Spotted fever group rickettsioses (SFGR), typhus group rickettsioses (TGR), scrub typhus (caused by Orientia tsutsugamushi), ehrlichiosis, and anaplasmosis often present as undifferentiated fever but are not treated by agents (penicillins and cephalosporins) typically used for acute febrile illness. Inability to diagnose these infections when the patient is acutely ill leads to excess morbidity and mortality. Failure to confirm these infections retrospectively if a convalescent blood sample is not obtained also impairs epidemiologic and clinical research.
KEYWORDS: spotted fever and typhus group rickettsioses, scrub typhus, ehrlichioses, anaplasmosis, Orientia spp., Rickettsia spp., Ehrlichia chaffeensis, Anaplasma phagocytophilum, Rickettsiales, ticks, diagnostics, etiology of fever studies
ABSTRACT
Spotted fever group rickettsioses (SFGR), typhus group rickettsioses (TGR), scrub typhus (caused by Orientia tsutsugamushi), ehrlichiosis, and anaplasmosis often present as undifferentiated fever but are not treated by agents (penicillins and cephalosporins) typically used for acute febrile illness. Inability to diagnose these infections when the patient is acutely ill leads to excess morbidity and mortality. Failure to confirm these infections retrospectively if a convalescent blood sample is not obtained also impairs epidemiologic and clinical research. We designed a multiplex real-time quantitative PCR (qPCR) assay to detect SFGR, TGR, O. tsutsugamushi, and infections caused by Anaplasma phagocytophilum and Ehrlichia chaffeensis with the ompA, 17-kDa surface antigen gene, tsa56, msp2 (p44), and vlpt gene targets, respectively. Analytical sensitivity was ≥2 copies/μl (linear range, 2 to 2 × 105) and specificity was 100%. Clinical sensitivities for SFGR, TGR, and O. tsutsugamushi were 25%, 20%, and 27%, respectively, and specificities were 98%, 99%, and 100%, respectively. Clinical sensitivities for A. phagocytophilum and E. chaffeensis were 93% and 84%, respectively, and specificities were 99% and 98%, respectively. This multiplex qPCR assay could support early clinical diagnosis and treatment, confirm acute infections in the absence of a convalescent-phase serum sample, and provide the high-throughput testing required to support large clinical and epidemiologic studies. Because replication of SFGR and TGR in endothelial cells results in very low bacteremia, optimal sensitivity of qPCR for these rickettsioses will require use of larger volumes of input DNA, which could be achieved by improved extraction of DNA from blood and/or extraction of DNA from a larger initial volume of blood.
INTRODUCTION
The burden and clinical impact of rickettsioses and related infections is increasingly recognized (1–7). Spotted fever group rickettsioses (SFGR), typhus group rickettsioses (TGR), and human granulocytic anaplasmosis (HGA) are causes of acute febrile illness worldwide. Human monocytic ehrlichiosis (HME), caused by Ehrlichia chaffeensis, also results in acute febrile illness across the Americas, whereas scrub typhus has been found only in the Asia-Pacific triangle until recently (3, 8). Recent data suggest both increasing incidence and significant underreporting (<25%) of rickettsioses (3, 9–13). Little is known about anaplasmosis in tropical regions (14–20). The current diagnostic reference standard for agents responsible for rickettsioses (SFGR and TGR), scrub typhus (Orientia tsutsugamushi), and ehrlichioses (HGA and HME) is a 4-fold rise in IgG antibody titer between paired acute- and convalescent-phase serum samples by indirect immunofluorescence (IFA), which is laborious, subjective, and intrinsically retrospective. Furthermore, IFA cannot absolutely distinguish between SFGR and TGR or between Anaplasma phagocytophilum and E. chaffeensis without testing for both agents of rickettsioses and ehrlichioses because of serologic cross-reactions. Rapid sensitive and specific detection of rickettsial agents could avert needless morbidity and mortality (21, 22) related to failure to prescribe needed doxycycline (5). High-throughput assays are needed to support large epidemiologic and clinical studies to define the global burden, distribution, and clinical features of rickettsioses and related infections, including the pathogenicity of different species of SFGR (23, 24). To allow high-throughput simultaneous assessment of multiple potential pathogens, including coinfections, we developed a quantitative, multiplex 5′-nuclease quantitative real-time PCR (qPCR) assay to rapidly detect and distinguish SFGR, TGR, O. tsutsugamushi, A. phagocytophilum, and E. chaffeensis.
MATERIALS AND METHODS
Samples.
We used specimens from patients with acute febrile illness in whom rickettsioses were independently confirmed by ≥1 reference method as follows: 4-fold IgG increase in antibody titer, PCR using different primers, culture, or observation of morulae in blood leukocytes (for A. phagocytophilum and E. chaffeensis). These patients included 20 with SFGR, 30 with TGR, 30 with O. tsutsugamushi infection, 15 with A. phagocytophilum infection, and 45 with E. chaffeensis infection. Control samples were from patients with other etiologies of acute febrile illness (including 17 with bloodstream infections, 33 with blood smear and PCR-confirmed malaria [28 caused by Plasmodium falciparum and 5 caused by Plasmodium vivax], and 6 with PCR-confirmed Ehrlichia ewingii infection) or those from patients convalescent from A. phagocytophilum (1) or E. chaffeensis infections (2) (Table 1). Depending upon origin, sample types included EDTA-anticoagulated blood, buffy coat, mononuclear cells, and DNA prepared from whole blood or buffy coat. For blood and buffy coat samples, DNA was prepared using the QIAamp DNA blood minikit (Qiagen), starting with 200 μl and resuspended into 200 μl buffer AE or water as recommended. For samples received as DNA, methods included the QIAamp blood minikit, QIAsymphony DNA midikit, Qiagen DNA extraction kit (Qiagen), the Wizard SV genomic DNA purification system (Promega, Madison, WI, USA), and IsoQuick extraction kit (ORCA Research, Bothell, WA).
TABLE 1.
Specimens used for evaluation of multiplex PCR
| Organism (no.) | Laboratory (no. of samples; location) | Specimen type (no.) | Reference assay (no. positive) | Reference(s) |
|---|---|---|---|---|
| SFGR (20) | National Institute of Health, Ricardo Jorge (18 samples; Portugal); CDC (2 samples; USA) | Blood/buffy coat (18), DNA (2) | Conventional PCR (18), real-time PCR (2), serology (18), culture (18) | 41, 42 |
| TGR (30) | Mahidol Oxford Tropical Medicine Research Unit (20 samples; Thailand); Lao-Oxford-Mahosot Hospital-Wellcome Trust Research Unit (10 samples; Lao PDR) | Blood/buffy coat (10), DNA (20) | Real-time PCR (30), serology (10), culture (10) | 35 |
| O. tsutsugamushi (30) | Mahidol Oxford Tropical Medicine Research Unit (20 samples; Thailand); Lao-Oxford-Mahosot Hospital-Wellcome Trust Research Unit (10 samples; Lao PDR) | Blood/buffy coat (10), DNA (20) | Real-time PCR (30), serology (10), culture (10) | 35 |
| E. chaffeensis (45) | Johns Hopkins University School of Medicine (USA); Mayo Clinic (USA); University of Texas Medical Branch (USA) | Blood (6), DNA (39) | Conventional PCR (12), real-time PCR (32), serology (10), morulae (1), culture (4) | 26, 43–45 |
| A. phagocytophilum (15) | Johns Hopkins University School of Medicine (USA) | Blood | Real-time PCR (15), serology (11), morulae (14), culture (4) | 26 |
| Bloodstream infections (17) | Johns Hopkins Hospital/University School of Medicine (USA) | Blood | Blood culture (17) | 26 |
| P. falciparum (28) | Johns Hopkins Hospital/University School of Medicine (USA) | Blood | Blood smear (28), real-time PCR (22) | 36 |
| P. vivax (5) | Johns Hopkins Hospital/University School of Medicine (USA) | Blood | Blood smear (5), real-time PCR (5) | 36 |
| E. chaffeensis (6) | USA reference laboratory (Mayo Clinic) | Blood | Real-time PCR (6) | 45 |
| Convalescent from E. chaffeensis or A. phagocytophilum (3) | Johns Hopkins University School of Medicine (USA) | Blood | Conventional PCR (3), serology (3), morulae (2), culture (3) | 26 |
Multiplex PCR assay development.
As previously reported (25, 26), we used AlleleID 6 (Premier Biosoft, Palo Alto, CA) software to design primers and probes. Since the conditions for amplification are standardized and the specific design of primers and probes identical, the assays were run similarly. However, there were a number of smaller changes, including the quantity of DNA input used, the range of standards and controls tested, and the total number of highly pedigreed samples tested. Our gene targets included a SFGR consensus (23 members of SFGR) ompA sequence, the genus-wide 17-kDa lipoprotein gene optimized for Rickettsia typhi (TGR), a consensus conserved region of the O. tsutsugamushi 56-kDa major outer membrane protein gene, the A. phagocytophilum msp2 (p44) gene, and the E. chaffeensis vlpt gene (sequences in Table 2).
TABLE 2.
Primers and probes used for multiplex PCR
| Target pathogen | Accession no. | Target gene | Forward primer | Reverse primer | Probe |
|---|---|---|---|---|---|
| SFGR | NC_009882.1 | SFGR ompA consensus | TTGTCAGGCTCTGAAGCTAAAC | AGCACCTGCCGTTGTGATATC | FAM-TAGCCGCAGTCCCTACAACACCGC-BHQ1 |
| TGR | M28481 | R. typhi 17-kDa antigen gene TGR optimized | ACTTGGTTCTCAATTCGGTCAC | CAGACTTGCACCGATTTGTCC | TXR-TGCCCCAAGTAATGCGCCTACACC-BHQ1 |
| O. tsutsugamushi | AY836148.1 | Orientia tsutsugamushi strain Kato 56-kDa antigen gene | GGTGGTAATGCTTTCGCTAATCAG | TGCTGCTTCTTGCGCCTGTAG | CY3/HEX-TGCTGCTGTTGCTGCCCTTGCC-BHQ1 |
| E. chaffeensis | AF181986 | vlpt | CTAATTCTGATTTACACGAGTCTTC | GCATCATCTTCGAATTGAACTTC | TET-TTGAGTTACCTGGTCC-BHQ1 |
| A. phagocytophilum | Many | msp2 (5’ conserved domain) | GAAGATGAWGCTGATACAGTA | CAACHGCCTTAGCAAACT | Cy5-TTATCAGTCTGTCCAGTAACA-BHQ1 |
The assay was run as either a pentaplex (Bio-Rad CFX384 PCR instrument) or separate triplex and duplex assays (Bio-Rad IQ5 PCR instrument). DNA from 200 μl of blood was reconstituted in buffer to 200 μl, and 1 to 3 μl of blood/buffy coat DNA was used for all PCR assays. Quantitative results were adjusted for input volume. Controls and standards included DNA from microscopically quantified bacterial cells or infected mammalian cells (positive controls), DNA obtained from the blood of healthy human study participants (negative controls), and no template controls. Plasmid-cloned amplicons were used to generate a standard curve for quantification (100 to 105 copies per reaction). Standards were accepted only when the curve’s R value was >0.90, the PCR efficiency was 85 to 115%, and the limit of detection was ≤10 copies per reaction. Samples run in duplicate or triplicate were accepted as positive only if ≥2 were positive. After initial experiences showed that triplicate was rarely contributory, we reduced the assay to duplicates.
Each measurement was adjusted for input volume or dilution/concentration effects of DNA elution to obtain a final measurement in bacteria per milliliter blood. We used the endpoint analysis program in the CFX Manager wherein the relative fluorescent units (RFUs) for each sample or control are calculated over the final 5 of 40 cycles to establish cutoffs. The positive cutoff value was calculated by identifying the average RFUs for the negative controls for each analyte/fluor and by adding a percentage of the range of RFUs on each plate for each analyte/fluor (highest to lowest RFU = range). We used data generated by testing 101 samples (14 A. phagocytophilum samples, 5 E. chaffeensis samples, 20 O. tsutsugamushi samples, 6 SFGR samples, 20 TGR samples, 3 convalescent samples of A. phagocytophilum and E. chaffeensis, 23 P. falciparum samples, and 20 negative controls) to compare the receiver operator characteristic (ROC) curves (x axis, 1 specificity; y axis, sensitivity at each cutoff) (27) for cutoffs set at 2.5, 3, 4, 5, 7.5, 10, 12.5, and 15% of the plate RFU range above the average negative sample RFUs for the analyte and plate. All final results were calculated using the cutoff selected for each pathogen analyte/fluor combination from the ROC curves. Multiple small informal comparisons suggested no differences in detection sensitivity between singleplex versus multiplex testing for each of the analytes (25). To formally determine this, DNA samples from blood obtained from 14 patients with A. phagocytophilum and 66 non-A. phagocytophilum “negative” controls (8 with SFGR, 20 with TGR, 7 with O. tsutsugamushi, 6 with E. chaffeensis, 1 convalescent from A. phagocytophilum and 2 from E. chaffeensis, and 22 with malaria) were run in a singleplex and compared to results using the multiplex assay.
Blood volume and DNA preparation in analytical sensitivity using rickettsiae-spiked blood.
To increase clinical sensitivity for vasculotropic rickettsiae (SFGR, TGR, and O. tsutsugamushi), the roles of starting blood volume, final DNA suspension volume, DNA preparation protocol, and pathogen DNA enrichment and isolation obtained with the MolYsis basic kit (Molzym GmbH & Co., Bremen, Germany) were studied using fresh human blood supplemented with spotted fever rickettsiae (Rickettsia parkeri Portsmouth strain)-infected human brain microvascular endothelial cells for which the quantity was determined by counting the proportion of infected cells among 200 cells and the average quantity of R. parkeri bacteria per infected cell in LeukoStat-stained cytofuged samples. Based on this calculation, an aliquot of 5 × 107 bacteria in endothelial cells was prepared by centrifugation and the pellet suspended in 5 ml of fresh EDTA-anticoagulated human blood. This 5-ml blood sample was then serially diluted (10-fold to 100 bacteria/ml) in human EDTA-anticoagulated blood, and 1 ml from each dilution was used to prepare buffy coat. The buffy coat and residual spiked blood were used for DNA preparation as below.
Effect of input volume of blood and output volume of resuspended DNA.
To examine the role of input blood volume used for extraction of DNA and output volume of DNA (resuspension volume), different methods were employed as follows: (i) DiaSorin/Arrow DNA extraction kit on the DiaSorin (NorDiag) Arrow nucleic acid extraction instrument, (ii) the QIAamp DNA blood minikit (Qiagen, Germantown, MD, USA), and (iii) the MolYsis basic kit (Molzym, Bremen, Germany). For the Arrow method, 500 and 100 μl of rickettsia-spiked blood were resuspended into a final volume of 150 μl and 100 μl buffer, respectively. For the QIAamp DNA blood minikit, 100 μl blood was extracted into 200 μl buffer. For the MolYsis kit, the pellet from 1 ml of blood was extracted using the Arrow protocol and resuspended into 100 μl buffer. Each DNA preparation was then used in the SFGR qPCR protocol.
Whole blood versus buffy coat.
Since buffy coat should increase sensitivity by enriching for host cell-associated rickettsiae, the sensitivity of qPCR using whole blood versus buffy coat was also compared. Using preparations supplemented with R. parkeri-infected endothelial cells as above, blood (200 μl starting volume) or buffy coat from 1 ml of blood (∼200 μl buffy coat after centrifugation) was used to prepare DNA (QIAamp DNA blood minikit), and both DNAs were resuspended in 200 μl DNA buffer. These preparations were then subjected to qPCR.
Ethics.
The study was reviewed by the ethics committee of the Johns Hopkins School of Medicine. Institutional review board (IRB) approval was granted to use archived discarded deidentified samples since consent was deemed both impractical and unnecessary (JHM protocol NA_00021376).
RESULTS
Singleplex versus multiplex qPCR.
Samples from 14 patients with A. phagocytophilum infection, 6 with E. chaffeensis infection, and 60 additional samples from patients with other rickettsial and nonrickettsial infections were tested using both the singleplex assay for A. phagocytophilum and our multiplex assay. We found no decrement in sensitivity with multiplexing. The singleplex assay for A. phagocytophilum detected 13 of 14 A. phagocytophilum infections (sensitivity, 93%) and detected 0 of 6 E. chaffeensis infections and 0 of 60 other infections (specificity, 100%). The multiplex assay correctly identified 12 of the 14 A. phagocytophilum infections (sensitivity, 86%) and 5 of 6 E. chaffeensis infections (sensitivity, 83%); specificity was 100% (66 and 74 other rickettsial and nonrickettsial samples, respectively, were negative). These findings confirmed multiple prior informal comparisons showing no differences in detection sensitivity between singleplex and multiplex assays as also described by others (25, 28).
Multiplex qPCR.
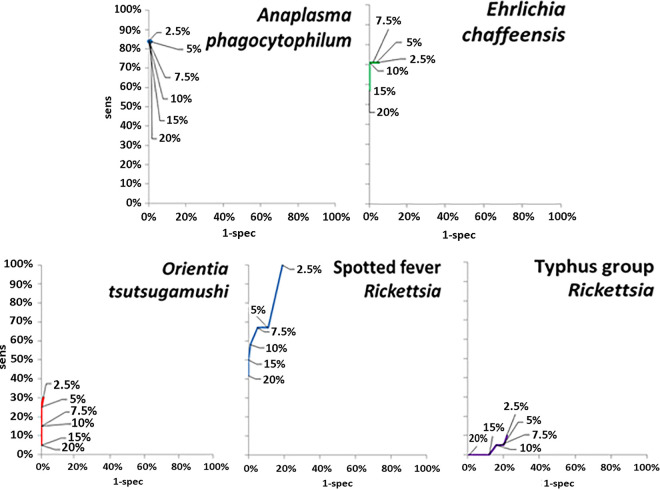
For each run in the pilot studies, the sensitivity and specificity of the multiplex qPCR assay were determined at a range of cutoffs and plotted on ROC curves. As shown in Fig. 1, the point of inflection on the ROC curve at which the highest sensitivity and specificity was achieved (of plate RFU range above the average of negative controls) was at 7.5% for SFGR (sensitivity, 67%; specificity, 95%), 2.5% for TGR (sensitivity, 10%; specificity, 78%), 5% for O. tsutsugamushi (sensitivity, 25%; specificity, 100%), 5% for A. phagocytophilum (sensitivity, 84%; specificity, 100%), and 10% for E. chaffeensis (sensitivity, 71%; specificity, 100%). These values were used to calculate the specific cutoffs for each 96- or 384-well multiplex plate assay for that specific analyte.
FIG 1.
ROC curves used to establish positive cutoffs for multiplex qPCR assays for rickettsioses. The data labels represent the percent of the total RFU range per run added to the average of negative control RFUs in order to establish a cutoff as described in Materials and Methods.
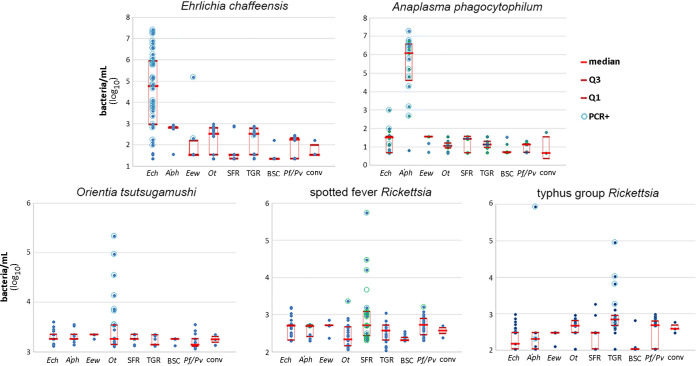
The established cutoffs were used in the full optimization study, from which final clinical sensitivity and specificity for each analyte could be estimated. Using the quantitation derived by comparing average final RFUs for each sample with the standard curve for that pathogen analyte/fluor and adjusting for DNA preparation dilution/concentration and DNA qPCR assay input volume, quantitative multiplex results were obtained (Fig. 2). The clinical diagnostic performance of the multiplex qPCR for the detection of rickettsial agents in the blood of humans with acute febrile illness is shown in Table 3.
FIG 2.
Quantitative multiplex results for the entire cohort by pathogen analyte (graph title). The x axis denotes the true identity of the individual sample tested (Ech, E. chaffeensis; Aph, A. phagocytophilum; Eew, E. ewingii; Ot, O. tsutsugamushi; SFR, spotted fever Rickettsia; TGR, typhus group Rickettsia; BSC, blood culture-positive sample controls; Pf/Pv, P. falciparum/P. vivax; conv, convalescent from E. chaffeensis or A. phagocytophilum infection), and the y axis is the bacterial quantity as discerned from the specific standard curve applied to the plate on which the sample was tested. Each individual solid circle denotes a single sample; open circles denote samples that were PCR positive for that analyte; the median for each group is denoted by the central red bar within the 1st and 3rd quartiles that are denoted by the top and bottom of the red box.
TABLE 3.
Performance of multiplex quantitative PCR for detection of acute rickettsial infections in patients with acute febrile illness
| Bacterium | Gene target | No. true positives | No. test positives | Sensitivitya | True negatives | Test negatives | Specificitya | PPVb | NPVb |
|---|---|---|---|---|---|---|---|---|---|
| Anaplasma phagocytophilum | msp2 (p44) | 14 | 14 | 0.93 | 184 | 182 | 0.99 | 0.88 | 0.99 |
| Ehrlichia chaffeensis | vlpt | 45 | 38 | 0.84 | 154 | 151 | 0.98 | 0.93 | 0.96 |
| Orientia tsutsugamushi | tsa56 (56-kDa surface antigen) | 30 | 8 | 0.27 | 169 | 169 | 1.0 | 1.0 | 0.88 |
| Spotted fever Rickettsia | ompA | 20 | 5 | 0.25 | 176 | 173 | 0.98 | 0.63 | 0.92 |
| Typhus group Rickettsia | 17-kDa surface antigena | 30 | 6 | 0.2 | 167 | 165 | 0.99 | 0.75 | 0.87 |
Sensitivity is the proportion of true positives with a positive test result (test positive/all with disease = test positives plus false negatives); specificity is the proportion of true negatives (nondiseased) with a negative test result (test negative/all without disease = test negatives plus false positives).
PPV, positive predictive value; NPV, negative predictive value.
In light of limited clinical sensitivity but high analytical sensitivity of the multiplex for vasculotropic rickettsioses (spotted fever and typhus group Rickettsia and Orientia tsutsugamushi), we examined the preanalytical role of input blood volume for DNA preparation versus final output of DNA suspension. We hypothesized that using a larger input volume of blood and smaller elution volume of extracted DNA would improve clinical sensitivity of qPCR. As shown in Table 4, input blood volume and elution DNA resuspension volume were correlated with sensitivity of qPCR; the limit of detection was 5 × 103 bacteria/ml when 100 μl of blood was resuspended into 200 μl final buffer (1:2 dilution from blood) but 1 × 102 bacteria/ml when using a method to obtain a 10:1 concentration of DNA from blood (1 ml of blood resuspended into 100 μl buffer). Regardless of method used to prepare DNA from blood, the final concentration of bacteria per microliter of eluted DNA at the limit of detection varied by 10-fold or less (from 0.3 bacteria/μl to 5/μl), which confirmed the excellent analytical sensitivity of the assay and suggested that concentration of blood DNA could further advance clinical sensitivity. Since larger starting blood volumes and smaller final DNA resuspension buffer volumes improved sensitivity, we finally employed the Molzym MolYsis basic kit to enable an input volume of 1 ml of blood with output volume of 100 μl buffer (theoretical 10:1 concentration from blood), which resulted in analytical sensitivity (5 × 102 bacteria/ml) comparable to that obtained with other methods that yield similar blood DNA concentration. Therefore, we next compared using the standard 200-μl input volume of blood versus 200 μl of buffy coat prepared from 1 ml blood with DNA in both cases eluted into 200 μl buffer. The use of buffy coat blood resulted in a 50-fold improvement in analytical sensitivity from 5 × 103 to 102 bacteria/ml blood.
TABLE 4.
The effect of starting blood volume and DNA concentration methods on detection sensitivity in Rickettsia-spiked whole-blood and buffy coat samples
| Method | Sample type | Starting blood vol (μl) | Elution vol (μl) | Concn factor | LODa Rickettsia parkeri/ml in blood | LOD Rickettsia parkeri/μl in eluate |
|---|---|---|---|---|---|---|
| Qiagen blood minikit | Blood | 100 | 200 | 0.5 | 5,000 | 5.0 |
| Arrow v2 | Blood | 100 | 100 | 1.0 | 1,000 | 1.0 |
| Arrow v1 | Blood | 500 | 150 | 3.3 | 333 | 0.3 |
| MolYsis basic/Arrow v1 | Blood | 1,000 | 100 | 10.0 | 500 | 5.0 |
| QIAsymphony | Blood | 1,000 | 100 | 10.0 | 100 | 1.0 |
| Qiagen blood minikit | Blood | 200 | 200 | 1.0 | 5,000 | 0.2 |
| Qiagen blood minikit | Buffy coat | 1,000 | 200 | 5.0 | 100 | 0.1 |
LOD, limit of detection in Rickettsia parkeri Portsmouth strain.
DISCUSSION
New diagnostic approaches are essential to reduce morbidity and mortality from rickettsioses, scrub typhus, and ehrlichiosis worldwide and to support large epidemiologic studies that define the global burden of these infections, including emerging species and ecologic niches of SFGR. Detection of bacteremia due to SFGR, TGR, and O. tsutsugamushi is inherently difficult compared with that due to A. phagocytophilum and E. chaffeensis because endothelial cells are infected rather than circulating leukocytes, which results in very low rickettsemia (29–32). The few molecular assays described to date, which include multiplexing and optimization using highly pedigreed samples, are difficult to compare because of lack of a uniform diagnostic standard comparator and different sample types and storage conditions (33, 34). In general, analytical sensitivity is lower for conventional PCR than for nested PCR and real-time PCR (1,000 to 10 ,000 and <100 to 5,000 genome equivalents/ml of blood DNA, respectively) (33) but heavily dependent on stage and severity of illness (30). A real-time multiplex qPCR assay to detect SFGR, TGR, O. tsutsugamushi, A. phagocytophilum, and E. chaffeensis would be ideal since there is great clinical, epidemiologic, and geographic overlap among them, and well-designed multiplex qPCR assays have similar sensitivity to singleplex assays (28). Finally, any assay with adequate analytical sensitivity requires clinical validation.
Paris et al. (35) described a multiplex qPCR assay for SFGR rickettsiae, TGR, and Orientia using ompB, gltA, and 47-kDa (unique to Orientia) gene targets. The limit of detection by multiplex qPCR was 1 copy/μl for SFGR and TGR and 24 copies/μl for O. tsutsugamushi. Clinical samples evaluated included 12 buffy coat samples from patients with suspected acute rickettsial infections on the basis of IgM- and IgG-based rapid immunochromatographic tests with or without IgM or IgG detection by IFA using paired serum samples. Of the 12 (3 SFGR, 2 TGR, and 7 O. tsutsugamushi samples), 6 were PCR positive (1 for TGR and 5 for O. tsutsugamushi); however, only 2 had a 4-fold rise in IgM and/or IgG antibody titer (1 TGR sample and 1 O. tsutsugamushi sample). A recent review (33), which included studies of >10 patients published since 2013 evaluated with serology and PCR, found that the median clinical sensitivity of real-time PCR for the detection of SFGR and TGR in blood was 18% overall, with SFGR improved (42%) versus TGR (3%). Tshokey et al. (37) evaluated 1,004 febrile patients in Bhutan for acute rickettsial infections, defining acute infection as a single high IgM titer or positive qPCR. Of 1,044 patients, 46 (4.4%), 4 (0.4%), and 70 (6.7%) patients had acute SFGR, TGR, and O. tsutsugamushi infection, respectively; however, only 7 were positive by qPCR for O. tsutsugamushi (4 PCR positive only and 3 qPCR and single-serum sample IFA positive).
To address the unmet need for improved detection of globally-distributed rickettsioses, we previously described development of a multiplex triplex qPCR to detect SFGR, TGR, and O. tsutsugamushi (25), in which analytical sensitivity and specificity were similar to that of Paris et al. (35), as well as development and limited clinical validation of a real-time duplex assay for Ehrlichia and Anaplasma (26). The primary strength of the current study is clinical validation of a 5-target real-time multiplex qPCR assay to detect and distinguish all 5 major rickettsioses and related infections worldwide using a large panel of specimens for which rigorous reference standard testing was completed. We found excellent clinical sensitivity for A. phagocytophilum and E. chaffeensis and clinical sensitivity for SFGR, TGR, and O. tsutsugamushi comparable to that of other reports with many fewer clinical samples and/or unclear confirmatory testing. We do not think that the low sensitivity of our assay for O. tsutsugamushi is due to a choice of antigen gene target since the original primers used in this study were established via identification of highly conserved regions of the 56-kDa antigen gene from 101 sequences deposited into GenBank using the AlleleID algorithms. Pilot studies examining amplification efficacy across the Kato, Karp, and Gilliam strains showed equivalence. Furthermore, the combination of primers and probes, when subjected to a BLAST search against the NCBI RefSeq Genome Database (Orientia [taxid 69474]), identified appropriate targets for amplification in a range of geographically distinct whole genomes, including those from Korea, Japan, Thailand, and even Orientia chuto from Dubai. The sensitivity of our assay is indeed very similar to that observed in other published studies, including those that target the O. tsutsugamushi 47-kDa gene (32–34, 38). It is recognized that the limited clinical sensitivity results from low-level bacteremia (38). Moreover, the 56-kDa antigen gene is a preferred target owing to its specificity for O. tsutsugamushi (31, 33); PCR positivity for this target provides strong evidence of pathogen DNA (33). Although the clinical sensitivities for SFGR, TGR, and O. tsutsugamushi were low (25%, 20%, and 27%, respectively), specificity was excellent; therefore, a positive result confirms acute infection when treatment decisions must be made (need for doxycycline) and when a convalescent-phase serum sample is not available (typical case and fatal cases). Furthermore, the high-throughput (384-well plate) platform of our multiplex PCR assay supports large clinical and epidemiological studies. We found no decrement in sensitivity with multiplexing and experimentally showed that increasing effective input blood volume and decreasing elution volume increased analytical sensitivity for detecting rickettsial DNA under experimental circumstances. Removing host DNA and concentrating microbial DNA from blood, an approach used to increase sensitivity of PCR for Mycobacterium tuberculosis (39), did not increase sensitivity of PCR for the spotted fever group rickettsia R. parkeri beyond that already obtained with other DNA concentration methods that lacked removal of host cell DNA.
In summary, our real-time multiplex qPCR assay showed high clinical specificity for all 5 rickettsial targets but higher clinical sensitivity for leukocytic rickettsiae versus vasculotropic rickettsiae. Further clinical validation of the assay, optimally using buffy coat and/or another method to concentrate nucleic acids (DNA ± RNA) from a larger volume of blood, is needed to yield a limit of detection of 101 to 103 bacteria/ml, the median rickettsemia observed in vivo during vasculotropic rickettsial infections in humans (29, 30, 40).
ACKNOWLEDGMENTS
M.E.R. was supported by a Johns Hopkins Center for Global Health Junior Faculty Grant, a Clinician Scientist Career Development Award from Johns Hopkins School of Medicine, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (K23AIO83931). The work was supported in part by NIAID R01AI44102, R01AI41213, and R21AI080911 grants to J.S.D.
The opinions expressed herein are those of the author(s) and are not necessarily representative of those of the Uniformed Services University of the Health Sciences (USUHS), the Department of Defense (DOD), or the United States Army, Navy, or Air Force.
We thank those who provided blood samples or blood DNA from patients with confirmed rickettsial infections; we specifically thank Johan Bakken (University of Minnesota, Duluth, MN), Gary Wormser (New York Medical College, Valhalla, NY), Daniel Paris (Swiss Tropical Medicine Institute, Basel, Switzerland, and Mahidol Oxford Research Unit, Bangkok, Thailand), Paul Newton (Oxford University, Laos Oxford Mahasot Research Unit, Vientiane, Laos), Juan Olano (University of Texas Medical Branch, Galveston, TX), Marina Eremeeva (Georgia State University, Statesboro, GA/Centers for Disease Control and Prevention, Atlanta, GA), and Rita DeSousa (Portuguese National Institute of Health, Lisbon, Portugal). We also thank Thomas Spahr (Johns Hopkins Hospital Clinical Microbiology Laboratory-Parasitology Laboratory), David Sullivan (The Johns Hopkins University Bloomberg School of Public Health and Malaria Research Center), Peggy Althaus (Duke University, Durham, NC), and Bobby Pritt (Mayo Clinic, Rochester, MN) for control specimens. We also thank the technical support team for outstanding dedication and expertise, including Meg Lichay, Emily Clemens, and Cindy Chen.
The work was jointly conceived, interpreted, and written by M.E.R. and J.S.D. The analytical approach, specific reagents, and analysis of multiplex results was done by J.S.D. and M.E.R. M.E.R. and J.S.D. conducted the final analysis and drafts of the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, Hinckley AF, Mead PS, Paz-Bailey G, Waterman SH, Drexler NA, Kersh GJ, Hooks H, Partridge SK, Visser SN, Beard CB, Petersen LR. 2018. Vital signs: trends in reported vectorborne disease cases - United States and territories, 2004–2016. MMWR Morb Mortal Wkly Rep 67:496–501. doi: 10.15585/mmwr.mm6717e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules CI, Marston HD, Bloom ME, Fauci AS. 2018. Tickborne diseases—confronting a growing threat. N Engl J Med 379:701–703. doi: 10.1056/NEJMp1807870. [DOI] [PubMed] [Google Scholar]
- 3.Xu G, Walker DH, Jupiter D, Melby PC, Arcari CM. 2017. A review of the global epidemiology of scrub typhus. PLoS Negl Trop Dis 11:e0006062. doi: 10.1371/journal.pntd.0006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonell A, Lubell Y, Newton PN, Crump JA, Paris DH. 2017. Estimating the burden of scrub typhus: a systematic review. PLoS Negl Trop Dis 11:e0005838. doi: 10.1371/journal.pntd.0005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsioutis C, Zafeiri M, Avramopoulos A, Prousali E, Miligkos M, Karageorgos SA. 2017. Clinical and laboratory characteristics, epidemiology, and outcomes of murine typhus: a systematic review. Acta Trop 166:16–24. doi: 10.1016/j.actatropica.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Chikeka I, Dumler JS. 2015. Neglected bacterial zoonoses. Clin Microbiol Infect 21:404–415. doi: 10.1016/j.cmi.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AJ, Paris DH, Newton PN. 2015. A systematic review of mortality from untreated scrub typhus (Orientia tsutsugamushi). PLoS Negl Trop Dis 9:e0003971. doi: 10.1371/journal.pntd.0003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ismail N, McBride JW. 2017. Tick-borne emerging infections: ehrlichiosis and anaplasmosis. Clin Lab Med 37:317–340. doi: 10.1016/j.cll.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Marshall GS, Tick-Borne Infections in Children Study Group, Stout GG, Jacobs RF, Schutze GE, Paxton H, Buckingham SC, DeVincenzo JP, Jackson MA, San Joaquin VH, Standaert SM, Woods CR. 2003. Antibodies reactive to Rickettsia rickettsii among children living in the southeast and south central regions of the United States. Arch Pediatr Adolesc Med 157:443–448. doi: 10.1001/archpedi.157.5.443. [DOI] [PubMed] [Google Scholar]
- 10.Marshall GS, Jacobs RF, Schutze GE, Paxton H, Buckingham SC, DeVincenzo JP, Jackson MA, San Joaquin VH, Standaert SM, Woods CR. 2002. Ehrlichia chaffeensis seroprevalence among children in the southeast and south-central regions of the United States. Arch Pediatr Adolesc Med 156:166–170. doi: 10.1001/archpedi.156.2.166. [DOI] [PubMed] [Google Scholar]
- 11.Bakken JS, Dumler JS. 2015. Human granulocytic anaplasmosis. Infect Dis Clin North Am 29:341–355. doi: 10.1016/j.idc.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N-Y, Huang P-Y, Leu H-S, Chiang P-C, Huang C-T. 2008. Clinical prediction of endemic rickettsioses in northern Taiwan—relevance of peripheral blood atypical lymphocytes. J Microbiol Immunol Infect 41:362–368. [PubMed] [Google Scholar]
- 13.Acestor N, Cooksey R, Newton PN, Menard D, Guerin PJ, Nakagawa J, Christophel E, Gonzalez IJ, Bell D. 2012. Mapping the aetiology of non-malarial febrile illness in Southeast Asia through a systematic review—terra incognita impairing treatment policies. PLoS One 7:e44269. doi: 10.1371/journal.pone.0044269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhu M, Nicholson WL, Roche AJ, Kersh GJ, Fitzpatrick KA, Oliver LD, Massung RF, Morrissey AB, Bartlett JA, Onyango JJ, Maro VP, Kinabo GD, Saganda W, Crump JA. 2011. Q fever, spotted fever group, and typhus group rickettsioses among hospitalized febrile patients in northern Tanzania. Clin Infect Dis 53:e8–e15. doi: 10.1093/cid/cir411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reller ME, Chikeka I, Miles JJ, Dumler JS, Woods CW, Mayorga O, Matute AJ. 2016. First identification and description of rickettsioses and Q fever as causes of acute febrile illness in Nicaragua. PLoS Negl Trop Dis 10:e0005185. doi: 10.1371/journal.pntd.0005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chikeka I, Matute AJ, Dumler JS, Woods CW, Mayorga O, Reller ME. 2016. Use of peptide-based enzyme-linked immunosorbent assay followed by immunofluorescence assay to document Ehrlichia chaffeensis as a cause of febrile illness in Nicaragua. J Clin Microbiol 54:1581–1585. doi: 10.1128/JCM.03331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reller ME, Bodinayake C, Nagahawatte A, Devasiri V, Kodikara-Arachichi W, Strouse JJ, Flom JE, Ostbye T, Woods CW, Dumler JS. 2012. Unsuspected rickettsioses among patients with acute febrile illness, Sri Lanka, 2007. Emerg Infect Dis 18:825–829. doi: 10.3201/eid1805.111563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blacksell SD, Kantipong P, Watthanaworawit W, Turner C, Tanganuchitcharnchai A, Jintawon S, Laongnuanutit A, Nosten FH, Day NP, Paris DH, Richards AL. 2015. Underrecognized arthropod-borne and zoonotic pathogens in northern and northwestern Thailand: serological evidence and opportunities for awareness. Vector Borne Zoonotic Dis 15:285–290. doi: 10.1089/vbz.2015.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh FX, Kho KL, Kisomi MG, Wong LP, Bulgiba A, Tan PE, Lim YAL, Nizam QNH, Panchadcharam C, Tay ST. 2018. Ehrlichia and Anaplasma infections: serological evidence and tick surveillance in Peninsular Malaysia. J Med Entomol 55:269–276. doi: 10.1093/jme/tjx204. [DOI] [PubMed] [Google Scholar]
- 20.Bezerra MCF, Melo ALT, Taques I, Aguiar DM, Pacheco RC, Slhessarenko RD. 2017. Seropositivity for Rickettsia spp. and Ehrlichia spp. in the human population of Mato Grosso, Central-Western Brazil. Rev Soc Bras Med Trop 50:399–403. doi: 10.1590/0037-8682-0318-2016. [DOI] [PubMed] [Google Scholar]
- 21.Murdoch DR, Woods CW, Zimmerman MD, Dull PM, Belbase RH, Keenan AJ, Scott RM, Basnyat B, Archibald LK, Reller LB. 2004. The etiology of febrile illness in adults presenting to Patan hospital in Kathmandu, Nepal. Am J Trop Med Hyg 70:670–675. doi: 10.4269/ajtmh.2004.70.670. [DOI] [PubMed] [Google Scholar]
- 22.Phongmany S, Rolain JM, Phetsouvanh R, Blacksell SD, Soukkhaseum V, Rasachack B, Phiasakha K, Soukkhaseum S, Frichithavong K, Chu V, Keolouangkhot V, Martinez-Aussel B, Chang K, Darasavath C, Rattanavong O, Sisouphone S, Mayxay M, Vidamaly S, Parola P, Thammavong C, Heuangvongsy M, Syhavong B, Raoult D, White NJ, Newton PN. 2006. Rickettsial infections and fever, Vientiane, Laos. Emerg Infect Dis 12:256–262. doi: 10.3201/eid1202.050900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alirol E, Horie NS, Barbe B, Lejon V, Verdonck K, Gillet P, Jacobs J, Buscher P, Kanal B, Bhattarai NR, El Safi S, Phe T, Lim K, Leng L, Lutumba P, Mukendi D, Bottieau E, Boelaert M, Rijal S, Chappuis F. 2016. Diagnosis of persistent fever in the tropics: set of standard operating procedures used in the NIDIAG febrile syndrome study. PLoS Negl Trop Dis 10:e0004749. doi: 10.1371/journal.pntd.0004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouqui P, European Network for Surveillance of Tick-Borne Diseases, Bacellar F, Baranton G, Birtles RJ, Bjoersdorff A, Blanco JR, Caruso G, Cinco M, Fournier PE, Francavilla E, Jensenius M, Kazar J, Laferl H, Lakos A, Lotric Furlan S, Maurin M, Oteo JA, Parola P, Perez-Eid C, Peter O, Postic D, Raoult D, Tellez A, Tselentis Y, Wilske B. 2004. Guidelines for the diagnosis of tick-borne bacterial diseases in Europe. Clin Microbiol Infect 10:1108–1132. doi: 10.1111/j.1469-0691.2004.01019.x. [DOI] [PubMed] [Google Scholar]
- 25.Prakash JAJ, Reller ME, Barat N, Dumler JS. 2009. Assessment of a quantitative multiplex 5’ nuclease real-time PCR for spotted fever and typhus group rickettsioses and Orientia tsutsugamushi. Clin Microbiol Infect 15:292–293. doi: 10.1111/j.1469-0691.2008.02242.x. [DOI] [PubMed] [Google Scholar]
- 26.Reller ME, Dumler JS. 2018. Development and clinical validation of a multiplex real-time quantitative PCR assay for human infection by Anaplasma phagocytophilum and Ehrlichia chaffeensis. Trop Med Infect Dis 3:14. doi: 10.3390/tropicalmed3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unal I. 2017. Defining an optimal cut-point value in ROC analysis: an alternative approach. Comput Math Methods Med 2017:3762651. doi: 10.1155/2017/3762651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hockman D, Dong M, Zheng H, Kumar S, Huff MD, Grigorenko E, Beanan M, Duncan R. 2017. Comparison of multiplex PCR hybridization-based and singleplex real-time PCR-based assays for detection of low prevalence pathogens in spiked samples. J Microbiol Methods 132:76–82. doi: 10.1016/j.mimet.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplowitz LG, Lange JV, Fischer JJ, Walker DH. 1983. Correlation of rickettsial titers, circulating endotoxin, and clinical features in Rocky Mountain spotted fever. Arch Intern Med 143:1149–1151. doi: 10.1001/archinte.1983.00350060073012. [DOI] [PubMed] [Google Scholar]
- 30.Kato C, Chung I, Paddock C. 2016. Estimation of Rickettsia rickettsii copy number in the blood of patients with Rocky Mountain spotted fever suggests cyclic diurnal trends in bacteraemia. Clin Microbiol Infect 22:394–396. doi: 10.1016/j.cmi.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Luce-Fedrow A, Mullins K, Kostik AP, St John HK, Jiang J, Richards AL. 2015. Strategies for detecting rickettsiae and diagnosing rickettsial diseases. Future Microbiol 10:537–564. doi: 10.2217/fmb.14.141. [DOI] [PubMed] [Google Scholar]
- 32.Watthanaworawit W, Turner P, Turner C, Tanganuchitcharnchai A, Richards AL, Bourzac KM, Blacksell SD, Nosten F. 2013. A prospective evaluation of real-time PCR assays for the detection of Orientia tsutsugamushi and Rickettsia spp. for early diagnosis of rickettsial infections during the acute phase of undifferentiated febrile illness. Am J Trop Med Hyg 89:308–310. doi: 10.4269/ajtmh.12-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paris DH, Dumler JS. 2016. State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus. Curr Opin Infect Dis 29:433–439. doi: 10.1097/QCO.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh S, Cuong NC, Hamaguchi S, Thuy PT, Cuong DD, Anh LK, Anh NTH, Anh DD, Sando E, Suzuki M, Fujita H, Yasunami M, Yoshihara K, Yoshida L-M, Paris DH, Ariyoshi K. 2019. Challenges in diagnosing scrub typhus among hospitalized patients with undifferentiated fever at a national tertiary hospital in northern Vietnam. PLoS Negl Trop Dis 13:e0007928. doi: 10.1371/journal.pntd.0007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paris DH, Blacksell SD, Stenos J, Graves SR, Unsworth NB, Phetsouvanh R, Newton PN, Day NP. 2008. Real-time multiplex PCR assay for detection and differentiation of rickettsiae and orientiae. Trans R Soc Trop Med Hyg 102:186–193. doi: 10.1016/j.trstmh.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Reller ME, Chen WH, Dalton J, Lichay MA, Dumler JS. 2013. Multiplex 5’ nuclease quantitative real-time PCR for clinical diagnosis of malaria and species-level identification and epidemiologic evaluation of malaria-causing parasites, including Plasmodium knowlesi. J Clin Microbiol 51:2931–2938. doi: 10.1128/JCM.00958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tshokey T, Stenos J, Durrheim DN, Eastwood K, Nguyen C, Vincent G, Graves SR. 2018. Rickettsial infections and Q fever amongst febrile patients in Bhutan. Trop Med Infect Dis 3:12. doi: 10.3390/tropicalmed3010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonthayanon P, Chierakul W, Wuthiekanun V, Phimda K, Pukrittayakamee S, Day NP, Peacock SJ. 2009. Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. J Clin Microbiol 47:430–434. doi: 10.1128/JCM.01927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bwanga F, Disque C, Lorenz MG, Allerheiligen V, Worodria W, Luyombya A, Najjingo I, Weizenegger M. 2015. Higher blood volumes improve the sensitivity of direct PCR diagnosis of blood stream tuberculosis among HIV-positive patients: an observation study. BMC Infect Dis 15:48. doi: 10.1186/s12879-015-0785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tantibhedhyangkul W, Wongsawat E, Silpasakorn S, Waywa D, Saenyasiri N, Suesuay J, Thipmontree W, Suputtamongkol Y. 2017. Use of multiplex real-time PCR to diagnose scrub typhus. J Clin Microbiol 55:1377–1387. doi: 10.1128/JCM.02181-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Sousa R, Franca A, Doria Nobrega S, Belo A, Amaro M, Abreu T, Pocas J, Proenca P, Vaz J, Torgal J, Bacellar F, Ismail N, Walker DH. 2008. Host- and microbe-related risk factors for and pathophysiology of fatal Rickettsia conorii infection in Portuguese patients. J Infect Dis 198:576–585. doi: 10.1086/590211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demma LJ, Traeger MS, Nicholson WL, Paddock CD, Blau DM, Eremeeva ME, Dasch GA, Levin ML, Singleton J Jr, Zaki SR, Cheek JE, Swerdlow DL, McQuiston JH. 2005. Rocky Mountain spotted fever from an unexpected tick vector in Arizona. N Engl J Med 353:587–594. doi: 10.1056/NEJMoa050043. [DOI] [PubMed] [Google Scholar]
- 43.Olano JP, Masters E, Hogrefe W, Walker DH. 2003. Human monocytotropic ehrlichiosis, Missouri. Emerg Infect Dis 9:1579–1586. doi: 10.3201/eid0912.020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dierberg KL, Dumler JS. 2006. Lymph node hemophagocytosis in rickettsial diseases: a pathogenetic role for CD8 T lymphocytes in human monocytic ehrlichiosis (HME)? BMC Infect Dis 6:121. doi: 10.1186/1471-2334-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell CA, Patel R. 2005. A real-time combined polymerase chain reaction assay for the rapid detection and differentiation of Anaplasma phagocytophilum, Ehrlichia chaffeensis, and Ehrlichia ewingii. Diagn Microbiol Infect Dis 53:301–306. doi: 10.1016/j.diagmicrobio.2005.06.019. [DOI] [PubMed] [Google Scholar]