INTRODUCTION
Cancers arising from the urothelium, the specialized epithelial lining of the urinary system, include urothelial carcinoma of the bladder (UCB) and upper tract urothelial carcinoma (UTUC).1 In the United States, an estimated 80,000 new UCB cases were diagnosed in 2019, representing the fourth most common cancer diagnosis in men and the sixth most common cancer diagnosis overall.1,2 UTUC is less common, with approximately 4,000 diagnoses in 2019.2 Prognosis and management of urothelial carcinoma is largely dictated by the grade (high v low) and stage (degree of invasion) found on cystoscopic/ureteroscopic biopsy or resection.3 The vast major-ity of cases are diagnosed when disease remains confined to the urinary tract,4-6 yet even localized disease causes substantial morbidity and bears significant risk for progression to lethal disease.7 Urine-based biomarkers have long been sought to improve diagnosis, disease monitoring, and treatment stratification.8-14 Although progress has been made, cytologic evaluation of the urine and cystoscopic and ureteroscopic evaluation of the urinary tract remain the gold standard for diagnosis and monitoring. However, not only is this invasive, it is also associated with the highest cost from diagnosis to death among all cancers in the United States.6,15,16 New opportunities for urine-based biomarker analysis have arisen because of better genomic characterization of urothelial carcinoma and technologic improvements for sensitive detection of tumor DNA on the basis of identification of cancer-related mutations and copy number changes.12,17-19 After initial proof-of-concept studies established the presence of tumor DNA in urine from patients with urothelial carcinoma, more recent studies have evaluated potential clinical applications in better-defined contexts.11-13,20-22 Many questions remain to be addressed, but these studies support the exciting possibility that urine-based analysis could decrease the need for invasive evaluations and aid in treatment selection. Here we discuss possible applications and barriers to implementing urinary tumor DNA (utDNA) analysis for patients with UCB and UTUC and translating these findings to improve clinical care.
CONTEXT
Key Objective
Can urinary tumor DNA (utDNA) be used as a biomarker for the diagnosis and surveillance of bladder cancer?
Knowledge Generated
We comprehensively reviewed modern utDNA technologies tested in the clinical setting. Technologies discussed include UroSEEK, Urine Cancer Personalized Profiling by deep Sequencing (uCAPP-Seq), Tagged-Amplicon Sequencing (TAm-Seq), digital droplet polymerase chain reaction (ddPCR), and shallow whole-genome sequencing (sWGS).
Relevance
Single-institutional studies suggest that utDNA analysis can facilitate early detection of bladder cancer in the diagnostic and surveillance settings. Larger prospective studies will need to be performed to determine clinical utility.
CHALLENGES IN UROTHELIAL CARCINOMA DIAGNOSIS AND CLINICAL MANAGEMENT
Diagnosis
Approximately 80% of patients diagnosed with urothelial cancer undergo evaluation after presenting with hematuria, either microscopic or gross.4 However, microscopic hematuria is a common finding in the general population and is usually transient and benign.23,24 For example, in a prospective study of 1,930 patients with hematuria, only 12% were diagnosed with UCB, and 0.7% were diagnosed with UTUC. In 61% of cases, no cause for their hematuria was established.23 Another study evaluated 292 patients with asymptomatic microscopic hematuria. Only 5.4% of patients were found to have urological malignancies, whereas the majority of patients converted to a negative urinalysis and remained cancer free, with a mean follow-up of 13 years.24 Although the diagnostic yield is low, endoscopic evaluation of hematuria remains necessary because of the lack currently of sufficiently sensitive noninvasive approaches and the ramifications of missing a diagnosis of malignancy.25
Non–Muscle-Invasive Bladder Cancer
Non–muscle-invasive bladder cancer (NMIBC), extending no deeper than the subepithelial lamina propria, accounts for 70% of newly diagnosed UCB.4 NMIBC is treated with transurethral resection alone or in conjunction with intravesical therapy (immunotherapy or chemotherapy). Five-year survival rates are 93.9% for carcinoma in situ (CIS) or Ta lesions and 84.2% for T1 disease26; however, 50%-70% will recur, and approximately 10%-45% will progress to muscle-invasive bladder cancer (MIBC).4,27 Patients are therefore closely monitored post-treatment, undergoing cystoscopy and urine cytology every 3 to 6 months for at least 1 to 2 years, followed by annual cystoscopy and urine cytology.3 In the United States, the cost of bladder cancer care from diagnosis to death was reported to be US$102,700 per patient in 2013, and an overall annual cost of US$5.25 billion dollars is expected in 2020.15
MIBC
Bladder cancer extending beyond the lamina propria, termed MIBC, accounts for approximately 25% of newly diagnosed UCB cases.28 Standard treatment of MIBC involves 3 to 6 cycles of neoadjuvant chemotherapy (NAC) followed by radical cystectomy (RC). However, 5-year survival is still only approximately 50%, and many patients likely derive no significant benefit from the addition of chemotherapy.29-31 It is challenging to assess response to chemotherapy in patients with MIBC in real time because of the need for cystoscopic evaluation for direct tumor visualization. Early identification of patients not responding to neoadjuvant chemotherapy might enable them to be immediately directed toward surgical intervention, preventing unnecessary surgical delay and chemotherapy-related toxicity.22 Conversely, identification of patients with profound sensitivity to neoadjuvant therapy as assessed by both pathologic and molecular assays might enable some patients to be spared the morbidity of subsequent RC.
In carefully selected patients, a trimodality approach of maximal transurethral resection, chemotherapy, and radiation may lead to similar oncologic outcomes as NAC plus RC.32,33 Although this treatment strategy spares many patients from the morbidity of RC, in trials assessing bladder-sparing approaches, the reported risk of salvage RC at 10 years was 21%-31%, as some patients still needed to undergo RC because of recurrent or persistent disease or functional decline of their bladder.34,35 Bladder-sparing management of MIBC therefore still necessitates frequent cystoscopic monitoring for recurrence.6,36 With a growing number of systemic treatment options that effectively treat UCB (immune checkpoint inhibitors, antibody drug conjugates, and kinase inhibitors), neoadjuvant and bladder-sparing treatment paradigms are poised to evolve.31,32,37 Thus, the value of being able to monitor MIBC treatment responses will likely grow and inform personalized treatment algorithms on the basis of dynamic monitoring.
UTUC
Diagnosis and treatment of UTUC generally parallels that for UCB, although there is greater controversy regarding optimal treatments, largely due to fewer clinical studies focused on this rare disease entity.38,39 If initial diagnostic imaging suggests a filling defect in the upper urinary tract, direct visualization is generally performed via ureteroscopy. UTUC is more challenging to biopsy endoscopically, particularly limiting evaluation of stage, and thus more prone to sampling error.39,40 UTUC treatment may include purely endoscopic management in carefully selected patients but often includes complete removal of the affected kidney, ureter, and a portion of the bladder (nephroureterectomy), with the role and optimal timing of perioperative chemotherapy remaining incompletely defined.41,42 After surgical management, a risk associated with UTUC diagnosis is the chance of developing a secondary UCB. In a series of 82 patients diagnosed with UTUC who were treated with curative intent with nephroureterectomy or segmental distal ureterectomy, 36 (44%) later developed UCB.41 Thus, current recommendations are to perform ureteroscopy, cystoscopy, and upper tract imaging every 3 months for 1 year, then at increasing intervals for several years.36 Sensitive noninvasive methods that reduce the need for direct tumor visualization could thus greatly decrease the morbidity of UTUC surveillance.
URINE BIOMARKERS FOR UROTHELIAL CARCINOMA DETECTION AND MONITORING
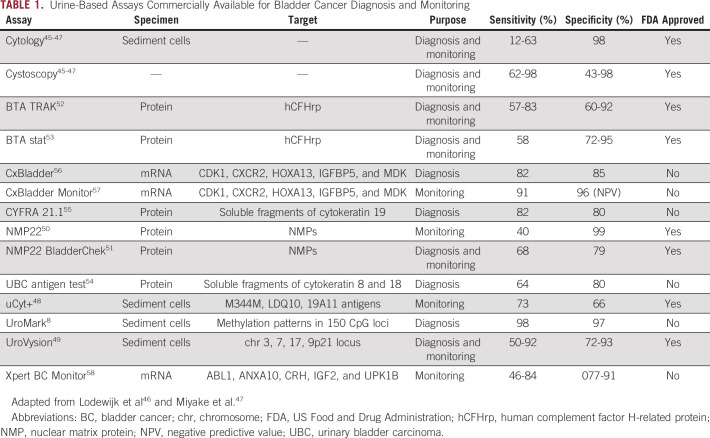
Cytology and cystoscopy are the gold standard methods for the detection and surveillance of bladder cancer.43,44 When combined, these tests confer high diagnostic sensitivity and specificity (Table 1).45-47 Nonetheless, they are invasive and costly, and their sensitivity to detect early disease is less than optimal.9,47 To overcome the shortcomings of cytology, several urinary biomarkers have been evaluated. These urine tests analyze: sediment cells (uCyt+,48 UroVysion,49 UroMark8), proteins (NMP22,50 NMP22 BladderChek,51 BTA TRAK,52 BTA stat,53 UBC test,54 CYFRA 21.155), or mRNA (CxBladder,56 CxBladder Monitor,57 Xpert Bladder Cancer Monitor58; Table 1). Generally, these tests have lower sensitivity than cystoscopy for the detection of high-grade lesions, and specificity is lower than urine cytology, explaining their lack of adoption into clinical practice.
TABLE 1.
Urine-Based Assays Commercially Available for Bladder Cancer Diagnosis and Monitoring
One test that may be comparable to the standard-of-care work-up is the UroMark assay, a targeted bisulfite next-generation sequencing (NGS) assay that evaluates urine sediment. The assay evaluates 150 CpG loci first identified to have potentially predictive value in a training set of urine sediment samples from 86 patients with MIBC and 30 tumor-free controls. In a validation cohort with 167 healthy controls and 107 bladder cancer cases, it performed with 98% sensitivity and 97% specificity. The UroMark assay is currently under investigation in a large observational study to assess its performance diagnosing new and recurrent cases of bladder cancer (ClinicalTrials.gov identifier: NCT02781428) and is not yet commercially available.59
GENOMICS OF BLADDER CANCER
Genomic Landscapes of MIBC and NMIBC
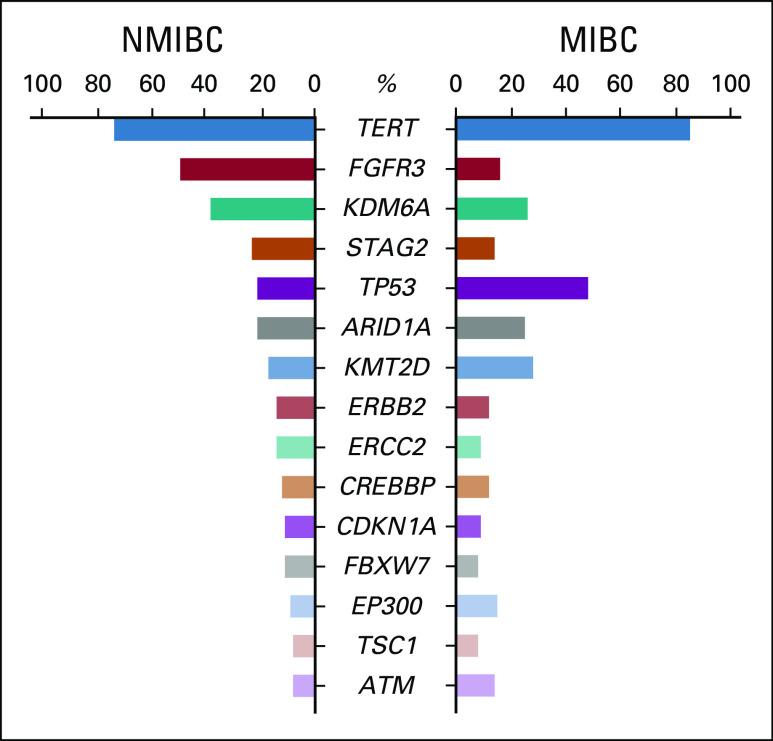
Several sequencing studies have reported on the incidence of genomic alterations in both NMIBC and MIBC specimens.17-19,30,60,61 MIBC has been more extensively evaluated with whole-exome sequencing analysis of > 400 MIBC samples by The Cancer Genome Atlas (TCGA).18 This revealed that 23 genes were significantly mutated at a rate > 7% in MIBC cases.18 Consistent with other cancers strongly linked to smoking, the overall mutational burden in bladder cancer is high, averaging 12 mutations per megabase in MIBC and high-grade NMIBC.61 The most commonly mutated genes in MIBC and NMIBC as reported by TCGA and the Memorial Sloan Kettering (MSK) studies are listed in Figure 1.17-19
FIG 1.
Significantly mutated genes in bladder cancer. Bar graphs represent the frequencies of overlapping driver genes mutated in non–muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC) at rates > 7% as reported in Table S4 of the Memorial Sloan Kettering (MSK) 2017 study19 and Figure 1 of The Cancer Genome Atlas (TCGA) 2017 study,18 respectively. For the FGFR3 mutational frequency in MIBC, we summed the single-nucleotide variant/indel rate (14%) with the fusion rate (2%) as reported in TCGA 2017.18 For TERT promoter mutations, the data for both MIBC and NMIBC are from the MSK 2017 study,19 as the TERT promoter region was not sequenced by TCGA.
Many of the significantly mutated genes in MIBC overlap with those identified in NMIBC, but there are quantitative differences. For example, TP53 mutations were present in almost half of the MIBC samples analyzed in TCGA studies,17,18 but mutations in this gene were only seen in approximately 20% of the 105 NMIBC samples analyzed by the MSK group.19 On the other hand, FGFR3 mutations were detected in almost half of NMIBC tumors, compared to only 16% of MIBCs.18,19 Mutations in PIK3CA and ARID1A were present at similar rates in both MIBC and NMIBC patient samples, seen in approximately a quarter of cases.17,19 In addition to these coding mutations, the TERT promoter has been shown to be mutated in 60%-85% of both MIBC and NMIBC cases.62-65 In Appendix A, we provide a detailed discussion of the genomic landscape and clonal diversity in UCB. Of particular relevance to the application of utDNA analysis to UCB, it is notable that across 32 cancer types, UCBs harbor the second-highest average number of nonsilent mutations in cancer-associated consensus genes per sample (approximately 5), with > 95% of samples having a nonsilent mutation in at least 1 consensus gene.66 Thus, in the vast majori-ty of cases, targeted evaluation of defined consensus regions would enable identification of at least 1, and typically multiple, mutations for diagnostic and surveillance purposes.
Genomic Predictive Markers
Several somatic mutations with potentially predictive value in urothelial carcinoma have been described. DNA damage repair (DDR) gene alterations have been shown to be associated with improved chemoradiation and cisplatin-based chemotherapy outcomes in patients with MIBC.67,68,69a Defects in the nucleotide excision repair gene ERCC2 were the most common DDR gene alteration in both high-grade NMIBC and MIBC, occurring in approximately 20% of cases.19 ERCC2 mutation correlated strongly with complete pathologic response after NAC and led to improved overall survival,69b a finding that was validated in a second cohort of patients.67 Because cisplatin induces DNA adducts, defective excision repair likely sensitizes cells to cisplatin-induced cell death by the principle of synthetic lethality.67,69b Less frequent DDR gene alterations were identified in ATM, BRCA1, BRCA2, ERCC4, PALB2, CHECK2, FANCC, RB1, PRKDC, ATRX, and MSH6.19,61,68 Mutations in these genes may also predict MIBC response to cisplatin-based chemotherapy.68 Furthermore, DDR gene alterations were associated with higher tumor mutational burden in MIBC and high-grade NMIBC.19,61 These data suggest that it is important to monitor mutations in DDR genes, which can serve as predictive biomarkers of therapeutic response, especially for patients on cisplatin-based treatment.
In the MSK study of NMIBC, 62 high-grade samples were examined to identify somatic alterations associated with recurrence after Bacillus Calmette-Guerin (BCG) treatment.19 ARID1A mutations were significantly associated with an increased risk of recurrence after BCG treatment, a finding that held true after correcting for multiple comparisons. ARID1A mutations also remained associated with recurrence within the larger 100-patient cohort treated with transurethral resection of bladder tumor (TURBT) with or without adjuvant intravesical therapy.19 In contrast, ERBB2 and FGFR3 mutations and co-occurring mutations in TP53 and MDM2 did not confer increased risk of recurrence after BCG.19 Finally, the authors did not find an association between mutational burden and recurrence after BCG.19 Although preliminary, these findings suggest that ARID1A mutations may serve as a specific biomarker for high-grade NMIBC resistance to BCG treatment.
Consistent with other disease types, activating mutations in receptor tyrosine kinases (FGFR3 or ERRB2) appear to predict responses to kinase inhibitors in metastatic urothelial carcinoma.70,71 An FGFR inhibitor, erdafitinib, has been approved by the US Food and Drug Administration for use in patients with locally advanced or metastatic disease, and is now being tested in recurrent high-risk NMIBC (ClinicalTrials.gov identifier: NCT04172675).71 Thus, the detection of certain actionable mutations can guide systemic treatments specifically targeting them.
POTENTIAL CLINICAL APPLICATIONS OF utDNA
Several studies have applied NGS-based liquid biopsy assays to urinary DNA to evaluate urothelial cancer noninvasively (assays summarized in Fig 2, with pre-analytic factors discussed in Appendix B). UroSEEK has been the most broadly tested and consists of 3 tests, all applied to urine sediment: SafeSeqS72 for multiplex PCR-based NGS mutational analysis of 10 genes associated with urothelial cancer, a separate TERT promoter SafeSeqS assay, and aneuploidy detection by FastSeqS.73 When applied to 570 patients who were at risk for developing bladder cancer, UroSEEK achieved a sensitivity of 83% for cancer detection, compared to 43% for cytology.11 Sensitivity increased to 95% when results from both UroSEEK and urine cytology were combined.11 UroSEEK positivity preceded the clinical diagnosis of bladder cancer by 2.3 months on average and by more than a year in 8 cases.11 Similarly, UroSEEK successfully identified UTUC; in urine collected before surgery from 56 patients with UTUC, 75% tested positive by UroSEEK, including 79% of those with noninvasive tumors. In contrast, urine cytology detected only 10% of UTUC cases.11 On evaluation of tumor tissue in both the bladder cancer and UTUC cohorts, mutations in genes assessed by UroSEEK were identified in 62%-75% of cases originally missed by urine testing.11 In a follow-up study of 527 bladder tumor samples, 92% were found to have a mutation identified by UroSEEK.11 Thus, the primary factor leading to a false-negative test result appears to be the assay’s limit of detection, rather than insufficient genomic breadth of the UroSEEK panel.
FIG 2.
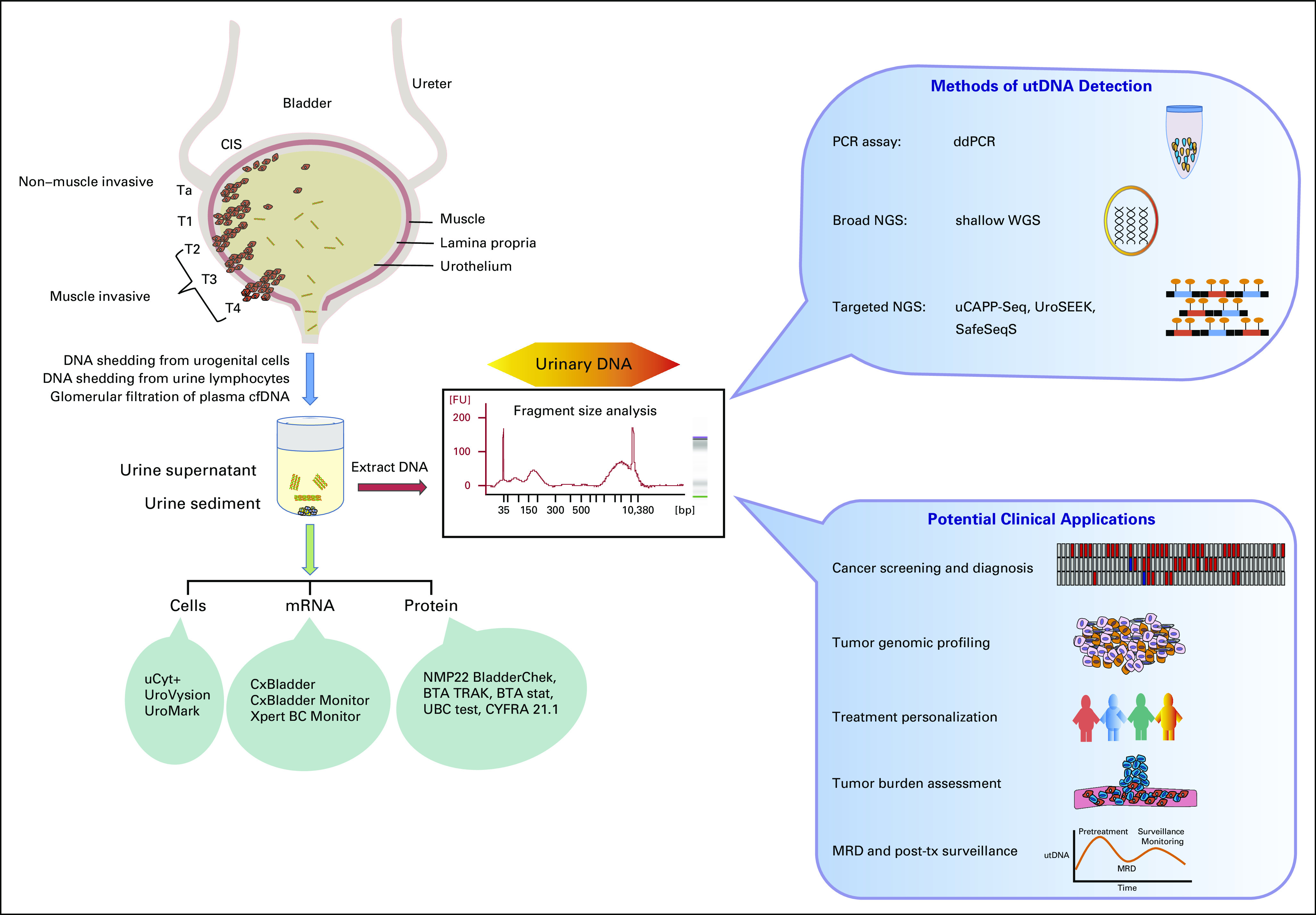
Origins of urinary DNA, methods of urinary tumor DNA (utDNA) detection, and potential clinical applications in bladder cancer. American Joint Committee on Cancer (ed 8)79 T staging is represented, with pictorial representations of carcinoma in situ (CIS), non–muscle-invasive bladder cancer (NMIBC), and muscle-invasive bladder cancer (MIBC). Commercial urine-based assays are also depicted, associated with the urine components they analyze. A representative electropherogram is shown of urinary DNA from a patient with cancer. Next-generation sequencing (NGS) and polymerase chain reaction (PCR)–based methods of utDNA detection are shown, as are potential clinical applications of utDNA analysis. cfDNA, cell-free DNA; ddPCR, digital droplet PCR; MRD, minimal residual disease; tx, treatment; uCAPP-Seq, urine Cancer Personalized Profiling by deep Sequencing; WGS, whole-genome sequencing.
UroSEEK was also investigated for post-treatment monitoring of patients who underwent tumor resection. Urine samples were collected from 322 patients whose tumors, based on evaluation of the resected tumor specimen, had a mutation in at least 1 of the genes (or the TERT promoter) queried by SafeSeqS.11 In these patients, UroSEEK identified recurrences with 68% sensitivity and 80% specificity.11 Relapse was detected by UroSEEK on average 7 months earlier than the clinical diagnosis. Surveillance cytology was also available for 196 patients, achieving a sensitivity of only 25%.11 Taken together, these results suggest that UroSEEK has the potential to improve our ability to detect bladder cancer in both the early diagnostic and recurrence settings. Although greater sensitivity will be needed to supplant cystoscopic evaluation, UroSEEK could be useful for developing risk-adapted protocols that reduce the need for invasive monitoring.
Dudley et al13 similarly used utDNA analysis to detect early-stage UCB and monitor post-treatment residual disease. They developed a hybrid capture-based NGS assay called Urine Cancer Personalized Profiling by Deep Sequencing (uCAPP-Seq), which evaluates genomic regions from 460 genes found to be recurrently mutated in MIBC by TCGA analysis,18 and applied it to urine supernatant.13 Comparing urine pretreatment samples from 54 patients with bladder cancer (CIS, pTa-T2) to samples from 34 healthy volunteers, they detected utDNA in 83% of cancer cases when blinded to tumor mutational status, with 97% specificity. Sensitivity of uCAPP-Seq improved to 93% with tumor-informed profiling, when the patient’s mutations were first identified by sequencing tumor biopsy tissue with the same gene panel and subsequently evaluated by uCAPP-Seq in urine specimens. They also evaluated a cohort of 64 patients undergoing surveillance after treatment of localized bladder cancer. uCAPP-Seq performed significantly better than standard urine cytology and cystoscopy, detecting 84% of patients who developed recurrence, whereas the combined sensitivity of cytology and cystoscopy was only 53%.13 Specificity of uCAPP-Seq remained high at 96%-100%, comparable to urine cytology. Detection of utDNA preceded clinical disease recurrence in 92% of patients by a median of 2.7 months, suggesting that uCAPP-Seq is a sensitive modality capable of robustly detecting UCB recurrence early.
Across their studies, Dudley et al identified a median of 6 mutations per patient in DNA isolated from urine supernatant. Concordance of mutations detected in paired tumor tissue and utDNA from 18 patients was reasonably high, with 67% of mutations detected in tumor tissue also detected in utDNA.13 Some discordance is to be expected, given intratumoral heterogeneity not fully captured by tumor biopsy sequencing and decreased probability of detecting subclonal mutations in cell-free DNA that are present subclonally at low levels in tumors.74-76
Digital droplet PCR (ddPCR) is another strategy that has been used for the detection of specific mutations in urine DNA.20 ddPCR separates DNA molecules into individual oil droplets with a target of 1 template molecule per droplet.77 PCR reactions are performed within individual droplets and the extent of amplification is digitized, yielding a binary result per droplet.77 In this way, ddPCR can be used to query specific mutations in a targeted fashion. In a retrospective pilot study, Birkenkamp-Demtröder et al14 examined 101 banked NMIBC urine samples from 12 patients who went on to develop recurrent or progressive/metastatic disease. They used germline and tumor sequencing to identify tumor-specific variants and then generated personalized ddPCR assays to query utDNA in 101 longitudinally collected urine samples. utDNA was detected in 50% of samples from patients with recurrent NMIBC and in 96% of samples from patients who developed MIBC or metastatic UCB. They also detected higher levels of utDNA in patients who developed progressive or metastatic disease, an average of 1,282 copies/mL, versus 31 copies/mL in patients who developed localized recurrence, suggesting that perhaps utDNA levels reflect invasive potential.14 Although the study was too small to delineate sensitivity and specificity, it established proof of concept for the use of personalized ddPCR assays to monitor utDNA. Such an approach may provide an alternative means of surveillance for the small proportion of patients who lack mutations in genes queried by NGS panel-based assays.
Rather than evaluating for specific mutations, Ge et al12 developed UCdetector to detect urothelial carcinoma through shallow whole-genome sequencing (sWGS) to identify copy number alterations in urine supernatant as well as urine sediment. Their study included urine analysis from 65 patients who had urothelial carcinoma. Genome-wide copy number changes in tumor had higher concordance with findings from urine supernatant (cell-free DNA) compared with sediment. Predicted tumor fractions were also significantly higher from urine supernatant than from urine sediment.12 The authors further developed a urine-based diagnostic classifier through machine learning to sensitively detect urothelial carcinoma on the basis of genome-wide copy number alteration features. Internal cross-validation revealed that the median clinical sensitivity was 86% and specificity was 95%, suggesting that urinary cell-free DNA sWGS could form the basis of noninvasive urothelial cancer detection. Using an independent validation cohort of 24 patients without tumors and 28 patients with urothelial carcinoma, UCdetector performed with a clinical sensitivity of 79% and specificity of 88%. The authors also performed pre- and post-operative analyses of urine samples from 7 patients undergoing TURBT. UCdetector identified disease in all 7 pretreatment samples and in 2 post-treatment samples.12 Follow-up and sample size, however, were too limited to ascertain the clinical significance of the post-operative findings.
Patel et al22 aimed to interrogate the longitudinal dynamics of biofluid-derived tumor DNA in patients undergoing platinum-based NAC for MIBC. They used a combination of tagged-amplicon sequencing (TAm-Seq) and sWGS to noninvasively evaluate single-nucleotide variants and copy number alterations in bladder cancer.17,60,78 They assessed tumor DNA serially in blood plasma, urine supernatant, and urine sediment. As part of this analysis, TAm-Seq was performed just prior to cycle 2 of NAC. Twelve patients who had single-nucleotide variants detected in their tumor tissue were included in this analysis. Mutant DNA was detected in a biofluid sample (either urine sediment, urine supernatant, or plasma) in 5 of the 6 patients whose disease recurred and in none of the 6 patients who remained recurrence-free, resulting in an overall sensitivity and specificity of 83% and 100% to predict recurrence, respectively.22 Mutant DNA detection before cycle 1 of NAC did not correlate with recurrence. These results suggest that the detection of mutant DNA from analysis of both plasma and urine may be used to monitor treatment response and serve as an early on-treatment predictive biomarker. The authors did not, however, report sensitivity or specificity using utDNA alone (without plasma) or of sWGS.
On serial time point analysis of patients undergoing NAC, Patel et al22 also noted a trend of decreasing utDNA levels over time, although there were examples of utDNA mutant allele fractions rising then falling or remaining persistently elevated in patients who experienced early recurrence. In addition, there was evidence of dynamic tumor evolution identified by biofluid analysis during NAC in 5 patients, including the emergence of new clonal driver mutations in a patient who recurred quickly, suggestive of chemotherapy resistance.22 Although small and not validated using an independent cohort, the study’s findings suggest that serial utDNA analysis could, in the future, help response-adapt treatment in patients with MIBC receiving NAC.
FUTURE DIRECTIONS
Urothelial carcinoma has a poor prognosis when recurrent or advanced.3,36,37 Early detection followed by definitive treatment when disease burden is minimal is critical for achieving long-term disease-free survival.6,26 It is also important to be able to monitor patients after treatment to identify relapse early. In addition, assessment while on neoadjuvant chemotherapy is important, especially when bladder-sparing approaches are being considered.22 Although cystoscopy and cytology are gold standard for the detection and surveillance of bladder cancer, there is a need for more-sensitive, less-invasive, and less-costly modalities.15,45 Analysis of utDNA is a noninvasive approach, with promising results suggesting diagnostic and surveillance capability. Although most studies testing utDNA as a surrogate for disease detection and monitoring have been conducted in small patient cohorts, the results are encouraging, demonstrating reasonable concordance between tissue and utDNA genotyping, higher sensitivity than cytology, and comparable specificity.13,14,20,22
To be validated as a cancer biomarker with clinical utility, large prospective clinical trials need to be performed to thoroughly test utDNA analysis for the diagnosis, surveillance, and management of urothelial carcinoma. Undoubtedly, the field will continue to refine utDNA detection assays to further enhance sensitivity and specificity, which will be needed to supplant cystoscopic monitoring in most scenarios. Although reducing the need for invasive monitoring will be a major priority, the potential for utDNA assays to improve outcomes should not be overlooked. For example, utDNA analysis during NAC might provide new opportunities for tailoring individualized treatments on the basis of dynamic molecular profiling. In light of an expanding armamentarium of agents active against urothelial carcinoma, opportunities are growing for treatment individualization on the basis of rapid response assessment. It is also notable that utDNA often detects localized disease before standard clinical approaches, raising the possibility that it can be used as a surrogate for minimal residual disease or perhaps persistence of premalignant cells. Thus, utDNA might also serve as a capable biomarker for guiding early treatment escalation to reduce the risk of recurrence and progression while disease burden is still minimal.
Summary
Urothelial cancer standard-of-care diagnosis and surveillance consists of invasive approaches associated with high costs.
DNA in urine arising from malignant cells is referred to as “urinary tumor DNA” (utDNA).
utDNA can be detected and quantified using NGS– or digital droplet PCR–based assays.
Single-institutional studies have shown that utDNA analysis can enable early detection of bladder cancer in the diagnostic and surveillance settings.
utDNA has potential as an early on-treatment biomarker for patients undergoing neoadjuvant chemotherapy.
Appendix A
Clonal Origin and Evolution of Bladder Cancer
Understanding the drivers of carcinogenesis in bladder cancer is key for the development of biomarkers that may enable the detection of subclinical or early disease.
To this purpose, different genomic analyses have been applied to identify founding clones in the carcinogenesis process. In 1992, Sidransky et al (N Engl J Med 326:737-740, 1992) analyzed the cystectomy specimens from 4 female patients who were diagnosed with multifocal bladder cancer to assess whether the tumors were derived from the same precursor cell. Using a combination of DNA gel electrophoresis and Southern blot, the authors investigated X-chromosome inactivation and allelic losses. Their analysis demonstrated that although normal bladder mucosa had a polyclonal pattern of X-inactivation, all analyzed tumors within a patient showed monoclonal X-inactivation with the same X-chromosome inactivated. In addition, 3 patients’ tumors were examined for somatic loss of chromosome 9q alleles, which showed loss of the same 9q allele in each tumor. In contrast, losses of chromosome 17p and 18q, later events in tumor progression, did not show clonality between tumors within a patient. Their findings corroborate the hypothesis that multifocal tumors arise from a single transformed cell and subsequently evolve to acquire new genetic alterations.
With advancements in genomic testing, more sophisticated techniques enabled a better understanding of the molecular changes involved in bladder carcinogenesis. Majewski et al (Cell Rep 26:2241-2256.e4, 2019) conducted a comprehensive analysis of a cystectomy specimen from a patient diagnosed with multifocal papillary bladder cancer. They hypothesized that epigenetic and/or genetic changes in histologically normal-appearing bladder mucosa show evidence of early cancer-initiating events, referred to as the “field effect”. Applying next-generation sequencing (NGS) to tumor and surrounding mucosal DNA, the authors analyzed mutations, copy number variations (CNVs), and methylation patterns. They identified widespread and uniform CNVs and methylation changes in normal-appearing mucosa on a background of highly heterogeneous low-allele fraction mutations, suggesting that the CNVs and methylation changes represented early cancer driver events. The authors also identified an inactivating mutation in the ACIN1 gene in normal mucosa, which demonstrated clonal expansion to invasive carcinoma along with mutations in 21 additional genes. Finally, an activating mutation in KRAS was identified as an important driver of progression to high-grade cancer in this patient. Although restricted to a single patient, these results support the clonal underpinnings of multifocal bladder carcinoma.
Another important factor to consider is the impact of chemotherapy driving clonal evolution in urothelial carcinoma (UC). To address this, Faltas et al (Nat Genet 48:1490-1499, 2016) performed whole-exome sequencing (WES)-based clonality analysis of 72 UC samples from 32 patients, including 16 matched sets of primary and metastatic UC and germline samples and 2 rapid autopsy cases. Interestingly, only 28.4% of mutations were shared pre- and post-chemotherapy, and even mutations in known driver genes, such as PIK3CA, KMT2D (MLL2), ATM, and TP53, were not consistently shared. Some post-chemotherapy tumors even developed different mutations in the same key gene. In an analysis of 21 sets of matched tumors, a pattern of early branching was observed where an ancestral clone gave rise to multiple cell populations evolving in parallel. The authors showed a significant increase in APOBEC signatures in chemotherapy-treated tumors, suggesting that APOBEC-induced mutagenesis could be contributing to UC clonal evolution in response to chemotherapy. These results suggest that UC undergoes extensive clonal evolution in response to chemotherapy, which could lead to chemotherapy resistance.
Bladder Cancer Genomic Landscape
Non–muscle-invasive bladder cancer genomic landscape.
Pietzak et al19 at Memorial Sloan Kettering (MSK) used the MSK-IMPACT targeted exome platform to sequence tumor and matched germline DNA from 105 patients with non–muscle-invasive bladder cancer (NMIBC). The most commonly identified mutations were in TERT promoter (73%), FGFR3 (49%), KDM6A (38%), PIK3CA (26%), STAG2 (23%), AR1D1A (21%), and TP53 (21%; Fig 1). Mutations in chromatin-modifying genes were highly prevalent, occurring in 69% of cases, most commonly involving KDM6A (38%) and AR1D1A (21%). Alterations in KDM6A and ARID1A did not correlate significantly with grade or stage. Truncating STAG2 mutations were associated with low-grade Ta tumors,19 although the association of STAG2 mutations with aggressive versus low-grade bladder cancer are conflicting in other studies (Balbás-Martínez C, et al: Nat Genet 45:1464-1469, 2013; Guo G, et al: Nat Genet 45:1459-1463, 2013; Solomon DA, et al: Science 333:1039-1043, 2011; Taylor CF, et al: Hum Mol Genet 23:1964-1974, 2014). The TERT promoter mutation was highly prevalent across different grades and stages, with a mutation rate of 61% in low-grade Ta tumors, 88% in high-grade Ta tumors, and 79% in high-grade T1 tumors.19 FGFR3 mutations were associated with lower grade and stage. Alterations in the tyrosine kinase/phosphatidylinositol3-kinase (RTK-PIK3) pathway, which includes FGFR3, were overall present in 79% of NMIBC cases.
MIBC genomic landscape.
The Cancer Genome Atlas (TCGA) study is the largest one to date assessing the muscle-invasive bladder cancer (MIBC) genomic landscape.18 It included analysis of 412 MIBC tumor samples analyzed by WES. Fifty-eight genes were significantly mutated, and tumor mutational burden correlated with the APOBEC signature. Furthermore, several canonical signaling pathways were shown to be altered in MIBC (Fig 1). The authors observed an 89% rate of inactivation in the p53/cell-cycle pathway in MIBC tumor samples, with TP53 itself mutated in 48% of cases. Other relatively common inactivating mutations were seen in RB1 (17%), CDKN1A (11%), and CDKN2A (7%). TERT promoter mutations were commonly found in 85% of patients with MIBC in a different study performed at the MSK Cancer Center.19 Of note, TERT promoter mutations were not assessed by the TCGA study,17,18 because the WES method they applied only interrogated protein-coding regions. Other important findings in TCGA consisted of oncogenes that harbored recurrent hotspot mutations including FGFR3, PIK3CA, and RAS. FGFR3 mutations were present in 16% of cases and were more common in lower-stage tumors,18 consistent with their greater prevalence in NMIBC.19 PIK3CA mutations were commonly seen in the helical domain and were likely due to APOBEC-induced mutagenesis.18 ERBB2 mutations were common at serine 310 (S310) in the extracellular domain (42% of mutated cases), also likely from APOBEC-induced mutagenesis. PPARG was also altered in MIBC (primarily by amplification) in 17% of cases, and has been shown to interact with mutated RXRA to further promote urothelial growth (Halstead AM et al, eLife 6:e30862, 2017). Mutually exclusive alterations were seen between CDKN2A and TP53, CDKN2A and RB1, CDKN2A and E2F3, TP53 and MDM2, FGFR3 and E2F3, and FGFR3 and RB1.18 Co-occurring genomic alterations were commonly seen between TP53 and RB1, TP53 and E2F3, and FGFR3 and CDKN2A. FGFR3 mutations and CDKN2A copy number alterations were found to co-occur in 7% of cases, possibly evidence of MIBC arising from progression of NMIBC tumors (Rebouissou S, et al: J Pathol 227:315-324, 2012).
APOBEC mutational signature.
The APOBEC mutational pattern is linked to increased activity of APOBEC cytidine deaminases, which have been shown to induce mutations in several cancer types, including MIBC (Roberts SA, et al: Nat Genet 45:970-976, 2013).61 In MIBC, 2 different variants of the APOBEC signature, APOBEC-a and APOBEC-b, were described in the TCGA study.18 These 2 APOBEC signatures accounted for 67% of all detected single-nucleotide variants and were strongly associated with hypermutation, increased PD-1 expression, and improved overall survival.18,66 APOBEC signatures were also identified in a study from the Dana Farber Cancer Institute that conducted targeted exome sequencing in 472 UC specimens spanning a range of grades and anatomic sites.61 The authors demonstrated that although APOBEC signature mutations were observed across UC types and grades, they were significantly more frequent in MIBCs and high-grade NMIBCs and less frequent in high-grade upper tract urothelial carcinomas and low-grade NMIBCs. Overall, APOBEC appears to play an important role in driving mutagenesis in UC, especially for MIBC and potentially high-grade NMIBC.
Appendix B
Pre-analytic Factors in Urinary Tumor DNA Testing
Sources of tumor DNA in the urine.
Tumor DNA is found in both the urine supernatant cell-free DNA (cfDNA) and in the cellular pellet (urine sediment). cfDNA reaches the urine via direct shedding from cells in the urogenital tract and from lymphocytes present in the urine (Fig 2; Botezatu I, et al: Clin Chem 46:1078-1084, 2000; Panagopoulou M, et al: J Cell Physiol 234:14079-14089, 2019; Su YH, et al: Ann N Y Acad Sci 1022:81-89, 2004; Chang HW, et al: Int J Biol Markers 22:287-294, 2007; Su YH, et al: J Mol Diagn 6:101-107, 2004). cfDNA can also reach the urine through glomerular filtration of plasma-derived cfDNA (Botezatu I, et al: Clin Chem 46:1078-1084, 2000). Urinary cfDNA can be divided based on length into low- and high-molecular-weight DNA: low-molecular-weight DNA ranges from 10 bp to 400 bp, and high-molecular-weight DNA measures at least 1 kbp. There is currently no widely adopted consensus on the use of urine supernatant versus sediment, although some studies suggest that the supernatant is enriched for tumor DNA relative to the sediment fraction (Togneri FS, et al: Eur J Hum Genet 24:1167-1174, 2016).12
Fragment size selection.
Fragment size selection of urinary cfDNA may not be necessary for bladder cancer urinary tumor DNA (utDNA) detection.13,64 Dudley et al13 found similar variant allele fractions of mutant DNA in both long (> 500 bp) and short (< 500 bp) urinary cfDNA fragments. Similar findings were demonstrated by Russo et al64 on the basis of detecting TERT promoter mutations. After using bead fractionation to separate DNA fragments from 8 patient cfDNA samples on the basis of size, it was found that the TERT mutant allele frequency detected by digital droplet polymerase chain reaction was similar for both long and short DNA fragments per sample.64
Specimen collection.
Although inter- and intra-donor variations in the quantity and quality of urinary cfDNA is inevitable (Johnson DJ, et al: J Forensic Sci 52:110-113, 2007), consideration of several factors can increase productive recovery. Amounts of cfDNA in a urine specimen depend on the volume of urine collected and the timing of collection. At our institution, we typically aim to collect up to 90 mL of urine, the size of most standard urinalysis cups. Urinary cfDNA concentrations, and unwanted particulate matter, tend to increase the longer urine remains inside the bladder (Brisuda A, et al: Urol Int 96:25-31, 2016). As such, first void morning samples usually contain higher amounts of DNA. However, these samples also harbor a higher content of debris and impurities; to decrease urinary debris contaminating cfDNA, the second urine void of the morning is preferred (Brisuda A, et al: Urol Int 96:25-31, 2016). Alternatively, urine samples can be collected at the time of cystoscopy or other urologic procedures, such as catheterization for intravesical therapy administration, which allows for more flexibility and convenience (Schmitz-Dräger C, et al: Urol Oncol 34:452-459, 2016). DNA concentrations have also been shown to vary during urination, with the highest concentration being at the beginning of the voiding process (Johnson DJ, et al: J Forensic Sci 52:110-113, 2007). Nonetheless, the first portion of voided urine may be contaminated by blood or urethral cellular debris; thus, midstream urine is a reasonable alternative, despite lower DNA concentrations (Vorsters A, et al: Eur J Clin Microbiol Infect Dis 33:2005-2014, 2014). Because patients with bladder cancer sometimes require the use of percutaneous nephrostomy tubes for medical reasons, attention should be paid to avoid evaluating urine from these sources, because it may have bypassed contact with the tumor. Interestingly, sex differences have been reported regarding DNA content in the urine. Female urine yields 3 to 4 times higher DNA concentrations than male urine, which may be related to genitourinary anatomic differences leading to differences in epithelial cell–derived DNA levels (Johnson DJ, et al: J Forensic Sci 52:110-113, 2007; Vu NT, et al: Forensic Sci Int 102:23-34, 1999).
Human urine is susceptible to nuclease activity, and for this reason cfDNA can be heterogeneous with respect to size and composition (Bryzgunova OE, et al: Ann N Y Acad Sci 1075:334-340, 2006). In an attempt to improve nucleic acid stability, freezing urine shortly after collection has been suggested (Ng HH, et al: Forensic Sci Int 287:36-39, 2018). However, this approach is not practical in the clinical setting, so other ways to stabilize urinary nucleic acids have been studied. Streck Laboratories and Norgen developed proprietary liquid reagents that stabilize cfDNA in urine specimens (Benhamou S, et al: BMC Cancer 16:837, 2016). The former reported that their reagent stabilizes DNA in urine samples at room temperature for 7 days. Another urine DNA preservation method that has been used is the addition of EDTA to urine samples before storage (Melkonyan HS, et al: Ann N Y Acad Sci 1137:73-81, 2008).13 Dudley et al found that when EDTA was added to urine samples to 0.5 mM, urinary cfDNA remained stable at 4°C for up to 1 week.13
Methods of urinary cfDNA isolation.
Methods of urinary cfDNA isolation can vary in their volume capacity, total cfDNA yield, and the size distribution of isolated DNA fragments (Streleckiene G, et al: Biotechniques 64:225-230, 2018). Column-based kits that have been used for cfDNA isolation from urine of patients with bladder cancer include the Qiagen Circulating Nucleic Acid Kit (Brisuda A, et al: Urol Int 96:25-31, 2016), QIAquick Gel Extraction Kit (Kim YH, et al: Investig Clin Urol 57:106-112, 2016), QIAamp DNA Mini Kit (Zancan M, et al: Int J Biol Markers 3:147-155, 2009), and QIAamp Viral RNA Mini Kit.63 These kits are generally volume-limited to 5 mL of urine supernatant, usually too low for utDNA analysis. Methods specifically catered to isolating cfDNA from urine for subsequent utDNA analysis can accommodate much larger volumes. For example, Dudley et al used a resin-based isolation method to isolate cfDNA from a median urine volume of 50 mL.13 Analysis of cfDNA fragment size from isolated urine samples revealed a distribution of low-molecular-weight DNA (< 500 bp), including ultra-short DNA (< 100 bp), and high-molecular-weight DNA (1-10 kbp). Magnetic bead–based DNA isolation approaches such as the MagMAX Cell-Free DNA Isolation Kit have also been used successfully to extract urinary cfDNA before utDNA analysis (Lee DH, et al: Sci Rep 8:14707, 2018).
SUPPORT
This work was supported by the Alvin J. Siteman Cancer Research Fund at Washington University in St Louis, MO (A.A.C.), the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (NCATS) under award number UL1TR002345 (Principal Investigator, Bradley Evanoff; A.A.C.), the NIH National Cancer Institute (NCI) under award number K08CA238711 (A.A.C.), the Cancer Research Foundation Young Investigator Award (A.A.C.), and the Damon Runyon Clinical Investigator Award (V.K.A). B.P. acknowledges the Washington University School of Medicine R25 STRENGTH Program (R25CA190190; Principal Investigator, Ramaswamy Govindan) for protected research time.
The views expressed in the submitted article are the authors’ views and not an official position of Washington University.
AUTHOR CONTRIBUTIONS
Conception and design: Aadel A. Chaudhuri, Vivek K. Arora
Financial support: Aadel A. Chaudhuri
Administrative support: Aadel A. Chaudhuri
Provision of study material or patients: Aadel A. Chaudhuri
Collection and assembly of data: Aadel A. Chaudhuri, Bruna Pellini, Nadja Pejovic, Pradeep S. Chauhan, Peter K. Harris, Vivek K. Arora
Data analysis and interpretation: Aadel A. Chaudhuri, Bruna Pellini, Jeffrey J. Szymanski, Zachary L. Smith, Vivek K. Arora
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Aadel A. Chaudhuri
Stock and Other Ownership Interests: Geneoscopy
Honoraria: Foundation Medicine, Roche
Consulting or Advisory Role: Geneoscopy, Roche, Fenix Group International, Tempus
Patents, Royalties, Other Intellectual Property: US Patent No. US8685727B2
Travel, Accommodations, Expenses: Roche, Foundation Medicine
Other Relationship: Roche
Vivek K. Arora
Stock and Other Ownership Interests: ORIC Pharmaceuticals
Consulting or Advisory Role: H3 Biomedicine, Bristol Myers Squibb, Seattle Genetics/Astellas
Research Funding: ORIC Pharmaceuticals
Travel, Accommodations, Expenses: H3 Biomedicine, Bristol Myers Squibb
No other potential conflicts of interest were reported.
REFERENCES
- 1.Scosyrev E, Yao J, Messing E. Urothelial carcinoma versus squamous cell carcinoma of bladder: Is survival different with stage adjustment? Urology. 2009;73:822–827. doi: 10.1016/j.urology.2008.11.042. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3. doi: 10.6004/jnccn.2017.0156. Spiess PE, Agarwal N, Bangs R, et al: Bladder cancer, version 5.2017: Clinical practice guidelines in oncology. J Natl Compr Cancer Netw 15:1240-1267, 2017. [DOI] [PubMed] [Google Scholar]
- 4.Kirkali Z, Chan T, Manoharan M, et al. Bladder cancer: Epidemiology, staging and grading, and diagnosis. Urology. 2005;66(suppl 1):4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 5.Schned AR, Andrew AS, Marsit CJ, et al. Histological classification and stage of newly diagnosed bladder cancer in a population-based study from the Northeastern United States. Scand J Urol Nephrol. 2008;42:237–242. doi: 10.1080/00365590801948166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang SS, Boorjian SA, Chou R, et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J Urol. 2016;196:1021–1029. doi: 10.1016/j.juro.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Prout GR, Jr, Barton BA, Griffin PP, et al. Treated history of noninvasive grade 1 transitional cell carcinoma. The National Cancer Group. J Urol. 1992;148:1413–1419. doi: 10.1016/s0022-5347(17)36924-0. [DOI] [PubMed] [Google Scholar]
- 8.Feber A, Dhami P, Dong L, et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics. 2017;9:8. doi: 10.1186/s13148-016-0303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yafi FA, Brimo F, Steinberg J, et al. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol. 2015;33:66.e25–66.e31. doi: 10.1016/j.urolonc.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Nisman B, Barak V, Shapiro A, et al. Evaluation of urine CYFRA 21-1 for the detection of primary and recurrent bladder carcinoma. Cancer. 2002;94:2914–2922. doi: 10.1002/cncr.10565. [DOI] [PubMed] [Google Scholar]
- 11.Springer SU, Chen C-HH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife. 2018;7:1–27. doi: 10.7554/eLife.32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. doi: 10.1373/clinchem.2019.309633. Ge G, Peng D, Guan B, et al: Urothelial carcinoma detection based on copy number profiles of urinary cell-free DNA by shallow whole-genome sequencing. Clin Chem 66:188-198, 2020. [DOI] [PubMed] [Google Scholar]
- 13.Dudley JC, Schroers-Martin J, Lazzareschi DV, et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. 2018;9:500–509. doi: 10.1158/2159-8290.CD-18-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol. 2016;70:75–82. doi: 10.1016/j.eururo.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Yeung C, Dinh T, Lee J. The health economics of bladder cancer: An updated review of the published literature. Pharmacoeconomics. 2014;32:1093–1104. doi: 10.1007/s40273-014-0194-2. [DOI] [PubMed] [Google Scholar]
- 16. Tan WS, Feber A, Dong L, et al: EAU–ESMO consensus statements on the management of advanced and variant bladder cancer—an international collaborative multi-stakeholder effort: Under the auspices of the EAU and ESMO Guidelines Committees. Eur Urol 7:311-331, 2017. [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Weinstein JN, Akbani R, et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1016/j.cell.2017.09.007. Robertson AG, Kim J, Al-Ahmadie H, et al: Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171:540-556.e25, 2017 [Erratum: Cell 174:1033, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietzak EJ, Bagrodia A, Cha EK, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol. 2017;72:952–959. doi: 10.1016/j.eururo.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen E, Birkenkamp-Demtröder K, Nordentoft I, et al. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur Urol. 2017;71:961–969. doi: 10.1016/j.eururo.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Birkenkamp-Demtröder K, Christensen E, Nordentoft I, et al. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur Urol. 2018;73:535–540. doi: 10.1016/j.eururo.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Patel KM, van der Vos KE, Smith CG, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep. 2017;7:5554. doi: 10.1038/s41598-017-05623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khadra MH, Pickard RS, Charlton M, et al. A prospective analysis of 1,930 patients with hematuria to evaluate current diagnostic practice. J Urol. 2000;163:524–527. [PubMed] [Google Scholar]
- 24.Mishriki SF, Nabi G, Cohen NP. Diagnosis of urologic malignancies in patients with asymptomatic dipstick hematuria: Prospective study with 13 years’ follow-up. Urology. 2008;71:13–16. doi: 10.1016/j.urology.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 25.Linder BJ, Bass EJ, Mostafid H, et al. Guideline of guidelines: Asymptomatic microscopic haematuria. BJU Int. 2018;121:176–183. doi: 10.1111/bju.14016. [DOI] [PubMed] [Google Scholar]
- 26.Howlader N, Noone AM, Krapcho M, et al. , (eds): SEER Cancer Statistics Review, 1975-2017, National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2017/
- 27.Sylvester RJ, van der Meijden APM, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–475, discussion 475-477. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of bladder cancer: A systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74:784–795. doi: 10.1016/j.eururo.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349:859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 30.McConkey DJ, Choi W, Shen Y, et al. A prognostic gene expression signature in the molecular classification of chemotherapy-naïve urothelial cancer is predictive of clinical outcomes from neoadjuvant chemotherapy: A phase 2 trial of dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin with bevacizumab in urothelial cancer. Eur Urol. 2016;69:855–862. doi: 10.1016/j.eururo.2015.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Advanced Bladder Cancer (ABC) Meta-analysis Collaboration Neoadjuvant chemotherapy in invasive bladder cancer: Update of a systematic review and meta-analysis of individual patient data advanced bladder cancer (ABC) meta-analysis collaboration. Eur Urol. 2005;48:202–205, discussion 205-206. doi: 10.1016/j.eururo.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Efstathiou JA, Spiegel DY, Shipley WU, et al. Long-term outcomes of selective bladder preservation by combined-modality therapy for invasive bladder cancer: The MGH experience. Eur Urol. 2012;61:705–711. doi: 10.1016/j.eururo.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 33.James ND, Hussain SA, Hall E, et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;366:1477–1488. doi: 10.1056/NEJMoa1106106. [DOI] [PubMed] [Google Scholar]
- 34.Mak RH, Hunt D, Shipley WU, et al. Long-term outcomes in patients with muscle-invasive bladder cancer after selective bladder-preserving combined-modality therapy: A pooled analysis of Radiation Therapy Oncology Group protocols 8802, 8903, 9506, 9706, 9906, and 0233. J Clin Oncol. 2014;32:3801–3809. doi: 10.1200/JCO.2014.57.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giacalone NJ, Shipley WU, Clayman RH, et al. Long-term outcomes after bladder-preserving tri-modality therapy for patients with muscle-invasive bladder cancer: An updated analysis of the Massachusetts General Hospital Experience. Eur Urol. 2017;71:952–960. doi: 10.1016/j.eururo.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 36.Flaig TW, Spiess PE, Agarwal N, et al. Bladder cancer, NCCN evidence blocks. Cancer Discov. 2018;23:311–331. [Google Scholar]
- 37.Balar A V, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien T, Ray E, Singh R, et al. Prevention of bladder tumours after nephroureterectomy for primary upper urinary tract urothelial carcinoma: A prospective, multicentre, randomised clinical trial of a single postoperative intravesical dose of mitomycin C (the ODMIT-C Trial) Eur Urol. 2011;60:703–710. doi: 10.1016/j.eururo.2011.05.064. [DOI] [PubMed] [Google Scholar]
- 39. Rouprêt M, Babjuk M, Burger M, et al: EAU Guidelines on Upper Urinary Tract Urothelial Carcinoma 2018. Arnhem, the Netherlands, European Association of Urology, 2018. [Google Scholar]
- 40.Bagley DH, Rivas D. Upper urinary tract filling defects: Flexible ureteroscopic diagnosis. J Urol. 1990;143:1196–1200. doi: 10.1016/s0022-5347(17)40223-0. [DOI] [PubMed] [Google Scholar]
- 41.Raman JD, Sosa RE, Vaughan ED, Jr, et al. Pathologic features of bladder tumors after nephroureterectomy or segmental ureterectomy for upper urinary tract transitional cell carcinoma. Urology. 2007;69:251–254. doi: 10.1016/j.urology.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 42. Reference deleted. [Google Scholar]
- 43.Babjuk M, Böhle A, Burger M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 44.Newman AM, Bratman S V., To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith ZL, Guzzo TJ. Urinary markers for bladder cancer. F1000Prime Rep. 2013;5:21. doi: 10.12703/P5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lodewijk I, Dueñas M, Rubio C, et al. Liquid biopsy biomarkers in bladder cancer: A current need for patient diagnosis and monitoring. Int J Mol Sci. 2018;19:1–34. doi: 10.3390/ijms19092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyake M, Owari T, Hori S, et al. Emerging biomarkers for the diagnosis and monitoring of urothelial carcinoma. Res Rep Urol. 2018;10:251–261. doi: 10.2147/RRU.S173027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He H, Han C, Hao L, et al. ImmunoCyt test compared to cytology in the diagnosis of bladder cancer: A meta-analysis. Oncol Lett. 2016;12:83–88. doi: 10.3892/ol.2016.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galván AB, Salido M, Espinet B, et al. A multicolor fluorescence in situ hybridization assay: A monitoring tool in the surveillance of patients with a history of non-muscle-invasive urothelial cell carcinoma: A prospective study. Cancer Cytopathol. 2011;119:395–403. doi: 10.1002/cncy.20168. [DOI] [PubMed] [Google Scholar]
- 50. doi: 10.3310/hta14040. Mowatt G, Zhu S, Kilonzo M, et al: Systematic review of the clinical effectiveness and cost-effectiveness of photodynamic diagnosis and urine biomarkers (FISH, ImmunoCyt, NMP22) and cytology for the detection and follow-up of bladder cancer. Heal Technol Assess 14:1-331, iii-iv, 2010. [DOI] [PubMed] [Google Scholar]
- 51.Hatzichristodoulou G, Kübler H, Schwaibold H, et al. Nuclear matrix protein 22 for bladder cancer detection: Comparative analysis of the BladderChek and ELISA. Anticancer Res. 2012;32:5093–5097. [PubMed] [Google Scholar]
- 52.Ellis WJ, Blumenstein BA, Ishak LM, et al. Clinical evaluation of the BTA TRAK assay and comparison to voided urine cytology and the Bard BTA test in patients with recurrent bladder tumors. The Multi Center Study Group. Urology. 1997;50:882–887. doi: 10.1016/s0090-4295(97)00508-6. [DOI] [PubMed] [Google Scholar]
- 53.Leyh H, Marberger M, Conort P, et al. Comparison of the BTA stat test with voided urine cytology and bladder wash cytology in the diagnosis and monitoring of bladder cancer. Eur Urol. 1999;35:52–56. doi: 10.1159/000019819. [DOI] [PubMed] [Google Scholar]
- 54.Ecke TH, Weiß S, Stephan C, et al. UBC Rapid Test for detection of carcinoma in situ for bladder cancer. Tumour Biol. 2017;39:1010428317701624. doi: 10.1177/1010428317701624. [DOI] [PubMed] [Google Scholar]
- 55.Huang YL, Chen J, Yan W, et al. Diagnostic accuracy of cytokeratin-19 fragment (CYFRA 21-1) for bladder cancer: A systematic review and meta-analysis. Tumour Biol. 2015;36:3137–3145. doi: 10.1007/s13277-015-3352-z. [DOI] [PubMed] [Google Scholar]
- 56.O’Sullivan P, Sharples K, Dalphin M, et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J Urol. 2012;188:741–747. doi: 10.1016/j.juro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Kavalieris L, O’Sullivan P, Frampton C, et al. Performance characteristics of a multigene urine biomarker test for monitoring for recurrent urothelial carcinoma in a multicenter study. J Urol. 2017;197:1419–1426. doi: 10.1016/j.juro.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 58.D’Elia C, Pycha A, Folchini DM, et al. Diagnostic predictive value of Xpert Bladder Cancer Monitor in the follow-up of patients affected by non-muscle invasive bladder cancer. J Clin Pathol. 2019;72:140–144. doi: 10.1136/jclinpath-2018-205393. [DOI] [PubMed] [Google Scholar]
- 59.Tan WS, Feber A, Dong L, et al. DETECT I & DETECT II: A study protocol for a prospective multicentre observational study to validate the UroMark assay for the detection of bladder cancer from urinary cells. BMC Cancer. 2017;17:767. doi: 10.1186/s12885-017-3758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forbes SA, Beare D, Bindal N, et al. COSMIC: High-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr Protoc Hum Genet. 2016;91:10.11.1–10.11.37. doi: 10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 61.Nassar AH, Umeton R, Kim J, et al. Mutational analysis of 472 urothelial carcinoma across grades and anatomic sites. Clin Cancer Res. 2019;25:2458–2470. doi: 10.1158/1078-0432.CCR-18-3147. [DOI] [PubMed] [Google Scholar]
- 62.Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stasik S, Salomo K, Heberling U, et al. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin Biochem. 2019;64:60–63. doi: 10.1016/j.clinbiochem.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Russo IJ, Ju Y, Gordon NS, et al. Toward personalised liquid biopsies for urothelial carcinoma: Characterisation of ddPCR and urinary cfDNA for the detection of the TERT 228 G>A/T mutation. Bladder Cancer. 2018;4:41–48. doi: 10.3233/BLC-170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hurst CD, Platt FM, Knowles MA. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur Urol. 2014;65:367–369. doi: 10.1016/j.eururo.2013.08.057. [DOI] [PubMed] [Google Scholar]
- 66. doi: 10.1016/j.cell.2018.07.034. Bailey MH, Tokheim C, Porta-Pardo E, et al: Comprehensive characterization of cancer driver genes and mutations. Cell 173:371-385.e18, 2018 [Erratum: Cell 174:1034-1035, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu D, Plimack ER, Hoffman-Censits J, et al. Clinical validation of chemotherapy response biomarker ERCC2 in muscle-invasive urothelial bladder carcinoma. JAMA Oncol. 2016;2:1094–1096. doi: 10.1001/jamaoncol.2016.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur Urol. 2015;68:959–967. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69a.Desai NB, Scott SN, Zabor EC, et al. Genomic characterization of response to chemoradiation in urothelial bladder cancer. Cancer. 2016;122:3715–3723. doi: 10.1002/cncr.30219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69b.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siefker-Radtke AO, Mellado B, Decaestecker K, et al. A phase 2 study of JNJ-42756493, a pan-FGFR tyrosine kinase inhibitor, in patients (pts) with metastatic or unresectable urothelial cancer (UC) harboring FGFR gene alterations. J Clin Oncol. 2016;34(15_suppl; abstr TPS4575) [Google Scholar]
- 71.Loriot Y, Necchi A, Park SH, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381:338–348. doi: 10.1056/NEJMoa1817323. [DOI] [PubMed] [Google Scholar]
- 72.Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinde I, Papadopoulos N, Kinzler KW, et al. FAST-SeqS: A simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS One. 2012;7:e41162. doi: 10.1371/journal.pone.0041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Azad TD, Chaudhuri AA, Fang P, et al: Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology 158:495-505.e6, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chin R-II, Chen K, Usmani A, et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA) Mol Diagn Ther. 2019;23:311–331. doi: 10.1007/s40291-019-00390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 77.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 79.Bochner BH, Hansel DE, Efstathiou JA, et al. Definitions, in Amin MB, Edge SB, Greene FL, et al (eds): AJCC Cancer Staging Manual (ed 8). Chicago, IL, American College of Surgeons, pp 765-773, 2017. [Google Scholar]