Abstract
Regeneration is the process by which organisms replace lost or damaged tissue, and regenerative capacity is highly context-dependent. Tissue regeneration shares certain hallmarks of embryonic development in that lineage-specific factors can be repurposed upon injury to initiate morphogenesis; however, many differences exist between regeneration and embryogenesis. Recent studies of regenerating tissues in laboratory model organisms such as acoel worms, frogs, fish, and mice have revealed that chromatin structure, dedicated enhancers and transcriptional networks are regulated in a context-specific manner to control key gene expression programs. A deeper mechanistic understanding of the gene regulatory networks of regeneration pathways might ultimately enable their targeted reactivation to treat human injuries and degenerative diseases. In this Review, we consider regeneration of body parts across a range of tissues and species to explore common themes and potentially exploitable elements.
ToC blurb
The capacity to regenerate tissue varies across different species and tissue types. The poor regenerative capacity of organs such as the heart and nervous system contribute to the etiology of a number of serious diseases such as heart failure and Alzheimer’s disease, respectively. In this Review, Goldman and Poss discuss how genetic programs of regeneration are regulated, and how these control mechanisms might be adapted to treat human disease
Introduction
Regeneration is the replacement of tissue in homeostasis or following trauma. Across the animal kingdom, there is remarkable diversity in the capacity for tissue regeneration. Many non-mammalian vertebrates — in particular, teleost fish and urodele amphibians such as the axolotl — possess a generally elevated regenerative potential and can regenerate whole limbs, large pieces of heart, or even a fully transected spinal cord1. Some invertebrate animals, such as Hydra and planarians, are so effective at regeneration that entire organisms can regenerate from tiny body fragments1. In mammalian species, including humans, some tissues have high regenerative potential throughout life, including skeletal muscle, liver, intestinal epithelium, skin, and blood, up to a certain threshold of damage or loss2–5. However, several organ systems, including the brain, spinal cord, heart, and joints, possess minimal regenerative capacity. In humans, this lack of regenerative ability contributes to the etiology of debilitating diseases such as heart failure and neurodegeneration; therefore, understanding the mechanisms and diverse phenotypes of regeneration may lead to novel treatments for these diseases.
Organs and appendages are complex tissue structures composed of sometimes hundreds of cell populations and subpopulations. Successful regeneration therefore requires programs of repatterning [G] to rebuild complex tissue structures from unique primordia. Some tissues like blood, skeletal muscle, and hair follicles of skin utilize a specialized stem cell to regenerate new cells, typically affiliated with a niche that maintains quiescence until homeostatic or injury-based division and differentiation. Other cell types like the heart muscle cells of adult zebrafish or neonatal mice, hepatocytes, and the endothelial cells that line blood vessels, self-duplicate after injury. Thus, the genetic control mechanisms involved in regeneration will differ for each cell type within an organ and for its corresponding mode of regeneration (Fig. 1).
Figure 1. A comparison between patterning in embryogenesis and regeneration events.
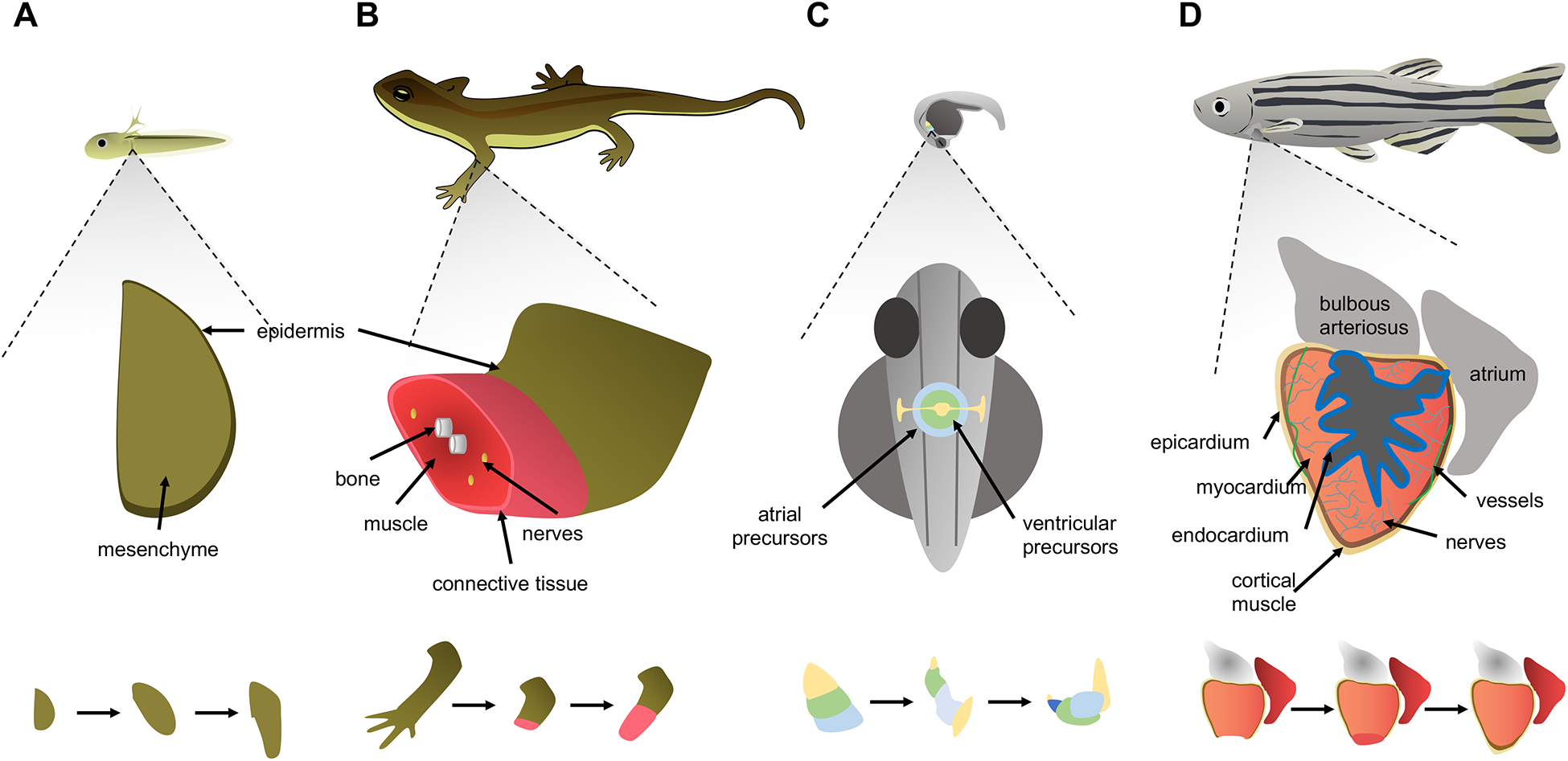
A. Growth of forelimb buds in the larval newt begins with an early bud stage before skeletal elements form, indicating a simple developmental primordium that grows and becomes patterned as the entire organism grows. Amputation stimulates the formation of a primordium called a blastema from remaining cells, which similar to a limb bud, grows and is patterned into mature skeletal elements, connective tissue, glands, nerves, skeletal muscle and epidermis. B. During zebrafish embryogenesis, mesoderm-derived cells destined to form the cardiac atrium and ventricle are arranged in a ring or cone pattern, prior to patterning into chambers and acquiring contractile activity. An adult zebrafish ventricle is comprised of multiple types of innervated and vascularized heart muscle surrounded by an outer layer of epicardial cells and coated internally by a layer of endocardial cells. After resection of the ventricular apex, spared cardiomyocytes divide and non-muscle tissues are re-established. Injury is indicated by a lightning bolt icon.
Over the past 10–15 years, new techniques for purifying and transplanting cells, and the ability to permanently label and map the fate of cells in situ have led to a number of critical breakthroughs in regeneration research. Recent work has defined the cellular sources of regenerating tissues, showing that distinct stem cell and non-stem cell populations are involved in regeneration in a tissue-specific and context-specific manner6–10. Further work has elucidated molecular mechanisms for regeneration that are adapted from, or distinct from, pathways used in the initial construction of the corresponding embryonic tissue. Molecular ‘triggers’ and ‘brakes’ for normal regeneration are primary targets for manipulation, with the ultimate purpose of initiating new tissue growth in human organs with poor innate regenerative capacity. A major challenge in defining the molecular machinery behind regenerative events has been the lack of tools for conditionally manipulating specific genes in a tissue-specific manner in adult organisms. In recent years, new tools such as CRISPR that enable targeted genome editing have emerged to allow the manipulation of regeneration pathways in virtually every model system. A growing number of research groups are investigating regeneration and elucidating the crucial functions of essential molecular players, thus revealing elegant new concepts and mechanisms of regeneration. However, it is likely that the current mechanistic models for the regeneration of most tissues are incomplete.
In this Review, we discuss what is known regarding the molecular mechanisms controlling gene expression programs of regeneration. We consider the regeneration of body parts across a range of tissues and species, as these varied contexts and underlying mechanisms offer unique insights into the regeneration process (model organisms commonly used to study regeneration are outlined in Box 1). Concepts and mechanisms identified from experiments in key supporting tissues, such as vascular cells or cultured stem cells, are reviewed extensively elsewhere3,11–13. We discuss the DNA regulatory elements that interact with dynamic genes and the chromatin regulators and transcription factors (TFs) that instruct gene expression. We also explore novel gene regulatory mechanisms of regeneration, and conclude by considering future advances needed for clinical applications.
Box 1: Model organisms for regeneration.
Many animal model systems are employed to study tissue regeneration, each with advantages and disadvantages153. Certain planarian flatworms can regenerate all anterior and posterior structures from a tissue fragment, based on populations of stem cells called neoblasts. This is a robust model for interrogating stem cell involvement in regeneration and can accomodate loss-of-function experiments by RNAi. Stable transgenesis for planarians has been challenging to develop, but its arrival will enable researchers to visualize cell populations in live animals and perform gain-of-function studies. Hydra exhibit whole-body regeneration and offer similar advantages as planarians, albeit with less tissue complexity and with transgenesis techniques available. Drosophila researchers have any and all molecular genetic tools at their disposal, although the number of tissues with high regenerative capacity is lower than other invertebrate models.
Among vertebrates, anuran models like Xenopus laevis and Xenopus tropicalis enable visual and manipulatable studies of limb, tail, or spinal cord regeneration in tadpoles. Gene editing and transgenesis are available, though generation times in these models are several months long (~12 months in X. laevis and ~4 months in X. tropicalis). An interesting advantage of these models is the ability to study and manipulate the progressive loss of regenerative capacity during development – comparable to what happens in mammals. Zebrafish, a popular teleost fish model, can regenerate a whole host of adult tissues, including fins and scales, as well as those with therapeutic relevance such as the spinal cord, pancreatic β-cells, heart, kidney, liver, retinae and brain. With the advent of CRISPR, few genetic tools remain untapped for zebrafish, although the generation time of ~3 months is no shorter than that of mice. Salamanders are also highly regenerative. Newts and axolotls regenerate many of the same critical tissues as zebrafish and possess the spectacular ability to regenerate limbs. Genome sequences for certain salamander species are now available. Axolotls have the shortest generation times (12 months or less) and are most amenable to transgenesis and gene editing techniques. Finally, while limitations in mammalian regenerative capacity can challenge the ability to study innate regeneration in several key adult tissues, mice, like humans, are able to regenerate a large array of tissues, particularly young, healthy individuals. The diminution of regenerative capacity in tissues like the heart, skeletal muscle, and blood with development or aging provides new opportunities for comparison and intervention. The powerful toolset available in mice enables practically any question to be asked.
Expression programs of regeneration
Hundreds to thousands of genes change expression levels as tissues transition from an uninjured structure to various stages of regeneration. These rapid changes orchestrate injury responses such as inflammation and wound healing, followed by morphogenetic events such as source cell division and patterning [G]. As regeneration completes, these programs largely return to a pre-injury state.
Regeneration deploys genes that function in embryogenesis.
A dominant feature across regeneration is the activation of genes that are critical during embryogenesis and typically drive proliferative, migratory or morphogenetic functions. The activation of embryogenesis genes has been described for most regeneration models, including for heart and retinal regeneration in zebrafish6,14,15 (with cell sources of cardiomyocytes and Müller glial cells, respectively) and for mandibular (resident stem cells) and liver regeneration (hepatocytes) in mammals16. Moreover, gene expression programs in regeneration can share features with those seen in embryogenesis. As an example, adult cardiomyocytes involved in regeneration in zebrafish take on a gene expression profile akin to an embryonic cardiomyocyte, with a higher degree of glycolytic gene expression and glucose uptake, and with a lower expression of mitochondrial genes, than cardiomyocytes distant from a site of active regeneration (Fig. 2a)17. It should be noted that those conclusions may be skewed toward the most highly expressed transcripts — those involved with metabolism — and do necessarily represent the entirety of the RNA-seq profile.
Figure 2. Relationship between genetic programs of regeneration and embryogenesis.
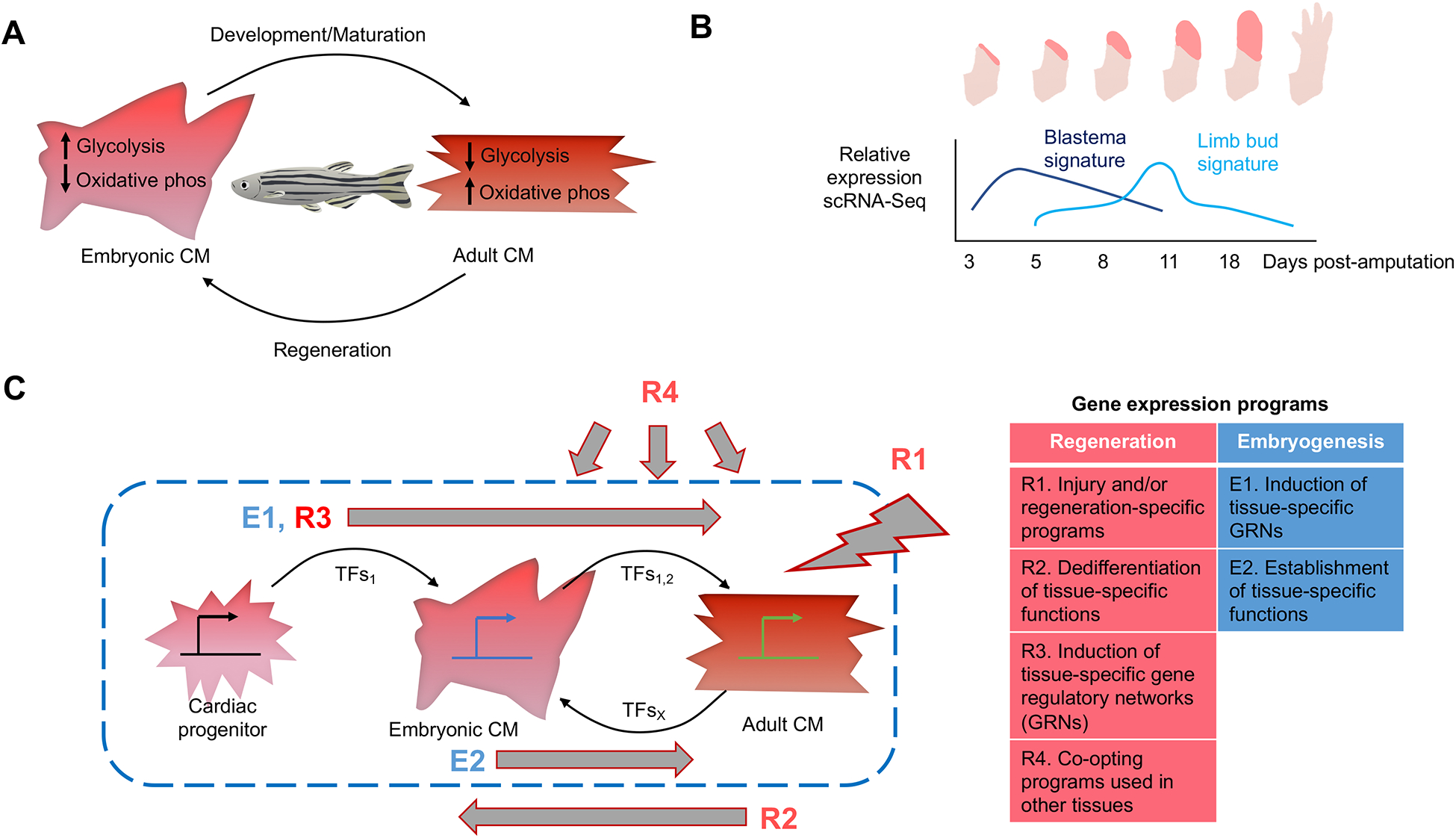
A. Heart regeneration in zebrafish is characterized by a return to certain embryonic gene expression programs17. Cardiomyocyte maturation from the embryo to the adult results in a switch from glycolytic metabolism to mitochondrial respiration (OXPHOS). During regeneration of cardiomyocytes, cells revert to a more glycolytic state through upregulating the expression of glycolytic genes B. Time series showing the biphasic gene expression response during regeneration of axolotl limbs after amputation22. Gene expression levels in sorted connective tissue cells indicate a distinct gene profile in early regenerates that is specific to blastemal generation (dark blue). A separate signature similar to that of embryonic limb buds is expressed in later regenerates (light blue). C. A model integrating gene expression programs relevant to regeneration, visualized for cardiac muscle cells. Embryonic gene programs utilize cascades of tissue-specific or tissue-preferred transcription factors, establishing tissue-specific functions. Regeneration involves defining injury signals, programs for de-differentiation of adult cell functions to approximate a less mature state, and employment of a tissue-specific gene regulatory network. Gene expression programs can also be co-opted from other tissue types or disease contexts. Transcription factors help establish and stabilize each state.
In contrast, only a few genes have been implicated solely in regeneration to date. One such gene is the salamander-specific factor Prod118–21. A reason why there are few regeneration-specific genes may be because adult regenerative events occurring after reproductive stages are not likely to be subject to natural selection pressures as strongly as those influencing ontogenetic [G] development.
Regeneration programmes have unique signatures.
Adult tissues are of a different scale and complexity than their embryonic primordia or early patterned form (Fig. 1), and homeostatic or injury-induced regeneration of tissues such as skeletal muscle, olfactory neurons or blood rely on adult stem cell populations not present in the embryo. Massive, rapid, genome-wide changes in gene expression from the basal gene expression program of an adult structure are required for an injured tissue to re-initiate growth and repatterning. Although genes employed for embryonic development are often repurposed for regeneration, evidence indicates that the underlying genetic programmes are nevertheless distinct.
Methods to collect and interpret changes in gene expression are rapidly evolving and increasing in their power to define regeneration programs. Recently, lineage tracing experiments were elegantly combined with single-cell RNA sequencing (scRNA-Seq) to assess the molecular trajectories of cell subpopulations in different regeneration contexts. In axolotl salamander limbs, which form a proliferative blastema [G] after amputation that acquires the pattern of the limb, connective tissue cells from heterogenous cell types dedifferentiate [G] into a homogeneous population of progenitors, with expression profiles resembling that of connective tissue cells of the early limb bud22. scRNA-Seq identified a gene expression signature for regeneration that was distinct from, and activated prior to, the emergence of the limb bud profile22. It is likely that this signature and the genes that it influences are necessary for forming an embryonic-like precursor (Fig. 2b). Regeneration-specific gene programs based on the assessment of gene expression profiles at multiple stages of regeneration were also reported in studies of tail regeneration in frog tadpoles23, sea star limb regeneration24, and in digit tip regeneration in mice25. This finding indicates that, although embryonic programs are often employed during regeneration, cells appear to transition through regeneration-specific stages.
Regeneration programs reflect features of other tissues and contexts.
A number of features differ among tissues and are expected to influence regeneration programs, including the inflammatory environment, distinct tissue tensions, and differences in vascularization, innervation, and exposure to circulating metabolites and hormones. Whereas one cell type might revert toward programs representative of embryonic development, the regeneration programs in other cell types may more closely resemble pathways found in diseased or in altogether distinct tissues. For example, TFs such as Homeobox protein Nkx-2.5, GATA4, HAND2, and T-box protein 5 (TBX5) that direct the specification and differentiation of early cardiac progenitors in the heart26 are induced in and/or can modulate heart muscle regeneration in zebrafish27,28, yet these or similar factors are induced in or are important for hypertrophy of muscle cells after cardiac injury in adult mammals29,30. Moreover, activation of early developmental programs is a hallmark of cancer31. Thus, although repurposing established gene expression programs to build new tissue in the adult is a feature of regeneration, it appears to be just one piece of the puzzle (Fig. 2c). These reused programs must be regulated in a context-specific manner and then integrated with specialized gene expression signatures and morphogenetic programs that are specific to adult cell types to enable regeneration.
DNA regulatory elements in regeneration
Regeneration programs are the combined output of the activity of DNA regulatory elements, TFs and chromatin regulators. Profiling of chromatin marks indicative of active gene regulation have revealed that changes in enhancer activity occur at thousands of sequence regions during regeneration. Thus, there is likely to be a huge number of active and potentially important regulatory sequences with differential activity during regeneration. Enhancer discovery [G] assays rely both on sufficiency and necessity experiments each with their caveats32. For example, a particular enhancer may function only with a select group of promoters, limiting ectopic reporter validation. Functional redundancy among several enhancers linked to a single gene can mask any in vivo role of a regulatory element upon genetic deletion33,34.
Enhancer elements dedicated to tissue regeneration.
It was recently demonstrated that gene expression during tissue regeneration is controlled by enhancers in a context-dependent manner35. By mapping dynamic histone modifications using chromatin immunoprecipitation sequencing (ChIP-seq) on samples from regenerating zebrafish hearts, a transcriptional enhancer that directs gene expression during regeneration, but that has no detectable activity in developing embryos or uninjured adults, was identified upstream of the gene encoding Leptin b (lepb)35.
A handful of studies have validated individual enhancers as preferential for or specific to regeneration using in vivo tests in transgenic animals. Investigations of such tissue regeneration enhancer elements (TREEs) in zebrafish heart regeneration35–37 have identified TREEs that direct gene expression only after injury and maintain expression for weeks during ongoing regeneration (Fig. 3). Other TREEs have been identified in studies of Drosophila melanogaster imaginal disc regeneration38 and many more have been inferred from profiles of the dynamic accessibility of chromatin during regeneration of worms, frogs and plants39–41. Recent loss-of-function experiments in mice revealed that separate intronic regulatory elements were required respectively for the expression of the haematopoietic transcription factors SAMD14 and GATA2 in red blood cell regeneration during anemia42,43. To our knowledge, only one enhancer that is both necessary and sufficient for a regeneration event has been described. The BVR-B element that drives wingless expression after injury to the imaginal disc is preferentially repressed during adulthood through repressive chromatin regulation, which is discussed in a forthcoming section44.
Figure 3. Discovering DNA regulatory elements involved in regeneration.
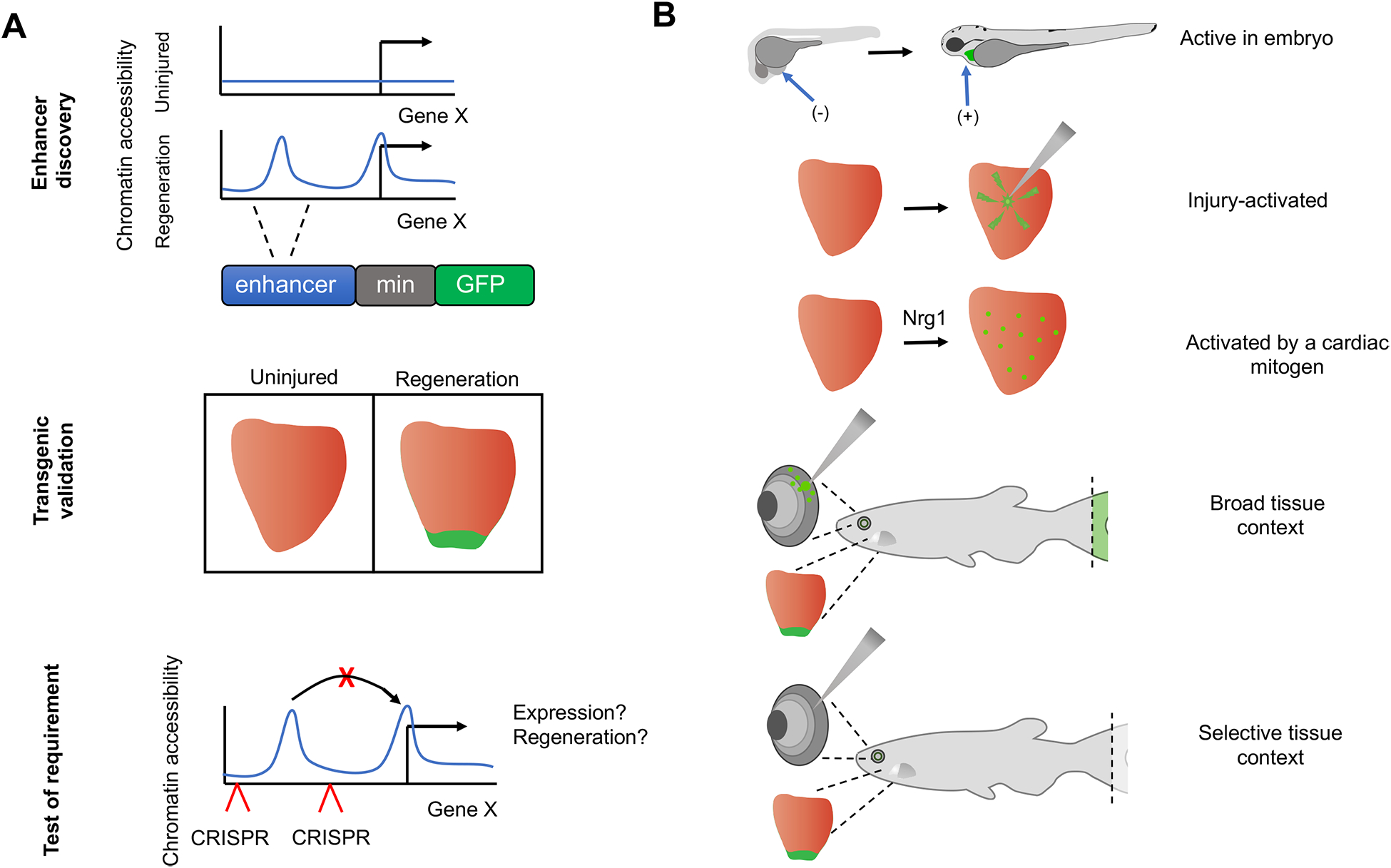
A. Tissue regeneration enhancer element (TREE) discovery typically begins with identification of a sequence region enriched for active or open chromatin marks in regenerating samples versus uninjured samples. Test DNA regions (blue) are subcloned upstream of a minimal promoter (gray) directing a reporter gene, such as GFP. In this example shown for a regenerating heart, stable transgenic lines only express GFP during regeneration. In vivo validation experiments delete the test region from the genome using CRISPR techniques and determine resulting changes in gene expression or regenerative events. B. TREEs validated in reporter experiments are activated in a context-specific manner. An enhancer might direct expression during embryonic or regenerative development. Enhancer activity can be induced by injury or by tissue creation, which can be discerned by testing expression in the presence of a mitogenic trigger such as Nrg1. Enhancers might direct expression in multiple regeneration contexts, for example regenerating fins, retinae, and heart, or instead might be tissue-specific. GFP reporter expression in represented in green.
Inputs and implications of TREEs.
There are many possible influences on and components of regeneration in complex tissues, such as mechanical tension, inflammation, interactions with the extracellular matrix (ECM) and growth factors, cell dedifferentiation, cell division, and tissue patterning, each of which is likely controlled by distinct TFs. These influences and components vary between tissue types, and therefore different tissues presumably engage different core TFs. Different enhancers appear to have different activation kinetics and stimuli, with some, for example, responsive only in certain tissue types, or with different responses to injury and cell proliferation36 (Fig. 3a). For instance, the lepb-linked enhancer in zebrafish is responsive to fin or heart injury through the activity of different adjacent sequences35. An enhancer linked to runx1 is also responsive to fin and heart injury and in addition can direct expression in the presence of a cardiomyocyte mitogen36 (Fig. 3b). We speculate that differences in regenerative capacity among species is somehow linked to differences in the catalog or placement of TREEs near key genes. The use of transgenic techniques through which enhancer elements are transferred between species could interrogate this idea. Other future studies must identify more regeneration-responsive enhancers, validate their role in regeneration contexts, and uncover their common features, including the TFs that interact with them.
Transcription factors in regeneration
Over 30 years ago, Weintraub and colleagues demonstrated that cell identities can be controlled by a single TF; they converted tissue culture fibroblasts into myoblasts by introducing MYOD145. Since then, studies have shown the TFs OCT4, SOX2 and KLF4 can act together to revert differentiated cell types into a pluripotent state46. Although the literature does not suggest that regeneration is likely to be driven by a central transcription factor or control node acting as a ‘master regulator’, recent studies have revealed TFs that act early in regeneration greatly influence gene expression and regenerative capacity through interaction with cis regulatory elements.
Gene regulatory networks in the response of flatworms to injury.
Many planarian flatworms and acoels can regenerate their entire head and tail. These organisms maintain mass under homeostatic conditions and regenerate upon injury, in planarians from the activity of somatic pluripotent stem cells called neoblasts7,47. Using assay for transposase-accessible chromatin using sequencing [G] (ATAC-seq) to profile chromatin accessibility, a recent study identified thousands of sequences in the acoel Hofstenia miamia that display increased accessibility during regeneration. These loci were highly enriched for a DNA sequence motif recognized by the transcription factor Egr39. Knockdown of egr expression by RNA interference (RNAi) dampened chromatin accessibility and transcriptional activity near several genes known to be induced during regeneration and consequently blocked head or tail regeneration. A similar effect was observed for egr RNAi in the planarian species Schimidtea mediterranea and might extend to deuterostomes such as sea stars48, in which egr expression is sharply induced during regeneration. Evidence suggests that Egr likely acts as a pioneer factor49 [G] in response to wounding, as the emerging regenerative program is dependent on its activity to recruit other key transcriptional regulators such as Runt-139. While the loss-of-function data suggest a master regulator of wound-induced genes based on these properties, gain-of-function studies to examine the ability of Egr to affect gene expression and morphogenesis at a ground state in the absence of injury are needed to support these findings. In mammals, TFs with master regulator properties might be found in certain tissue-specific adult stem cells50. For example, induced overexpression of either Hoxc4, Hoxc6, or Hoxc8 genes can ectopically regenerate hair follicles in the mouse51. Whereas the discovery of master regulators of regeneration may provide shortcuts to therapeutic applications, they will almost certainly be tissue-specific and context-specific.
Pluripotency TFs in regenerating neural tissue.
TFs can modulate the regenerative capacity of neurons in vertebrates. Zebrafish can regenerate damaged retinae through the proliferation and differentiation of Müller glia cells into photoreceptors and retina-associate neurons such as bipolar and amacrine cells52,53. Oryzias latipes, or the medaka, a teleost fish distantly related to the Zebrafish, seems to possess a more limited capacity to regenerate structures such as the retina and heart54, as retinal Müller glia only produce photoreceptors upon injury and not associated neurons55. Recently it was discovered that medaka have lower levels of sox2 expression than zebrafish. sox2 encodes Sox2, a TF important in the identity and activity of neural progenitor cell populations. Experimentally increasing Sox2 levels in medaka Müller glia cells using inducible transgenes, to approximate Sox2 levels in zebrafish Müller glia, restored retinal cell types such as retinal ganglion cells and amacrine cells in addition to photoreceptors in medaka55. Other studies identified a requirement for SOX2 during the regeneration of mouse olfactory neurons56, which are slowly renewed by a stem cell population located in the olfactory epithelium. KLF4, a binding partner of SOX2, is also required for the regeneration of murine retinal ganglion neurons57. SOX2 and KLF4 are part of a small group of TFs that can revert differentiated cells into a pluripotent state46. SOX2 function during the development of the nervous system can also inhibit neural cell production by maintaining a plastic precursor state58. Thus, it appears that SOX2 is a master regulator of neural precursor cells, which assists regeneration by maintaining a plastic state or reverting cells to a more developmentally potent cell depending on the context. It will be important to determine what regulates the transcription of SOX2 expression itself in these cells and whether the pioneer activities of SOX2 and KLF449 mediate partial reprogramming [G] of chromatin during regeneration, analogous to that observed during the reprogramming of somatic cells to induced pluripotent stem cells.
TFs that control cardiac regenerative capacity.
TFs can limit regeneration programs, for example by governing terminal differentiation states. This fact is evident from recent studies of cardiac regeneration, which is measurable in early neonatal mammals but minimal in late neonates and adults59,60. For example, a negative correlation between thyroid hormone levels and the capacity for heart regeneration was reported in both mice61 and frogs62. Furthermore, conditional inhibition of the TF thyroid hormone receptor α (THRA) in heart muscle increased the percentage of diploid and mononucleate adult cardiomyocytes, a phenotype associated with a more robust regenerative response to myocardial infarction63 (Box 2). The authors of these studies speculated that regulation of thyroid hormone levels and signaling is a potential target of natural selection, affecting the DNA content and regenerative capacity of cardiomyocytes61. Other TFs such as the homeobox protein Meis1 have been reported to limit proliferation of heart muscle cells through repression of cell-cycle regulator p2164. The TF Yes-associated protein (YAP) is a potent activator of cell division and growth, modulated through multiple branches of regulation but primarily controlled by Hippo kinase signaling, which restricts YAP from entering the nucleus65. Recent studies have pointed to the negative control of YAP by the Hippo pathway as a key constraint on heart regeneration66–69. Nuclear YAP is normally depleted in cardiomyocytes during postnatal development, but a mutant YAP that retains high nuclear levels has the potential to drive cardiomyocyte proliferation, even in adult mice70. Thus, signaling cascades that ultimately affect TF function can inhibit the heart’s ability to regenerate in response to injury.
Box 2. Single-cell RNA sequencing in regeneration.
Batch analysis of gene expression changes in whole tissues has been useful for identifying gene expression changes during regeneration. However, the responses of key cells such as adult stem cells, which can represent a tiny percentage of whole tissue (for example, <5% of skeletal muscle157, and ~.004% of hematopoietic tissue158) can be masked using this approach. Cell sorting may not be effective for isolating stem cells if only a small subset of the sorted cells within a particular tissue type are actively participating in regeneration. Transgenic methods for isolating regenerating cells are promising; a fluorescent reporter under the control of a promoter or enhancer that is activated during regeneration can be used either to mark regenerating tissue for fluorescence-activated cell sorting (FACS) or to direct expression of an affinity-tagged, regeneration-associated factor usable for profiling36. However, it is unclear if isolating cells using these known promoters and enhancers will produce a physiologically-relevant cell subpopulation.
Single-cell RNA sequencing (scRNA-Seq) can sample the gene expression signature of thousands of single cells or nuclei within a complex tissue and is therefore able to identify rare cell types and infer molecular trajectories and potential developmental transitions from a timecourse sampled from regenerating tissue. Recent scRNA-Seq studies have found potentially new cell types required for regeneration, for example Pdgfra-expressing endoneurial mesenchymal cells during the regeneration of amputated mouse sciatic nerves and digit tips159. Contributions of these Pdgfra-positive cells to regenerated nerves, skin and bone were validated using both lineage-tracing and transplantation, and the new tissue was reported to be derived from neural crest cells that persist after development. The molecular basis for the regenerative capacity of these cells has not been clarified; however their emergence required tissue innervation – a general theme in complex tissue regeneration160,161. Comparisons of scRNA-Seq datasets between early-stage tadpoles that successfully regenerate their tails and later-stage, regeneration-defective tadpoles identified unique gene expression programs23; based on subsequent transgenic ablation experiments, an epithelial cell type called regenerating-organizing-cells (ROC) was described that is required for regeneration162. Other notable studies have characterized diverse populations of regenerating stem cells from planarians163 and hydra164 as well as in the mammalian gut165.
A major drawback of scRNA-Seq is that sequencing depth [G] for a given cell is limited to accommodate the data from thousands of individual cells. This results in a bias against detection of poorly expressed genes that are not abundant enough to compete for more limited reads, including many if not most genes that encode TFs and other gene regulatory proteins.
Regeneration and chromatin remodelling
For TFs to bind regulatory elements and activate the gene expression programs of regeneration, they must be able to bind to open, accessible chromatin. Several major families of chromatin regulatory complexes, such as Polycomb Group proteins (PcG) and ATP-dependent chromatin remodelers, are well-studied for their roles in modulating genome accessibility during cell fate transitions71,72. In each case, nucleosomes directly impact gene expression as a response to modification of their histone components and/or three-dimensional structure73. Regenerative programs involve both gene activation and gene silencing36,74, and each can be achieved through a variety of chromatin regulatory mechanisms75.
Differences between embryogenesis and regeneration.
Chromatin regulatory events during regeneration are unlikely to replicate analogous events during initial embryonic development, even if the gene expression outcomes are the same. As embryogenesis proceeds, cell genomes are progressively condensed in a tailored manner that promotes or restricts distinct cell identities76,77. The chromatin regulation necessary to produce cardiac precursors from early mesoderm cells is likely to be distinct from chromatin changes that are associated with dedifferentiation of a mature cardiomyocyte to a more proliferative state6,78,79. For example, the p300/CBP transactivation complex acetylates histone H3 at lysine 27 (H3K27Ac)80, which reflects an open chromatin state more amenable to transcriptional gene activation. This activity is well-studied in early embryonic events; for example, in the formation of the cardiac fields in mice, where p300/CBP directly interacts with and facilitates the transcriptional activity of the critical cardiogenic TFs Gata4 and Hand281. Using a histone variant to map open chromatin during zebrafish heart regeneration showed most loci were not linked to H3K27Ac modifications36. Thus, the p300/CBP-mediated pathway of chromatin activation, which is critical during embryonic heart development, might only play a minor role in preparing the genome during regeneration. Other marks and pathways stimulating open chromatin observed in other regenerative contexts are potentially more critical16,41,75.
Reversing the repressive effects of chromatin.
In several organisms, the ability to regenerate is confined to specific developmental stages82; for instance, D. melanogaster larvae can regenerate damaged wing discs, whereas adult wings have no regenerative capacity83. Repressive chromatin marks narrow the developmental window of regenerative capacity in imaginal wing discs. The trimethylation of lysine 27 on histone H3 (H3K27me3), a highly repressive chromatin modification associated with chromatin compaction84, increases at sequences near the wingless gene over time, an effect that limits wingless signaling and restricts regeneration. Similarly, the cardiac regenerative response of neonatal mice59 is restricted by a reduction in the accessibility of chromatin by H3K27me3 around genes associated with cell proliferation85. Chromatin packaging has been shown in many species and developmental contexts to control cell proliferation more generally, such as regulation of the INK4a/ARF tumor-suppressor gene locus86 by the Polycomb complex protein BMI-1, or by phosphorylation of the chromatin remodeling complex Swi/Snf, a process that precedes mitosis87. Recent studies indicate that chromatin also regulates regeneration by controlling the expression of proliferation-associated genes85,88.
Repressive chromatin marks are strategically removed to permit gene activation during regeneration. During regeneration of Xenopus laevis kidneys, the histone demethylase arid3a is recruited to an enhancer to remove the repressive H3K9me3 mark and allow binding of lhx1, a TF that promotes kidney development40. Similarly, chromatin silencing must be reversed during the regeneration of utricle-supporting cells in the cochlea to activate the expression of genes that are required for the development of inner ear hair cells in mouse embryos89. This phenomenon is observed extensively across kingdoms, including in plants, where removal of H3K4me2 by the histone demethylase LDL3 in Arabidopsis thaliana allows for activation of genes required for callus formation in regenerating shoots41. It is not clear whether the removal of repressive chromatin marks is a genome-wide feature of cell lineage changes or whether removal only occurs at key regulatory nodes. However, the ability of a cell to reverse repressive chromatin marks is likely to be a key component that determines regenerative capacity.
Modulation of Polycomb activity influences regeneration.
PcGs were originally described as stable repressors of gene expression that can maintain the silent state by compacting chromatin after TFs disengage from their binding sites71,73. The ability of PcGs to control differentiation states by toggling the expression of critical developmental TFs has led to the suggestion that they may represent a master, repressive controller of regeneration90–92. Under this model, inhibition of PcG might de-repress the expression of developmentally potent TFs that in turn trigger large programs of gene expression. A signature of PcG silencing is the H3K27me3 modification84, which is erased from enhancers that direct expression during regeneration38,93 (Fig. 4a). Pharmacological inhibition of EZH2, the PcG enzyme responsible for the H3K27me3 mark, caused a modest but measurable increase in regeneration of mouse olfactory neurons after transgenic ablation by an undescribed mechanism94. However, we are not aware of studies showing that direct manipulation of PcG repression alone can confer regenerative capacity to tissues.
Figure 4. Chromatin dynamics in regeneration.
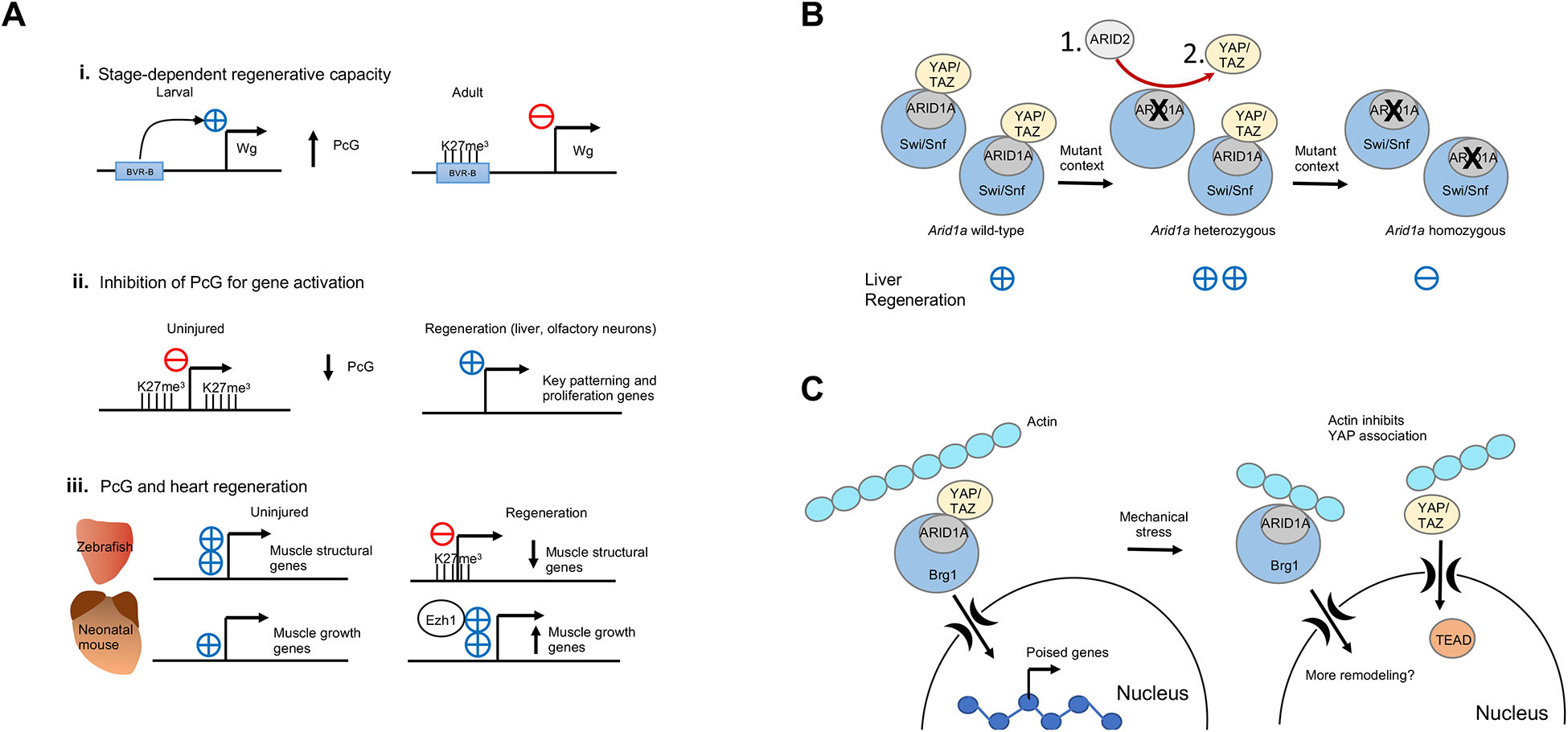
A. The role of Polycomb regulation in regeneration. Polycomb signaling can silence pro-regenerative enhancers, restricting regeneration, as shown here for the BRV-B enhancer of the wingless gene during imaginal disc regeneration in Drosophila. Polycomb silencing is sometimes inhibited in order to activate pro-regeneration genes, such as cell-cycle genes, in the case of regeneration of liver or olfactory neurons. Polycomb has different functions during heart regeneration depending on the model organism; in adult zebrafish, Polycomb silences highly expressed muscle structural genes that encode central components of the sarcomere such as titin and the myosin heavy and light chains (cmlc1, ttna, ttnb, vmhcl). Alternatively, in neonatal mice, Polycomb subunit Ezh1 acts as a transcriptional activator for genes involved in tissue morphogenesis, muscle structure and metabolism such as Bmp7, Irx2 and Tnni1.
B. Heterozygosity of Arid1a promotes liver regeneration. Some possible mechanisms include exchange of Arid1a subunits with Arid1b or Arid107 (1), a decrease in Hippo signaling via YAP/TAZ (2), or the fine tuning of gene expression to restrict antagonistic pathways (not pictured).
C. ATP-dependent chromatin remodeling complex Swi/Snf promotes regeneration after mechanical stress. In the uninjured setting, the ARID1A subunit of Swi/Snf sequesters YAP/TAZ and prevents it from binding its co-factor, TEAD-1. Swi/Snf remodels chromatin to poise pro-regeneration genes for activation in the liver. Upon mechanical stress, actin binds to ARID1A and releases YAP/TAZ, which can then associate with its active cofactor TEAD-1 and activate regeneration. Swi/Snf itself may then also further remodel chromatin102.
PcG function can also be inhibited through indirect methods (Fig. 4a). The Uhrf1 gene encodes a protein with multiple functional domains, which recruits DNA methyltransferase DNMT1 to hypomethylated DNA. Reducing DNA methylation by depleting Uhrf1 from mouse hepatocytes was expected to de-repress expression of transposons genome-wide, as DNA methylation is known to be the primary mechanism to silence transcription of transposons95. Instead, transposons were silenced by PcG proteins, which were redistributed from their usual targets to transposons, likely as a mechanism to protect genome integrity. Interestingly, this indirect mode of PcG inhibition, where PcG proteins are sequestered by transposons, led to increased expression levels of cell cycle genes, rather than developmental TFs, promoting regeneration of hepatocytes in the livers of Uhrf1 mutants. Thus, inhibition of Polycomb silencing proteins may be important in regeneration as well as initial development.
PcGs can also promote regeneration directly in some models (Fig. 4a). Recently, it was reported that expression of the embryonic variant of Ezh, Ezh2, was upregulated during zebrafish heart regeneration at the expense of the adult counterpart Ezh174. During regeneration, there was a concomitant increase of the H3K27me3 mark at genes encoding sarcomeric proteins, coinciding with dedifferentiation of cardiomyocytes. Overexpression of a mutant histone (H3.3K27M) known to inhibit the methylation activity of Polycomb Repressive Complex PRC2 impaired heart regeneration in zebrafish74. Although inhibiting Polycomb activity to release gene repression may be a common theme in regeneration, in the zebrafish heart, PRC2 activity itself reduces the expression of lineage-specific genes and promotes dedifferentiation. Similarly, the mouse orthologue of the Ezh2 gene is upregulated during regeneration of the pancreas in mice96,97, where it regulates the proliferation of insulin-producing cells. In contrast, Ezh2 was found to be dispensable as part of the cardiac regenerative response of neonatal mice98; however, the alternative catalytic subunit of PRC2, Ezh1, was implicated unexpectedly as a transcriptional activator of cardiac growth genes98. Interpreting Polycomb function is notoriously complicated; methods to control PcG activity in specific cell types and with fine temporal precision may be necessary to optimize its impact on regeneration for every given context.
Other chromatin regulatory pathways.
The gene EGR1, involved in whole-body regeneration of the acoel worm, has been well-characterized as an immediate-early gene [G] (IEG). These genes can be activated in minutes by TFs already present in the cell, avoiding the need for translation99,100. The chromatin around such genes is perpetually accessible and heavily acetylated, leaving the gene poised for activation99. Upon initiation of a stimulus – such as a signaling pathway or a steroid hormone - expression of IEGs is rapidly induced. This initial signaling cascade often leads to expression of secondary response genes that are regulated by the protein products of IEGs, such as EGR1. It is unclear if other regeneration-related genes are IEGs. Perpetually “open” regions of chromatin associated with regeneration are likely to be overlooked using current methods for chromatin analysis, as their accessibility would be similar in samples from uninjured and regenerating tissue.
Swi/Snf, a counterpart and antagonist of PcG that exposes DNA around nucleosomes, has been reported in some contexts to preferentially function upstream of pro-regeneration genes. ARID1A, an integral component of Swi/Snf, is required for liver regeneration and the formation of injury-induced progenitor-like cells101,102. Without ARID1A, chromatin accessibility is reduced at genes key to establishing hepatic progenitor cells in embryos, suggesting that the Swi/Snf complex poises these genes for activation during regeneration102. A screen in patients with liver cirrhosis identified mutations in several genes that were clonally expanded in diseased livers103, including ARID1A and the gene KMT2D, which encodes a protein homolog of the D. melanogaster PcG antagonist Trithorax-related and is part of the Mixed-lineage leukemia (MLL) complex104. These mutations improved the regenerative response to liver injury in mice when heterozygous, but other studies have shown that they are deleterious when homozygous94,101. How the dosage of gene-activating complexes such as MLL contribute to regeneration is unclear, but these findings suggest that in addition to activating pro-regenerative genes, Swi/Snf and MLL may regulate antagonistic pathways in parallel, and the outcome is therefore based on the concentration of gene products (Fig. 4b). Swi/Snf may do more during regeneration than remodel nucleosomes; studies in human cells have shown ARID1A can sequester the Hippo pathway factors YAP and TAZ, preventing them from binding and activating TEAD-1, the key TF that trans-activates Hippo pathway target genes105 (Fig. 4b). Hippo signaling has been implicated in several settings of regeneration, including following autotomy [G] in lizards and in the intestine, heart, liver, and skin of mammals69,105. Under high mechanical stress, a potential feature of regeneration106,107, actin inhibits the interaction between YAP/TAZ and ARID1A, allowing YAP/TAZ to bind to TEAD-1, which in turn mediates Hippo pathway gene activation108. Chromatin remodeling of YAP targets during liver regeneration suggests that Swi/Snf may also have a dual role in activating this crucial pathway, connecting chromatin regulation with perturbations of the cytoskeleton102 (Fig. 4c). Over-expression of constitutively-active YAP in mouse hearts resulted in changes in chromatin accessibility that coincided with increased cardiomyocyte proliferation70. It will be interesting to note for the future if this is a consequence of an altered interaction of Swi/Snf factors with the mutated YAP protein.
Emerging modes of gene regulation
Novel pathways for regulating gene expression are consistently being discovered, opening up new paradigms of what may constitute a regeneration ‘trigger’ or a ‘brake’. Recent literature describes the role of new classes of regulatory molecules such as long non-coding RNAs (lncRNAs), as well as new mechanisms by which signaling pathways can be integrated to control transcriptional events and the processes that may underlie forms of regeneration.
Membrane proteins signal in the nucleus.
Other signaling pathways involved in regeneration are typically directed by secreted or membrane-bound ligands with key roles in embryonic tissue patterning3,109,110. These signals can act on context-specific enhancer elements engaged by downstream TFs, or they may act in non-traditional ways to control transcription. For instance, it was recently reported that in mouse livers the insulin receptor – traditionally considered a standard receptor tyrosine kinase – translocates to the nucleus and binds to RNA polymerase II (Pol II) at insulin-responsive genes111. Insulin-like growth factor signaling has been implicated in promoting the regeneration of heart, skeletal muscle, liver, and nerves in various ways, and it is possible that the ability to directly regulate transcription is part of its function during these events112–116.
Non-coding RNAs regulate regeneration genes.
The regulation of gene expression by lncRNAs is a well-known gene regulatory paradigm117. lncRNAs can induce cis-regulated expression of linked genes through relaxing chromatin at enhancers and promoters as they are transcribed117. Additionally, lncRNAs can recruit chromatin remodeling factors to the genome and can form complexes with proteins that promote transcription. Recently, it was reported that lncRNAs expressed within the coding region of each gene of the protocadherin gene cluster in humans and mouse are required for DNA demethylation at the linked promoter as a mechanism to regulate stochastic expression of individual genes118.
Several recent studies have described roles for lncRNAs in regeneration. The long noncoding RNA Falcor fine-tunes expression of the adjacent Foxa2 gene by an unknown mechanism in adult mice, an interaction that maintains lung epithelial homeostasis and promotes regeneration119. FOXA2 is a pioneer factor required for development of endoderm and maintenance of secretory cell identity in the lung epithelium. Similarly, knockout of Silc1 lncRNA delays sciatic nerve regeneration in mice through cis-upregulation of the Sox11 gene, which encodes a key TF that regulates the specification and identity of cortico-spinal neurons during development120 and lies over 200 kilobases away from Silc1121. In regenerating chicken feathers, several lncRNAs modulate regeneration in a negative feedback loop in trans with WNT signaling122.
lncRNAs may also restrict regeneration programs. In mice, expression of an lncRNA called cardiomyocyte proliferation regulator or CPR increases into adulthood, when it recruits the DNA methyltransferase DNMT3A to promoter sequences of MCM388, a gene encoding a DNA helicase that is required for DNA replication. This regulatory mechanism contributes to one of several ‘transcriptional brakes’ on regeneration mentioned previously, including YAP sequestration108 and MEIS1-based control of cell cycle inhibitors64.
Trauma byproducts that regulate gene expression.
Stress, in the form of tissue tension or the release of cellular contents that change the local environment, is one of the first signals cells experience before activating regeneration. Recent studies have found that stress responses can directly integrate with global transcription programs123,124. In the heart, abnormal excitation–contraction coupling, a result of increased demands during injury, leads to cleavage of the cardiomyocyte structural protein junctophilin-2 (JP-2). A cleaved fragment of the N-terminus of JP-2 called JP2NT translocates to the nucleus to compete with the key cardiogenic TF MEF2 at its binding sites, which can reprogram transcription and protect against pathologic hypertrophy125. In another example, stress within the gut as a result of aging, infection, or injury stimulates regeneration of adult stem cells50. Transgenic ablation of midgut intestinal stem cells in D. melanogaster caused the release of reactive oxygen species from apoptotic cells at the ablated site, which stimulated stressed enterocytes126 to release the Unpaired cytokines; Upd, Upd2 and Upd3. These cytokines promoted the proliferation of intestinal stem cells by stimulating the JAK–STAT pathway and subsequent tissue regeneration. Recently, a Drosophila TF named Ets21C was found to function downstream of this stress-mediated feed forward loop and, interestingly, it regulates different cohorts of genes in enterocytes and intestinal stem cells127. It is likely there are other, as yet unknown novel signaling systems that can promote regeneration in stressed tissue.
Extreme injury regulates differentiation states.
Massive injuries can revert cells directly or indirectly to a stem-like state, where they can adopt alternative fates in a mechanism called adaptive cellular reprogramming128. Prominent examples of this are conversion of α-endocrine cells to β-cells in the mouse pancreas after genetic ablation of most of the pancreatic β-cell population129, and also in mice, the repair of severed axons by Schwann cells130. Cell conversion events of this sort suggest marked chromatin reprogramming, similar to that found during the generation of induced pluripotent cells131. A recent study compared healing after jawbone fracture to mandibular distraction osteogenesis, in which the bones are separated by a gap and require new bone growth to close the wound16. During jaw distraction [G], resident tissue stem cells showed evidence of reversion to a neural crest cell (NCC)-like state, with extensive genome-wide reprogramming, including expression of transposons mirroring early NCC signatures. In D. melanogaster, a protein called Taranis prevents maladaptive ectopic switching of cell fates during wing imaginal disc regeneration132. Vertebrate Taranis orthologues have not been identified; however, recent work implied that the conserved transcriptional co-repressor CtBP1 has a similar function to Taranis133.
It is possible that adaptive cellular reprogramming in regenerating tissue might involve TF complexes undergoing phase transitions. Many non-membrane-bound cell components are compartmentalized based on their physical properties, a concept known as phase separation. Phase separation can be visualized by the idea of oil and water. Differences in hydrophobicity drive the separation of these liquids; in the case of proteins in cells, protein–protein or protein–nucleic acid interactions may dictate the separation or transition between different phases134. Silent genes and active genes may represent different phases, as active genes may associate with a number of transcription factors, co-activators and Pol II, leading to the formation of multi-molecular nuclear condensates that are phase-separated from the karyolymph [G]. Transition between silent-gene and active-gene liquid phases is therefore governed by a collection of TFs, chromatin marks, and RNA135. Pluripotent cells such as embryonic stem cells have few nuclear condensates, suggesting that adaptive cellular reprogramming is likely to be concurrent with a general loss of nuclear condensates and a transition to a more homogenous phase (Fig. 5). In this model, upon massive injury, signaling cascades are likely to impact the number and distribution of epitopes for binding, thus regulating the fluidity of chromatin states and, possibly, genome plasticity. For example, following injury, phosphorylation events, including on histones tails themselves, may perturb the binding of histone covalent modifications on adjacent residues to the network of proteins forming the aggregate90,136,137. Thus, the binding reactions supporting condensates in differentiated cells are dismantled within the nucleus and the genome is reset into a more plastic state. A recent study reported that regulation by YAP can compartmentalize TEAD-1, open chromatin regions, and loci encoding proliferation factors into hubs of active transcription through phase transitions138. It will be interesting to see how our emerging understanding of the regulation of phase transitions will uncover new paradigms regarding dedifferentiation, reprogramming and gene regulation during regeneration.
Figure 5. Phase transitions and injury-induced regeneration programs.
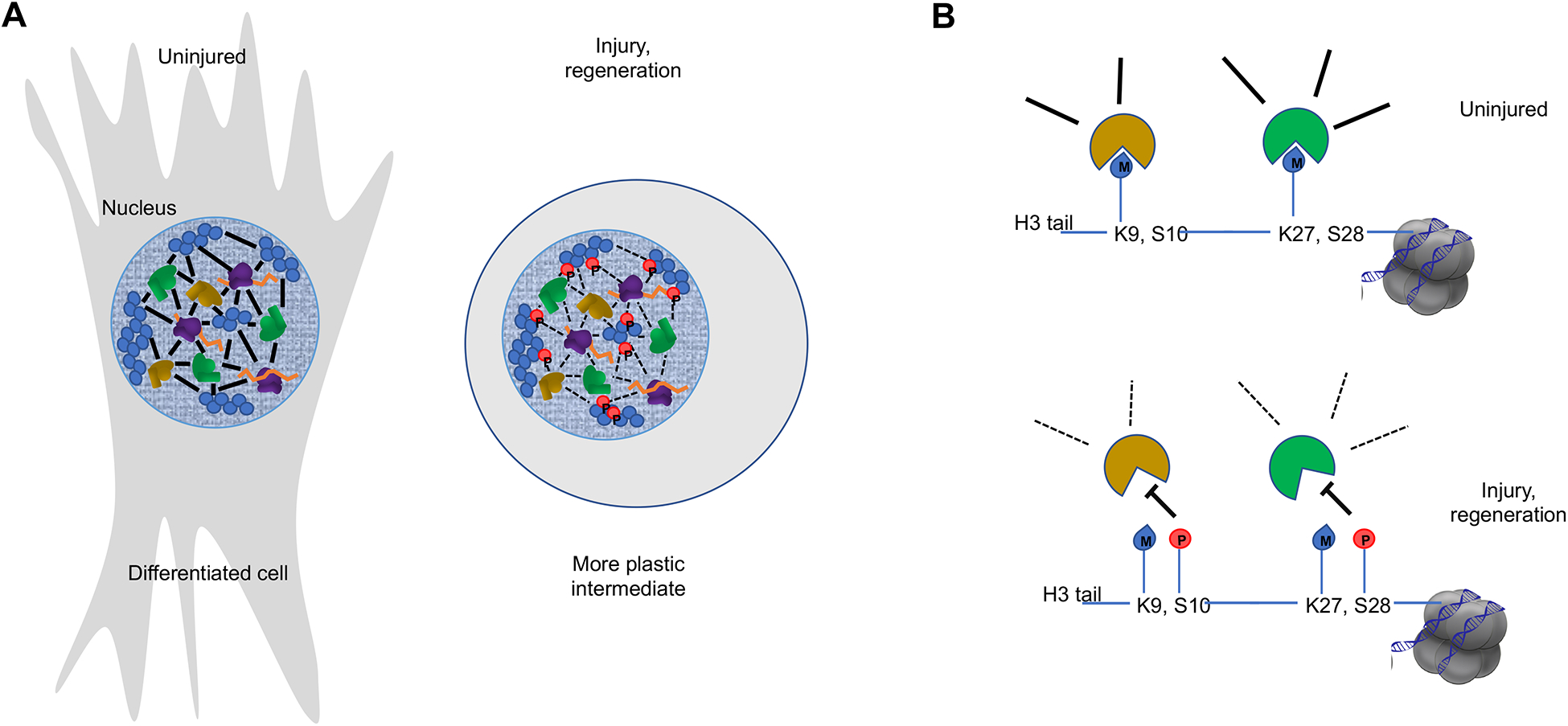
A. A model by which major phase transitions in chromatin might underlie sharp phenotypic changes during adaptive cell reprogramming. In differentiated cells, networks of transcription factors (brown), nucleosomes (blue), chromatin proteins (green), RNA (orange), and covalent modifications (not picture) coalesce into compartments of active and repressed genes. During regeneration, numerous signaling pathways are activated to help dismantle such aggregates, for example phosphorylation events (red) that inhibit binding reactions. Thus, nuclei are reset to an embryonic-like state where the genome organization is loosened, allowing for changes in cell differentiation and proliferative capacity. Chromatin binding proteins that recognize covalent modifications on histone tails, such as methylations of H3K9 and H3K27, are integral components of binding interactions that constitute a membrane-less organelle. Phosphorylation of Ser residues adjacent to the methylated Lys disrupts binding of these proteins to chromatin and could lead to phase transitions where previously structured repressed domains are released, leading to an opening of the genome.
Future perspectives
Advances in understanding regenerative biology are occurring through critical technology improvements such as scRNA-seq, CRISPR and high-throughput sequencing. Further advances are changing our ability to detect, visualize and manipulate gene expression, with possible therapeutic applications.
Novel technologies.
Novel methods are emerging to visualize gene expression during morphogenetic change and in three dimensions. Spatial information regarding gene expression domains can be recreated using Tomo-seq, which involves high-throughput sequencing of serial tissue sections139. Newer techniques make use of a scRNA-seq variant called Slide-seq to get near single-cell resolution140. In Slide-seq, rather than isolating single cells together with an individually barcoded bead, the beads are arranged in an ordered lattice that is overlaid with tissue sections. Subsequent sequencing of tissue associated with each bead can then be correlated with three-dimensional space. A second method, called sequential fluorescence in situ hybridization (seqFISH+), makes use of barcoding and serial fluorescent in situ hybridizations in tissue that remains intact141. All of these technologies are unlikely to detect poorly expressed transcripts, similar to standard scRNA-seq, as hybridization of rare transcripts can also be challenging. In the future, it would be interesting to adapt these techniques to visualize gene expression changes in a tissue as it is regenerating. The greatest boost for all of these technologies will be the ever-expanding capabilities of high-throughput sequencing, which may improve sequencing depth to the point that rare transcripts can be identified.
To understand the chromatin dynamics that underlie programmatic changes, there is an unmet need to identify the earliest-acting TFs and genomic loci. This challenging goal might be achieved by using proximity labeling, where a tag appended to a critical TF can mark associating proteins and enable their purification142. Transcriptional enhancer elements have been identified that direct gene expression during regeneration; however, further in vivo profiling is needed to identify enhancer elements that act as locus control regions. Continued evolution of CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) techniques that employ sequence-specific targeting of transcription control proteins for activation and inhibition, respectively, will help dissect the functions of regulatory sequences143,144. Methods to assess chromatin state at single-cell level should also expand the identification of active regulatory elements and concepts in rare and transitioning cell types145–147. This should apply to enhancers as well as silencer elements, which are more challenging to discover but could be of critical importance to elucidate regenerative mechanisms.
Looking beyond transcription.
Tissue regeneration is expected to be regulated at the levels of translation and post-translation. Beyond transcription, processes such as mRNA transport, degradation, or translation may provide more precise means of control without an intervening transcriptional event. Proteome and ribosome foot-printing technologies are opening the door to studies of protein levels that will give a more complete picture of gene expression148. Innovations in proximity labeling and mass spectrometry will help to further describe important signaling interactions that underlie regeneration142.
Regenerative medicine.
Rapid, large-scale changes in chromatin structure and gene expression are integral to success during innate regeneration, and therapeutics to induce regenerative capacity in the tissue will likely need to induce notable chromatin remodeling events. Critically, there must be inherent controls to ensure the epigenome transitions from a growth landscape toward one of patterning and completion of regeneration, for example, to avoid the risk of tissue overgrowth or cancer. Gene therapy as a method to virally deliver therapeutic proteins in humans is nearing its full potential149,150. Advances in gene therapy vectors - to evade the human immune system, to carry heavier cargoes, and to target potent cargoes to affected tissues – are moving forward in step with our understanding of regeneration pathways151–155. These therapies will be especially useful when stem cell transplantation is impractical or ineffective.
Conclusions
Dynamic changes in gene expression are a central and defining component of regeneration, enabling cells to respond to injury, toggle between differentiation states, and carry out tissue growth and patterning. There have been many informative and valuable insights into tissue regeneration from the investigation of model species, and broadly available techniques for genome-wide profiling and molecular genetics have helped turn attention toward gene regulatory mechanisms. Cell-by-cell assessment of gene expression programs has generated overviews of how features of the embryonic state can be revisited during regeneration. Dedicated enhancer elements for regeneration are being revealed and represent anchoring points for identification of DNA-protein complexes that turn on gene expression. It will also be important to understand how activity at these control regions is tempered as regenerative events conclude. Advances in ways to interpret and potentially manipulate chromatin states will help to interpret and potentially exploit regenerative pathways for clinical use.
Box 3: Polyploidy and regeneration.
Cell division requires sequential steps of karyokinesis [G] and cytokinesis. In some mammalian tissues, endoreplication [G] and binucleation — a polyploid [G] state — is more common than successful division. Like the cardiomyocytes of the mammalian heart, a high percentage of hepatocytes in the liver are polyploid, with a high frequency of binucleation. The liver can heal through hypertrophic growth or through division of existing diploid cells; still, there is evidence that some binucleated hepatocytes are able to divide in response to injury. A recent study using multi-color, Brainbow-based lineage tracing of mouse hepatocytes was able to distinguish between single-colour diploid cells and multi-coloured polyploid cells, and found that polyploid cells, even those above 4N, are able to proliferate166. In most cases, cell-division was reductive, and daughter cells became diploid. Instances of proliferation of polyploid cells themselves were also observed. Interestingly, many of the diploid cells quickly returned to polyploidy after a round of endoreplication, which suggests a mechanism to prepare for further rounds of proliferation166.
The binucleation status of cardiomyocytes has been inversely correlated with the ability of cardiomyocytes to divide63,167. For example, in zebrafish, elegant genetic manipulations to induce cardiomyocyte binucleation are able to disrupt heart regeneration167. Why binucleation might be more of an impediment to division for the key cells of the heart rather than the liver may reflect in part these cells’ differences in structure and function. A transgene expressing mutated YAP in the mouse heart appears to cause cardiomyocyte hyperplasia even in the adult mammalian state, suggesting a dominant polyploid organ can still be converted to a regenerative phenotype by strong developmental rewiring70. Moreover, polyploidy is being described as a natural regenerative response in a growing number of tissues in addition to the liver, including intestinal tissue in fruitflies168 and epicardial tissue in zebrafish169, and it has long been know that skeletal muscle regeneration is achieved by fusion of mononucleated myoblasts with other myoblasts and existing myofibers5.
Acknowledgments
We thank Nutishia Lee for contributions to the artwork in this review. We thank A. Burghes, B. Blaser and C. Lepper for discussions, and J. Kang, V. Cigliola, and M. Mokalled for comments on the manuscript. We apologize to colleagues in the field if the depth of the discussion of their work was limited as a result of space constraints. J.A.G. acknowledges funding from the American Heart Association (AHA)(17SDG33660922) and Ohio State University. K.D.P. acknowledges funding from the National Institutes of Health, AHA, and Fondation Leducq.
GLOSSARY
- Repatterning
the recapitulation of previous arrangements of cell types during regrowth
- Patterning
the developmental process by which cells acquire their identities, depending on their relative spatial positions within the embryo
- Ontogenetic
relating to the developmental history of an organism from fertilization to adulthood
- Blastema
a mass of proliferative cells that forms at the salamander limb stump after amputation, ultimately giving rise to the new limb structures. Additional regeneration contexts in other species and tissues similarly invoke a blastema
- Enhancer Discovery
enhancers are classically validated using assays where the test region is removed from its normal location and engineered into a heterologous context using reporters. Measures of endogenous enhancer activity have centered on scanning the accessibility of their chromatin, but CRISPR technology has made loss-of-function experiments feasible in virtually all lab model systems, enabling new definitions of enhancers based on function in vivo
- Dedifferentiate
a process where cells lose their lineage-restricted characteristics and may adopt a more developmentally primitive status
- Assay for transposase-accessible chromatin using sequencing
(ATAC-seq) an in vitro assay for chromatin accessibility that measures the ability of a transposase to access the underlying DNA
- Pioneer factor
a transcription factor that directly binds nucleosomes and can therefore initiate gene regulation from a previously silenced state
- Reprogramming
the process of conversion from a committed cell type to a different cell type
- Immediate-early gene
(IEG) a gene that is activated rapidly because it does not require the need of an intervening transcription event to produce already-poised activating factors
- Autotomy
regulated removal of a body part as a defense mechanism, for example a lizard losing its tail to escape a predator
- Jaw distraction
a surgical method for lengthening the jaw by cutting bone and resetting more distally
- Karyolymph
the contents of the nucleus including chromatin, nuclear fluid and particulate condensates such as the nucleolus
- Sequencing depth
the number of high-throughput sequencing reads per given sample indicative of the abundance of a transcript or chromatin feature
- Karyokinesis
the division of nuclei after mitosis to compartmentalize the two daughter genomes
- Endoreplication
an incomplete form of mitosis where the genome is replicated but the daughter cells never physically separate, leading to polyploidy
- Polyploid
a state where cells have an increasing number of paired chromosomes beyond the normal 2N (eg. 4N, 8N, etc.).
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Genetics thanks I. Hariharan and the other, anonymous reviewers for their contribution to the peer review of this work.
References
- 1.Poss KD Advances in understanding tissue regenerative capacity and mechanisms in animals. Nature Publishing Group 11, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokalled MH & Poss KD A Regeneration Toolkit. Developmental Cell 47, 267–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rafii S, Butler JM & Ding B-S Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells JM & Watt FM Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wosczyna MN & Rando TA A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Developmental Cell 46, 135–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuchi K et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner DE, Wang IE & Reddien PW Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kragl M et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Dor Y, Brown J, Martinez OI & Melton DA Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Knopf F et al. Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Developmental Cell 20, 713–724 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Salinero JM & Rafii S Endothelial cell adaptation in regeneration. Science 362, 1116–1117 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Naik S, Larsen SB, Cowley CJ & Fuchs E Two to Tango: Dialog between Immunity and Stem Cells in Health and Disease. Cell 175, 908–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens KR & Murry CE Human Pluripotent Stem Cell-Derived Engineered Tissues: Clinical Considerations. Cell Stem Cell 22, 294–297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepilina A et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Fausett BV & Goldman D A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J. Neurosci 26, 6303–6313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ransom RC et al. Mechanoresponsive stem cells acquire neural crest fate in jaw regeneration. Nature 563, 514–521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honkoop H et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 8, 98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Gates PB & Brockes JP Positional identity of adult stem cells in salamander limb regeneration. C. R. Biol 330, 485–490 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Nowoshilow S et al. The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 1–20 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Blassberg RA, Garza-Garcia A, Janmohamed A, Gates PB & Brockes JP Functional convergence of signalling by GPI-anchored and anchorless forms of a salamander protein implicated in limb regeneration. Journal of Cell Science 124, 47–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elewa A et al. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nature Communications 8, 2286–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerber T et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used single-cell transcriptome sequencing of connective tissue in regenerating Axolotl limbs to detect a regeneration gene signature that precedes the emergence of an embryonic-like gene expression program.
- 23.Aztekin C et al. Identification of a regeneration-organizing cell in the Xenopus tail. Science 364, 653–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a time series between regenerative and non-regenerative stages in Xenopus, Aztekin et. al. uncovered a regeneration-specific gene expression signature.
- 24.Oulhen N et al. Regeneration in bipinnaria larvae of the bat star Patiria miniata induces rapid and broad new gene expression. Mech. Dev 142, 10–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storer MA et al. Acquisition of a Unique Mesenchymal Precursor-like Blastema State Underlies Successful Adult Mammalian Digit Tip Regeneration. Developmental Cell (2019). doi: 10.1016/j.devcel.2019.12.004 [DOI] [PubMed] [Google Scholar]
- 26.Olson EN Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta V et al. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol 23, 1221–1227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindler YL et al. Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141, 3112–3122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka T, Xu J & Molkentin JD Re-employment of developmental transcription factors in adult heart disease. Semin. Cell Dev. Biol 18, 117–131 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisping E et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc. Natl. Acad. Sci. U.S.A 103, 14471–14476 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong AY & Whited JL Parallels between wound healing, epimorphic regeneration and solid tumors. Development 147, dev181636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catarino RR & Stark A Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes & Development 32, 202–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterwalder M et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 1–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farley EK et al. Suboptimization of developmental enhancers. Science 350, 325–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang J et al. Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper revealed regeneration-responsive transcriptional enhancers and showed they can be adapted to deliver pro-regenerative factors.
- 36.Goldman JA et al. Resolving Heart Regeneration by Replacement Histone Profiling. Developmental Cell 40, 392–404.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used a transgenic histone-tagging approach to identify and validate many regeneration-responsive enhancer elements relevant to cardiomyocyte regeneration.
- 37.Pfefferli C & Jaźwińska A The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nature Communications 8, 15151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcaya-Molina E et al. Damage-responsive elements in Drosophila regeneration. Genome Research 28, 1852–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work profiled enhancers from Drosophila imaginal discs that are active during regeneration and compared them to those in embryonic development and from other regeneration models.
- 39.Gehrke AR et al. Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 363, eaau6173 (2019). [DOI] [PubMed] [Google Scholar]; This manuscript describes egr1 as a pioneer transcription factor and possible master regulator required for whole body regeneration in the acoel worm, Hofstenia miamia.
- 40.Suzuki N et al. Arid3a regulates nephric tubule regeneration via evolutionarily conserved regeneration signal-response enhancers. Elife 8, 455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihara H et al. Primed histone demethylation regulates shoot regenerative competency. Nature Communications 10, 1786 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hewitt KJ et al. GATA Factor-Regulated Samd14 Enhancer Confers Red Blood Cell Regeneration and Survival in Severe Anemia. Developmental Cell 42, 213–225.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soukup AA et al. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J. Clin. Invest 129, 1180–1192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris RE, Setiawan L, Saul J & Hariharan IK Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. Elife 5, 49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript describes Polycomb-mediated decommissioning of an embryonic enhancer limiting regenerative capacity in Drosophila imaginal discs.
- 45.Davis RL, Weintraub H & Lassar AB Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K & Yamanaka S Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Ivankovic M et al. Model systems for regeneration: planarians. Development 146, dev167684 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Cary GA, Wolff A, Zueva O, Pattinato J & Hinman VF Analysis of sea star larval regeneration reveals conserved processes of whole-body regeneration across the metazoa. BMC Biol 17, 16–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwafuchi-Doi M & Zaret KS Cell fate control by pioneer transcription factors. Development 143, 1833–1837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keyes BE & Fuchs E Stem cells: Aging and transcriptional fingerprints. The Journal of Cell Biology 217, 79–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Z et al. Hoxc-Dependent Mesenchymal Niche Heterogeneity Drives Regional Hair Follicle Regeneration. Cell Stem Cell 23, 487–500.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Bernardos RL, Barthel LK, Meyers JR & Raymond PA Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J. Neurosci 27, 7028–7040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramachandran R, Reifler A, Parent JM & Goldman D Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J. Comp. Neurol 518, 4196–4212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito K et al. Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Dev. Dyn 243, 1106–1115 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Lust K & Wittbrodt J Activating the regenerative potential of Müller glia cells in a regeneration-deficient retina. Elife 7, 7028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gadye L et al. Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell 21, 775–790.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocha-Martins M et al. De novo genesis of retinal ganglion cells by targeted expression of Klf4 in vivo. Development 146, dev176586 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Bylund M, Andersson E, Novitch BG & Muhr J Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat Neurosci 6, 1162–1168 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Porrello ER et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porrello ER et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. U.S.A 110, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirose K et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall LN et al. Stage-dependent cardiac regeneration in Xenopus is regulated by thyroid hormone availability. Proceedings of the National Academy of Sciences 116, 3614–3623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patterson M et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nature Publishing Group 49, 1346–1353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahmoud AI et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249–253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu F-X & Guan K-L The Hippo pathway: regulators and regulations. Genes & Development 27, 355–371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xin M et al. Hippo pathway effector Yap promotes cardiac regeneration. Proceedings of the National Academy of Sciences 110, 13839–13844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leach JP et al. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gise, von A et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. U.S.A 109, 2394–2399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Liu S, Heallen T & Martin JF The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol 15, 1–684 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Monroe TO et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Developmental Cell 48, 765–779.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; The combined work of Chang et al., Li et al., Sun et al. and Monroe et al. suggest a model where mechanical transduction initiates chromatin reprogramming by the combined action of Swi/Snf and YAP signaling during liver and heart regeneration.
- 71.Schuettengruber B & Cavalli G Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Narlikar GJ, Fan H-Y & Kingston RE Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Lau MS et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 355, 1081–1084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Yair R et al. H3K27me3-mediated silencing of structural genes is required for zebrafish heart regeneration. Development 146, dev178632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work showed that Polycomb activity can be pro-regenerative by promoting dedifferentiation through silencing sarcomeric genes in muscle.
- 75.Maki N, Tsonis PA & Agata K Changes in global histone modifications during dedifferentiation in newt lens regeneration. Mol. Vis 16, 1893–1897 (2010). [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmad K & Henikoff S Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104, 839–847 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Meshorer E et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental Cell 10, 105–116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jopling C et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan X et al. Dynamic Alterations to α-Actinin Accompanying Sarcomere Disassembly and Reassembly during Cardiomyocyte Mitosis. PLoS ONE 10, e0129176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rada-Iglesias A et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai Y-S, Cserjesi P, Markham BE & Molkentin JD The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem 277, 24390–24398 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Samarajeewa A et al. Transcriptional response to Wnt activation regulates the regenerative capacity of the mammalian cochlea. Development 145, dev166579–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hariharan IK & Serras F Imaginal disc regeneration takes flight. Current Opinion in Cell Biology 48, 10–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pengelly AR, Copur Ö, Jäckle H, Herzig A & Müller J A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Quaife-Ryan GA et al. Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation 136, 1123–1139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bracken AP et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Development 21, 525–530 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sif S, Stukenberg PT, Kirschner MW & Kingston RE Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Development 12, 2842–2851 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ponnusamy M et al. The Long Non-Coding RNA CPR Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 331, 1078–44 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Jen H-I et al. Transcriptomic and epigenetic regulation of hair cell regeneration in the mouse utricle and its potentiation by Atoh1. Elife 8, W3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palacios D et al. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7, 455–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katsuyama T & Paro R Epigenetic reprogramming during tissue regeneration. FEBS LETTERS 585, 1617–1624 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Shaw T & Martin P Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep 10, 881–886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stewart S, Tsun Z-Y & Izpisua Belmonte JC A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. U.S.A 106, 19889–19894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin B et al. Injury Induces Endogenous Reprogramming and Dedifferentiation of Neuronal Progenitors to Multipotency. Cell Stem Cell 21, 761–774.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang S et al. Epigenetic Compensation Promotes Liver Regeneration. Developmental Cell 50, 43–56.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work showed that disruption of DNA methylation by the knockout of Uhrf1 in mice inhibits normal Polycomb function by redistributing it to compensate for silencing of transposons.
- 96.Mallen-St Clair J et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes & Development 26, 439–444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen H et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes & Development 23, 975–985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ai S et al. Divergent Requirements for EZH1 in Heart Development Versus Regeneration. Circ Res 121, 106–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fowler T, Sen R & Roy AL Regulation of primary response genes. Molecular Cell 44, 348–360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Balamotis MA et al. Complexity in transcription control at the activation domain-mediator interface. Sci Signal 2, ra20–ra20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun X et al. Suppression of the SWI/SNF Component Arid1a Promotes Mammalian Regeneration. Stem Cell 18, 456–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li W et al. A Homeostatic Arid1a-Dependent Permissive Chromatin State Licenses Hepatocyte Responsiveness to Liver-Injury-Associated YAP Signaling. Stem Cell 25, 54–68.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Zhu M et al. Somatic Mutations Increase Hepatic Clonal Fitness and Regeneration in Chronic Liver Disease. Cell 177, 608–621.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that heterozygous mutations in chromatin-activating complexes can foster regeneration in mammalian livers.
- 104.Smith E, Lin C & Shilatifard A The super elongation complex (SEC) and MLL in development and disease. Genes & Development 25, 661–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moya IM & Halder G Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 20, 1–16 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Benham-Pyle BW, Pruitt BL & Nelson WJ Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao J et al. Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Developmental Cell 42, 600–615.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chang L et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 563, 265–269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tanaka EM The Molecular and Cellular Choreography of Appendage Regeneration. Cell 165, 1598–1608 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Wehner D & Weidinger G Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. 31, 336–343 (2015). [DOI] [PubMed] [Google Scholar]
- 111.Hancock ML et al. Insulin Receptor Associates with Promoters Genome-wide and Regulates Gene Expression. Cell 177, 722–736.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Apel PJ et al. Effect of locally delivered IGF-1 on nerve regeneration during aging: an experimental study in rats. Muscle Nerve 41, 335–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barton ER, Morris L, Musaro A, Rosenthal N & Sweeney HL Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. The Journal of Cell Biology 157, 137–148 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chablais F & Jaźwińska A IGF signaling between blastema and wound epidermis is required for fin regeneration. Development 137, 871–879 (2010). [DOI] [PubMed] [Google Scholar]
- 115.Choi W-Y et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660–666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Desbois-Mouthon C et al. Hepatocyte proliferation during liver regeneration is impaired in mice with liver-specific IGF-1R knockout. FASEB J. 20, 773–775 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Kopp F & Mendell JT Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Canzio D et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 177, 639–653.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Swarr DT et al. The long noncoding RNA Falcor regulates Foxa2 expression to maintain lung epithelial homeostasis and promote regeneration. Genes & Development (2019). doi: 10.1101/gad.320523.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shim S, Kwan KY, Li M, Lefebvre V & Sestan N Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perry RB-T, Hezroni H, Goldrich MJ & Ulitsky I Regulation of Neuroregeneration by Long Noncoding RNAs. Molecular Cell 72, 553–567.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin X et al. Long non-coding RNAs regulate Wnt signaling during feather regeneration. Development 145, dev162388 (2018). [DOI] [PubMed] [Google Scholar]
- 123.Boos F et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nature Cell Biology 21, 442–451 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Eming SA, Wynn TA & Martin P Inflammation and metabolism in tissue repair and regeneration. Science 356, 1026–1030 (2017). [DOI] [PubMed] [Google Scholar]
- 125.Guo A et al. E-C coupling structural protein junctophilin-2 encodes a stress-adaptive transcription regulator. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jiang H et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mundorf J, Donohoe CD, McClure CD, Southall TD & Uhlirova M Ets21c Governs Tissue Renewal, Stress Tolerance, and Aging in the Drosophila Intestine. Cell Rep 27, 3019–3033.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jessen KR, Mirsky R & Arthur-Farraj P The Role of Cell Plasticity in Tissue Repair: Adaptive Cellular Reprogramming. Developmental Cell 34, 613–620 (2015). [DOI] [PubMed] [Google Scholar]
- 129.Thorel F et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Arthur-Farraj PJ et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 75, 633–647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Papp B & Plath K Epigenetics of Reprogramming to Induced Pluripotency. Cell 152, 1324–1343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schuster KJ & Smith-Bolton RK Taranis Protects Regenerating Tissue from Fate Changes Induced by the Wound Response in Drosophila. Developmental Cell 34, 119–128 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Worley MI, Alexander LA & Hariharan IK CtBP impedes JNK- and Upd/STAT-driven cell fate misspecifications in regenerating Drosophila imaginal discs. Elife 7, 487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hyman AA, Weber CA & Jülicher F Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 135.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gehani SS et al. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Molecular Cell 39, 886–900 (2010). [DOI] [PubMed] [Google Scholar]
- 137.Lau PNI & Cheung P Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. U.S.A 108, 2801–2806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cai D et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nature Cell Biology 21, 1578–1589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu C-C et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Developmental Cell 36, 36–49 (2016). [DOI] [PubMed] [Google Scholar]
- 140.Rodriques SG et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rhee H-W et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xie S, Duan J, Li B, Zhou P & Hon GC Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Molecular Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 144.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hainer SJ, Bošković A, McCannell KN, Rando OJ & Fazzio TG Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177, 1319–1329.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cusanovich DA et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309–1324.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cao J et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.van Heesch S et al. The Translational Landscape of the Human Heart. Cell 178, 242–260.e29 (2019). [DOI] [PubMed] [Google Scholar]
- 149.Anguela XM & High KA Entering the Modern Era of Gene Therapy. Annu. Rev. Med 70, 273–288 (2019). [DOI] [PubMed] [Google Scholar]
- 150.Mendell JR et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med 377, 1713–1722 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Chicoine LG et al. Plasmapheresis Eliminates the Negative Impact of AAV Antibodies on Microdystrophin Gene Expression Following Vascular Delivery. Molecular Therapy 22, 338–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Recino A et al. Immunosuppression overcomes insulin- and vector-specific immune responses that limit efficacy of AAV2/8-mediated insulin gene therapy in NOD mice. Gene Ther. 26, 40–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Trapani: Adeno-Associated Viral Vectors as a Tool for Large Gene Delivery to the Retina Genes 10, 287–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liao H-K et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Davidsson M et al. A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism. Proceedings of the National Academy of Sciences 116, 27053–27062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bracken AP, Brien GL & Verrijzer CP Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes & Development 33, 936–959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Venable JH Constant cell populations in normal, testosterone-deprived and testosterone-stimulated levator ani muscles. Am. J. Anat 119, 263–270 (1966). [DOI] [PubMed] [Google Scholar]
- 158.Wilson A et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Carr MJ et al. Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell 24, 240–256.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 160.Mahmoud AI et al. Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Developmental Cell 34, 387–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kumar A, Godwin JW, Gates PB, Garza-Garcia AA & Brockes JP Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.THORNTON CS The effect of apical cap removal on limb regeneration in Amblystoma larvae. J. Exp. Zool 134, 357–381 (1957). [DOI] [PubMed] [Google Scholar]
- 163.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM & Reddien PW Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Siebert S et al. Stem cell differentiation trajectories in Hydraresolved at single-cell resolution. Science 365, eaav9314–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ayyaz A et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125 (2019). [DOI] [PubMed] [Google Scholar]
- 166.Matsumoto T, Wakefield L, Tarlow BD & Grompe M In Vivo Lineage Tracing of Polyploid Hepatocytes Reveals Extensive Proliferation during Liver Regeneration. Cell Stem Cell 26, 34–47.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.González-Rosa JM et al. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Developmental Cell 44, 433–446.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lucchetta EM & Ohlstein B Amitosis of Polyploid Cells Regenerates Functional Stem Cells in the Drosophila Intestine. Cell Stem Cell 20, 609–620.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


