Abstract
Lipidated small GTP-binding proteins of the Arf family interact with multiple cellular partners and with membranes to regulate intracellular traffic and organelle structure. Here, we focus on the ADP-ribosylation factor 1 (Arf1), which interacts with numerous proteins in the Arf pathway, such as the ArfGAP ASAP1 that is highly expressed and activated in several cancer cell lines and associated with enhanced migration, invasiveness, and poor prognosis. Understanding the molecular and mechanistic details of Arf1 regulation at the membrane via structural and biophysical studies requires large quantities of fully functional protein bound to lipid bilayers. Here, we report on the production of a functional human Arf1 membrane platform on nanodiscs for biophysical studies. Large scale bacterial production of highly pure, N-myristoylated human Arf1 has been achieved, including complex isotopic labeling for NMR studies, and the myr-Arf1 can be readily assembled in small nanoscale lipid bilayers (nanodiscs). It is determined that myr-Arf1 requires a minimum binding surface in the nanodiscs of approximately 20 lipids. Fluorescence and NMR were used to establish nucleotide exchange and ArfGAP-stimulated GTP hydrolysis at the membrane, indicating that phophoinositide stimulation of the activity of the ArfGAP ASAP1 is ≧ 2000-fold. Differences in nonhydrolyzable GTP analogues are observed, and GMPPCP is found to be the most stable. Combined, these observations establish a functional environment for biophysical studies of Arf1-effectors and interactions at the membrane.
Graphical Abstract
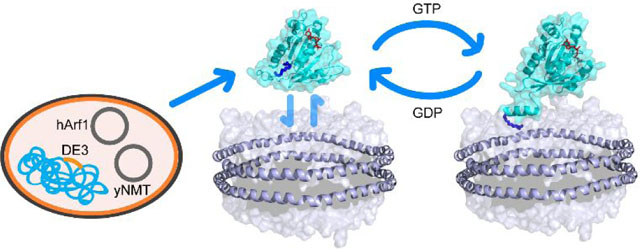
ADP-ribosylation factor-1 (myr-Arf1) is a small myristoylated GTPase which belongs to the Ras superfamily. Like other Ras-related GTP-binding proteins, Arf1 cycles between an inactive GDP-bound form and an active GTP-bound form. Nucleotide exchange is regulated by Sec7-domain containing guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP to GTP, and GTPase-activating proteins (ArfGAPs), which stimulate Arf1 GTPase activity1, 2. Arf1 activation by GDP/GTP exchange is coupled to its recruitment to membranes via its myristoylated N-terminal amphipathic helix3, 4. The exchange of GDP to GTP triggers a conformational switch in Arf1, where the myristoylated N-terminal α-helix dissociates from a surface groove on Arf1 and binds to the membrane surface3–6. When bound to GTP, myr-Arf1 recruits diverse effectors to the membrane to regulate membrane trafficking, organelle structures and actin1, 2. Arf-dependent membrane traffic and actin remodeling are necessary for cell motility and signaling that contribute to invasion and metastasis of cancer cells. ArfGAPs, which bind directly to Arf1, have been found to be abnormally expressed in different cancer cell types and human cancers and is associated with invasion, metastasis and poor prognosis7–17 .
Most structural work on Arf1, apart from an NMR study on yeast Arf1 bound to lipid bicelles5, 6, has been done on a truncated protein missing the myristoylated N-terminal amphipathic helix18–21. Therefore, an understanding of how human myr-Arf1 interacts at the membrane surface with its GEFs and ArfGAPs has remained elusive. Over the last few years, small nanoscale lipid bilayers, or nanodiscs (ND), have emerged as a prevailing lipid bilayer mimicking environment22, 23 , their size, homogeneity and the ability to modulate the lipid composition of the lipid bilayer have allowed the investigation of important structural information on membrane proteins using a variety of biophysical techniques24–28.
Here, we report on the formation and characterization of a fully functional membrane-bound myr-Arf1 platform on nanodiscs for structural and biophysical studies. We show (i) that inducible co-expression of human Arf1 with yeast NMT in E. coli produces milligram quantities of either unlabeled human myr-Arf1 for biochemical studies or isotopically labeled myr-Arf1 for NMR studies, (ii) that the myr-Arf1 protein is fully functional for nucleotide exchange- triggered membrane binding and ArfGAP-stimulated GTP hydrolysis, and (iii) a suitably stable myr-Arf:arfGAP complex can be established at the membrane surface.
Materials and Methods
Construction of expression vectors.
The human Arf1 (hArf1; Uniprot P84077) gene was synthesized (Integrated DNA Technologies, Inc) as a codon-optimized gBlocks fragment, and subcloned (in our lab) into a pET3a vector under T7 promoter control. The clone was subsequently modified by adding a -GSGSHHHHHH-tag at the c-terminus using conventional PCR-cloning techniques. The yeast N-myristoyltransferase (yNMT; Uniprot P14743) gene was synthesized in the same manner (Integrated DNA Technologies, Inc) and subcloned into a pET9b vector, also under T7 promoter control. Both genes were inserted between NdeI and BamHI restriction sites.
For production of the myr-Arf1 protein, hArf1/pET3a and yNMT/pET9b plasmids were co-transformed into the E. coli competent cells BL21 star (Invitrogen). Positive cotransformants were selected on minimal media (M9) agar plates containing both carbenicillin (100 mg/L) and kanamycin (50 mg/L).
Production of the myr-Arf1-HIS protein.
For the production of unlabeled myr-Arf1-HIS, co-transformed BL21 (star) cells were cultured in M9 media [reference] at 37°C until OD600 = 0.8. Sodium myristate (Sigma-Aldrich Cat. No. M8005) was added to a final concentration of 100 μM per liter. Coenzyme-A was added as one-half of a CoA supplement capsule (Coenzyme-A Technologies™) per liter of culture. Following an additional 30 min of growth, 1 mM IPTG was added to induce protein expression, and the cells were incubated for another 4 h at 37°C. The cells were pelleted by centrifugation at 7000 RCF for 30 min. The cell pellet was collected and frozen at −80°C. Production of isotope labelled myr-Arf1-HIS for NMR experiments is as described in the Supplemental section S1.
Purification of Arf1 (Arf1s) from the supernatant of cell lysis.
After the disruption of the frozen cells (Supplemental section S2). The supernatant from lysed cells was collected and purified on a prepacked 5ml Ni-NTA column which was pre-equilibrated by lysis buffer. The desired protein was separated by gradient elution using elution buffer (Tris 20 mM, NaCl 250 mM, MgSO4 1 mM, DTT 1 mM, Imidazole 300 mM, pH8.0 (T20N250M1D1I300)) from 0% to 100% in 15 column volumes. The eluted component was pooled and concentrated to a 2 ml volume, using an Amicon centrifugal device with 10 kDa MWCO (Millipore). The sample was applied to a Superdex S75 16/60 size exclusion column (running buffer: Tris 20 mM, MgSO4 1 mM, DTT 1 mM, pH8.0 (T20M1D1)) for further purification and removal of imidazole and sodium chloride. The eluate was pooled as Arf1s.
Purification of Arf1 (Arf1p) from the pellets of cell lysis.
The pellets obtained following cell lysis were resuspended in resolubilization buffer Tris 20 mM, NaCl 250 mM, MgSO4 1 mM, DTT 1 mM, Imidazole 20 mM, Cholate 25 mM, pH 8.0 (T20N250M1D1I20C25)). The mixture was shaken on an orbital ‘shaker’ at 4°C for 1 day. The soluble fraction was collected after centrifugation at 48000 RCF for 30 min. The supernatant was then applied to a 5 ml Ni-NTA column, which was pre-equilibrated by resolubilization buffer. Gradient elution with elution buffer (T20N250M1D1I300C25, pH 8.0) was used to separate the desired protein. The eluate was collected and concentrated to approximately 2 ml volume. The sample was then loaded on a Superdex S75 16/60 size exclusion column (running buffer T20M1D1, pH8.0) to eliminate all NaCl, cholate and aggregated portion. The eluate was pooled as Arf1p.
Separation of the myr-Arf1-HIS from Arf1-HIS.
The Arf1s and Arf1p samples contained both myr-Arf1-HIS and Arf1-HIS. Arf1s and Arf1p were combined and concentrated to 0.5 ml volume. Ammonium sulfate powder was added to the sample to a final concentration of 0.6 M. After centrifugation at 48000 RCF for 10 minutes, the soluble fraction/supernatant was collected and applied to a pre-equilibrated Phenyl HP 5 ml hydrophobic interaction column using the running buffer T20M1D1 containing 0.6 M (NH4)2SO4. The non-myristoylated and myristoylated Arf1 separated under isocratic flow, wherein myr-Arf1-HIS eluted later than non-myristoylated Arf1-HIS. The myr-Arf1-HIS component was collected and buffer exchanged to buffer condition T20N150M0.5D1 pH 7.4 by dialysis. Samples from cell disruption and purification were analyzed by SDS-PAGE, Mass spectrometry and NMR.
Production of L8K-Arf1.
Production of isotope labeled L8K-Arf1 samples was described in Supplemental sections S3–4. L8K-Arf1 does not have a HIS tag.
Mass spectrometry measurement.
Mass spectra were obtained using Agilent Technologies 6100 Series Single Quadrupole LC/MS. Data deconvolution was performed using OpenLab CDS (Agilent).
Preparation of Membrane Scaffolding Protein (MSP) belt proteins.
The plasmids for MSPs MSP1D1, MSPΔH5, and MSPΔH4,5 were a generous gift of Drs. Franz Hagn and Gerhard Wagner, Harvard Medical School. The proteins were expressed and purified as described previously22.
Preparation of empty NDs.
All lipids were purchased from Avanti Polar Lipids and were mixed in chloroform solutions, then air-dried with nitrogen flow and re-solubilized with cholate in aqueous buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl and 75 mM sodium cholate). NDs were prepared using the phospholipids 16:0–18:1-PC (1-palmitoyl-2-oleoyl-snglycero-3-phosphocholine (POPC)), 16:0–18:1-PS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS)) and 18:1–18:1-PI(4,5)P2 (1-oleoyl-2-oleoyl-snphosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). NDs were assembled by mixing MSP1D1, MSPΔH5 or MSPΔH4,5 with solubilized lipids at a 1:65, 1:45 and 1:20 ratio, respectively, followed by removal of sodium cholate from the mixture with Bio-beads™ SM-2 resin (BIO-RAD). Assembled NDs were then purified via a Superdex-200 size exclusion column (GE Healthcare).
Preparation of myr-Arf1-HIS anchored MSPΔH5 NDs for NMR.
The membrane mimicking ND was formed using MSPΔH522 and phospholipids. To assemble the ND and load the particle with myr-Arf1-HIS, established protocols29 were employed and modified. MSPΔH5 and cholate- solubilized lipids (POPC:POPS:PI(4,5)P2 = 7:2:1) were incubated with myr-Arf1-HIS in Tris 20 mM, NaCl 150 mM, MgSO4 0.5 mM, DTT 1 mM, EDTA 1 mM and GTPγS 2 mM (T20N150M0.5D1E1GTPγS2), pH7.4 with the final cholate concentration of 18 mM. The lipids: MSPΔH5 ratio is 45:1, and the myr-Arf1-HIS:MSPΔH5 ratio was kept at 1:1. After incubation at 4°C for 2 h, 1 g of washed Bio-Beads resin (BIO-RAD) was added to the mixture per ml, and the suspension was rotated at 4°C for another 12 h. The supernatant was then pooled and purified on a pre-equilibrated S200 100/300 size exclusion column using the buffer T20N150, pH 7.4. The main peak was collected and concentrated.
NMR measurements.
NMR samples were prepared as described in Supplemental section S5. All NMR spectra were collected at 25°C on Bruker Avance III 700 and 850 MHz spectrometers equipped with TCI triple-resonance cryoprobes. The 1H-15N HSQC and 1H-13C ILV methyl HMQC spectra were collected for the GDP-bound forms of myr-Arf1-HIS and Arf1-HIS and ND-anchored, GTP-analogue-bound forms of myr-Arf1-HIS. The same experiments were acquired for GDP and GTPγS bound L8K-Arf1. All spectra were processed using TopSpin and NMRPipe30 and analyzed and illustrated by NMRFAM-Sparky31.
NMR diffusion measurements were made using the bipolar gradient LED sequence32, 33 while monitoring the lipid choline methyl resonance of the ND. The diffusion time was 300 msec and gradients were varied from 5 to 45 G2/cm2. Measurements were made at 25°C on a Bruker Avance III 800 MHz spectrometer equipped with a TCI triple-resonance cryoprobe.
Backbone NH and ILV methyl assignments of L8K-Arf1:GTPγS.
The backbone NH resonances of L8K-Arf1:GTPγS were assigned by using a standard set of triple resonance experiments (HNCACB, HN(CO)CACB, HNCA, HN(CO)CA, HNCO, HN(CA)CO). The ILV methyl resonances were assigned by using (H)CC(CO)NH (20 ms mixing time at 30°C) and verified using a 4D methyl – methyl NOESY (200 ms mixing time) spectrum and known crystal structure (PDB ID: 1J2J).
Fluorescence assays.
All fluorescence experiments measurements were performed with a Horiba Fluoromax-4 spectrofluorometer in a 120 μL quartz cell. The sample (140 μL) was thermostated at 22°C. The time constant of the fluorometer was set according to the kinetic studied, down to 500 ms for fast kinetic measurements. The excitation λexc and emission wavelength λem were 297 and 337 nm, respectively. The excitation and emission bandwidth were set to 4 and 10 nm, respectively.
Nucleotide exchange assays.
Nucleotide exchange of purified myr-Arf1-HIS (5 μM) was assessed by monitoring the change in tryptophan fluorescence following addition of EDTA (2 mM) in the presence of 20 μM of GTP or GTPγ-S, which takes advantage of the large difference in fluorescence between the GDP- and GTP-bound forms of Arf proteins4. The reaction was stopped by addition of 1 mM MgCl2.
GAP assays.
ASAP1-PZA (Uniprot Q9ULH1) is a GTPase activating protein (GAP). Induction of hydrolysis of myr-Arf1-HIS•GTP to myr-Arf1-HIS•GDP was determined by following the change in tryptophan fluorescence after addition of 1mM MgCl2, as described previously34. ASAP1-PZA was titrated into the reaction containing ND-bound myr-Arf1-HIS•GTP as a substrate.
Results and Discussion
Expression and purification of isotopically myr-Arf1-HIS.
N-myristoylation of Arf1 is essential for its recruitment to subcellular membranes and its effect on membrane trafficking . N-myristoylation is catalyzed by N-myristoyltransferases (NMTs) and generally occurs cotranslationally after the initial methionine has been cleaved. Previous studies reporting the production of myristoylated proteins generally include co-expression of yeast NMT with the target protein35. This approach produces relatively small amounts of myr-Arf1 (usually 5~15%) relative to Arf1. In order to produce milligram quantities of the myr-Arf1-HIS protein, the genes of human Arf1 (carrying a C-terminal 6-His tag) and yeast NMT (yNMT) were synthesized with optimized E.coli codons and inserted into two separate plasmids, wherein each protein expression is inducible. yNMT was constructed in pET9 with kanamycin resistance, and human Arf1 was cloned in pET19b selected by ampicillin. Both constructs are under control of the T7 promoter, inducible by IPTG. ESI-MS analysis (Figures S1 and S2) identified three Arf1 components with molecular mass of 21886 Da (Component A, theoretical mass 21884 Da), 21805 Da (Component B, theoretical mass 21806 Da) and 21675 Da (component C, theoretical mass 21674 Da). The Δm of 211 ± 2 Da between components A and C is similar to the monoisotopic and average delta masses expected for N-terminal myristoylation (210.1984 and 210.3598 Da, respectively) indicating that component A is myr-Arf1 carrying a 6-His tag (myr-Arf1-HIS) and component C corresponds to the Arf1 processed by methionyl aminopeptidase but not myristoylated. Component B corresponds to a small fraction of Arf1 protein where cleavage of the initiator methionine did not occur. Typically, we found the level of myristoylation to be ≥ 50% of the total Arf1 expression. Interestingly, after cell lysis and centrifugation, the protein partitions equally in the soluble and the pellet fraction. Treatment of the pellet with cholate allowed the recovery of 30% of that sub-fraction. After purification by successive Ni-NTA, Size Exclusion (SEC) and Phenyl HP chromatography, which was inspired by the previous protocol35 (see Materials and Methods and Chart S1 in the supporting information), we typically obtained 4 to 6 mg of myr-Arf1-HIS per liter of culture. 15N and/or Isoleucine, Leucine and Valine 13CH3 methyl sidechain isotopic labeling in a protonated background36, 37 yielded similar amounts of myr-Arf1-HIS. Preparation of deuterated myr-Arf1-HIS reduced the yield by up to 90%. Further work is in progress to improve this labeling protocol. The purified myr-Arf1-HIS is entirely in the GDP-bound form based on NMR spectra.
Myr-Arf1 is fully functional in biochemical assays.
Myr-Arf1 cannot switch to the active GTP-bound state in the absence of membrane. This reflects the fact that the GTP-bound form is stable only when associated to membrane lipids. To assess if myr-Arf1-HIS is fully functional on ND, we followed GDP/GTP exchange and GTP hydrolysis by fluorescence. The experiments take advantage of the difference in tryptophan fluorescence between the GDP- and GTP-bound forms of Arf proteins38 . First, we compared the rate of nucleotide exchange in the presence of ND or lipid vesicles for myr-Arf1-HIS and for the HIS-free protein. Normally, myr-Arf1-HIS would first engage with the membrane in a GDP-bound state, via the myristoyl moiety, followed by an encounter with its GEF and displacement of the nucleotide. Here, nucleotide exchange was triggered by addition of EDTA in the presence of an excess of GTP. As for other GTP-binding proteins of the Ras superfamily, the chelation of magnesium by EDTA accelerates the dissociation of GDP from Arf and, thus, facilitates its replacement by GTP3. Figure 1A shows the tryptophan fluorescence kinetic trace of GDP/GTP exchange for myr-Arf1-HIS in the presence of 500 μM of exposed lipids presented as ND formed with a truncated version of the belt protein MSP1D1 missing helix 5 (MSPΔH5)22. The ND (diameter ~8.2 nm) contain a total of ~70 lipids (~35 lipids per leaflet) as measured by 31P NMR. Kinetic traces obtained in the presence of large unilamellar vesicles (LUVs) were superimposable with those in the presence of ND (data not shown). Analysis of the traces yielded a t1/2 of 80 ±5 s, showing that the membrane surface of a ND does support nucleotide exchange and protein binding equivalent to that of LUVs. In addition, the influence of the C-terminal 6-His tag on the rate of GDP/GTP exchange was investigated by comparing kinetic traces for the tagged and untagged protein in the presence of ND. Indeed, it was previously shown that modifications of Arf1 C-terminal end can affect nucleotide exchange kinetics and interactions with its regulators39. It is observed that addition of the C-terminal 6-His tag only minimally alter the rate of GDP/GTP exchange in vitro (t1/2 =105 ±5 s for the untagged myr-Arf1, data not shown), demonstrating that any interaction between the Arf1 and the membrane is not significantly altered by C-terminal 6-HIS tag. Additional GDP/GTP exchange experiments were conducted in the presence of larger or smaller ND formed with wild type MSP1D1 (110 lipids per ND, 55 lipids per leaflet, diameter ~9.5 nm) or MSP missing helices 4,5 and 6 (MSPΔH4–6, 30 lipids per ND, 15 lipids per leaflet, diameter 6.5 nm), respectively22. Interestingly, while the kinetic trace measured for the larger ND was essentially indistinguishable from ND formed with MSPΔH5, the addition of EDTA in the presence of the smallest ND triggered a much smaller fluorescence increase (~10%), which was unstable. This suggests that stable binding of myr-Arf•GTP to lipid bilayers requires a membrane surface area of 20 lipids (~ 20 nm2) or more. Association of myr-Arf1•GTP to nanodiscs was further characterized by 1H NMR translational diffusion experiments as described in “Material and Methods” (Figure 1B). The diffusion of myr-Arf1-HIS loaded ND is slower than ND alone, indicating anchoring and association of myr-Arf1-HIS with the ND.
Figure 1.
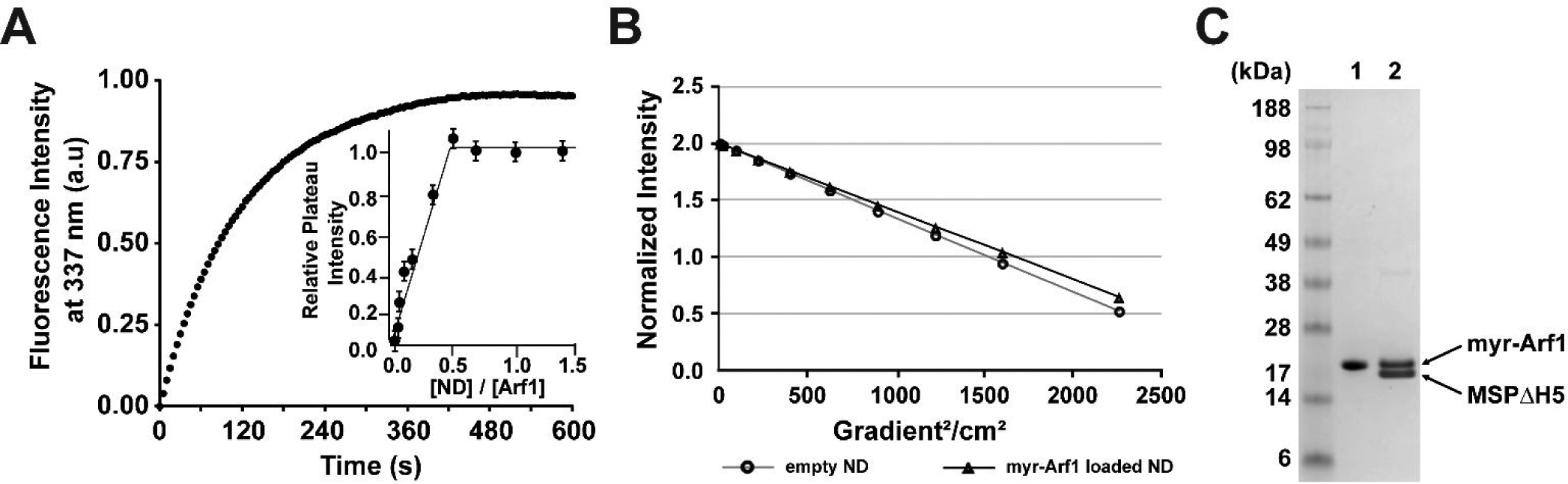
Functional activity and characterization of ND-anchored myr-Arf1. (A). Nucleotide exchange of myr-Arf1•GDP. Representative tryptophan fluorescence kinetics trace of GDP/GTP exchange of myr-Arf1•GDP (5 μM) in the presence of ND containing 0.5 mM of accessible lipids. Nucleotide exchange was triggered by the addition of 2 mM EDTA in the presence of 20 μM GTPγS, at 22°C. Inset: Relative fluorescence intensity of the plateau as a function of the ND: myr-Arf1 ratio. This shows that each ND can bind two myr-Arf1•GTP. (B). 1H NMR translational diffusion measurements on empty (●) and myr-Arf1 loaded (▲) ND. (C). Coomassie stained SDS-PAGE gel of (lane 1) pure myr-Arf1 and (lane 2) MSPΔH5 ND-anchored myr-Arf1 where the myrArf1: ND ratio is 2:1.
We performed additional experiments to probe the number of Arf molecules per ND. To do so, we followed the amplitude of the fluorescence plateau after nucleotide exchange as a function of ND concentration, using MSPΔH5 NDs (diameter ~8.2 nm, 35 lipids per leaflet). Figure 1A, inset shows the relative intensity of the plateau upon titration of ND. At ND/Arf1 ratio of 0.5 and higher, corresponding to two Arf per ND or less, the intensity of the plateau was independent of the ND/Arf ratio indicating that the available membrane surface is enough to support full binding of myr-Arf1-HIS•GTP. Below 0.5, intensity decreased linearly with the ND concentration suggesting that binding is limited by the available lipid area. Since both monolayers of a ND are accessible for interaction, this shows that one MSPΔH5 ND can bind on average two myr-Arf1•GTP, most likely one on each monolayer. The average of two myr-Arf1-HIS to one ND particle stoichiometry was confirmed by Coomassie blue stained SDS-PAGE (Figure 1C). The same experiment repeated with MSP1D1 ND (diameter ~9.5 nm, ~55 lipids per monolayer), showed that 3 to 4 proteins could bind to the larger membrane surface area of those ND (data not shown). These data illustrate that, through control of the ND size and myr-Arf1 ratio, it is possible to construct ND with a single myr-Arf1 per leaflet/side of the ND.
In order to probe if the restricted surface area of a ND allows simultaneous binding of myr-Arf1•GTP and its ArfGAP, we monitored GTP hydrolysis activated by ASAP1 using fluorescence. ASAP1 is an Arf GTPase-activating protein that catalyzes the conversion of Arf•GTP to Arf•GDP. ASAP1 is structurally complex, containing BAR, PH, Arf GAP, Ank repeat, Proline rich and SH3 domains13. Here, we used a recombinant protein composed of the PH, Arf GAP and Ankyrin repeat domains of ASAP1 ([325–724]ASAP1), which is called PZA for PH, Zinc binding (the Arf GAP domain is a zinc binding motif) and Ankyrin repeats. These domains are necessary and sufficient for maximum GAP activity13, 40. We used NDs formed with the belt protein MSPΔH5, purified with size exclusion chromatography, and the size homogeneity was checked with dynamic light scattering. We compared the rate of hydrolysis for the HIS-tagged and untagged myr-Arf1. Figure 2 shows the tryptophan fluorescence kinetic trace of GTP hydrolysis following addition of 10 nM of ASAP1-PZA in the presence of 5 μM of myr-Arf1-HIS•GTP bound to PI(4, 5)P2-containing NDs. ASAP1–stimulated GTP hydrolysis requires phosphatidylinositol 4,5-bisphosphate (PI(4, 5)P2) binding to the cognate PH domain41 . Addition of ASAP1-PZA triggers a decrease of fluorescence indicating hydrolysis of myr-Arf1-HIS•GTP into myr-Arf1-HIS•GDP on the minute time scale. We tested the influence of the presence of PI(4,5)P2 in the nanodisc lipid bilayer on GTP hydrolysis. Fig 2, inset shows the percentage of GTP hydrolyzed after 3 minutes as a function of ASAP1-PZA concentration using NDs with or without PI(4,5)P2. The amount of ASAP1-PZA necessary to hydrolyze 50% of the GTP bound to Arf in the reaction (C50) was estimated. In the absence of PI(4,5)P2, 2.5 ± 0.25 μM of ASAP1-PZA was required. The presence of PI(4,5)P2 increased GAP activity by 2000-fold (C50 = 2 ± 1 nM). The C-terminal 6-His tag did not significantly affect the rate of GTP hydrolysis (C50 = 1.5 ± 1 nM for the untagged protein), indicating that the C-terminal 6-HIS tag does not significantly alter the energetics of myr-Arf1/ASAP1-PZA interactions in a model membrane system. However, while PI(4,5)P2 clearly stimulates GAP activity on NDs, the stimulation is approximately one order of magnitude lower than what was reported in vesicles41. This slower rate of GTP hydrolysis in ND may be the result of a slower rate of complex formation at the ND surface since ASAP1-PZA has to dissociate from ND and reassociate with another ND, compared to lateral diffusion on a surface of a LUV. Alternatively, bound Arf may reduce the effective concentration of PI(4,5)P2 by sequestration, altering the binding of ASAP1. Indeed, it was recently observed that the complex between myr-Arf1 and Brag2 forms interactions with multiple PIP2 lipids, with lipids binding to both partners42. Finally, optimal on- and off- rates may also subtly depend on lipid composition. Elucidation of these mechanisms requires further study.
Figure 2.
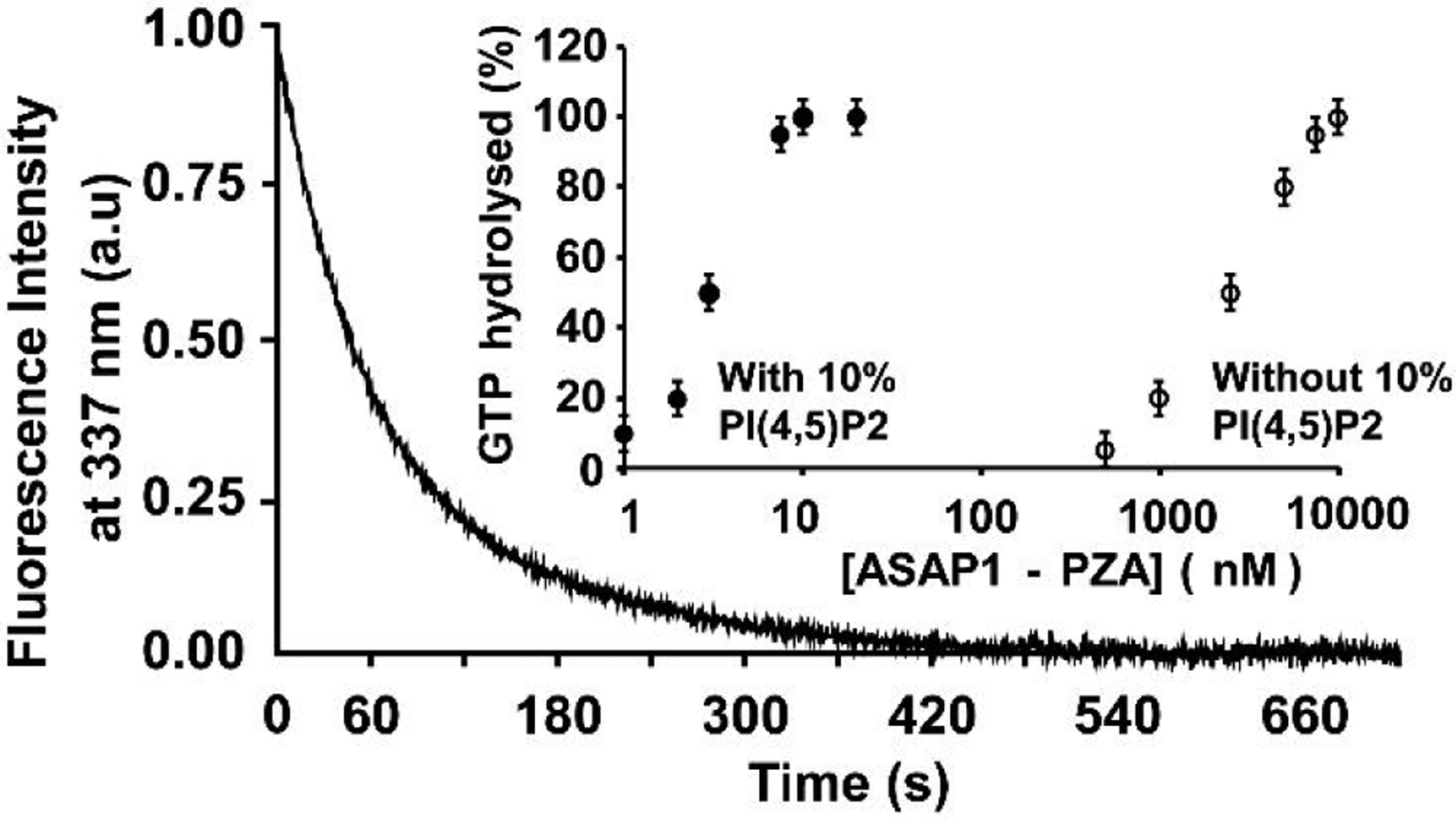
Functional activity of ND-anchored myr-Arf1. GTP hydrolysis of ND-anchored myr-Arf1•GTP by ASAP1-PZA. Representative tryptophan fluorescence kinetic trace of GTP hydrolysis of myr-Arf1•GTP (5μM) by ASAP1-PZA (9 nM). Inset: Effect of PtdIns(4,5)P2 on the relative activity of ASAP1-PZA. PZA was titrated into a reaction containing 5 μM myr-Arf1•GTP as a substrate and ND containing 16:0–18:1 PC only (o) or doped with 10 mol% of 18:1–18:1 PI(4,5)P2 (●). The total concentration of exposed lipids was 500 μM. The percentage of GTP bound to myr-Arf1 hydrolyzed in 3 min is plotted against ASAP1-PZA concentration.
Characterization of Arf1, nucleotide exchange and ASAP1-stimulated hydrolysis by NMR.
Chemical shift changes measured by NMR are a powerful tool to monitor ligand binding and enzymatic reactions with atomic resolution. Here, we used NMR to (i) determine completion of the nucleotide exchange reaction in the presence of ND, (ii) monitor the formation of a functional complex between myr-Arf1-HIS•GTP and ASAP1-PZA at the surface of the ND particle and (iii) characterize the lifetime of the complex as a function of the nucleotide analog. ND-anchored myr-Arf1-HIS•GTP was prepared as described in the material and methods. The MSPΔH5 ND particle, loaded with myr-Arf1-HIS, represents a particle of approximately 110 kD. As anticipated for a molecular system of this size, [1H-15N] TROSY NMR measurements performed on uniformly 15N-labelled protein at 800 MHz and 25°C produced spectra in which virtually all signals were absent. This observation is in contrast to reports for Rheb GTPase domain tethered to NDs43. The differential behavior indicates that the myr-Arf1-HIS is immobilized at the surface compared to the Rheb domain. It is anticipated that overcoming the limitations related to perdeuteration of myr-Arf1-HIS will enable at least partial examination of 15N resonances. However, it is also well known that specific 13C-labeling of the sidechain methyl groups of Isoleucine, Leucine, and Valine can be combined with [13C-1H]-methyl-HMQC NMR methods (also referred to as methyl-TROSY)44 to observe the resonances of these methyl groups in very large biomolecular particles, even for particles >300 kDa. Mazhab-Jafari et al. demonstrated that high quality 13C-HMQC spectra could be obtained for KRAS4B involved in a complex on the surface of a ND45. The methyl-TROSY method takes advantage of the favorable relaxation properties of sidechain methyl groups, and, while it is optimal when combined with perdeuteration, it does function reasonably well in protonated systems. NDs containing 70:20:10 POPC:PS:PI(4,5)P2 and protonated ILV-13CH3-labeled myr-Arf1-HIS•GTPγS yielded high quality 13C-HMQC spectra (Figure 3). At magnetic field strengths corresponding to 1H frequencies of 700 MHz and higher, signals were observed for all 11 Isoleucine (9 were assigned) and for 29 out of 33 leucine and valine residues.
Figure 3.
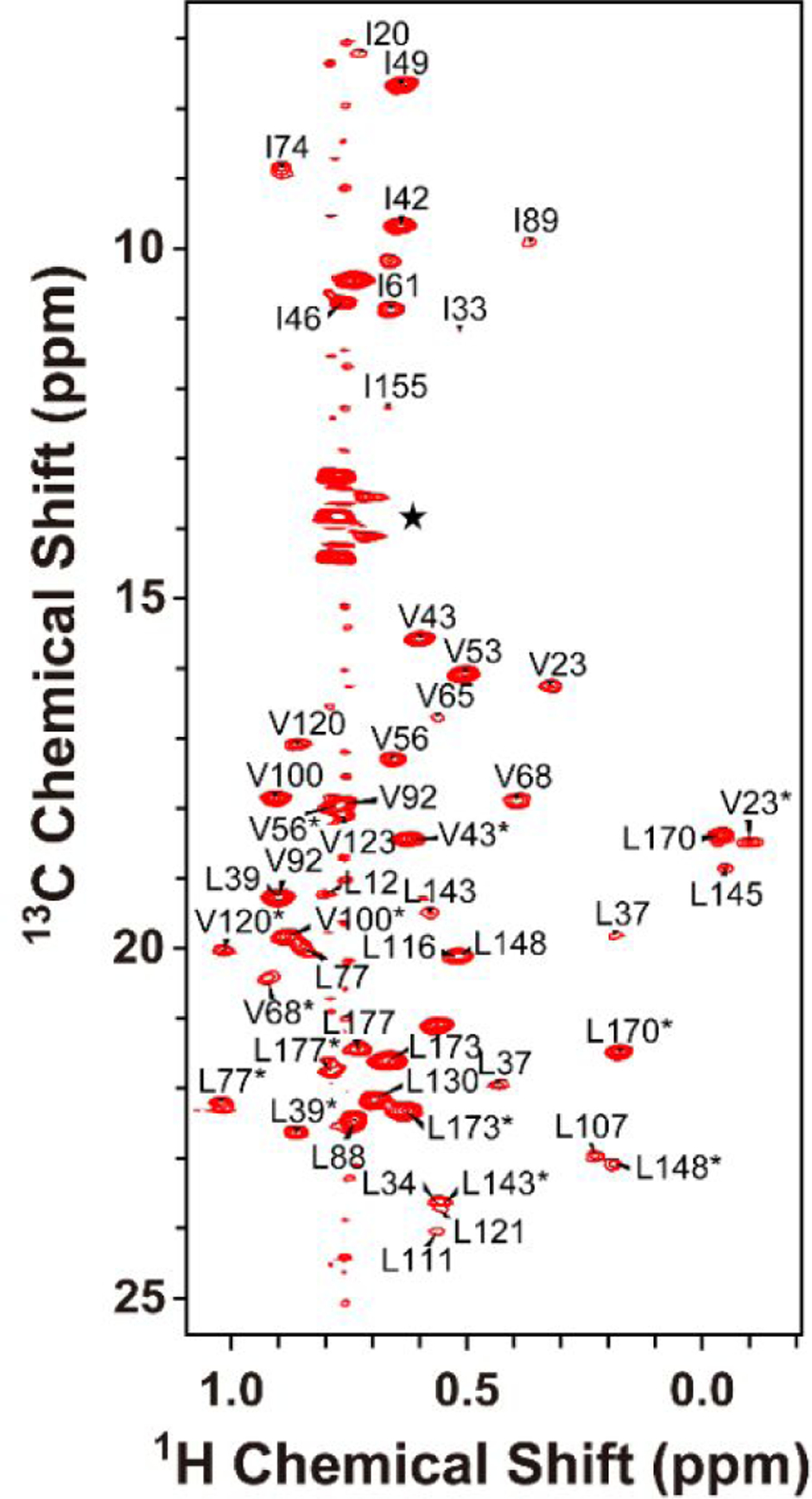
Two-dimensional 1H-13C ILV methyl HMQC spectra of GTPγS bound and ND-anchored protonated-myrArf1: ND. The peaks at approximately 13.5–14.1 ppm (marked with ) arise from natural abundance 13C signals of the terminal methyl moiety of the lipid acyl chains. The Leu and Val resonance assignments are stereospecifically ambiguous and are marked with *.
In order to assign the methyl resonances of myr-Arf1-HIS, we utilized a soluble, non-myristoylated L8K-mutant of Arf1 (L8K-Arf1), which differs from wild type Arf1 in being soluble while bound to either GDP or GTP and analogues of GTP such as GTPγS46. Both forms of L8K-Arf1 yield high quality 1H-15N HSQC spectra. Backbone resonance assignments (Fig 4A) were made for GTPγS-bound L8K-Arf1, using standard triple-resonance methods. The sidechain methyl resonances of Leu, Ile, and Val residues were assigned for deuterated L8K-Arf1 bound to GTPγS (Fig. 4B) (BMRB 27726). We observed that the sidechain resonances, particularly the methyl resonances of Ile, Leu, and Val residues, do not differ significantly between L8K-Arf1•GTPγS and myr-Arf1-HIS•GTPγS forms. Hence, the methyl NMR assignments of L8K-Arf1 enabled transfer and assignment of the methyl resonances of myr-Arf1-HIS•GTPγS when anchored to ND.
Figure 4.
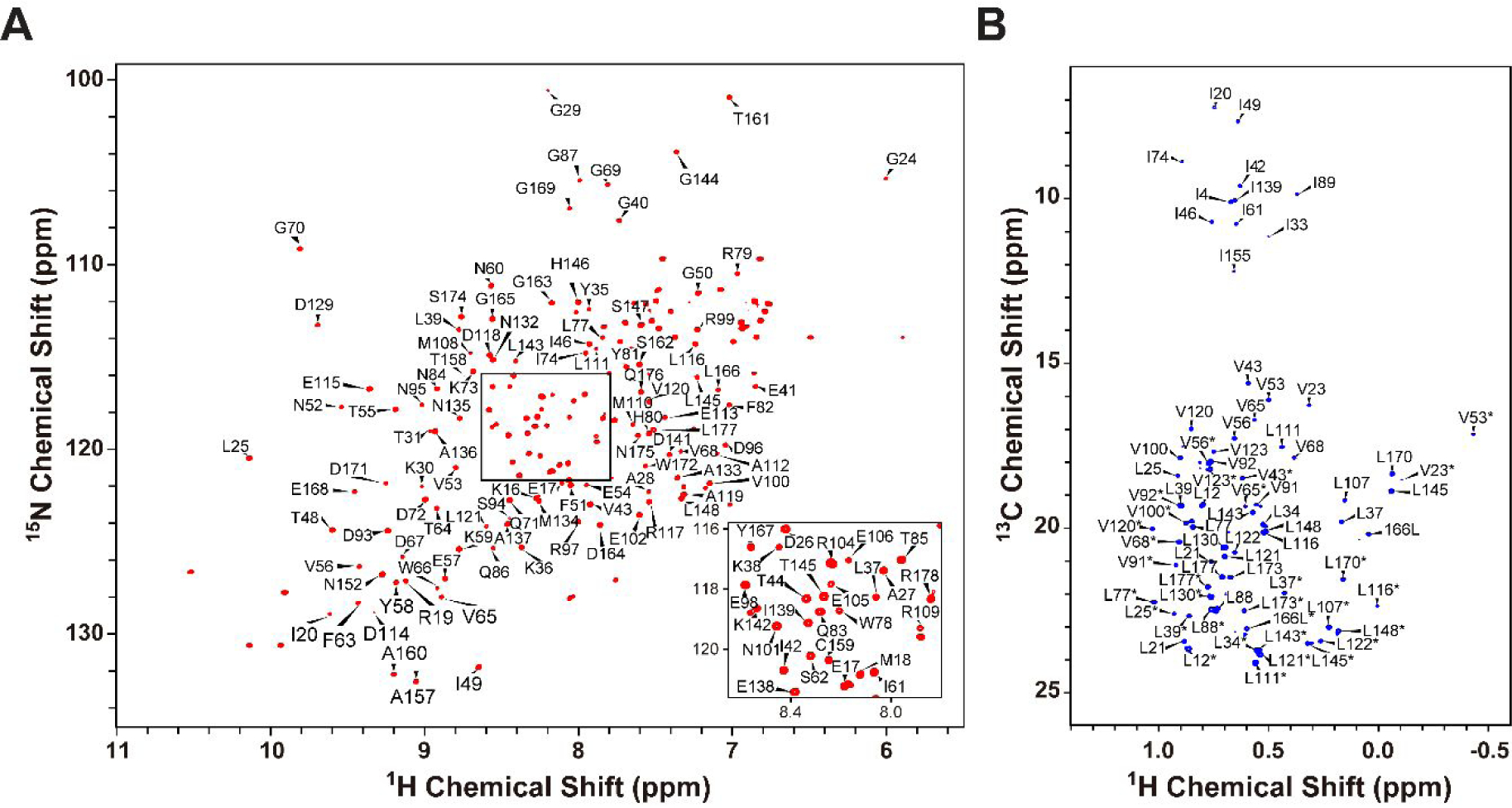
NMR spectra of the GTPγS-bound form of L8K-Arf1. (A) 1H-15N HSQC (B) 1H-13C methyl HMQC. Residue assignments are annotated. The Leu and Val resonances assignments are stereospecifically ambiguous and marked with *.
Methyl-TROSY spectra were recorded for L8K-Arf1 and ND-anchored myr-Arf1 loaded with either GDP, GTPγS, or GMPPCP, a non-hydrolysable GTP analog. The Ile region shows especially well resolved signals (Fig. 5A), which provide clear reporters for the state of bound nucleotide and the kinetics of hydrolysis. The peak corresponding to isoleucine 74 (I74), which is part of switch 2 in Arf1, was used as a reporter for nucleotide exchange. Figure 5B shows expansions from [1H-13C]-methyl-TROSY 2D experiments focused on I74 of ND-anchored myr-Arf1-HIS in the presence of GDP (5B, blue), GTPγS (5B, red) and GMPPCP (5B, green). It is important to note (i) that not only does the position of the peak shift with nucleotide exchange and chemical form of the GTP-analogue, but also (ii) that a single peak is observed for each nucleotide-bound form. These characteristics indicate complete nucleotide exchange and a single homogeneous protein population.
Figure 5.
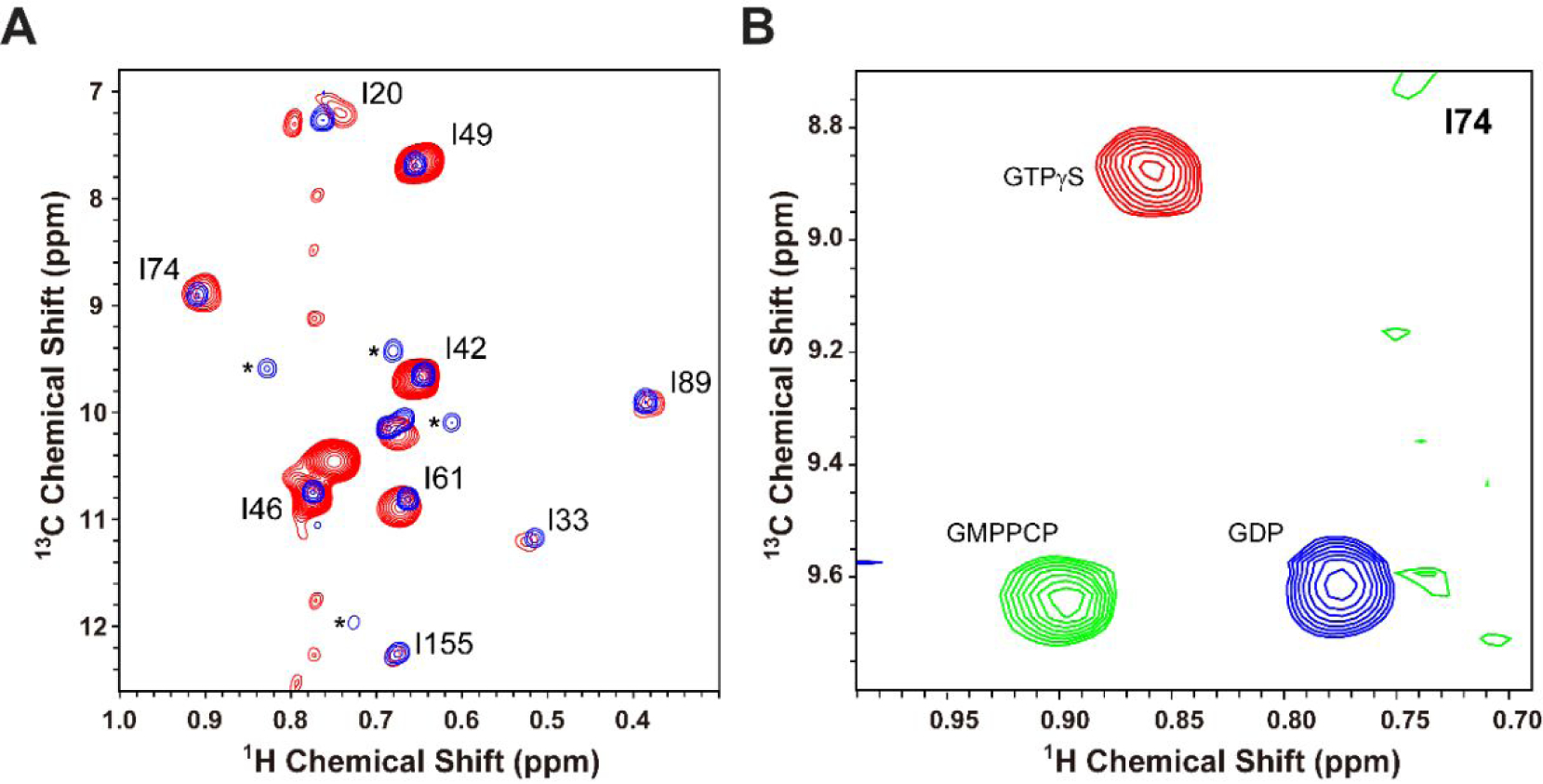
(A) Overlay of the 2D 1H-13C methyl ILV HMQC spectra of GTPγS bound forms of deuterated L8K-Arf1 (blue) and ND-anchored protonated-myrArf1-HIS:ND (red). The peaks marked by * belong to a small residual of the GDP bound form of L8K-Arf1. (B) Overlay of I74 Ile region of the 2D 1H-13C HMQC spectra for ND-anchored myrArf1 with either GTPγS bound (red), GMPPCP bound (green) or GDP bound (blue).
For structural studies of stoichiometric complexes of myr-Arf1:ASAP1-PZA, it is not only critical to know the state of nucleotide-bound myr-Arf1 but also its lifetime. NMR was utilized to monitor nucleotide hydrolysis in situ. The methyl chemical shifts of ND-anchored myr-Arf1-HIS•GTPγS (~200 μM) were monitored over the anticipated time scale of NMR experiments required for complexes (hours), in the presence of a catalytic concentration of ASAP1 (~3 μM for a myr-Arf1-HIS:ASAP1 ratio of ~70:1) (Fig 6). The complex was formed using GTPγS instead of GTP, since the rate of GTPγS hydrolysis was anticipated to be many-fold slower than for GTP. Figure 6 A–C shows spectra centered on I74 collected at time points of 0, 12 h and 36 h. At time 0, only one peak is observed, corresponding to the chemical shift of myr-Arf1-GTPγS, as described previously. At time 12 h, a second peak appeared which corresponds to chemical shift of the GDP-bound form. Finally, at time 36 h, only the peak corresponding to the GDP bound form is observed. These data confirm that a functional complex between myr-Arf1-HIS•GTPγS and ASAP1-PZA forms on the NDs. Complete hydrolysis is achieved within a day. However, this time period is short relative to that required for detailed structural studies. On the contrary, utilization of a GMPPCP analog in similar time course experiments, Figure 6 (D and E), illustrates that there is no hydrolysis of this GTP-analogue following 72 h, even at a higher myr-Arf1-HIS:ASAP1-PZA (10:1). These data indicate that the lifetime of a complex between myrArf1-HIS●GMPPCP:ASAP1-PZA on the membrane surface of NDs is long enough to support structural studies. Structural studies remain extremely challenging due to the large particle size of the complex and will require a range of novel labeling schemes and experimental ingenuity to reveal the nature of the inter-protein interactions and conformational shifts that form the mechanism of action for ArfGAP function.
Figure 6.
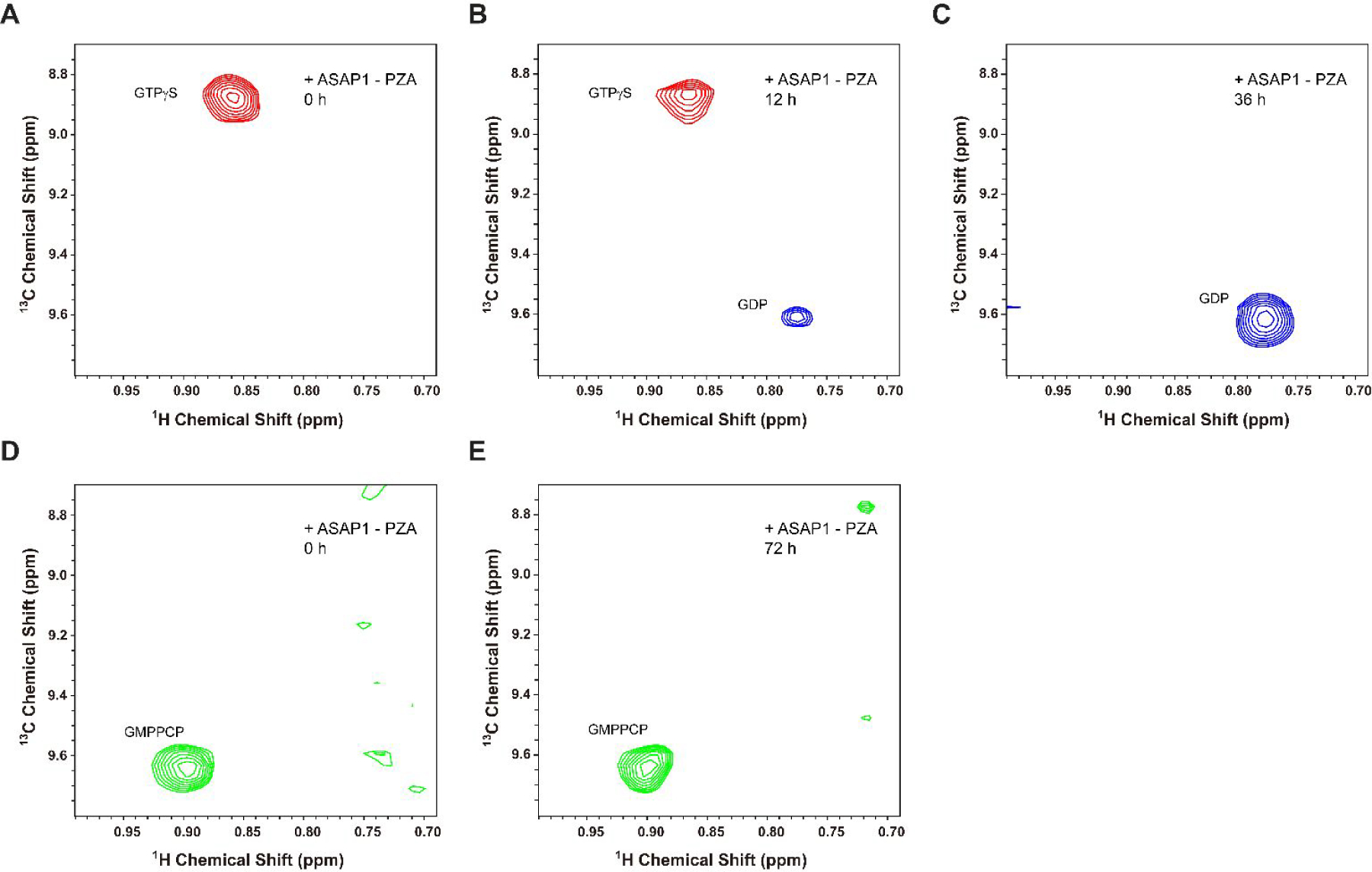
The 2D 1H-13C Ile HMQC spectra of I74 in ND-anchored myrArf1-HIS:GTP bound forms. (A-C) Spectra monitoring the conversion of GTPγS to GDP using a 200 μM myrArf1ND sample buffered at 1 mM Mg2+. Data were collected before (A), 12 h (B), and 36 h (C) after 3 μM ASAP1-PZA was added. (D, E) The 2D 1H-13C Ile HMQC spectra of I74 in ND-anchored myrArf1-HIS:GMPPCP bound form buffered at 1 mM Mg2+. Using a 200 μM myrArf1-HIS:ND sample, spectra were collected before (D) and 72 h (E) after 20 μM ASAP1-PZA was added.
Conclusion
In this work, we describe the efficient production and purification of myristoylated human Arf1. The high expression and extent of myristoylation was achieved by combining (i) inducible over-expression of both human Arf1 and yeast NMT with E.coli codon optimization, (ii) a high level of sodium myristate, and (iii) addition of supplemental coenzyme-A (CoA) in the media. Purification procedures were optimized to include recovery of myr-Arf1 from cell lysis pellets via resolubilization and hydrophobic interaction chromatography, which resulted in high purity and maximum overall yield of human myr-Arf1. The myr-Arf1 is shown to be biologically active and to form a fully functional membrane-bound myr-Arf1 platform on NDs. The minimal context required to establish a functional complex on ND membrane mimetics has been established. NMR spectroscopy has enabled the observation of various forms of myr-Arf1, including the GDP-bound form and various ND-anchored GTP/GTP-analogue bound forms. Methyl-TROSY NMR has enabled monitoring of ASAP1-mediated GTP hydrolysis at the ND surface. These NMR and biophysical methods allow optimization of lipid composition, concentrations and ratios, and ND size to form stable myr-Arf1:ASAP1-PZA complexes. These studies enable the pursuit of structural and interaction studies of myr-Arf1 with its cognate ArfGAP ASAP1 at the surface of a membrane.
Supplementary Material
ACKNOWLEDGMENT
The authors are very grateful for the assistance of Ms. Marzena Dyba and Dr. Sergey Tarasov of the Structural Biophysics Laboratory’s Biophysics Resource.
Funding Sources:
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (Project number ZIA BC011419).
ABBREVIATIONS
- ArfGAP
Arf GTPase-activating protein
- GMPPCP
βγ-Methyleneguanosine 5′-triphosphate
- coA
Coenzyme A
- ESI-MS
Electro Spray Ionization - Mass Spectrometry
- EDTA
Ethylenediaminetetraacetic acid
- GAP
GTPase-activating protein
- IPTG
isopropyl β-D-1-thiogalactopyranoside: IPTG
- LUV
Large unilamellar vesicles
- MSP
Membrane scaffolding protein
- myr-Arf1
N-myristoylated human Arf1
- NMR
Nuclear Magnetic Resonance
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- yNMT
yeast N-myristoyltransferase
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Expanded material and methods sections describing experimental protocols and purification schemes is available as a PDF file.
The NMR assignments for L8K-Arf1 have been deposited in the BioMagResBank (BMRB) as accession number 27726.
REFERENCES
- [1].Gillingham AK, and Munro S (2007) The small G proteins of the Arf family and their regulators, Annu Rev Cell Dev Biol 23, 579–611. [DOI] [PubMed] [Google Scholar]
- [2].Donaldson JG, and Jackson CL (2011) ARF family G proteins and their regulators: roles in membrane transport, development and disease, Nat Rev Mol Cell Biol 12, 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, and Kahn RA (1995) The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch, J Biol Chem 270, 14809–14815. [DOI] [PubMed] [Google Scholar]
- [4].Antonny B, Beraud-Dufour S, Chardin P, and Chabre M (1997) N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange, Biochemistry 36, 4675–4684. [DOI] [PubMed] [Google Scholar]
- [5].Liu Y, Kahn RA, and Prestegard JH (2010) Dynamic structure of membrane-anchored Arf*GTP, Nature structural & molecular biology 17, 876–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu Y, Kahn RA, and Prestegard JH (2009) Structure and membrane interaction of myristoylated ARF1, Structure 17, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T, Manabe T, Wada H, Matsuura N, and Sabe H (2005) Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities, EMBO J 24, 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sato H, Hatanaka KC, Hatanaka Y, Hatakeyama H, Hashimoto A, Matsuno Y, Fukuda S, and Sabe H (2014) High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients, Cell Commun Signal 12, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, Miura K, Yano H, and Onodera Y (2009) The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis, Traffic 10, 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Onodera Y, Nam JM, Hashimoto A, Norman JC, Shirato H, Hashimoto S, and Sabe H (2012) Rab5c promotes AMAP1-PRKD2 complex formation to enhance beta1 integrin recycling in EGF-induced cancer invasion, J Cell Biol 197, 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kahn RA, Bruford E, Inoue H, Logsdon JM Jr., Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, and Cassel D (2008) Consensus nomenclature for the human ArfGAP domain-containing proteins, J Cell Biol 182, 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ha VL, Luo R, Nie Z, and Randazzo PA (2008) Contribution of AZAP-Type Arf GAPs to cancer cell migration and invasion, Adv Cancer Res 101, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, and Randazzo PA (1998) ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src, Mol Cell Biol 18, 7038–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Randazzo PA, Andrade J, Miura K, Brown MT, Long YQ, Stauffer S, Roller P, and Cooper JA (2000) The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton, Proc Natl Acad Sci U S A 97, 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Muller T, Stein U, Poletti A, Garzia L, Rothley M, Plaumann D, Thiele W, Bauer M, Galasso A, Schlag P, Pankratz M, Zollo M, and Sleeman JP (2010) ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients, Oncogene 29, 2393–2403. [DOI] [PubMed] [Google Scholar]
- [16].Hou T, Yang C, Tong C, Zhang H, Xiao J, and Li J (2014) Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer, Int J Clin Exp Pathol 7, 280–287. [PMC free article] [PubMed] [Google Scholar]
- [17].Ehlers JP, Worley L, Onken MD, and Harbour JW (2008) Integrative genomic analysis of aneuploidy in uveal melanoma, Clin Cancer Res 14, 115–122. [DOI] [PubMed] [Google Scholar]
- [18].Amor JC, Harrison DH, Kahn RA, and Ringe D (1994) Structure of the human ADP-ribosylation factor 1 complexed with GDP, Nature 372, 704–708. [DOI] [PubMed] [Google Scholar]
- [19].Goldberg J (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching, Cell 95, 237–248. [DOI] [PubMed] [Google Scholar]
- [20].Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, and Wakatsuki S (2003) Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport, Nat Struct Biol 10, 386–393. [DOI] [PubMed] [Google Scholar]
- [21].Amor JC, Horton JR, Zhu X, Wang Y, Sullards C, Ringe D, Cheng X, and Kahn RA (2001) Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases, J Biol Chem 276, 42477–42484. [DOI] [PubMed] [Google Scholar]
- [22].Hagn F, Etzkorn M, Raschle T, and Wagner G (2013) Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins, J Am Chem Soc 135, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Denisov IG, and Sligar SG (2017) Nanodiscs in Membrane Biochemistry and Biophysics, Chem Rev 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao Y, Cao E, Julius D, and Cheng Y (2016) TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action, Nature 534, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bayburt TH, Leitz AJ, Xie G, Oprian DD, and Sligar SG (2007) Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins, J Biol Chem 282, 14875–14881. [DOI] [PubMed] [Google Scholar]
- [26].Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, and Tidow H (2018) Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs, Structure 26, 1072–1079 e1074. [DOI] [PubMed] [Google Scholar]
- [27].Rouck JE, Krapf JE, Roy J, Huff HC, and Das A (2017) Recent advances in nanodisc technology for membrane protein studies (2012–2017), FEBS Lett 591, 2057–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barnaba C, and Ramamoorthy A (2018) Picturing the Membrane-assisted Choreography of Cytochrome P450 with Lipid Nanodiscs, Chemphyschem 19, 2603–2613. [DOI] [PubMed] [Google Scholar]
- [29].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, and Sligar SG (2009) Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs, Methods Enzymol 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- [31].Lee W, Tonelli M, and Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Altieri AS, Hinton DP, and Byrd RA (1995) Association of Biomolecular Systems via Pulsed Field Gradient NMR Self-Diffusion Measurements, Journal of the American Chemical Society 117, 7566–7567. [Google Scholar]
- [33].Wu DH, Chen AD, and Johnson CS (1995) An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses, Journal of Magnetic Resonance, Series A 115, 260–264. [Google Scholar]
- [34].Che MM, Nie Z, and Randazzo PA (2005) Assays and properties of the Arf GAPs AGAP1, ASAP1, and Arf GAP1, Methods Enzymol 404, 147–163. [DOI] [PubMed] [Google Scholar]
- [35].Ha VL, Thomas GM, Stauffer S, and Randazzo PA (2005) Preparation of myristoylated Arf1 and Arf6, Methods Enzymol 404, 164–174. [DOI] [PubMed] [Google Scholar]
- [36].Tugarinov V, and Kay LE (2005) Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins, Chembiochem 6, 1567–1577. [DOI] [PubMed] [Google Scholar]
- [37].Tugarinov V, Kanelis V, and Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy, Nat Protoc 1, 749–754. [DOI] [PubMed] [Google Scholar]
- [38].Jian X, Brown P, Schuck P, Gruschus JM, Balbo A, Hinshaw JE, and Randazzo PA (2009) Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1, J Biol Chem 284, 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jian X, Cavenagh M, Gruschus JM, Randazzo PA, and Kahn RA (2010) Modifications to the C-terminus of Arf1 alter cell functions and protein interactions, Traffic 11, 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kam JL, Miura K, Jackson TR, Gruschus J, Roller P, Stauffer S, Clark J, Aneja R, and Randazzo PA (2000) Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. Evidence for the pleckstrin homology domain functioning as an allosteric site, J Biol Chem 275, 9653–9663. [DOI] [PubMed] [Google Scholar]
- [41].Jian X, Tang WK, Zhai P, Roy NS, Luo R, Gruschus JM, Yohe ME, Chen PW, Li Y, Byrd RA, Xia D, and Randazzo PA (2015) Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1, Structure 23, 1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Karandur D, Nawrotek A, Kuriyan J, and Cherfils J (2017) Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane, Proc Natl Acad Sci U S A 114, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mazhab-Jafari MT, Marshall CB, Stathopulos PB, Kobashigawa Y, Stambolic V, Kay LE, Inagaki F, and Ikura M (2013) Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc, J Am Chem Soc 135, 3367–3370. [DOI] [PubMed] [Google Scholar]
- [44].Kay LE (2011) Solution NMR spectroscopy of supra-molecular systems, why bother? A methyl-TROSY view, J Magn Reson 210, 159–170. [DOI] [PubMed] [Google Scholar]
- [45].Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, and Ikura M (2015) Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site, Proc Natl Acad Sci U S A 112, 6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Luo R, Jacques K, Ahvazi B, Stauffer S, Premont RT, and Randazzo PA (2005) Mutational analysis of the Arf1*GTP/Arf GAP interface reveals an Arf1 mutant that selectively affects the Arf GAP ASAP1, Current biology : CB 15, 2164–2169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


