Abstract
Background
New strategies are urgently needed to deal with the growing problem of multidrug-resistant bacterial pathogens. As the natural viruses against bacteria, recently, bacteriophages have received particular attention. Here, we identified and characterized a novel peptidoglycan hydrolase named MMPphg by decoding the complete genome sequence of Meiothermus bacteriophage MMP17, which was isolated in Tengchong hot spring in China and contains a circular genome of 33,172 bp in size and a GC content of 63.4%.
Findings
We cloned the MMPphg gene, overproduced and purified the phage lytic protein, which contains a highly conserved M23 metallopeptidase domain and can be activated by Mg2+ and Zn2+. MMPphg is capable of withstanding temperatures up to 70 °C, and preserved more than 80% of its activity after a 30 min treatment between 35 and 65 °C. More interestingly, by disrupting bacterial cells, MMPphg exhibits surprising antimicrobial activity against both Gram-negative and Gram-positive pathogenic bacteria, especially antibiotic-resistant strains such as Escherichia coli O157, Staphylococcus aureus and Klebsiella pneumonia.
Conclusions
In the current age of mounting antibiotic resistance, these results suggest the great potential of MMPphg, the gene product of bacteriophage MMP17, in combating bacterial infections and shed light on bacteriophage-based strategies to develop alternatives to conventional antibiotics for human or veterinary applications.
Keywords: Meiothermus bacteriophage, Genome sequencing, Endolysin, Antibiotic-resistant bacteria, Antimicrobial activity
Main text
With the uncontrolled and inappropriate use of antibiotics, multidrug-resistant bacterial pathogens are becoming increasingly common, which is one of the major concerns in combating serious bacterial infections, presenting great challenge for clinical therapy [1, 2]. To deal with this problem of antibiotic resistance, there is an urgent need to develop alternative therapeutic methods to replace or supplement antibiotics [3].
Bacteriophages (also known as phages) are natural viruses of microbes that specifically infect only host bacterial cells without affecting other microflora and exist in nearly every environment [4, 5]. They were found to be natural agents against bacteria in the early part of the twentieth century [6]. Recently, phage lytic enzymes that can destroy the cell wall of bacteria and thus cause host cell lysis have also gained new ground as alternative antibacterial agents because of their safety and high efficiency [7, 8].
Meiothermus bacteria belong to the family Thermaceae of the phylum Deinococcus-Thermus, which are moderately thermophilic strains and grow at temperatures of 35–68 °C [9]. Phages infecting Meiothermus bacteria are particularly interesting because they are sources of valuable agents resistant to denaturation at high temperatures. So far, however, studies on Meiothermus phages and their genome are scarce. Previously, we have isolated a Meiothermus phage named MMP17 from Tengchong hot spring in Yunnan Province of China [10]. In this study, we further decoded its complete genome sequence, and present the antimicrobial potential of MMPphg, an interesting gene product of Meiothermus phage MMP17.
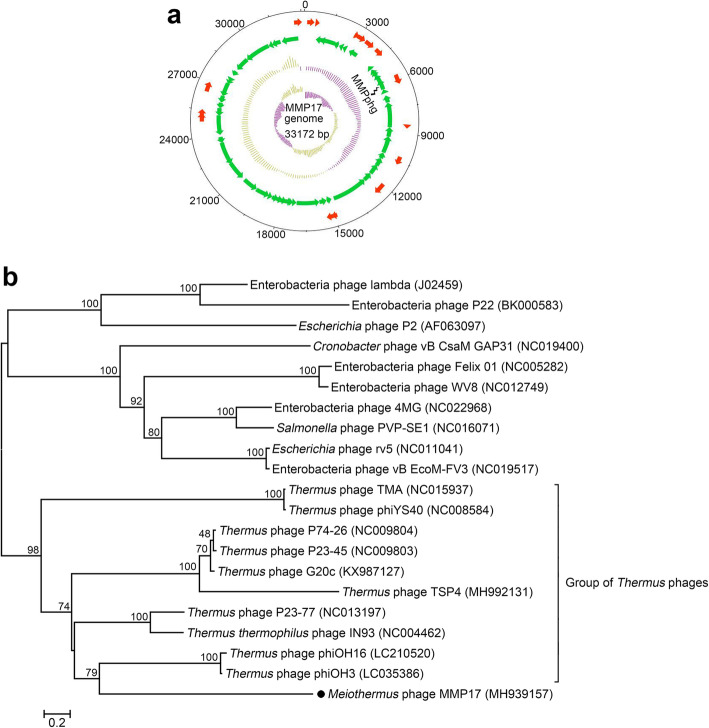
The genomic DNA of phage MMP17 was extracted using the phenol-chloroform method as previously described [11] and sequenced in the majorbio company (Shanghai, China) by Sanger sequencing. As shown in Fig. 1a, MMP17 has a small genome of 33,172 bp in size and a GC content of 63.4%; the high GC content of phage MMP17 is consistent with its growth temperature (63 °C). Based on reads overlapping features in the assembly, MMP17 has a circular genome. To validate this, the restriction enzymes digestion of MMP17 genome DNA was performed. As expected, the NcoI and HindIII (both have a single cutting site in the genome) digestion results were in line with the circular topology of MMP17 genome (Additional file 1: Fig. S1). Similar to many other thermophilic phage genomes such as P23–77 (GenBank accession: NC_013197.1) and IN93 (GenBank accession: NC_004462.2), this circular organization contributes to genome stability and may help the survival of phage MMP17 under extreme environment. This genome project has been deposited in GenBank under the accession number MH939157.
Fig. 1.
Genome map of phage MMP17 and whole-genome based neighbor-joining phylogenetic analysis. a Genome map of MMP17, from inside to outside: circle 1 demonstrates the GC skew [(G-C)/(G + C)]; circle 2 denotes the GC percentage plot; circles 3 and 4 show the ORFs on the minus strand (green) and plus strand (red) respectively; circle 5 displays the numbered scale with an interval of 3 kb. b Whole-genome-based phylogenetic tree of phage MMP17, which was created with MEGA6 program using the neighbor-joining method. The values at the nodes indicate bootstrap probabilities that were calculated based on 1000 replicates. Numbers in parentheses are the GenBank accession numbers of phage genomes
The BLASTn analysis further indicated that, at the nucleotide level, MMP17 exhibits no significant similarity to publicly available phage genomes in the GenBank database, suggesting its novelty. The closest neighbor of MMP17 is Thermus aquaticus Y51MC23 (GenBank accession: NZ_CP010822.1), but with only 1% sequence coverage and 82.9% identity, followed by Meiothermus ruber DSM1279 (GenBank accession: CP005385.1), with 1% sequence coverage and 80.4% identity. Open reading frame (ORF) prediction and annotation of MMP17 genome were performed with ORF finder on NCBI website and further refined with BLASTp program. Through these analyses, we found 86 ORFs in the MMP17 genome but there is no tRNA or rRNA gene in it. Of the 86 protein-encoding genes, only 26.7% (23/86) were assigned predicted functions (Additional file 3: Table S1). Consistent with current taxonomic results [9, 12], whole-genome-based neighbor-joining phylogenetic analysis demonstrated that there is a close evolutionary relationship between Meiothermus phage MMP17 and representatives of Thermus phages (Fig. 1b). In the phylogenetic tree, the most intimate relatives of MMP17 are Thermus phage phiOH16 (GenBank accession: LC210520) and Thermus phage phiOH3 (GenBank accession: LC035386). However, comparisons at the protein level by employing CoreGenes 3.5 [13] showed that only 2/86 (2.3%) proteins of phage MMP17 are homologous to those of both phiOH16 and phiOH3. Together, these results suggest that MMP17 indeed represents a novel phage.
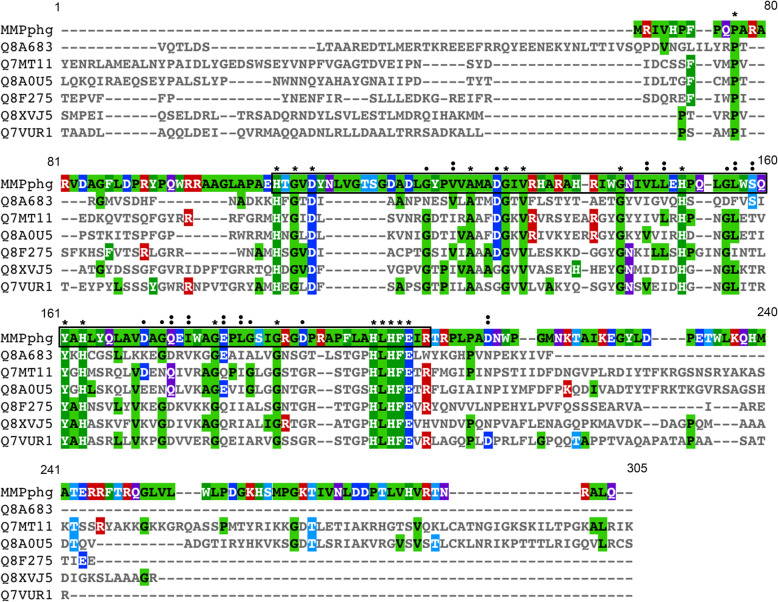
Among all the protein-encoding genes in MMP17 genome, ORF20 with a length of 633 bp is very interesting as it encodes a M23 family metallopeptidase which is composed of 210 aa (GenBank accession: QAY18044.1). The used initiation codon and termination codon of ORF20 are ATG and TAG, respectively. For convenience, we named ORF20 as MMPphg (peptidoglycan hydrolytic gene of MMP17) in this study. Next, we carried out the conserved domain analysis of MMPphg against NCBI Conserved Domain Database (CDD v3.17, last update: 2019-04-03) [14]. The results revealed that it contains a conserved M23 peptidase domain (NCBI domain architecture ID: 10480195) and is highly similar to peptidase of M23 family (accession: pfam01551) (Additional file 2: Fig. S2). Further, multiple sequence alignment of MMPphg and other six representatives of M23 peptidase family was performed by using Clustal Omega (version 1.2.4) with the default settings [15]. As shown in Fig. 2, there are 18 fully conserved residues (P77, H105, G107, D109, A127, G131, V133, G144, H151, Y161, H163, G178, G185, H196, L197, H198, F199 and E200) in the amino acid sequence of MMPphg. Interestingly, we also found a fully conserved motif HLHFE at the C-terminus of MMPphg M23 domain [16], which might be involved in the peptidoglycan-degrading process of bacterial cell wall and deserves further study.
Fig. 2.
Multiple sequence alignment of MMPphg and other six representatives of M23 peptidase family. The alignment was carried out with CLUSTAL Omega (1.2.4). An asterisk (*) indicates positions that have a single, fully conserved residue, while colon (:) and period (.) denote conservation among groups of strong similarity and weak similarity, respectively. The amino acid residues of MMPphg within M23 peptidase domain were boxed. The UniProtKB/Swiss-Prot accession numbers Q8A683, Q7MT11, Q8A0U5, Q8F275, Q8XVJ5 and Q7VUR1 represent protein sequences of putative membrane peptidase from Bacteroides thetaiotaomicron, peptidase of M23/M37 family from Porphyromonas gingivalis, putative peptidase from Bacteroides thetaiotaomicron, peptidase of M23/M37 family from Leptospira interrogans, putative ipr002886 peptidase M23b family transmembrane protein from Ralstonia solanacearum and putative peptidase family M23/M37 protein from Bordetella pertussis, respectively. This figure was prepared with MView version 1.63
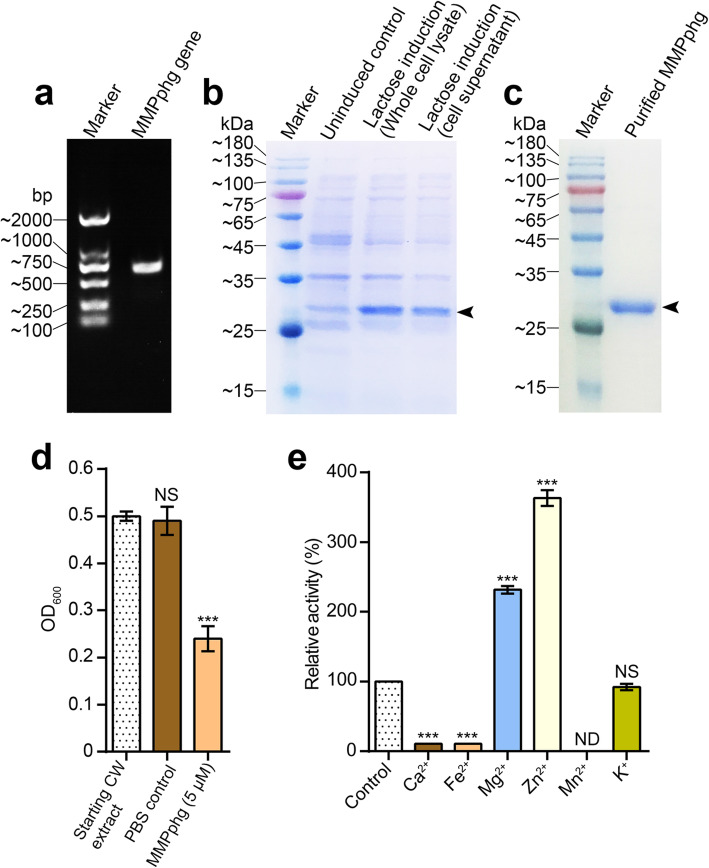
Next, the DNA sequence of the putative peptidoglycan hydrolase, MMPphg, was cloned by PCR amplification (Fig. 3a). To overproduce MMPphg, E. coli Rosetta cells containing the recombinant expression vector pET28a-MMPphg were cultivated at 37 °C in LB medium supplemented with Kanamycin (50 μg/mL), and lactose (1 g/L) was used for induction of T7 promoter in the vector (Fig. 3b). The purification step was performed using HisTrap affinity column according to the manufacturer’s instructions (GE Healthcare, USA), and the purity of recombinant MMPphg was confirmed by 12% SDS-PAGE (Fig. 3c). The details of PCR primers, regular gene cloning, recombinant protein expression and purification are presented in Additional file 4. Subsequently, the capability of MMPphg to digest cell wall obtained from Meiothermus sp. TG17 (GenBank accession: GU329951), the host bacterium for phage MMP17, was confirmed (Fig. 3d). It is known that members of M23 peptidase family are usually zinc-dependent metallopeptidases [17], to confirm this, the effects of metal ions (Mn2+, Ca2+, Mg2+, Zn2+, Fe2+ and K+) on the lytic activity of MMPphg against Meiothermus sp. TG17 cells were evaluated. It can be seen from Fig. 3e that both Zn2+ and Mg2+ showed positive effects on MMPphg lytic activity. Especially, with a concentration of 1 mM, Zn2+ can reconstitute MMPphg activity to 363.4% compared with untreated control, possibly because of increased interaction of MMPphg with the peptidoglycan backbone mediated by divalent zinc cations [18]. Together, these results are in line with the function of MMPphg as a peptidoglycan hydrolase belonging to M23 peptidase family.
Fig. 3.
Gene cloning, protein expression and purification of MMPphg, the gene product of Meiothermus phage MMP17, and functional analysis of MMPphg as a metallopeptidase. a PCR amplification of MMPphg gene. b Lactose (1 g/L) was used for induction to overproduce MMPphg. c SDS-PAGE analysis of the purity of recombinant MMPphg, which is at approximately 26 kDa as the black arrowhead indicates. d MMPphg is able to digest cell wall (CW) extracted from Meiothermus sp. TG17, the host bacterium for phage MMP17. e Effects of metal ions on the lytic activity of MMPphg. Relative activity of MMPphg against Meiothermus sp. TG17 cells was calculated as percentage in relation to the non-treated control. Each experiment was repeated in triplicate; error bars indicate the standard deviation. P values were determined using the Student’s t test. ***, P < 0.001; ND, non-detectable; NS, not significant
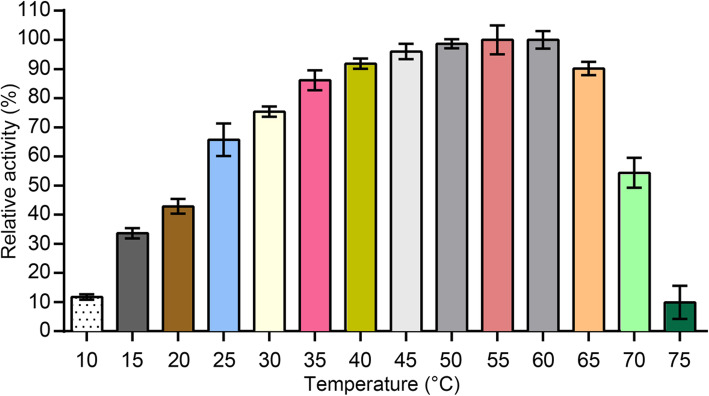
Furthermore, the thermostability of MMPphg was examined at different temperatures ranging from 10 to 75 °C. It is noteworthy that MMPphg retained more than 80% of its activity after a 30 min heat treatment between 35 and 65 °C, and still preserved more than 50% activity at 70 °C (Fig. 4), suggesting its considerable thermo-resistance to denaturation. Recently, a few lysins from thermophilic phages have been studied [19, 20]. For example, it was reported that an endolysin named PlyGVE2 from the deep-sea thermophilic Geobacillus phage GVE2 was relatively stable and active over a broad range of temperatures from 40 to 80 °C, and maintained 80% of its activity after a 30 min incubation at 55 °C [21, 22], which is similar to MMPphg. Moreover, the engineered chimeolysin of PlyGVE2, PlyGVE2CpCWB, could still preserve 57% activity after a treatment at 55 °C for 30 min [23]. More surprisingly, two endolysins with extremely high thermostability have also been reported [24, 25]. The first one is Ph2119 endolysin from the Thermus scotoductus MAT2119 bacteriophage Ph2119, and it retains approximately 86.7% of its initial activity after 6 h of incubation at 95 °C [24]. The other is Ts2631 endolysin from the Thermus scotoductus phage vB_Tsc2631. The Ts2631 endolysin has been demonstrated to be not only thermoresistant, maintaining 64.8% of its original activity after 2 h at 95 °C, but also highly thermodynamically stable, with a Tm of 99.8 °C [25]. By comparison, PlyC, an antimicrobial endolysin against Streptococcus pyogenes, completely lost its activity after a 50 °C heat treatment for 30 min [20, 26]. Therefore, we strongly believe that studies and development of these thermo-resistant phage proteins would be very helpful for biotechnological applications in humans or livestock.
Fig. 4.
Thermostability of MMPphg, which was first incubated at different temperatures (from 10 to 75 °C) for 30 min, and then its activity was determined by the standard turbidity reduction assay against Meiothermus sp. TG17 cells. All data were normalized to the maximal lytic activity among the dataset (= 100%) and presented as mean ± standard deviation of n = 3 experiments
Next, the bacteriolytic activity of MMPphg was tested at 37 °C in a turbidity reduction assay against Salmonella ser. Paratyphi B (without treatment by permeabilization agents), which is a Gram-negative pathogenic bacterium that can contaminate many food products especially those of animal origin, and cause diseases in poultry and pork farming [27, 28]. As shown in Fig. 5a, the bacteriolytic activity of MMPphg increased proportionally to its concentration, and the curve pattern exhibited rapid kinetics of bacterial cell lysis by MMPphg. For example, with the MMPphg concentration of 7.5 μM, an OD600 reduction of 0.34 could be achieved after a 20 min incubation of MMPphg with the bacteria at 37 °C. For bacterial colony counting of Salmonella ser. Paratyphi B after MMPphg treatment on LB agar plates, the dose-dependent effect was also observed (Fig. 5b). Finally, a log10 reduction by 5.05 ± 0.46 in bacterial counts was reached upon exposure of 107 CFU/mL of Salmonella ser. Paratyphi B cells to MMPphg (7.5 μM) at 37 °C for 1 h.
Fig. 5.
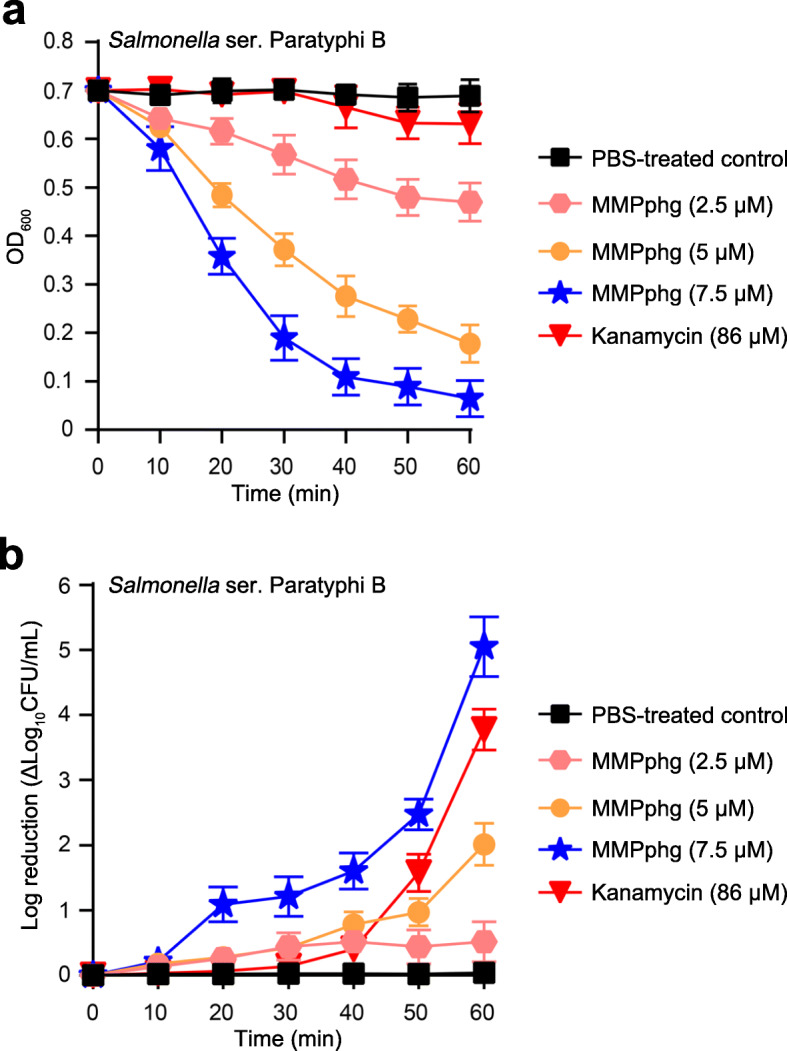
Dependence of MMPphg bacteriolytic and bactericidal activity on its concentration. a Turbidity reduction assays. Different concentrations of MMPphg were incubated with Salmonella ser. Paratyphi B at 37 °C, and the changes in OD600 were measured over time. b Bactericidal activity of MMPphg against Salmonella ser. Paratyphi B. After incubation at 37 °C over 60 min period (samples were taken at an interval of 10 min), aliquots were serially diluted and spread onto LB agar plates. Log kills were presented as ΔLog10 (CFU/mL) compared with bacterial counts before treatment. Kanamycin (50 μg/mL, approximately 86 μM) and PBS were used as the controls. Data were presented as mean ± SD, and all experiments were repeated in triplicate
Considering the potential antimicrobial applications of MMPphg, we further tested its bactericidal activity against various Gram-negative or Gram-positive bacteria (see Additional file 4 for the detailed information of bacterial strains and methods). As shown in Table 1 and Additional file 4: Table S2, MMPphg exhibits antimicrobial activity against both Gram-negative and Gram-positive bacterial pathogens, which is quite rare and very interesting as currently most reported phage lysins only have lytic activity against Gram-negative or Gram-positive strains [3, 18, 29]. Recently, Plotka et al. have demonstrated that the Ts2631 endolysin, a promising antimicrobial agent from the extremophilic Thermus scotoductus bacteriophage vB_Tsc2631 [30], has a unique 20-residue N-terminus with 7 positively charged amino acids. And this 20-residue N-terminus of Ts2631 endolysin mediates peptidoglycan binding and is crucial for the antibacterial activity of the protein [31]. The same functional features were also highlighted in the case of the lytic enzyme LysC from Clostridium intestinale URNW [32], and it is noteworthy that the highly positively charged N-terminal tail of LysC is of vital importance for antimicrobial activity of the enzyme, which sheds light on new strategies to develop antimicrobial agents with superb activity. Accordingly, we found that the C-terminal tail of M23 peptidase domain of MMPphg (GRGDPRAPFLAHLHFEIRTR) contains six positively charged residues (underlined) as well as a highly conserved motif HLHFE. We hypothesize that this special 20-residue tail could assist penetration of MMPphg through the outer membrane of Gram-negative bacteria by electrostatic interactions with negatively charged molecules on the bacterial surface [33, 34], and hence be responsible for the observed antibacterial activity of MMPphg. Together, these clues suggest that MMPphg might be a multifunctional phage lytic protein with specific structure that is able to interfere with the outer membrane and hydrolyze bacterial peptidoglycan [18], which warrants future structure-based studies.
Table 1.
Antimicrobial activity of MMPphg on various Gram-negative or Gram-positive bacteria. The significant log reduction units observed (≥ 1 log kill) are marked in bold
| Strain | Antibiotic resistance a | Antimicrobial activity (Log10 reduction) b |
|---|---|---|
| Escherichia coli | ||
| CMCC(B)44102 | No | 3.23 ± 0.15 (***) |
| Staphylococcus aureus | ||
| ATCC6538 | No | 1.31 ± 0.03 (***) |
| KMUST1606BL1486 | Yes | 1.04 ± 0.11 (***) |
| Salmonella ser. Enteritidis | ||
| CMCC(B)50335 | No | 1.08 ± 0.03 (***) |
| Salmonella ser. Typhi | ||
| CGMCC1.1190 | No | 0.92 ± 0.01 |
| Salmonella ser. Paratyphi B | ||
| CMCC(B)50094 | No | 3.42 ± 0.15 (***) |
| Shigella dysenteriae | ||
| KMUSTDS8 | Yes | 0.96 ± 0.01 |
| KMUSTDS6 | Yes | 1.01 ± 0.01 (***) |
| Klebsiella pneumoniae | ||
| 13A14165 | Yes | 3.02 ± 0.01 (***) |
| 13A14918 | Yes | 1.47 ± 0.03 (***) |
| 13A15188 | Yes | 2.49 ± 0.07 (***) |
| 13A15382 | Yes | 1.59 ± 0.03 (***) |
| 1412SP0200 | Yes | 1.24 ± 0.02 (***) |
| 1412SP0057 | Yes | 2.26 ± 0.10 (***) |
| 13 V1837 | Yes | 0.97 ± 0.03 |
| 14 V0622 | Yes | 1.09 ± 0.02 (***) |
| 1501SP0134 | Yes | 2.57 ± 0.07 (***) |
aDetailed antibiotic-resistant information for the relevant strains is shown in Additional file 4: Table S2; bValues represent mean ± standard deviation. For all tests, the antimicrobial activity was measured after incubating 106 CFU/mL of the bacterial cells with 6 μM MMPphg at 37 °C for 1 h, and six independent replicates were run in each reaction. ***, P < 0.001 for MMPphg-treated cells versus PBS-treated controls
In addition, we found that MMPphg also has broad antimicrobial activity against different multidrug-resistant strains of Klebsiella pneumoniae (Table 1); these Gram-negative strains belong to the ESKAPE group pathogens and are very difficult to treat in clinic care because of their antibiotic resistance and virulence [35]. To further investigate the detailed view of bacterial cell alteration after MMPphg treatment, scanning electron microscope (Quanta 200, FEI, Holland) analysis was performed according to the manufacturer’s instructions. As shown in Fig. 6, exogenous MMPphg treatment (5 μM) at 37 °C for 1 h could severely promote bacteria destruction of both Gram-negative (E. coli O157) and Gram-positive (S. aureus) cells, resulting in release of their intracellular components. These findings are consistent with the aforementioned activity of MMPphg and further confirmed the role of MMPphg as a phage-derived lytic enzyme with surprising antimicrobial activity against both Gram-negative and Gram-positive bacteria.
Fig. 6.
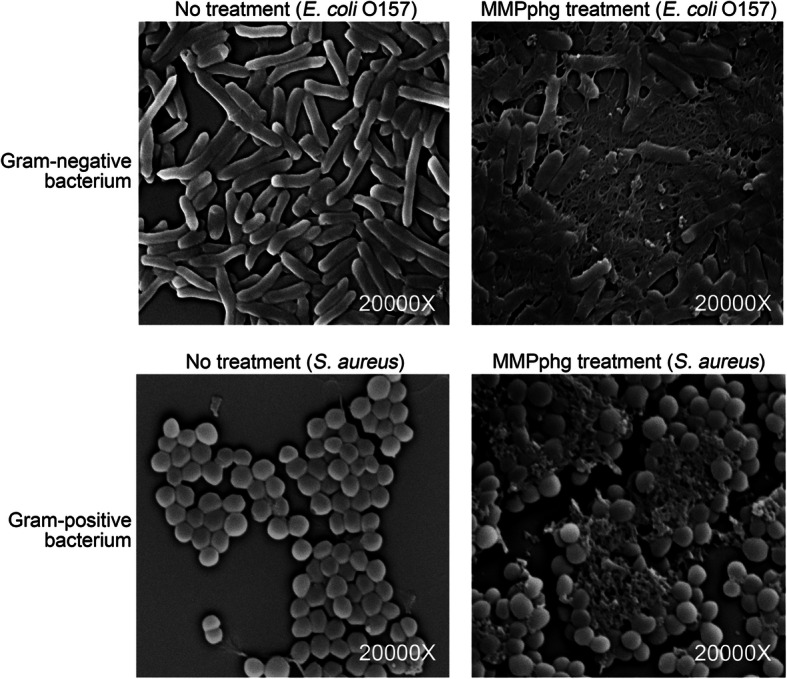
Scanning electron microscope showing the effects of exogenous MMPphg treatment on bacteria destruction of both Gram-negative (E. coli O157) and Gram-positive (S. aureus) cells. For all tests, 105 CFU/mL of bacterial cells were treated with or without 5 μM MMPphg at 37 °C for 1 h. The magnification is 20,000 times the original size
Conclusions
Summarily, we report here the complete genome sequence of Meiothermus phage MMP17, and by annotating it, we identified and characterized a novel peptidoglycan hydrolase, MMPphg, which has antimicrobial activity against both Gram-negative and Gram-positive bacteria, especially multidrug-resistant pathogenic strains. In the current age of mounting antibiotic resistance, these results suggest the potential applications of MMPphg as an antimicrobial agent to fight against bacterial infections and provide insights into bacteriophage-based technologies to develop novel alternatives to antibiotics for human or veterinary applications. Still, future optimization and structure-based studies are needed to further increase the efficiency of MMPphg and delineate the underlying mechanism more precisely.
Supplementary information
Additional file 1: Fig. S1. Patterns of restriction enzymes digestion of phage MMP17 genome DNA. a The positions of two restriction enzymes, NcoI and HindIII. Both have a single cutting site in the genome of phage MMP17. b Restriction enzymes cutting results of MMP17 genome DNA.
Additional file 2: Fig. S2. The conserved domain analysis of MMPphg at both nucleotide and protein levels.
Additional file 3: Table S1. Genetic features of open reading frames in the Meiothermus phage MMP17 genome. Protein sequences of the predicted ORFs of Meiothermus phage MMP17 were subjected to BLASTp program to analyze their best known matches on the NCBI website (https://blast.ncbi.nlm.nih.gov). The NCBI non-redundant database (nrdb) was used as the reference database, with the cutoff E-value set at 1E-05.
Additional file 4: Table S2 and additional methods. Table S2. Detailed antibiotic-resistant information for the bacteria used in this study.
Acknowledgements
We thank Prof. Xueshan Xia and Dr. Yuzhu Song from the Research Center of Molecular Medicine of Yunnan Province, Kunming University of Science and Technology for kindly providing the strains of Klebsiella pneumoniae.
Abbreviations
- MMPphg
Peptidoglycan hydrolytic gene of MMP17
- MMP17
Meiothermus Myoviridae Phage 17
- ORF
Open reading frame
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ESKAPE
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species
- SEM
Scanning electron microscope
- CFU
Colony-forming unit
- PBS
Phosphate-buffered saline
- LB
Luria Broth
- CW
Cell wall
- OD
Optical density
- SD
Standard deviation
Authors’ contributions
FW and LL conceived and designed the study. JH isolated the bacteriophage MMP17 and prepared its DNA samples for genome sequencing. YAO, JH and FW performed bioinformatic analyses. YAN carried out the experiments. FW and YAN analyzed the data. FW wrote the manuscript. XD and LL were involved in discussion. All authors read and approved the final manuscript.
Funding
This study was funded by the Natural Science Foundation of China under Grant No. 31760042 to LL and Grant No. 31760338 to FW, and the Start-up Grant from Kunming University of Science and Technology for the Introduction of Talent Research Project under Grant No. KKSY201626003 to FW. These funding sources had no role in the design of the study; the collection, analysis, and interpretation of data; or in writing of this manuscript.
Availability of data and materials
The complete genome sequence of Meiothermus bacteriophage MMP17 generated in this study was deposited in GenBank with the accession number MH939157.1 (https://www.ncbi.nlm.nih.gov/nuccore/MH939157.1/). All the other materials are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Feng Wang, Yan Xiong and Yao Xiao contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12985-020-01403-0.
References
- 1.Luepke KH, Suda KJ, Boucher H, Russo RL, Bonney MW, Hunt TD, et al. Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy. 2017;37:71–84. doi: 10.1002/phar.1868. [DOI] [PubMed] [Google Scholar]
- 2.Manohar P, Nachimuthu R, Lopes BS. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018;18:97. doi: 10.1186/s12866-018-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Love MJ, Bhandari D, Dobson RCJ, Billington C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics. 2018;7:E17. doi: 10.3390/antibiotics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abedon ST, Garcia P, Mullany P, Aminov R. Editorial: phage therapy: past, present and future. Front Microbiol. 2017;8:981. doi: 10.3389/fmicb.2017.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trudil D. Phage lytic enzymes: a history. Virol Sin. 2015;30:26–32. doi: 10.1007/s12250-014-3549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7:1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raposo P, Viver T, Albuquerque L, Froufe H, Barroso C, Egas C, et al. Transfer of Meiothermus chliarophilus (Tenreiro et al.1995) Nobre et al. 1996, Meiothermus roseus Ming et al. 2016, Meiothermus terrae Yu et al. 2014 and Meiothermus timidus Pires et al. 2005, to Calidithermus gen. nov., as Calidithermus chliarophilus comb. nov., Calidithermus roseus comb. nov., Calidithermus terrae comb. nov. and Calidithermus timidus comb. nov., respectively, and emended description of the genus Meiothermus. Int J Syst Evol Microbiol. 2019;69:1060–1069. doi: 10.1099/ijsem.0.003270. [DOI] [PubMed] [Google Scholar]
- 10.Lin L, Han J, Ji X, Hong W, Huang L, Wei Y. Isolation and characterization of a new bacteriophage MMP17 from Meiothermus. Extremophiles. 2011;15:253–258. doi: 10.1007/s00792-010-0354-z. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz JE, Schuch R, Fischetti VA. Identifying active phage lysins through functional viral metagenomics. Appl Environ Microbiol. 2010;76:7181–7187. doi: 10.1128/aem.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albuquerque L, Rainey FA, Nobre MF, da Costa MS. Meiothermus granaticius sp. nov., a new slightly thermophilic red-pigmented species from the Azores. Syst Appl Microbiol. 2010;33:243–246. doi: 10.1016/j.syapm.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Turner D, Reynolds D, Seto D, Mahadevan P. CoreGenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res Notes. 2013;6:140. doi: 10.1186/1756-0500-6-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D2D3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sievers F, Higgins DG. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamford CV, Francescutti T, Cameron CE, Jenkinson HF, Dymock D. Characterization of a novel family of fibronectin-binding proteins with M23 peptidase domains from Treponema denticola. Mol Oral Microbiol. 2010;25:369–383. doi: 10.1111/j.2041-1014.2010.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabowska M, Jagielska E, Czapinska H, Bochtler M, Sabala I. High resolution structure of an M23 peptidase with a substrate analogue. Sci Rep. 2015;5:14833. doi: 10.1038/srep14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briers Y, Lavigne R. Breaking barriers: expansion of the use of endolysins as novel antibacterials against gram-negative bacteria. Future Microbiol. 2015;10:377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 19.São-José C. Engineering of phage-derived lytic enzymes: improving their potential as antimicrobials. Antibiotics. 2018;7:29. doi: 10.3390/antibiotics7020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heselpoth RD, Nelson DC. A new screening method for the directed evolution of thermostable bacteriolytic enzymes. J Vis Exp. 2012;69:4216. doi: 10.3791/4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye T, Zhang X. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl Microbiol Biotechnol. 2008;78:635–641. doi: 10.1007/s00253-008-1353-1. [DOI] [PubMed] [Google Scholar]
- 22.Jin M, Ye T, Zhang X. Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology. 2013;159:1597–1605. doi: 10.1099/mic.0.067611-0. [DOI] [PubMed] [Google Scholar]
- 23.Swift SM, Seal BS, Garrish JK, Oakley BB, Hiett K, Yeh H-Y, et al. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-Clostridium antimicrobial with improved thermostability. Viruses. 2015;7:3019–3034. doi: 10.3390/v7062758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plotka M, Kaczorowska AK, Stefanska A, Morzywolek A, Fridjonsson OH, Dunin-Horkawicz S, et al. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl Environ Microbiol. 2014;80:886–895. doi: 10.1128/AEM.03074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plotka M, Kaczorowska AK, Morzywolek A, Makowska J, Kozlowski LP, Thorisdottir A, et al. Biochemical characterization and validation of a catalytic site of a highly thermostable Ts2631 endolysin from the Thermus scotoductus phage vB_Tsc2631. PLoS One. 2015;10:E0137374. doi: 10.1371/journal.pone.0137374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wernicki A, Nowaczek A, Urban-Chmiel R. Bacteriophage therapy to combat bacterial infections in poultry. Virol J. 2017;14:179. doi: 10.1186/s12985-017-0849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gand M, Mattheus W, Saltykova A, Roosens N, Dierick K, Marchal K, et al. Development of a real-time PCR method for the genoserotyping of Salmonella Paratyphi B variant Java. Appl Microbiol Biotechnol. 2019;103:4987–4996. doi: 10.1007/s00253-019-09854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen HK, Trachsel J, Looft T, Casey TA. Finding alternatives to antibiotics. Ann N Y Acad Sci. 2014;1323:91–100. doi: 10.1111/nyas.12468. [DOI] [PubMed] [Google Scholar]
- 30.Plotka M, Kapusta M, Dorawa S, Kaczorowska AK, Kaczorowski T. Ts2631 endolysin from the extremophilic Thermus scotoductus bacteriophage vB_Tsc2631 as an antimicrobial agent against gram-negative multidrug-resistant bacteria. Viruses. 2019;11:E657. doi: 10.3390/v11070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plotka M, Sancho-Vaello E, Dorawa S, Kaczorowska A-K, Kozlowski LP, Kaczorowski T, et al. Structure and function of the Ts2631 endolysin of Thermus scotoductus phage vB_Tsc2631 with unique N-terminal extension used for peptidoglycan binding. Sci Rep. 2019;9:1261. doi: 10.1038/s41598-018-37417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plotka M, Szadkowska M, Håkansson M, Kovačič R, Al-Karadaghi S, Walse B, et al. Molecular characterization of a novel lytic enzyme LysC from clostridium intestinale URNW and its antibacterial activity mediated by positively charged N-terminal extension. Int J Mol Sci. 2020;21:E4894. doi: 10.3390/ijms21144894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihajlovic M, Lazaridis T. Antimicrobial peptides in toroidal and cylindrical pores. Biochim Biophys Acta. 1798;2010:1485–1493. doi: 10.1016/j.bbamem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2015;59:1329–1333. doi: 10.1128/AAC.03937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Patterns of restriction enzymes digestion of phage MMP17 genome DNA. a The positions of two restriction enzymes, NcoI and HindIII. Both have a single cutting site in the genome of phage MMP17. b Restriction enzymes cutting results of MMP17 genome DNA.
Additional file 2: Fig. S2. The conserved domain analysis of MMPphg at both nucleotide and protein levels.
Additional file 3: Table S1. Genetic features of open reading frames in the Meiothermus phage MMP17 genome. Protein sequences of the predicted ORFs of Meiothermus phage MMP17 were subjected to BLASTp program to analyze their best known matches on the NCBI website (https://blast.ncbi.nlm.nih.gov). The NCBI non-redundant database (nrdb) was used as the reference database, with the cutoff E-value set at 1E-05.
Additional file 4: Table S2 and additional methods. Table S2. Detailed antibiotic-resistant information for the bacteria used in this study.
Data Availability Statement
The complete genome sequence of Meiothermus bacteriophage MMP17 generated in this study was deposited in GenBank with the accession number MH939157.1 (https://www.ncbi.nlm.nih.gov/nuccore/MH939157.1/). All the other materials are available from the corresponding author on reasonable request.