Key Points
The ELN2017 classification distinguishes 3 distinct risk groups after allogeneic HSCT consolidation in patients with AML.
Patients with AML with TP53 mutation, monosomal karyotype, or MRD positivity at transplantation have poor prognoses after allogeneic HSCT.
Abstract
In 2017, an updated European LeukemiaNet (ELN) risk classification was published allocating patients with acute myeloid leukemia (AML) to 3 risk groups on the basis of certain cytogenetic and molecular aberrations. To date, studies of the prognostic significance of the ELN2017 risk classification in the context of an allogeneic hematopoietic stem cell transplantation (HSCT) are lacking. We performed risk stratification according to the ELN2017 classification in 234 patients with AML who underwent allogeneic HSCT as a consolidation therapy. In our cohort, the risk of 39.7% of the patients was classified as favorable, that of 12.8% as intermediate, and that of 47.4% as adverse. In the context of allogeneic HSCT, the assignment to the 3 ELN2017 risk groups retained its prognostic significance, with patients with favorable risk having the best prognosis and those with adverse risk having the worst one. Subgroup analyses showed that patients with a monosomal karyotype or TP53 mutation had considerably increased relapse rates, even in the adverse-risk group. When we analyzed the impact of digital droplet PCR–based measurable residual disease (MRD) before allogeneic HSCT, MRD+ patients had impaired prognoses, with cumulative incidence of relapse and overall survival comparable to those of patients classified as having an ELN2017 adverse genetic risk. This study is the first to demonstrate that the ELN2017 classification distinguishes the 3 risk groups with significantly distinct prognoses, even after allogeneic HSCT, and emphasizes the dismal prognosis of patients with AML with TP53 mutations, monosomal karyotype, or MRD positivity after allogeneic HSCT.
Visual Abstract
Introduction
Acute myeloid leukemia (AML) is a highly genetically heterogeneous disease. During recent years, its molecular landscape has been redefined greatly with the advent of high-throughput sequencing techniques.1-3
In 2010, an international expert panel came together on behalf of the European LeukemiaNet (ELN) to establish a risk classification based on cytogenetic and molecular aberrations.4 The definitions of 4 AML risk groups (favorable, intermediate-I, intermediate-II, and adverse) were shown to have significant value in predicting outcomes, mainly in patients treated with consolidation chemotherapy regimens.5-7 In patients who undergo allogeneic hematopoietic stem cell transplantation (HSCT) as a consolidation therapy, the prognostic impact of the ELN2010 classification was reduced, because patients with favorable risk have significantly longer survival than the other risk groups.8 In 2017, an updated ELN risk classification was released that incorporated the most recent insights into the molecular architecture of AML and their prognostic significance.9 Compared with the ELN2010 classification, in the ELN2017 classification, only 3 risk groups were defined: favorable, intermediate, and adverse. Based on studies showing that only patients with biallelic CEBPA mutations had significantly improved survival,10-15 the entity “biallelic mutated CEBPA” was introduced, to define patients with favorable risk. Because the negative prognostic impact of fms-related receptor tyrosine kinase-internal tandem duplications (FLT3-ITDs) depends on a high ITD/wild-type ratio (≥0.5),16-18 the FLT3-ITD allelic ratio is now applied, to stratify patients with AML into the 3 risk groups in the context of nucleophosmin-1 (NPM1) mutation status. Because the presence of a monosomal karyotype and mutations in the genes TP53, ASXL1, and RUNX1 were found to be associated with particularly poor prognoses, they were added to the ELN2017 classification to define patients with adverse risk.3,19-33 In a study analyzing the prognostic impact of the ELN2017 classification in 3 independent AML cohorts, the allocation to the genetic-risk groups distinguished significantly different overall survival groups in at least 2 of the 3 cohorts, mainly among those who underwent consolidation chemotherapy.34 Boddu et al showed that the ELN2017 risk group in which patients were classified correlated significantly with overall survival (OS) in younger patients with AML (<60 years) who received chemotherapy-based consolidation, with a caveat regarding the lack of data on the CEBPA mutation status of the entire cohort.35 In their study, the Japan Adult Leukemia Study Group demonstrated improved prognostic accuracy of the ELN2017 classification compared with that of the ELN2010.36 Another study recently demonstrated that the ELN2017 genetic-risk classification could be combined with monitoring of the peritransplantation measurable residual disease (MRD) to refine the prediction of relapse after allogeneic HSCT.37 The importance of MRD monitoring was also emphasized by the introduction in the updated ELN2017 classification of the new response category “complete remission (CR) without MRD.”9
In the context of allogeneic HSCT, the prognostic efficacy of the updated ELN2017 classification has not been evaluated. To our knowledge, ours is the first study that has been undertaken to investigate the impact of the allocation to the 3 ELN2017 risk groups on prediction of relapse and survival in an AML cohort that homogeneously underwent allogeneic HSCT as a consolidation therapy.
Methods
Patients and treatment
The cohort consisted of 234 patients with AML (median age, 59.9 years; range, 14.3-75.8) who underwent allogeneic HSCT at the University Hospital Leipzig from September 1998 through October 2018. All patients were treated with standard cytarabine-based chemotherapy and underwent consolidation treatment with allogeneic HSCT after achieving CR (n = 170; 74.9%), CR with incomplete peripheral recovery (CRi; n = 34, 15.0%), or CR with partial response (PR; n = 23; 10.1%). In our cohort, patients received consolidation therapy in the form of 1 of the following conditioning protocols: myeloablative (MAC; n = 49; 21.5%),38 reduced-intensity (RIC; n = 4; 1.8%), and nonmyeloablative (NMA; n = 162; 71.1%).39,40 Patients with insufficient response received a conditioning regimen according to investigators’ discretion.41-43 The decision to perform allogeneic HSCT was based on institutional protocol or the patient’s request and was dependent on the biological and clinical information available at the time of HSCT. Written informed consent for participation in the study was obtained in accordance with the Declaration of Helsinki. For detailed information on treatment regimens, please refer to the supplemental Data.
Genetic-risk groups were defined according to the ELN2017 recommendations based on the molecular and cytogenetic aberrations identified in bone marrow samples at diagnosis.9 Basic clinical characteristics of the patients are shown in Table 1. The median follow-up of patients who remained alive was 4.7 years after allogeneic HSCT.
Table 1.
Clinical characteristics, according to ELN2017 genetic-risk group, of patients with AML who underwent allogeneic HSCT
| Characteristics | All (N = 234) | ELN2017 favorable (n = 93) | ELN2017 intermediate (n = 30) | ELN2017 adverse (n = 111) | P* |
|---|---|---|---|---|---|
| Age at diagnosis, y | .01 | ||||
| Median | 59.9 | 55.1 | 60.8 | 62.7 | |
| Range | 14.3-75.8 | 14.3-73.9 | 19.2-72.2 | 20.0-75.8 | |
| Sex, n (%) | .33 | ||||
| Female | 116 (49.6) | 48 (51.6) | 11 (36.7) | 57 (51.4) | |
| Male | 118 (50.4) | 45 (48.4) | 19 (63.3) | 54 (48.6) | |
| WBC at diagnosis, ×109/L | <.001 | ||||
| Median | 6.9 | 15.8 | 2.5 | 4.7 | |
| Range | 0.7-385 | 1-324 | 0.7-146 | 0.7-385 | |
| Platelets at diagnosis, ×109/L | .49 | ||||
| Median | 66.5 | 75 | 77 | 60.5 | |
| Range | 3-950 | 3-238 | 10-268 | 3-950 | |
| Hemoglobin at diagnosis, g/dL | .43 | ||||
| Median | 8.8 | 8.9 | 9.2 | 8.2 | |
| Range | 4.2-14.9 | 4.3-14.7 | 5.2-13.4 | 4.2-14.9 | |
| Peripheral blasts at diagnosis, % | .32 | ||||
| Median | 26 | 32 | 14 | 22 | |
| Range | 0-97 | 2-97 | 1-96 | 0-97 | |
| Bone marrow blasts at diagnosis, % | .73 | ||||
| Median | 50 | 53.7 | 51 | 50 | |
| Range | 3-100 | 20-100 | 21-95 | 3-95 | |
| NPM1 mutation at diagnosis, n (%) | <.001 | ||||
| Absent | 162 (71.4) | 25 (28.7) | 27 (90.0) | 110 (100) | |
| Present | 65 (28.6) | 62 (71.3) | 3 (10.0) | 0 (0) | |
| FLT3-ITD mutation at diagnosis, n (%) | .41 | ||||
| Absent | 191 (84.1) | 73 (83.9) | 23 (76.7) | 95 (86.4) | |
| Present | 36 (15.9) | 14 (16.1) | 7 (23.3) | 15 (13.6) | |
| DNMT3A mutation at diagnosis, n (%) | .07 | ||||
| Absent | 112 (84.8) | 36 (75.0) | 25 (89.3) | 51 (91.1) | |
| Present | 20 (15.2) | 12 (25.0) | 3 (10.7) | 5 (8.9) | |
| FLT3-TKD mutation at diagnosis, n (%) | .09 | ||||
| Absent | 195 (89.0) | 69 (83.1) | 29 (96.7) | 97 (91.5) | |
| Present | 24 (11.0) | 14 (16.9) | 1 (0.3) | 9 (8.5) | |
| JAK2 mutation at diagnosis, n (%) | .008 | ||||
| Absent | 98 (92.5) | 26 (100.0) | 23 (79.3) | 49 (96.1) | |
| Present | 8 (7.5) | 0 (0.0) | 6 (20.7) | 2 (1.9) | |
| EVI1 expression at diagnosis, n (%) | <.001 | ||||
| Absent | 98 (77.8) | 44 (95.7) | 15 (71.4) | 39 (66.1) | |
| Present | 28 (22.2) | 2 (4.3) | 6 (28.6) | 20 (33.9) | |
| Remission status at HSCT, n (%) | .03 | ||||
| CR | 170 (74.9) | 73 (80.2) | 25 (83.3) | 72 (67.9) | |
| CR, MRD− | 47 (27.6) | 27 (37.0) | 7 (28.0) | 13 (18.1) | |
| CR, MRD+ | 24 (14.1) | 16 (21.9) | 4 (16.0) | 4 (5.6) | |
| CR, no information on MRD | 99 (58.2) | 30 (41.1) | 14 (56.0) | 55 (76.4) | |
| CRi | 34 (15.0) | 14 (15.4) | 1 (3.3) | 19 (17.9) | |
| PR | 23 (10.1) | 4 (4.4) | 4 (13.3) | 15 (14.2) | |
| Chemotherapy cycles before HSCT, n (%) | .09 | ||||
| 1 | 44 (25.6) | 12 (17.6) | 6 (23.1) | 26 (33.3) | |
| 2 | 97 (56.4) | 38 (55.9) | 16 (61.5) | 43 (55.1) | |
| ≥3 | 31 (18.0) | 18 (26.5) | 4 (15.4) | 9 (11.5) |
EVI1, ecotropic viral integration site-1; TKD, tyrosine kinase domain; WBC, white blood cell.
P values are from Fisher’s exact or Kruskal-Wallis test and compare the 3 ELN2017 genetic-risk groups.
Cytogenetic and mutation analyses of NPM1, FLT3, and CEBPA
Pretreatment bone marrow cytogenetics were determined using standard techniques for banding and fluorescence in situ hybridization. Genomic DNA of pretreatment samples was screened for the presence of the FLT3-ITD and NPM1 mutations, as previously described.8,44,45 The NPM1 mutation type was determined by applying Sanger sequencing of exon 12, as published before.8 We analyzed the presence of biallelic CEBPA mutations in diagnostic samples, applying the sequencing approach described by Benthaus et al.46
Next-generation sequencing of pretreatment bone marrow samples
In patients with adequate sample available, we performed targeted amplicon sequencing, using the TruSight Myeloid Sequencing Panel (Illumina, San Diego, CA). This panel covers 568 amplicons across 54 genes recurrently mutated in myeloid malignancies, including the genes TP53, ASXL1, and RUNX1. Next-generation sequencing was performed on the MiSeq platform (Illumina). We performed alignment and variant calling, as previously described.47 ASXL1 mutations at codon 646 were validated by applying a proofreading polymerase-based Sanger sequencing approach.29
MRD detection
In patients with NPM1-mutated AML at diagnosis and sufficient sample available (n = 45), we performed NPM1-mutated MRD assessment before allogeneic HSCT by applying a digital droplet PCR assay, as previously described.48 In addition, MRD was determined based on the expression of meningioma-1 (MN1; n = 71) and brain and acute leukemia, cytoplasmic (BAALC; n = 72) genes in pretransplant samples, as published before.49,50 Patients with only PR before allogeneic HSCT were excluded from MRD assessment.
Statistical analyses
Statistical analyses were performed using the R statistical software platform (version 3.4.3). Further details on statistical analyses, clinical end points, and multivariate analyses are provided in the supplemental Data.
Results
Associations between the ELN2017 risk groups and clinical and biological characteristics
In the AML cohort, 93 patients were classified as favorable (39.7%), 30 as intermediate (12.8%), and 111 as adverse (47.4%). The most frequently observed genetic aberrations were mutated NPM1 without FLT3-ITD or with a low FLT3-ITD allelic ratio in the favorable-risk group (66.7%), wild-type NPM1 without FLT3-ITD or with low FLT3/ITD allelic ratio in the intermediate-risk group (46.7%), and a complex karyotype in the adverse-risk group (21.5%). A detailed list of genetic characteristics according to the ELN2017 risk classification is provided in supplemental Table 1.
When we analyzed clinical associations, we observed that patients classified as favorable according to the ELN2017 risk classification were younger (P = .01) and had significantly higher white blood cell count at diagnosis than that of patients in whom risk was intermediate or adverse (P < .001; Table 1). Patients in the adverse-risk group were less likely to receive allogeneic HSCT after attaining CR (67.9%), compared with those with favorable or intermediate risk (80.2% and 83.3% respectively; P = .03, Table 1). The different genetic-risk groups were associated with the presence of additional somatic mutations that are not included in the ELN2017 risk classification. Those with favorable risk were more likely to harbor DNA methyltransferase-3α (DNMT3A; P = .07) and FLT3-tyrosine kinase domain mutations by trend (P = .09), whereas 75.0% of the identified JAK2 mutations were observed in patients with intermediate risk (P = .008; Table1). Expression of EVI1 was rarely seen in patients with favorable risk (4.3%), but it occurred in approximately one third of patients with intermediate or adverse risk (28.6% and 33.9%, respectively; P < .001; Table 1).
Prognostic impact of the ELN2017 classification of patients with AML who undergo allogeneic HSCT
In our cohort of patients with AML who underwent allogeneic HSCT, the cumulative incidence of relapse (CIR) was 35.8% (95% confidence interval [CI], 29.6-42.1), and OS was 57.3% (95% CI; 51.1-64.4) 2 years after allogeneic HSCT (supplemental Figure 1).
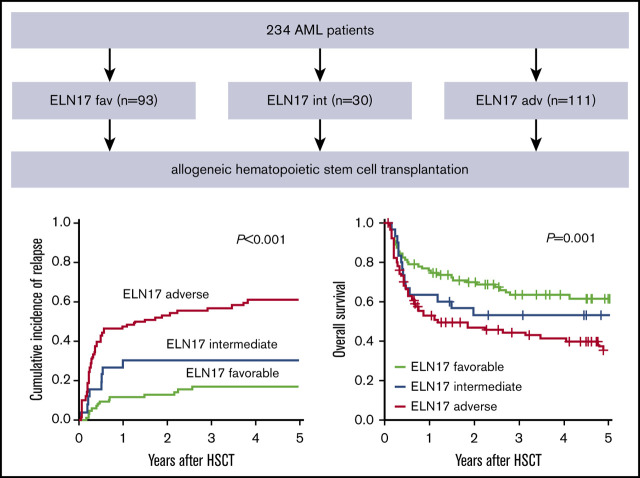
The allocation to the 3 ELN2017 risk groups separated outcome groups after transplantation (CIR, P < .001; OS, P = .001; Figure 1A-B). In a comparison of the 3 risk groups performed 2 years after allogeneic HSCT, patients classified as favorable had the lowest CIR (12.8%; 95% CI, 6.7-20.8), compared with patients in the intermediate (30.7%; 95% CI, 14.3-49.0) and adverse (54.0%; 95% CI, 43.0-63.9) risk groups. In the same comparison, these results translated into the longest OS for patients with favorable risk (69.9%; 95% CI, 61.0-80.1), whereas patients with adverse risk had the shortest OS (47.0%; 95% CI, 37.9-58.2). The OS of patients with intermediate risk ranged between those of the favorable- and adverse-risk groups (52.9%; 95% CI, 37.7-74.4).
Figure 1.
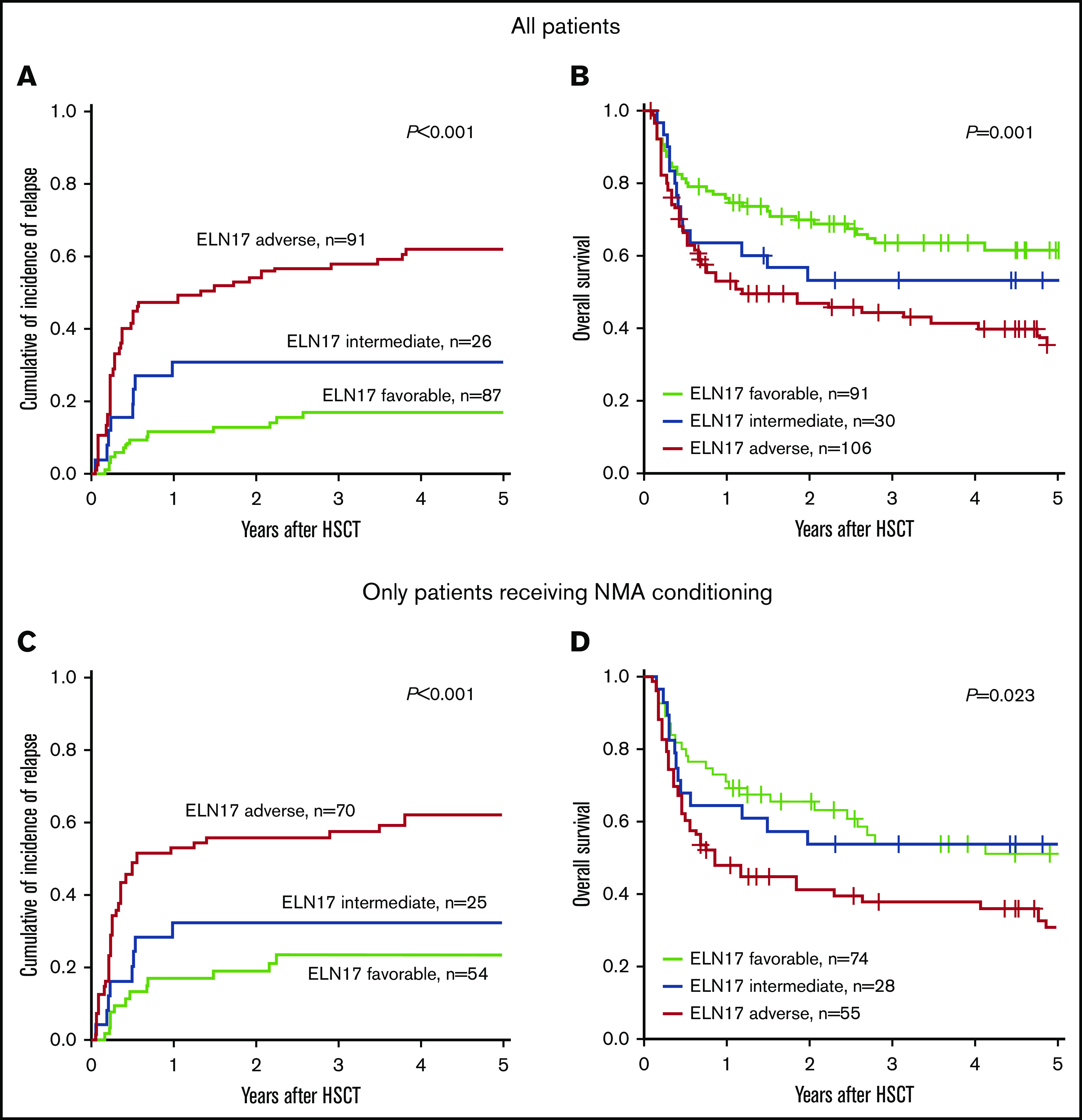
Prognostic impact of the ELN2017 risk classification in patients with AML who undergo allogeneic HSCT. Allocation to the 3 risk groups according to the ELN2017 classification predicted CIR (A) and OS (B) in patients with AML who undergo allogeneic HSCT. The prognostic impact, CIR (C) and OS (D) was retained in patients who underwent allogeneic HSCT after NMA conditioning.
In multivariate analyses, the ELN2017 risk classification remained a significant prognostic factor for CIR and OS after adjustment for age at allogeneic HSCT, hemoglobin level, platelets, and blast count in peripheral blood and bone marrow at diagnosis (Table 2; supplemental Figure 7).
Table 2.
Multivariate analyses of patients with AML who underwent allogeneic HSCT
| Variable | CIR | OS | ||
|---|---|---|---|---|
| HR (95% CI)* | P | OR (95% CI)† | P | |
| ELN2017 genetic-risk group (adverse vs intermediate vs favorable) | 2.95 (2.09-4.18) | <0.001 | 0.69 (0.54-0.90) | .005 |
| Age at HSCT, y | — | — | .95 (0.93-0.97) | <.001 |
| Hemoglobin at diagnosis, g/dL | — | — | 1.13 (1.0-1.26) | .04 |
| Platelets at diagnosis, ×109/L | — | — | 1.00 (1.00-1.01) | .06 |
| Bone marrow blasts at diagnosis, % | — | — | 1.02 (1.00-1.03) | .01 |
| Peripheral blasts at diagnosis, % | — | — | 0.99 (0.98-1.00) | .07 |
Variables considered in the models were those significant at α = 0.20 in univariate analyses. For the CIR end point, the variables were ELN2017 genetic-risk group (adverse vs intermediate vs favorable), hemoglobin at diagnosis, bone marrow blasts at diagnosis, age at HSCT, disease origin (secondary vs de novo), and remission status at HSCT (complete remission vs no complete remission). For the OS end point, the variables were ELN2017 genetic-risk group, hemoglobin at diagnosis, platelets at diagnosis, bone marrow blasts at diagnosis, peripheral blood blasts at diagnosis, age at HSCT, disease origin (secondary vs de novo), remission status at HSCT (complete remission vs no complete remission), HLA antigen match (antigen match vs mismatch), and application of the MAC regimen (MAC vs RIC vs NMA).
EVI1, ecotropic viral integration site-1; HR, hazard ratio.
Hazard ratio, <1 (>1) indicates a lower (higher) risk of relapse for the first category of dichotomous variable.
Odds ratio, <1 (>1) indicates a lower (higher) chance of survival for the first category of dichotomous variable.
When we restricted our 2-year postallogeneic HSCT analyses to patients with AML who underwent NMA-HSCT, the ELN2017 risk classification retained its prognostic impact on CIR (favorable, 18.8%; intermediate, 32.0%; and adverse; 55.7%; P < .001; Figure 1C) and OS (favorable, 65.1%; intermediate, 53.6%; and adverse, 41.1%; P = .02; Figure 1D). A prognostic significance of the ELN2017 risk classifications was also observed when we restricted our analyses to patients who underwent allogeneic HSCT after attaining CR1 (CIR, P < .001; OS, P = .03; supplemental Figure 2), patients who underwent HSCT after attaining CR without detectable MRD (CIR, P < .001; OS, P = .09; supplemental Figure 3), and older patients with AML (≥60 years at diagnosis; CIR, P < .001; OS, P = .03; supplemental Figure 4).
Prognostic impact of distinct mutations in the context of the ELN2017 risk classification
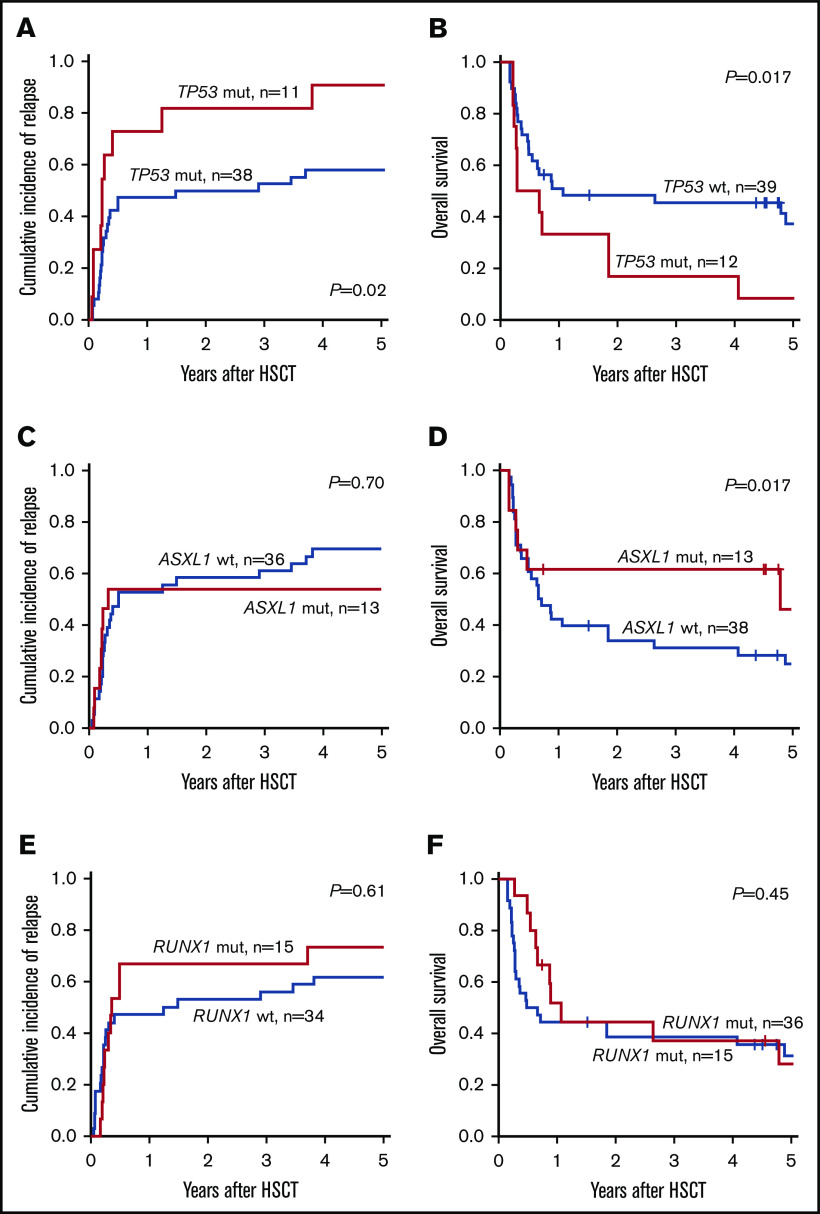
In the updated ELN2017 risk classification, TP53, RUNX1, and ASXL1 mutations were added to the definition of patients with AML with adverse prognosis in the absence of favorable risk aberrations. When we analyzed patients classified as adverse according to ELN2017 classification, the subgroup of patients harboring TP53 mutations had significantly impaired outcomes. Two years after allogeneic HSCT, patients with the TP53 mutation had higher CIR (81.8% vs 50.0%; P = .02; Figure 2A) and shorter OS (16.7% vs 48.4%; P = .017; Figure 2B) compared with patients who had adverse risk without TP53 mutations. In contrast, the presence of ASXL1 or RUNX1 mutation was not associated with a worse prognosis compared with patients adverse risk who lacked those mutations (ASXL1: CIR, P = .70; Figure 2C and OS, P = .17; Figure 2D; RUNX1: CIR, P = .61; Figure 2E and OS, P = .45; Figure 2F).
Figure 2.
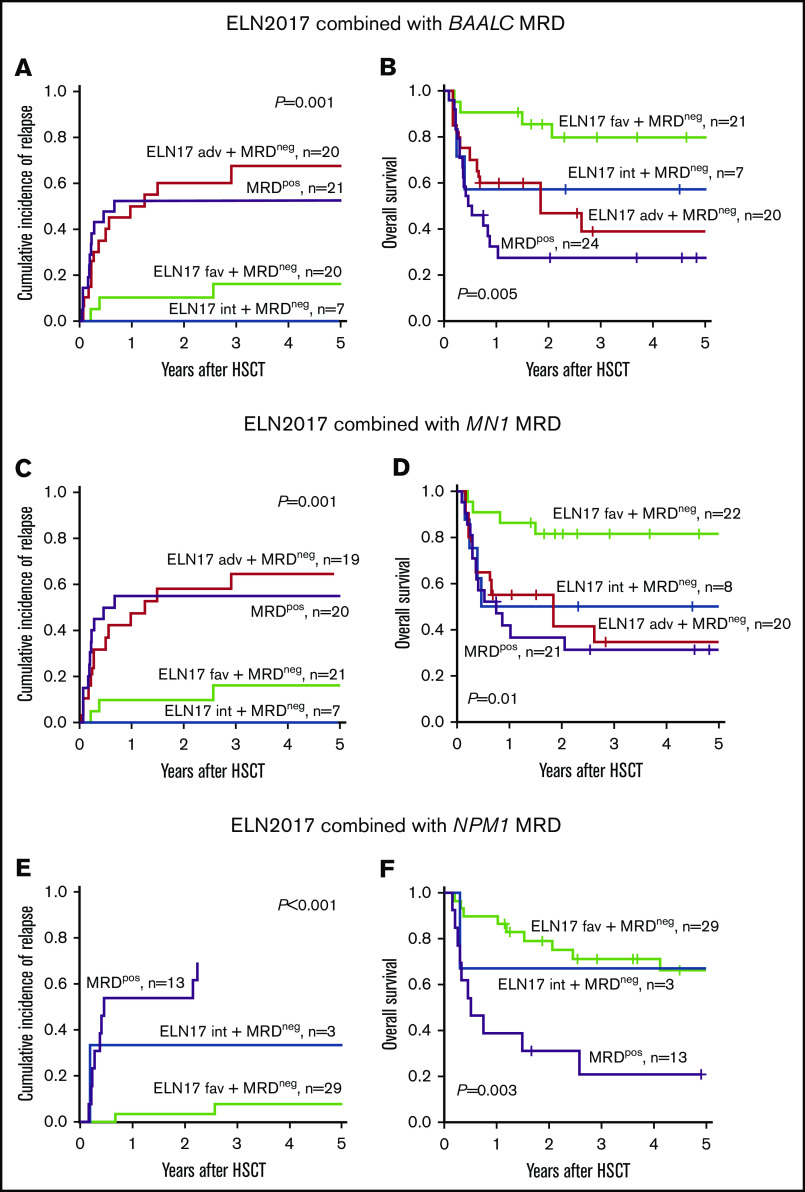
Impact of MRD on the outcome of patients with AML in the context of ELN2017 risk classification. CIR and OS of patients who were MRD− categorized according to the ELN2017 classification compared with BAALC MRD+ (A-B) and MN1 MRD+ (C-D) patients or those with NPM1-mutated MRD (E-F).
In addition, we analyzed the impact of the newly introduced FLT3-ITD allelic ratio across all 3 ELN2017 genetic-risk groups in our cohort consisting exclusively of patients with AML whose consolidation therapy was allogeneic HSCT. In this relatively small subset of patients with FLT3-ITD and available outcome data (n = 33), we observed no significant difference in CIR or OS between patients with a low (<0.5) or high (≥0.5) FLT3-ITD allelic ratio (CIR, P = .45; OS, P = .73; supplemental Figure 5). However, these findings should be validated in larger transplantation cohorts.
Association of the ELN2017 classification and presence of MRD before allogeneic HSCT
We also sought to determine whether assignment to a specific ELN2017 risk group correlates with the presence of MRD in patients in CR or CRi before undergoing allogeneic HSCT. There was no association between the presence of MRD, based on the expression of BAALC and MN1 or detectable NPM1 mutation and ELN2017 risk group (BAALC, P = .99; MN1, P = .99; mutated NPM1, P = .55; Table 3). Regarding outcome, patients who were BAALC MRD+ before allogeneic HSCT had unfavorable outcomes 2 years after allogeneic HSCT, irrespective of the ELN2017 risk category, comparable to patients allocated to the ELN2017 adverse-risk group (CIR, 52.4% vs 60.0%; Figure 3A; OS, 27.5% vs 46.7%; Figure 3B). Patients in the MN1 MRD+ and the ELN2017 adverse-risk groups consistently showed comparably impaired outcomes 2 years after allogeneic HSCT (CIR, 55.0% vs 57.9%; Figure 3C; OS, 36.7% vs 41.2%; Figure 3D). Outcomes of patients with NPM1-mutated MRD before allogeneic HSCT were considerably worse at the 2-year follow-up, compared with the patients who were MRD− with NPM1 mutations (CIR, 53.8% vs 36.8%; Figure 3E; OS, 30.8% vs 72.8%; Figure 3F), which is in line with previously published data.9,48
Table 3.
Association of ELN2017 genetic-risk group and the presence of MRD in patients with AML before allogeneic HSCT
| Characteristics | All (N = 234) | ELN2017 favorable (n = 93) | ELN2017 intermediate (n = 30) | ELN2017 adverse (n = 111) | P* |
|---|---|---|---|---|---|
| BAALC MRD at HSCT, n (%) | .99 | ||||
| Negative | 49 (68.1) | 21 (67.7) | 8 (66.7) | 20 (69.0) | |
| Positive | 23 (31.9) | 10 (32.3) | 4 (33.3) | 9 (31.0) | |
| MN1 MRD at HSCT, n (%) | .99 | ||||
| Negative | 50 (70.4) | 22 (71.0) | 8 (66.7) | 20 (71.4) | |
| Positive | 21 (29.6) | 9 (29.0) | 4 (33.3) | 8 (28.6) | |
| Mutated NPM1 MRD at HSCT, n (%) | .55 | ||||
| Negative | 32 (71.1) | 29 (69.0) | 3 (100.0) | — | |
| Positive | 13 (28.9) | 13 (31.0) | 0 (0.0) | — |
Fisher’s exact test, comparing the 3 ELN2017 genetic-risk groups.
Figure 3.
Prognostic impact of TP53, ASXL1, and RUNX1 mutations in the ELN2017 adverse-risk group. In patients with adverse risk, the presence of a TP53 mutation associated with higher CIR (A) and shorter OS (B). In contrast, patients who harbored mutations in ASXL1 (C-D) or RUNX1 (E-F) had CIR and OS comparable to that of patients with adverse risk who lacked these mutations.
Discussion
With the introduction of high-throughput sequencing, the molecular landscape of AML is evolving into a complex network with distinct mutation patterns, and the identification of significant novel genetic aberrations is continuous. Therefore, a reliable disease classification based on the AML genetic background is needed to stratify patients into clinical risk groups and to subsequently identify the optimal treatment of individual patients. The updated ELN2017 risk classification includes novel findings on cytogenetic and molecular aberrations.9 However, its prognostic value, especially in the context of an allogeneic HSCT, remains to be fully investigated. We present, to our knowledge, the first study to examine the prognostic impact of the ELN2017 classification in an AML cohort homogeneously treated with allogeneic HSCT.
In our cohort of 234 patients with AML, the ELN2017 classification distinguished 3 risk groups, with patients with favorable risk having the lowest relapse and longest survival rates and those with adverse risk having significantly impaired outcomes, even after undergoing allogeneic HSCT. In addition, in multivariate analyses, the allocation to the 3 ELN2017 risk groups was an independent predictor of CIR and OS after allogeneic HSCT. We previously examined the prognostic impact of the ELN2010 risk classification in an AML cohort homogeneously treated with allogeneic HSCT after NMA conditioning. In this study Bill et al found reduced prognostic significance of the ELN2010 risk classification after allogeneic HSCT, and CIR only trended toward significance in their 4 risk groups.8 For the ELN2017 classification, we observed a significant association of the allocation to the 3 risk groups and CIR after allogeneic HSCT, which may indicate an improved prognostic influence of the updated ELN classification in the context of an allogeneic HSCT. This result was supported by the lower Bayesian information criterion of the ELN2017, compared with the ELN2010 classification for the prediction of CIR (872.7 vs 882.9). In our cohort, most patients received NMA conditioning (71.1%) which may bias the statistical analyses and subsequent conclusions. However, when we excluded all NMA-conditioned patients, the ELN2017 classification retained its prognostic impact on CIR after allogeneic HSCT (P < .001). In addition, the conditioning regimen was not an independent predictor of CIR or OS in multivariate analyses of this AML set.
Comparable to our results, the Japan Adult Leukemia Study Group demonstrated improved prognostic significance of the ELN2017 risk classification compared with the ELN2010 version in a heterogeneously consolidated cohort.36 In line with our findings, the outcome of patients with low and high FLT3-ITD allelic ratio did not differ significantly in this cohort.36 Similarly, Boddu et al reported a caveat related to the lack of data on the presence of biallelic CEBPA mutations, with significantly different outcomes according to allocation to the 3 ELN2017 risk groups, but no significant impact of the FLT3-ITD allelic ratio on survival in the context of the NPM1 mutation status in this cohort of mainly younger patients with AML (median age, 51 years).35 This finding was attributed mainly to the high percentage of patients who underwent allogeneic HSCT in the subgroup with NPM1 wild-type and high FLT3-ITD allelic ratios.35 In another study, Sakaguchi et al also questioned the prognostic impact of the newly introduced FLT3-ITD allelic ratio and showed that patients with the NPM1 mutation and a low FLT3-ITD allelic ratio and, although classified as favorable according to ELN2017 recommendations, had significantly improved survival when they underwent allogeneic HSCT after attaining CR1.51 Straube et al validated the prognostic significance of the ELN2017 classification in 3 independent AML cohorts (2 consisting of adult patients and 1 of pediatric patients). They also analyzed the prognostic impact of the FLT3-ITD allelic ratio and reported that patients with NPM1 mutation and low FLT3-ITD allelic ratio seemed to have impaired prognoses compared with patients with NPM1 mutation and wild-type FLT3, especially in the subset of younger patients (<60 years).34 However, only a small number of patients who underwent allogeneic HSCT were included, preventing a further subgroup analyses based on the application of consolidation therapy. With the drawback of a low sample of patients, patients with high and low FLT3-ITD allelic ratio had a comparable outcome after allogeneic HSCT in our cohort (supplemental Figure 5), which is in context with other publications and suggests that a potentially negative prognostic impact of a high FLT3-ITD allelic ratio may be overcome by allogeneic HSCT.
When we analyzed the subgroups in the ELN2017 adverse-risk group based on the newly introduced aberrations, we noted that patients with TP53 mutations or a monosomal karyotype had particularly impaired outcomes compared with others in the group (Figure 2A-B; supplemental Figure 6). This observation indicates the lack of sufficient treatment strategies for this subgroup of patients, nearly all of whom had relapsed at 2 years after allogeneic HSCT (TP53-mutated CIR, 81.8%; monosomal karyotype CIR, 77.3%).
We also analyzed whether the ELN2017 risk classification correlates with the presence of MRD before allogeneic HSCT. For the available MRD data in our cohort consisting of mutated NPM1 MRD and MRD based on the expression of the genes BAALC and MN1 we did not observe significant differences in the presence of MRD pre-HSCT for the 3 risk groups. Regarding outcomes, patients who had detectable NPM1, BAALC, or MN1 MRD at allogeneic HSCT had unfavorable prognoses, irrespective of the allocated ELN2017 risk group, comparable to patients with AML ELN2017 classified as having an adverse genetic risk. These findings demonstrate that detectable MRD in AML is a negative predictor,52,53 and that the MRD status, if available, should be considered in addition to the genetic risk at diagnosis and closely monitored during the disease course to further optimize treatment decisions. The impaired prognosis of MRD+ patients with AML, even after allogeneic HSCT, also raises the question of whether these patients may benefit from treatment intensification, such as additional therapy before or additional maintenance therapy after allogeneic HSCT. However, prospective randomized data on how MRD could guide treatment decisions are scarce, preventing the routine use of MRD-guided therapy in clinical practice, to date.
In this study, we are the first, to our knowledge, to validate the prognostic impact of the ELN2017 genetic-risk classification in an AML cohort that homogeneously underwent consolidation therapy with allogeneic HSCT. Our data emphasize the exceptional adverse prognosis of patients with the TP53 mutation, monosomal karyotype, or MRD present before allogeneic HSCT. In addition to determining the ELN2017 genetic risk, detection of MRD in AML should be further standardized in the near future to help guide treatment decisions.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Laura Kloss, Janet Bogardt, Annette Jilo, Dagmar Cron, Ines Kovacs, Kathrin Wildenberger, Scarlett Schwabe, Christine Günther, Daniela Bretschneider, Evelin Hennig, and Christel Müller for their assistance.
This work was supported by the Deutsche José-Carreras-Stiftung (grants 04R/2016 and PS15/05) (J.G.), Verein Zusammen gegen den Krebs eV, and Ein Herz für Kinder eV.
Footnotes
Original data are available by e-mail request to the corresponding author (sebastian.schwind@medizin.uni-leipzig.de).
Authorship
Contribution: J.G., M.J., M.B., K.G., and J.S. performed the experiments; J.G., M.J., and S.S. analyzed the results; J.G. prepared the figures; J.G. and S.S. designed the research and wrote the paper; D.N., U.P., and S.S. provided administrative support; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sebastian Schwind, Medical Clinic and Policlinic 1, Hematology and Cellular Therapy University Hospital Leipzig, Liebigstr 22; Haus 7, 04103 Leipzig, Germany; e-mail: sebastian.schwind@medizin.uni-leipzig.de.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136-1152. [DOI] [PubMed] [Google Scholar]
- 2.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döhner H, Estey EH, Amadori S, et al. ; European LeukemiaNet . Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453-474. [DOI] [PubMed] [Google Scholar]
- 5.Röllig C, Bornhäuser M, Thiede C, et al. Long-term prognosis of acute myeloid leukemia according to the new genetic risk classification of the European LeukemiaNet recommendations: evaluation of the proposed reporting system. J Clin Oncol. 2011;29(20):2758-2765. [DOI] [PubMed] [Google Scholar]
- 6.Mrózek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alpermann T, Kern W, Schnittger S, et al. Evaluation of the proposed reporting system of the European LeukemiaNet and recommendations for prognosis of acute myeloid leukemia. Leuk Res. 2013;37(2):197-200. [DOI] [PubMed] [Google Scholar]
- 8.Bill M, Jentzsch M, Grimm J, et al. Prognostic impact of the European LeukemiaNet standardized reporting system in older AML patients receiving stem cell transplantation after non-myeloablative conditioning. Bone Marrow Transplant. 2017;52(6):932-935. [DOI] [PubMed] [Google Scholar]
- 9.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufour A, Schneider F, Metzeler KH, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28(4):570-577. [DOI] [PubMed] [Google Scholar]
- 11.Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol. 2010;28(16):2739-2747. [DOI] [PubMed] [Google Scholar]
- 12.Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100(8):1343-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlenk RF, Taskesen E, van Norden Y, et al. The value of allogeneic and autologous hematopoietic stem cell transplantation in prognostically favorable acute myeloid leukemia with double mutant CEBPA. Blood. 2013;122(9):1576-1582. [DOI] [PubMed] [Google Scholar]
- 14.Taskesen E, Bullinger L, Corbacioglu A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469-2475. [DOI] [PubMed] [Google Scholar]
- 15.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CAJ, van Putten WLJ, Valk PJM, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale RE, Green C, Allen C, et al. ; Medical Research Council Adult Leukaemia Working Party . The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111(5):2776-2784. [DOI] [PubMed] [Google Scholar]
- 17.Pratcorona M, Brunet S, Nomdedéu J, et al. ; Grupo Cooperativo Para el Estudio y Tratamiento de las Leucemias Agudas Mieloblásticas . Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121(14):2734-2738. [DOI] [PubMed] [Google Scholar]
- 18.Schlenk RF, Kayser S, Bullinger L, et al. ; German-Austrian AML Study Group . Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014;124(23):3441-3449. [DOI] [PubMed] [Google Scholar]
- 19.Breems DA, Van Putten WLJ, De Greef GE, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26(29):4791-4797. [DOI] [PubMed] [Google Scholar]
- 20.Cornelissen JJ, Breems D, van Putten WLJ, et al. Comparative analysis of the value of allogeneic hematopoietic stem-cell transplantation in acute myeloid leukemia with monosomal karyotype versus other cytogenetic risk categories. J Clin Oncol. 2012;30(17):2140-2146. [DOI] [PubMed] [Google Scholar]
- 21.Kayser S, Zucknick M, Döhner K, et al. ; German-Austrian AML Study Group . Monosomal karyotype in adult acute myeloid leukemia: prognostic impact and outcome after different treatment strategies. Blood. 2012;119(2):551-558. [DOI] [PubMed] [Google Scholar]
- 22.Medeiros BC, Othus M, Fang M, Roulston D, Appelbaum FR. Prognostic impact of monosomal karyotype in young adult and elderly acute myeloid leukemia: the Southwest Oncology Group (SWOG) experience. Blood. 2010;116(13):2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middeke JM, Herold S, Rücker-Braun E, et al. ; Study Alliance Leukaemia (SAL) . TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2016;172(6):914-922. [DOI] [PubMed] [Google Scholar]
- 24.Bowen D, Groves MJ, Burnett AK, et al. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia. 2009;23(1):203-206. [DOI] [PubMed] [Google Scholar]
- 25.Gaidzik VI, Teleanu V, Papaemmanuil E, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features [published correction appears in Leukemia. 2016;30(11):2282]. Leukemia. 2016;30(11):2160-2168. [DOI] [PubMed] [Google Scholar]
- 26.Gaidzik VI, Bullinger L, Schlenk RF, et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011;29(10):1364-1372. [DOI] [PubMed] [Google Scholar]
- 27.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22(8):1539-1541. [DOI] [PubMed] [Google Scholar]
- 28.Mendler JH, Maharry K, Radmacher MD, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol. 2012;30(25):3109-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschka P, Schlenk RF, Gaidzik VI, et al. ASXL1 mutations in younger adult patients with acute myeloid leukemia: a study by the German-Austrian Acute Myeloid Leukemia Study Group. Haematologica. 2015;100(3):324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratcorona M, Abbas S, Sanders MA, et al. Acquired mutations in ASXL1 in acute myeloid leukemia: prevalence and prognostic value. Haematologica. 2012;97(3):388-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnittger S, Eder C, Jeromin S, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27(1):82-91. [DOI] [PubMed] [Google Scholar]
- 33.Tang J-L, Hou H-A, Chen C-Y, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114(26):5352-5361. [DOI] [PubMed] [Google Scholar]
- 34.Straube J, Ling VY, Hill GR, Lane SW. The impact of age, NPM1mut, and FLT3ITD allelic ratio in patients with acute myeloid leukemia. Blood. 2018;131(10):1148-1153. [DOI] [PubMed] [Google Scholar]
- 35.Boddu PC, Kadia TM, Garcia-Manero G, et al. Validation of the 2017 European LeukemiaNet classification for acute myeloid leukemia with NPM1 and FLT3-internal tandem duplication genotypes. Cancer. 2019;125(7):1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada Y, Nagata Y, Kihara R, et al. ; Japan Adult Leukemia Study Group JALSG . Prognostic analysis according to the 2017 ELN risk stratification by genetics in adult acute myeloid leukemia patients treated in the Japan Adult Leukemia Study Group (JALSG) AML201 study. Leuk Res. 2018;66:20-27. [DOI] [PubMed] [Google Scholar]
- 37.Shah MV, Jorgensen JL, Saliba RM, et al. Early Post-Transplant Minimal Residual Disease Assessment Improves Risk Stratification in Acute Myeloid Leukemia. Biol Blood Marrow Transplant. 2018;24(7):1514-1520. [DOI] [PubMed] [Google Scholar]
- 38.Thomas ED, Clift RA, Hersman J, et al. Marrow transplantation for acute nonlymphoblastic leukemic in first remission using fractionated or single-dose irradiation. Int J Radiat Oncol Biol Phys. 1982;8(5):817-821. [DOI] [PubMed] [Google Scholar]
- 39.McSweeney PA, Niederwieser D, Shizuru JA, et al. Hematopoietic cell transplantation in older patients with hematologic malignancies: replacing high-dose cytotoxic therapy with graft-versus-tumor effects. Blood. 2001;97(11):3390-3400. [DOI] [PubMed] [Google Scholar]
- 40.Hegenbart U, Niederwieser D, Sandmaier BM, et al. Treatment for acute myelogenous leukemia by low-dose, total-body, irradiation-based conditioning and hematopoietic cell transplantation from related and unrelated donors. J Clin Oncol. 2006;24(3):444-453. [DOI] [PubMed] [Google Scholar]
- 41.Fleischhack G, Hasan C, Graf N, Mann G, Bode U. IDA-FLAG (idarubicin, fludarabine, cytarabine, G-CSF), an effective remission-induction therapy for poor-prognosis AML of childhood prior to allogeneic or autologous bone marrow transplantation: experiences of a phase II trial. Br J Haematol. 1998;102(3):647-655. [DOI] [PubMed] [Google Scholar]
- 42.Parker JE, Pagliuca A, Mijovic A, et al. Fludarabine, cytarabine, G-CSF and idarubicin (FLAG-IDA) for the treatment of poor-risk myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol. 1997;99(4):939-944. [DOI] [PubMed] [Google Scholar]
- 43.Schmid C, Schleuning M, Ledderose G, Tischer J, Kolb H-J. Sequential regimen of chemotherapy, reduced-intensity conditioning for allogeneic stem-cell transplantation, and prophylactic donor lymphocyte transfusion in high-risk acute myeloid leukemia and myelodysplastic syndrome. J Clin Oncol. 2005;23(24):5675-5687. [DOI] [PubMed] [Google Scholar]
- 44.Jentzsch M, Bill M, Nicolet D, et al. Prognostic impact of the CD34+/CD38- cell burden in patients with acute myeloid leukemia receiving allogeneic stem cell transplantation. Am J Hematol. 2017;92(4):388-396. [DOI] [PubMed] [Google Scholar]
- 45.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107(10):4011-4020. [DOI] [PubMed] [Google Scholar]
- 46.Benthaus T, Schneider F, Mellert G, et al. Rapid and sensitive screening for CEBPA mutations in acute myeloid leukaemia. Br J Haematol. 2008;143(2):230-239. [DOI] [PubMed] [Google Scholar]
- 47.Grimm J, Bill M, Jentzsch M, et al. Clinical impact of clonal hematopoiesis in acute myeloid leukemia patients receiving allogeneic transplantation. Bone Marrow Transplant. 2019;54(8):1189-1197. [DOI] [PubMed] [Google Scholar]
- 48.Bill M, Grimm J, Jentzsch M, et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann Hematol. 2018;97(10):1757-1765. [DOI] [PubMed] [Google Scholar]
- 49.Jentzsch M, Bill M, Grimm J, et al. Prognostic Impact of Blood MN1 Copy Numbers Before Allogeneic Stem Cell Transplantation in Patients With Acute Myeloid Leukemia. HemaSphere. 2019;3(1):e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jentzsch M, Bill M, Grimm J, et al. High BAALC copy numbers in peripheral blood prior to allogeneic transplantation predict early relapse in acute myeloid leukemia patients. Oncotarget. 2017;8(50):87944-87954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakaguchi M, Yamaguchi H, Najima Y, et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2(20):2744-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araki D, Wood BL, Othus M, et al. Allogeneic Hematopoietic Cell Transplantation for Acute Myeloid Leukemia: Time to Move Toward a Minimal Residual Disease-Based Definition of Complete Remission? J Clin Oncol. 2016;34(4):329-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N Engl J Med. 2018;378(13):1189-1199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.