480 cells was determined using a scratch wound healing assay. (D) Relative migratory rate of SW480 cells. All experiment were repeated three times independently (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a one-way ANOVA followed by a post hoc Tukey's test. *P<0.05, **P<0.01, ***P<0.001. shRNA, short hairpin RNA; NC, negative control; miR, microRNA.
miR-3619-5p inhibitor reverses the effects of LINC00958 silencing on the apoptosis of CRC cells
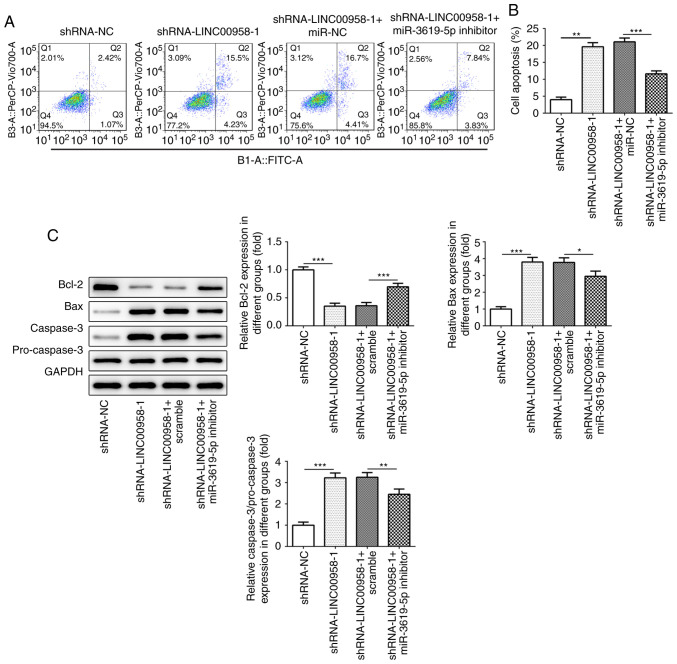
To confirm the regulatory effects of miR-3619-5p on the apoptosis of CRC cells, flow cytometry was performed. As presented in Fig. 8A and B, transfection with a miR-3619-5p inhibitor decreased the number of apoptotic SW480 cells compared with LINC00958 silencing alone. Meanwhile, the expression of apoptosis-related proteins was evaluated using western blot analysis. As shown in Fig. 8C, co-transfection with miR-3619-5p inhibitor and shRNA-LINC00958-1 notably enhanced the expression of Bcl-2, whereas the expression of Bax and casapase-3 was reduced in SW480 cells compared with shRNA-LINC00958-1 transfection alone. Taken together, the results indicated that the miR-3619-5p inhibitor reversed the effects of LINC00958 silencing on the apoptosis of CRC cells.
Figure 8.
miR-3619-5p inhibitor reverses the effects of LINC00958 silencing on the apoptosis of CRC cells. (A) Flow cytometry was used to measure the apoptotic rate of SW480 cells. (B) Cell apoptosis was quantified. (C) The expression levels of apoptosis-related proteins were detected using western blotting. The experiments were generated from three independent repeats (N=3). Data are expressed as mean ± standard deviation. Statistical comparisons were made by a one-way ANOVA followed by a post hoc Tukey's test. *P<0.05, **P<0.01, ***P<0.001. shRNA, short hairpin RNA; NC, negative control; miR, microRNA.
Discussion
Accumulating evidence has demonstrated that lncRNAs are abnormally expressed in colorectal cancer (CRC) and are involved in its progression (20,21). The present study aimed to investigate the roles of LINC00958 and miR-3619-5p in the proliferation, invasion and migration of CRC cells. The results demonstrated that LINC00958 was highly expressed in CRC cells and downregulation of LINC00958 inhibited CRC cell proliferation, invasion and migration by targeting miR-3619-5p.
An increasing amount of evidence suggests that dysregulation of lncRNAs are closely implicated in the pathogenesis, development and metastasis of CRC (22,23). Emerging evidence confirms that LINC00958 expression is notably overexpressed in cervical cancer cells, and LINC00958 contributes to the proliferation and metastasis of cervical cancer cells (24). In support, a previous study highlighted the importance of LINC00958 in the occurrence of glioma (25). Notably, accumulating evidence has demonstrated that CRC-related lncRNAs regulate the progression of CRC by inhibiting or promoting proliferation, invasion, apoptosis and metastasis of CRC cells (26,27). For example, lncRNA PCAT-1 can modulate cell proliferation, invasion, migration and apoptosis in CRC by targeting miR-149-5p (28). Downregulation of XLOC-010588 reduces CRC cell invasion and migration (29). In the present study, the expression of LINC00958 was markedly upregulated in CRC cells. Silencing of LINC00958 inhibited proliferation, invasion and migration, and promoted apoptosis of CRC cells. These findings demonstrated that LINC00958 silencing may suppress the progression of CRC.
lncRNAs act as competing endogenous (ce)RNAs to regulate the activity of miRNAs, which is an important mechanism in the regulation of tumor development (30,31). In the present study, the StarBase bioinformatics database predicted that LINC00958 is a potentially direct target of miR-3619-5p, which was verified by a luciferase reporter assay. miR-3619-5p was found to act as a tumor-suppressor gene, which can prevent proliferation and cisplatin resistance of skin squamous cell carcinoma cell via regulating the expression of KPNA4 (32). Moreover, a previous study reported that miR-3619-5p has the ability to inhibit the growth of prostate cancer cells (33). Compelling evidence indicates that miR-3619-5p could suppress β-catenin-mediated cancer growth and invasion in non-small cell lung cancer cells (34). In support of these findings, decreased miR-3619-5p expression has been detected in retinoblastoma cells, and transfection with miR-3619-5p mimic can improve the progression of retinoblastoma (35). In the present study, miR-3619-5p inhibitor reversed the effects of LINC00958 silencing on proliferation, invasion, migration and apoptosis of CRC cells. Thus, the results of the present study demonstrated that the downregulation of LINC00958 can suppress the progression of CRC cells by targeting miR-3619-5p.
In summary, to the best of our knowledge, the present study demonstrated for the first time that downregulation of LINC00958 suppresses CRC cell proliferation, invasion and migration, and promotes apoptosis by targeting miR-3619-5p, suggesting a potential novel biomarker and target for the diagnosis and treatment of CRC. However, the lack of studies of LINC00958 in tumor tissue from CRC patients, the absence of experiments concerning the miR-3619-5p effect on endogenous LINC00958, the characteristics of SW480 cells and how this could impact the extrapolation of our data to in vivo tumors are limitations of the present research and therefore, a comprehensive analysis is required in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YS designed the experiments. YS, SH and YL performed the experiments. YL, RW and PH analyzed the data. YC, LC and YS wrote the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zheng Y, Nie P, Xu S. Long noncoding RNA CASC21 exerts an oncogenic role in colorectal cancer through regulating miR-7-5p/YAP1 axis. Biomed Pharmacother. 2020;121:109628. doi: 10.1016/j.biopha.2019.109628. [DOI] [PubMed] [Google Scholar]
- 2.Meng S, Jian Z, Yan X, Li J, Zhang R. lncRNA SNHG6 inhibits cell proliferation and metastasis by targeting ETS1 via the PI3K/AKT/mTOR pathway in colorectal cancer. Mol Med Rep. 2019;20:2541–2548. doi: 10.3892/mmr.2019.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 5.van der Werf A, Arthey K, Hiesmayr M, Sulz I, Schindler K, Laviano A, Langius J, de van der Schueren M. The determinants of reduced dietary intake in hospitalised colorectal cancer patients. Support Care Cancer. 2018;26:2039–2047. doi: 10.1007/s00520-018-4044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Q, Liu B. Identification of the anticancer effects of a novel proteasome inhibitor, ixazomib, on colorectal cancer using a combined method of microarray and bioinformatics analysis. OncoTargets Ther. 2017;10:3591–3606. doi: 10.2147/OTT.S139686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu X, Yang B, Liu F, Fang Q. lncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomed Pharmacother. 2020;121:109623. doi: 10.1016/j.biopha.2019.109623. [DOI] [PubMed] [Google Scholar]
- 8.Bao Y, Tang J, Qian Y, Sun T, Chen H, Chen Z, Sun D, Zhong M, Chen H, Hong J, et al. Long noncoding RNA BFAL1 mediates enterotoxigenic bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 2019;10:675. doi: 10.1038/s41419-019-1925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, Liu Y, Wei C, Yang Z, Chang W, Zhang X. lncRNA HULC promotes the progression of gastric cancer by regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 2020;121:109607. doi: 10.1016/j.biopha.2019.109607. [DOI] [PubMed] [Google Scholar]
- 10.Shi X, Zhang W, Nian X, Lu X, Li Y, Liu F, Wang F, He B, Zhao L, Zhu Y, et al. The previously uncharacterized lncRNA APP promotes prostate cancer progression by acting as a competing endogenous RNA. Int J Cancer. 2020;146:475–486. doi: 10.1002/ijc.32422. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, Gu Y, Han P, Li Z, Zhao JL, Gao MZ. Long noncoding RNA SBF2-AS1 promotes colorectal cancer proliferation and invasion by inhibiting miR-619-5p activity and facilitating HDAC3 expression. J Cell Physiol. 2019;234:18688–18696. doi: 10.1002/jcp.28509. [DOI] [PubMed] [Google Scholar]
- 12.Sun A, Xiao LJ, Zhao XR, Wang JP, Chang XM, Zhao EH. Effect of lncRNA AK093407 on colorectal cancer cell line HCT-116. Eur J Immunol. 2019;49:1535–1536. [Google Scholar]
- 13.Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, Avendaño-Félix M, Silva-Benítez E, López-Camarillo C, Ramos-Payán R. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front Oncol. 2019;9:1008. doi: 10.3389/fonc.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seitz AK, Christensen LL, Christensen E, Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM, Palmfeldt J, Thomson M, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7:395. doi: 10.1038/s41598-017-00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Song ZJ, Wang Y, Zhong WF, Kang P, Yang Y. Elevated long non-coding RNA LINC00958 was associated with metastasis and unfavorable prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:598–603. doi: 10.26355/eurrev_201901_16872. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Liu M, Yu Y, Sun Y, Li J, Hu W, Wang X, Tong D. LINC00958 regulated miR-627-5p/YBX2 axis to facilitate cell proliferation and migration in oral squamous cell carcinoma. Cancer Biol Ther. 2019;20:1270–1280. doi: 10.1080/15384047.2019.1617571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL, Wang YD. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446:49–61. doi: 10.1016/j.canlet.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Peng J, Liu F, Zheng H, Wu Q, Liu S. Long noncoding RNA ZFAS1 promotes tumorigenesis and metastasis in nasopharyngeal carcinoma by sponging miR-892b to up-regulate LPAR1 expression. J Cell Mol Med. 2020;24:1437–1450. doi: 10.1111/jcmm.14823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Damas ND, Marcatti M, Côme C, Christensen LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF, Seemann SE, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Liu T, Zhang Y, Li Q, Jin LK. lncRNA-ZDHHC8P1 promotes the progression and metastasis of colorectal cancer by targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23:1476–1486. doi: 10.26355/eurrev_201902_17105. [DOI] [PubMed] [Google Scholar]
- 22.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388. [PubMed] [Google Scholar]
- 23.Wang X, Mo FM, Bo H, Xiao L, Chen GY, Zeng PW, Huang YN, Lei Z, Yuan WJ, Chen ZH. Upregulated expression of long non-coding RNA, LINC00460, suppresses proliferation of colorectal cancer. J Cancer. 2018;9:2834–2843. doi: 10.7150/jca.26046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia Z, Xu J, Xu L. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J Cell Biochem. 2020;121:2500–2509. doi: 10.1002/jcb.29472. [DOI] [PubMed] [Google Scholar]
- 25.Guo E, Liang C, He X, Song G, Liu H, Lv Z, Guan J, Yang D, Zheng J. Long noncoding RNA LINC00958 accelerates gliomagenesis through regulating miR-203/CDK2. DNA Cell Biol. 2018;37:465–472. doi: 10.1089/dna.2018.4163. [DOI] [PubMed] [Google Scholar]
- 26.Lin H, Guo Q, Lu S, Chen J, Li X, Gong M, Tang L, Wen J. lncRNA SUMO1P3 promotes proliferation and inhibits apoptosis in colorectal cancer by epigenetically silencing CPEB3. Biochem Biophys Res Commun. 2019;511:239–245. doi: 10.1016/j.bbrc.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Song Y, Yang C, Wu X. Overexpression of lncRNA TUSC7 reduces cell migration and invasion in colorectal cancer. Oncol Rep. 2019;41:3386–3392. doi: 10.3892/or.2019.7106. [DOI] [PubMed] [Google Scholar]
- 28.Wang AH, Fan WJ, Fu L, Wang XT. lncRNA PCAT-1 regulated cell proliferation, invasion, migration and apoptosis in colorectal cancer through targeting miR-149-5p. Eur Rev Med Pharmacol Sci. 2019;23:8310–8320. doi: 10.26355/eurrev_201910_19142. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Kuang H, Xue J, Liao L, Yin F, Zhou X. lncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;93:1230–1237. doi: 10.1016/j.biopha.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, Schmidt J, Ziebolz D, Schmalz G. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9. doi: 10.1016/j.oraloncology.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Wan X, Ding X, Chen S, Song H, Jiang H, Fang Y, Li P, Guo J. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36:521–532. doi: 10.1007/s13277-015-3136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Luo H, Hui L. miR-3619-5p hampers proliferation and cisplatin resistance in cutaneous squamous-cell carcinoma via KPNA4. Biochem Biophys Res Commun. 2019;513:419–425. doi: 10.1016/j.bbrc.2019.03.203. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Wang C, Yu X, Wu H, Hu J, Wang S, Ye Z. miR-3619-5p inhibits prostate cancer cell growth by activating CDKN1A expression. Oncol Rep. 2017;37:241–248. doi: 10.3892/or.2016.5250. [DOI] [PubMed] [Google Scholar]
- 34.Niu X, Liu S, Jia L, Chen J. Role of miR-3619-5p in β-catenin-mediated non-small cell lung cancer growth and invasion. Cell Physiol Biochem. 2015;37:1527–1536. doi: 10.1159/000438520. [DOI] [PubMed] [Google Scholar]
- 35.Yan G, Su Y, Ma Z, Yu L, Chen N. Long noncoding RNA LINC00202 promotes tumor progression by sponging miR-3619-5p in retinoblastoma. Cell Struct Funct. 2019;44:51–60. doi: 10.1247/csf.18033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.