The Elizabethkingia genus has gained global attention in recent years as containing sporadic, worldwide, nosocomial pathogens. Elizabethkingia spp. are intrinsically multidrug resistant, primarily infect immunocompromised individuals, and are associated with high mortality (∼20 to 40%). As yet, gaps remain in our understanding of transmission, global strain relatedness, antimicrobial resistance, and effective therapy. Over a 16-year period, 22 clinical and 6 hospital environmental isolates were collected from Queensland, Australia.
KEYWORDS: Elizabethkingia, MDR, multidrug resistance, nosocomial, MIC, minimum inhibitory concentration, antimicrobial resistance, AMR, comparative genomics, nosocomial infection
ABSTRACT
The Elizabethkingia genus has gained global attention in recent years as containing sporadic, worldwide, nosocomial pathogens. Elizabethkingia spp. are intrinsically multidrug resistant, primarily infect immunocompromised individuals, and are associated with high mortality (∼20 to 40%). As yet, gaps remain in our understanding of transmission, global strain relatedness, antimicrobial resistance, and effective therapy. Over a 16-year period, 22 clinical and 6 hospital environmental isolates were collected from Queensland, Australia. Identification using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Vitek MS) and whole-genome sequencing was compared with a global strain data set. Phylogenomic reconstruction robustly identified 22 Elizabethkingia anophelis, 3 Elizabethkingia miricola, 2 Elizabethkingia meningoseptica, and 1 Elizabethkingia bruuniana isolates, most of which branched as unique lineages. Global analysis revealed that some Australian E. anophelis isolates are genetically closely related to strains from the United States, England, and Asia. Comparative genomics of clinical and environmental strains identified evidence of nosocomial transmission in patients, indicating probable infection from a hospital reservoir. Furthermore, broth microdilution against 39 antimicrobials revealed almost ubiquitous resistance to aminoglycosides, carbapenems, cephalosporins, and penicillins. Like other international strains, our isolates expressed susceptibility to minocycline and levofloxacin and the less common trimethoprim-sulfamethoxazole. Our study demonstrates important new insights into the genetic diversity, environmental persistence, and transmission of and potential effective therapy for Australian Elizabethkingia species.
INTRODUCTION
The genus Elizabethkingia (formerly Chryseobacterium) comprises a group of environmental bacteria that have traditionally been isolated from soil and water environments (1–4). As opportunistic pathogens, Elizabethkingia spp. can cause sporadic nosocomial outbreaks and infections in immunocompromised or at-risk individuals (1, 2, 5–8). Infections have been documented worldwide, such as those in the Central African Republic (9), Mauritius (10), Singapore (11), Taiwan (12), and the United States (6), suggesting a comprehensive global distribution that is yet to be fully described. To date, the largest outbreak was caused by community-acquired Elizabethkingia anophelis in Wisconsin, USA, from 2015 to 2016. A total of 66 individuals were infected, and the outbreak spread to the neighboring states of Illinois and Michigan (6). Comparative genomics characterized unique mutations by an integrative conjugative element (ICE) insertion in the mutY gene in all infecting strains, which may have accelerated the transmission of the outbreak clone. Additionally, a mutation in the mutS gene was identified in the only hypermutator strain, although the significance of this strain was unclear (6). Often, the source of Elizabethkingia species infection remains unclear, and routes of transmission are still to be defined (2, 6, 9, 12–16). However, previous investigations have suggested that shared water reservoirs within hospitals may be an overlooked reservoir of infection (1, 2, 17).
As an understudied pathogen, taxonomic assignment within the Elizabethkingia genus is ongoing. Recently, a formal taxonomic revision using whole-genome sequencing (WGS) has confirmed six Elizabethkingia species, consisting of E. anophelis, E. meningoseptica, E. miricola, E. bruuniana, E. ursingii, and E. occulta (3–5). It is also now recognized that E. anophelis, not E. meningoseptica, is the primary species causing human infection, with mortality rates currently estimated at 23 to 26% and 23 to 41%, respectively (4, 13, 18–20). Bacteremia, pneumoniae, sepsis, and meningitis are common clinical presentations with E. anophelis (7, 14, 18, 19). Similarly, E. meningoseptica infections present as neonatal meningitis and/or sepsis, but E. meningoseptica can also cause infections in most organ systems (8, 12). Risk factors associated with Elizabethkingia species infection consist of being male, having underlying chronic medical conditions, such as malignancy or diabetes mellitus, and admission to critical care or neonatal units (8, 12, 13, 18, 19, 21). The remaining members of the genus are thought to be much less prevalent in human disease; however, difficulties in accurately identifying E. miricola, E. bruuniana, E. ursingii, and E. occulta from clinical specimens have hindered appropriate recognition and characterization of these species (4).
Elizabethkingia species are considered resistant to carbapenems, cephalosporins, aminoglycosides, and most β-lactams even in combination with β-lactamase inhibitors (except for piperacillin-tazobactam), due to two unique metallo-β-lactamases (blaBlaB and blaGOB) and a unique extended-spectrum β-lactamase (ESBL) (blaCME). Minocycline, levofloxacin, trimethoprim-sulfamethoxazole, and piperacillin-tazobactam are the most common antimicrobials that have been tested, with most strains demonstrating susceptibility to at least one or to various combinations of these antimicrobials (4, 6, 19–22). Due to the variations in susceptibility and the severity of infection, the most effective empirical therapy is still not known, highlighting the need for further MIC profiling (7, 19–21).
This study aimed to perform one of the largest comparative genomic analyses of Elizabethkingia species isolates to date, including isolates from Australia, a geographic area whose Elizabethkingia population is previously undescribed, as well as to assess the accuracy of identification of Elizabethkingia spp. with the Vitek MS version 3.2 database and to determine the MICs of clinical Australian Elizabethkingia isolates across 39 antimicrobials.
MATERIALS AND METHODS
Ethics statement.
This project was reviewed by the chairperson of a National Health and Medical Research Council (NHMRC) and registered with The Royal Brisbane and Women’s Hospital Human Research Ethics Committee (HREC) (EC00172) and was deemed compliant with the NHMRC guidance “Ethical Considerations in Quality Assurance and Evaluation Activities” 2014 and exempt from HREC review.
Isolates and initial identification.
Twenty-two clinical Elizabethkingia species isolates collected in Queensland, Australia, over a 16-year period (2002 to 2018) were included in this study (Table 1). Isolates were collected by two methods. First, laboratory database storage records from multiple public and private laboratories in Queensland were searched for Elizabethkingia spp. or Chryseobacterium meningoseptica. Second, isolates identified by current laboratory identification systems as Elizabethkingia spp. were collected prospectively from both private and public pathology laboratories throughout the state of Queensland between January 2017 and October 2018. In both methods, isolates were stored at −80°C with low-temperature bead storage systems prior to being collected from storage and resurrected on 5% horse blood agar (Edwards Group MicroMedia, Narellan, NSW, Australia). Single colonies were then double passaged on 5% horse blood agar and subjected to identification via Vitek MS Knowledge Base version 3.2 (bioMérieux, Murarrie, QLD, Australia). This database includes and can identify E. anophelis, E. miricola, and E. meningoseptica.
TABLE 1.
Elizabethkingia species isolates and associated identification information included in the current study
| Isolate | Patient age (yrs) | Date collected | Sample type or collection site | Species identification using: |
|
|---|---|---|---|---|---|
| Vitek MS Knowledge Base 3.2 | Whole-genome sequencing | ||||
| EkQ1 | 1 | 2017 | Sputum | E. miricola | E. miricola |
| EkQ3 | 43 | 2017 | Sputum | E. anophelis | E. anophelis |
| EkQ4 | 78 | 2017 | Blood | E. meningoseptica | E. meningoseptica |
| EkQ5 | 59 | 2017 | Blood | E. anophelis | E. anophelis |
| EkQ6 | 17 | 2018 | Bronchoalveolar lavage fluid | E. anophelis | E. anophelis |
| EkQ7 | 69 | 2018 | Blood | E. anophelis | E. anophelis |
| EkQ8 | 0 | 2018 | Urine | E. anophelis | E. anophelis |
| EkQ10 | 34 | 2018 | Sputum | E. miricola | E. miricola |
| EkQ11a | 85 | 2018 | Blood | E. miricola | E. bruuniana |
| EkQ12 | 53 | 2018 | Blood | E. meningoseptica | E. meningoseptica |
| EkQ13 | 1 | 2011 | Sputum | E. miricola | E. miricola |
| EkQ15 | 16 | 2002 | Bronchoalveolar lavage fluid | E. anophelis | E. anophelis |
| EkQ16 | 82 | 2017 | Blood | E. anophelis | E. anophelis |
| EkQ17 | 66 | 2018 | Blood | E. anophelis | E. anophelis |
| EkM1 | Unknown | 2018 | Unknown | E. anophelis | E. anophelis |
| EkM2 | Unknown | 2018 | Unknown | E. anophelis | E. anophelis |
| EkM3 | Unknown | 2014 | Unknown | E. anophelis | E. anophelis |
| EkS1 | 80 | 2013 | Blood | E. anophelis | E. anophelis |
| EkS2 | 82 | 2015 | Blood | E. anophelis | E. anophelis |
| EkS3 | 74 | 2016 | Blood | E. anophelis | E. anophelis |
| EkS4 | 73 | 2012 | Blood | E. anophelis | E. anophelis |
| EkS5 | 66 | 2018 | Dialysis fluid | E. anophelis | E. anophelis |
| EK1 | NA | 2019 | Bathroom sink drain, oncology ward | E. anophelis | E. anophelis |
| EK2 | NA | 2019 | Corridor sink drain, infectious disease ward | E. anophelis | E. anophelis |
| EK3 | NA | 2019 | Hand-washing sink drain, oncology ward | E. anophelis | E. anophelis |
| EK4 | NA | 2019 | Hand-washing sink, transplant ward | E. anophelis | E. anophelis |
| EK5 | NA | 2019 | Bathroom handrail, transplant ward | E. anophelis | E. anophelis |
| EK6 | NA | 2019 | Bathroom sink, transplant ward | E. anophelis | E. anophelis |
Strain EkQ11, marked in boldface, represents a species identification error according to Vitek MS Knowledge Base version 3.2.
To investigate the epidemiology and transmission potential of E. anophelis, six environmental isolates were collected in 2019 from the Princess Alexandra Hospital, Brisbane, Australia, via swabbing various surfaces throughout the environment (Table 1). Specimens were plated onto 5% horse blood agar, and Elizabethkingia species colonies were double passaged to ensure purity and then subjected to identification via Vitek MS Knowledge Base version 3.2.
DNA extraction, whole-genome sequencing, and genome assembly.
DNA was extracted using the DNeasy UltraClean microbial extraction kit (Qiagen, Chadstone, VIC, Australia) according to the manufacturer’s instructions. Purified DNA was quantified using both the NanoDrop 3300 spectrophotometer and the Qubit 4 fluorometer (Thermo Fisher Scientific). Sequencing libraries were generated using the Nextera Flex DNA library preparation kit and sequenced on the MiniSeq system (Illumina, Inc., San Diego, CA, USA) on a high-output 300-cycle cartridge according to the manufacturer’s instructions. Comparative genomic analyses were performed across a large Elizabethkingia data set (n = 128) (Table S1 in the supplemental material), including the 28 Australian genomes generated in the current study (Table 1), to assign species and to assess intraspecific and geographical relationships among strains. Publicly available Elizabethkingia Illumina reads (n = 119) were downloaded from the NCBI Sequence Read Archive database (January 2019), and Elizabethkingia species assemblies were downloaded from the GenBank database (n = 109). Publicly available Illumina reads were quality filtered with Trimmomatic version 0.38 (23) and subjected to quality control assessments with FastQC (24), followed by downsizing to 40× coverage using Seqtk (25). For assemblies without accompanying Illumina data, synthetic paired-end reads were generated with ART MountRainier-2016.06.05 (26). Genomes were limited to one representative per strain, and only sequence reads that were of high quality according to FastQC were included, to avoid errors in phylogenomic reconstruction (n = 100) (Table S1). The genomes were assembled using SPAdes version 3.13.0 (27) and annotated with Prokka version 1.13 (28) (Table S2).
Phylogenomic reconstruction.
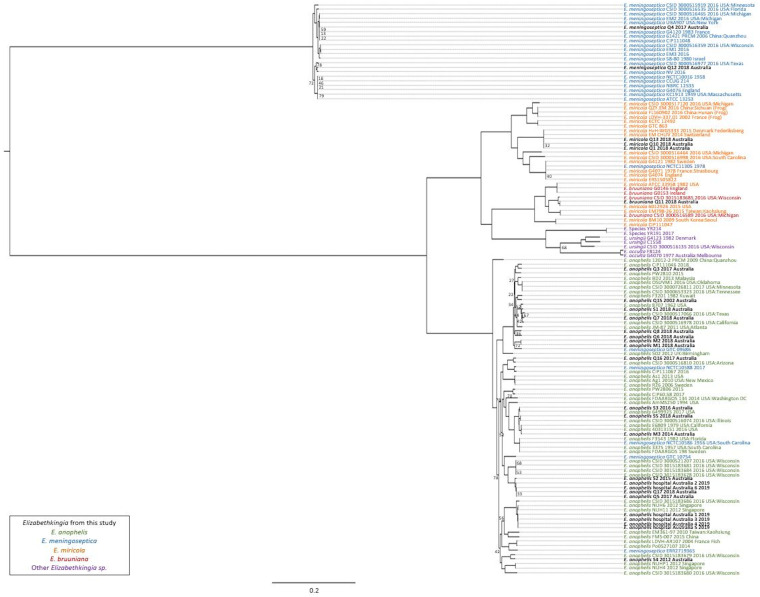
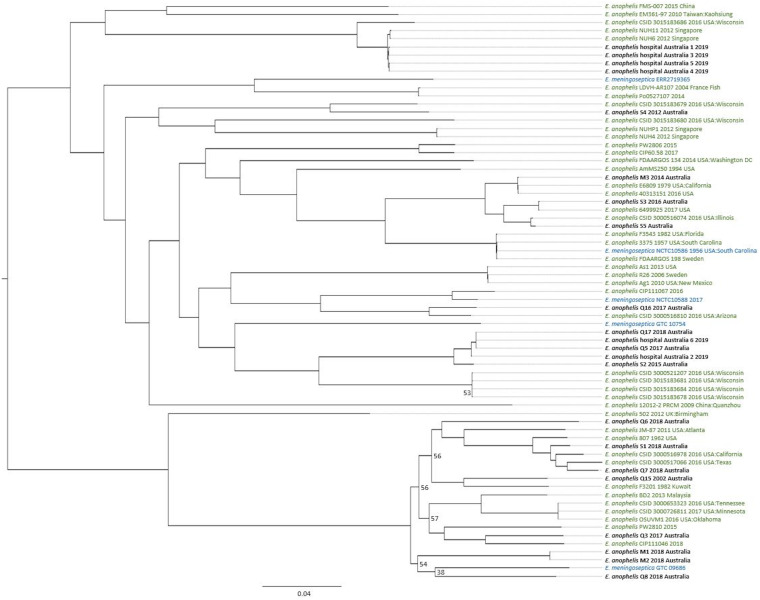
The comparative genomics pipeline SPANDx version 3.2 (29) was used under default settings to identify orthologous, biallelic, core-genome single-nucleotide polymorphism (SNP) and short insertion-deletion (indel) characters among the 128 Elizabethkingia genomes. E. anophelis strain NUHP1, E. miricola strain CSID_3000517120, E. meningoseptica strain G4120, and E. bruuniana strain G0146 (GenBank accession numbers NZ_CP007547.1, NZ_MAGX00000000.1, NZ_CP016378.1, and NZ_CP014337.1, respectively) were used as reference genomes for species-specific SPANDx read-mapping alignment. Reference genome E. anophelis NUHP1 was used as the reference strain for the genus read mapping and alignment. Outputs from SPANDx were used to generate maximum-likelihood trees under the GTR+G model determined by jModelTest 2 (30) in RAxML version 8.2.12 (31) and visualized in FigTree version 4.0 (http://tree.bio.ed.ac.uk/software/figtree). From the 128 genomes, 127,236 SNPs were used to construct the Elizabethkingia genus phylogeny (Fig. 1). Within-species phylogenies were also constructed, using 121,827 SNPs from 71 genomes for E. anophelis (Fig. 2), 135,087 SNPs from 18 genomes for E. miricola (Fig. S1), 61,500 SNPs from 22 genomes for E. meningoseptica (Fig. S2), and 82,680 SNPs from 10 genomes for E. bruuniana (Fig. S3). All phylogenies were statistically tested with 1,000 bootstrap replicates. Branch support of less than 0.8 is shown in figures. To assess SNP and indel differences among closely related strains, the earliest-collected strain was used as the reference in SPANDx, and SNP and indel variants that had passed quality filtering were visualized in Tablet 1.19.09.03 (32) and Geneious Prime 2019 2.1 (33) (Table 2). For clonal isolates, ratios of nonsynonymous to synonymous evolutionary changes (dN/dS ratios) were calculated with MEGAx (Table S3).
FIG 1.
Global phylogenomic analysis of Elizabethkingia species genomes. Maximum-likelihood midpoint-rooted phylogeny. Branches returning bootstrap support of <0.8 are labeled. This phylogeny was reconstructed using 127,236 bialleleic, orthologous single-nucleotide polymorphisms identified among the 128 Elizabethkingia genomes.
FIG 2.
Elizabethkingia anophelis species-specific phylogenomic analysis. Maximum-likelihood midpoint-rooted phylogeny was reconstructed using 121,827 bialleleic, orthologous single-nucleotide polymorphisms identified among the 71 E. anophelis genomes. E. anophelis genomes correctly identified to species level are colored green, Elizabethkingia meningoseptica genomes incorrectly identified to species level are colored blue, and new Elizabethkingia anophelis genomes generated in this study are colored black. Branches returning bootstrap support of <0.8 are labeled.
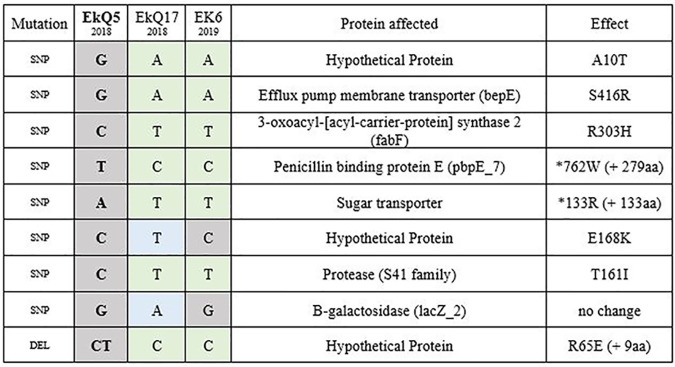
TABLE 2.
Single-nucleotide polymorphism and deletion differences between strains of the clonal cluster of clinical and environmental Elizabethkingia anophelis isolatesa
Single-nucleotide polymorphism and deletion differences between strains of the clonal cluster of clinical and environmental Elizabethkingia anophelis isolates. Clinical isolates EkQ5 (earliest-collected and reference strain) and EkQ17 were collected from two different transplant patients, while Ek6 was collected from a shared handwashing sink on the transplant ward. Gray shading shows no differences, green shows similarities between EkQ17 and EK6, and blue highlights unique changes. The proteins affected by each mutation and the resulting amino acid changes are also shown. SNP, single-nucleotide polymorphism; DEL, deletion.
MIC testing.
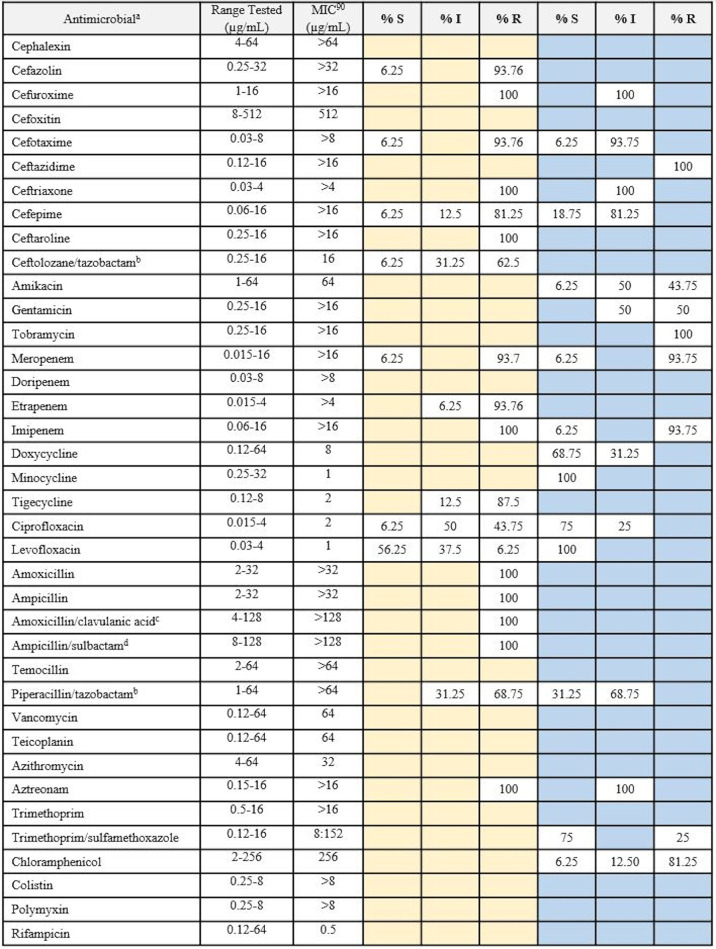
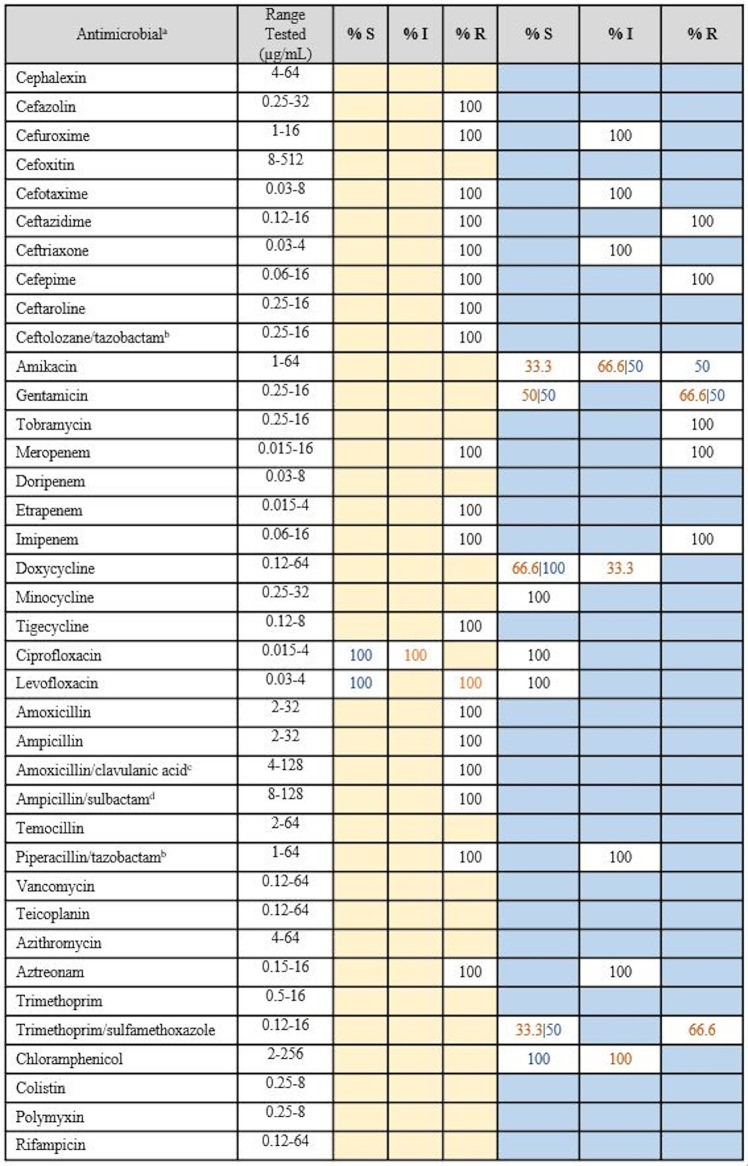
Elizabethkingia species clinical isolates were subjected to broth microdilution to determine the MICs of 39 clinically relevant antimicrobials, consistent with or complementary to previous Elizabethkingia studies (Tables 3 and 4) (12, 19, 22, 34). Custom Gram-negative Sensititre MIC plates (ThermoFisher Scientific, Scoresby, VIC, Australia) were used according to the manufacturer’s instructions. E. bruuniana isolate EkQ11 was excluded from MIC analyses due to poor growth. Elizabethkingia species isolates were compared against the European Committee on Antimicrobial Susceptibility Testing (EUCAST) pharmacokinetic-pharmacodynamic (PK-PD) nonspecies breakpoints (35) and the non-Enterobacteriaceae breakpoints according to the Clinical and Laboratory Standards Institute (CLSI) M100 guidelines (36–38). The MIC distributions for each antimicrobial are shown in Tables S4 and S5.
TABLE 3.
MIC data for Australian clinical Elizabethkingia anophelis isolates against clinically relevant antimicrobials
MIC data derived from broth microdilution testing of the 16 Australian clinical E. anophelis isolates against 39 clinically relevant antimicrobials. Pharmacokinetic-pharmacodynamic (non-species-specific) breakpoints applied from EUCAST clinical breakpoint tables (version 9.0) are shown in columns with yellow shading, and non-Enterobacteriaceae breakpoints applied from CLSI M100-29 (2019) are shown in columns with blue shading. Shaded blue and yellow cells indicate that no breakpoint is currently available for this antimicrobial within these schemes. S, susceptible; I, susceptible with high exposure (EUCAST definition) or intermediate (CLSI definition); R, resistant.
Tazobactam concentration fixed at 4 mg/liter.
Clavulanic acid concentration fixed at 2 mg/liter.
Sulbactam concentration fixed at 4 mg/liter.
TABLE 4.
MIC data for Australian clinical Elizabethkingia meningoseptica and Elizabethkingia miricola isolates against clinically relevant antimicrobials
MIC data derived from broth microdilution testing of the 2 E. meningoseptica (blue font) and 3 E. miricola (orange font) Australian clinical isolates against 39 clinically relevant antimicrobials. Pharmacokinetic-pharmacodynamic (non-species-specific) breakpoints applied from EUCAST Clinical Breakpoint Tables (version 9.0) are shown in columns with yellow shading, and non-Enterobacteriaceae breakpoints applied from CLSI M100-29 (2019) are shown in columns with blue shading. Shaded blue and yellow cells indicate that no breakpoint is currently available for this antimicrobial within these schemes. S, susceptible; I, susceptible with high exposure (EUCAST definition) or intermediate (CLSI definition); R, resistant.
Tazobactam concentration fixed at 4 mg/liter.
Clavulanic acid concentration fixed at 2 mg/liter.
Sulbactam concentration fixed at 4 mg/liter.
In silico antimicrobial resistance (AMR) gene predictions.
Clinical Elizabethkingia species WGS data were subjected to ABRicate using both the CARD and NCBI databases to predict AMR genes (https://github.com/tseemann/abricate) and RAST for an alternative confirmation (34, 39, 40). Geneious prime 2019.2.1 and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to generate single-protein sequence alignments (33).
Data availability.
Illumina sequence data for the 28 Elizabethkingia species genomes described in this study have been deposited in the NCBI in the SRA database under accession number SRP225137 and the BioProject database under accession number PRJNA576977 (BioSample accession numbers SAMN13016226 to SAMN13016247 and SAMN14081590 to SAMN14081595).
RESULTS
Elizabethkingia identification using comparative genomics versus mass spectrometry.
Phylogenomic reconstruction, including100 Elizabethkingia reference genomes collected internationally over the past 50 years, robustly identified the 22 clinical and 6 environmental Australian Elizabethkingia species isolates as E. anophelis (n = 22), E. miricola (n = 3), E. meningoseptica (n = 2), and E. bruuniana (n = 1) (Fig. 1; Table S1 in the supplemental material). Eleven identification errors were identified in the publicly available data set, including 2 identification errors within the E. anophelis clade, 5 within the E. bruuniana clade, and 1 within the E. miricola clade (Fig. 1). Additionally, comparison of Vitek MS Knowledge Base version 3.2 with genomic species assignments of the Australian isolates resulted in one identification error from the Vitek MS, incorrectly identifying an E. bruuniana isolate as E. miricola (Table 1).
Australian Elizabethkingia and global relatedness.
Australian Elizabethkingia spp. displayed no distinct phylogeographical signal within the genus phylogeny, as they disseminated across the phylogenetic tree (Fig. 1). No Australian Elizabethkingia isolate was identical to a previously described isolate, with those appearing to be nearly identical in the phylogenies separated by 16 to 284 SNPs (Fig. 1 and 2; Fig. S1 to S3). Australian E. anophelis isolates are not closely related to Wisconsin, USA, outbreak strains (Fig. 1 and 2). Clinical isolates EkQ17, EkQ5, and EkS2 and environmental isolates EK2 and EK6 branched off the Wisconsin, USA, outbreak cluster, diverging as a distantly related unique lineage separated by an estimated 20,400 SNPs and 500 indels using CSID_3015183681 as the reference strain. The truncations of the C termini of MutY and MutS, characteristic of the outbreak and hypermutator strains, respectively, were not identified in Australian strains from the amino acid alignment of these proteins. Furthermore, the 2019 hospital environmental isolates EK1, EK3, EK4, and EK5, collected from various wards’ hand-washing sinks or toilet environments, were from the same hospital as the EkQ5-EkQ17-EK6-EK2 clade. The EK hospital isolates from this study are separated from the 2012 Singaporean isolates NUH6 and NUH11 by 656 to 867 SNPs and 41 to 72 indels, respectively, and share a clade with 2016 outbreak isolate CSID_3015183686.
Evidence of E. anophelis nosocomial transmission.
Two instances of recent closely related Australian E. anophelis isolates were identified on two separate lineages by phylogenetic analysis (Fig. 2). In the first instance, two isolates, EkM1 and EkM2, were collected from the same patient 1 month apart, branching as a unique lineage with clinical isolate EkQ8 from a patient in a different hospital (Fig. 2).
In the second instance, diverging from the Wisconsin outbreak cluster in the E. anophelis phylogeny are five epidemiologically linked clinical isolates, EkQ5, EkQ17, EkS2, and hospital environmental isolates EK2 and EK6 (Fig. 2). SNP and indel comparisons between clinical strains EkQ5 and EkQ17, from two different patients admitted into the same transplant ward 9 months apart in 2018, revealed a difference of 8 SNPs and 1 indel. Epidemiologically, EkQ5, EkQ17, and EkS2 appear to be linked to a single environmental source within the transplant ward.
Mutational differences between EkQ5-EkQ17 and EK6 were mostly nonsynonymous in nature, consistent with adaptive evolution (dN/dS ratios presented in Table S3). Of the two SNPs separating EkQ17 and EK6, one resulted in a missense mutation (resulting in a change from E to K at position 168 [E168K]) in a hypothetical protein (Ek00046). Between EkQ5 and EK6, 4 SNPs resulted in missense mutations and 2 caused nonsense mutations in penicillin-binding protein E (PbpE) and a sugar transporter protein that increased protein length, likely leading to altered or lost protein function (Table 2). In addition, the indel mutation accrued by EkQ5 resulted in a frameshift mutation that elongated a hypothetical protein (Ek02802) by 9 residues, potentially altering its function.
Another hospital environmental isolate, EK2, was linked to the EkQ5-EkQ17-EK6 clade according to phylogenetic analysis, differing by 38 SNPs and 16 indels (Fig. 2). This isolate was collected in 2019 from a sink drain in the infectious disease ward adjacent to the transplant ward where EkQ5, EkQ17, and EK6 were isolated. A more distantly related clinical isolate, EkS2, also clustered within the same clade as the EkQ5-EkQ17-EK6-EK2 isolates but differed from these isolates by 3,552 SNPs and 120 indels. Consistent with the phylogenomic findings, EkS2 was not epidemiologically linked to the EkQ5-EkQ17-EK6-EK2 isolates, being isolated from a patient admitted to a different hospital in 2015.
MICs.
A total of 39 clinically relevant antimicrobials were tested across the 22 clinical E. anophelis, E. miricola, and E. meningoseptica isolates. Modal MICs were relatively consistent within and between species and predominantly sat on the higher end of the ranges tested (Tables S4 and S5). Elizabethkingia does not have a defined clinical breakpoint, and therefore, species were examined against the EUCAST nonspecies and CLSI non-Enterobacteriaceae PK-PD breakpoints. The EUCAST breakpoints suggest that Australian strains have the greatest resistance to cephalosporins, carbapenems, and penicillins, even in combination with β-lactamase inhibitors (amoxicillin-clavulanic acid, piperacillin-tazobactam, and ampicillin-sulbactam). Furthermore, the CLSI breakpoints suggest high levels of resistance to amikacin, gentamicin, tobramycin, and chloramphenicol. From the MICs (Tables 3 and 4), only a select few antimicrobials had modal MICs in the lower range, including tetracyclines (doxycycline, 2 μg/ml, and minocycline, 0.5 to 1 μg/ml), fluoroquinolones (ciprofloxacin, 0.25 μg/ml, and levofloxacin, 0.25 μg/ml) and trimethoprim-sulfamethoxazole (1 μg/ml) (Tables 3 and 4). Only minocycline achieved 100% susceptibility across all E. anophelis strains using the CLSI non-Enterobacteriaceae PK-PD breakpoints. Rifampin and azithromycin do not have corresponding EUCAST or CLSI PK-PD breakpoints; however, their respective modal MICs are also on the lower end of the ranges tested, suggesting the potential for susceptibility. Vancomycin also lacks corresponding EUCAST or CLSI PK-PD breakpoints, but based on the MICs observed (range, 8 to 64 μg/ml), it is expected the isolates are nonsusceptible. One E. anophelis isolate, EkQ6, was responsible for the low MICs observed across the antimicrobials tested, remaining susceptible to cephalosporins and carbapenems, in addition to the fluoroquinolones, tetracyclines, and trimethoprim-sulfamethoxazole.
In silico AMR gene analysis.
All 22 clinical Elizabethkingia species genomes carried all three previously described β-lactamases that are characteristic of Elizabethkingia. The chromosomal extended-spectrum β-lactamase blaCME encodes cephalosporin and β-lactamase activity, while metallo-β-lactams blaBlaB and blaGOB encode activity against carbapenemases and penicillin–β-lactamase combinations. The metallo-β-lactamase blaBlaB carried a missense mutation of blaBlaBΔT16A in EkQ6. Three E. miricola isolates and the E. bruuniana isolate carried an AmpC variant with 94 to 95% sequence similarity to AmpC identified in E. anophelis and E. miricola genomes (accession numbers CP006576, CP007547, and CP011059). All isolates carried a conserved AmpG, with three strains exhibiting 5′-end truncations AmpGM1_A243del and one strain exhibiting AmpGM1_A3del. All isolates also carried tetracycline resistance gene tet(X), chloramphenicol resistance gene catB, and aminoglycoside resistance gene aadS. Except for that of E. bruuniana EkQ11, all Australian Elizabethkingia species genomes carried the vancomycin resistance protein VanW.
DISCUSSION
Elizabethkingia spp. have caused serious nosocomial infections and outbreaks globally, and yet, they have received little attention to date. This study aimed to fill knowledge gaps surrounding the diversity, origin, and transmission events of clinical and environmental Elizabethkingia species isolates from Australia, a previously unstudied population.
Elizabethkingia identification using comparative genomics versus mass spectrometry.
The 28 Australian Elizabethkingia isolates were identified as E. anophelis, E. meningoseptica, E. miricola, and E. bruuniana, with E. anophelis as the primary infecting species in Australia, similar to recent global reports (7, 18, 21). Despite a previous review of identification failure using mass spectroscopy for species other than E. anophelis and E. meningoseptica (4), Vitek MS Knowledge Base version 3.2 performed reliably in this study, with 96.2% accuracy. E. bruuniana (one isolate) was the only species that could not be accurately identified, instead being identified as the sister species E. miricola. This could be due to the species not yet being present in the database, or perhaps to E. miricola and E. bruuniana being variations of the same species, as many previous identification errors were seen in the genus phylogeny (Fig. 1). Nevertheless, identification of E. miricola should be taken with caution until the database has been upgraded with the capabilities to differentiate between the sister species.
Australian Elizabethkingia and global relatedness.
Phylogenomic analyses of Australian clinical isolates revealed dispersal throughout the tree and unique lineages in some strains. Others branched with previously identified geographically diverse isolates from both clinical and environmental settings (Fig. 1). Recently, DNA-DNA hybridization and average nucleotide identity have allowed the reclassification of E. miricola strains ATCC 33958, BM10, and EM798-26 to E. bruuniana (3, 21, 41). Further to these corrections, using comparative genomics, we suggest the reclassification of E. miricola strains 6012926 and CIP111047 to E. bruuniana, E. meningoseptica strains NCTC10588 and NCTC10586 to E. anophelis, and lastly, E. meningoseptica NCTC11305 to E. miricola (Fig. 1). Evidence from past studies has described the structure of E. anophelis phylogenies as consisting of two and six major clades (6, 42); in this study, we identified six lineages, yet as sampling continues, this may expand (Fig. 2).
Several E. anophelis isolates from this study cluster phylogenetically with the Wisconsin outbreak strains from 2016, the most pathogenic Elizabethkingia outbreak to date (6). Outbreak and hypermutator strains have been characterized by their ICE insertions and truncations at the C terminus in both the MutS and MutY protein sequences, respectively (6). The MutS and MutY protein sequences in our clinical isolates aligned with few nonsynonymous amino acid changes and no truncations, and therefore, it is unlikely that the Australian clinical isolates would display the outbreak characteristics or phenotype suspected to be responsible for the increased pathogenesis of the Wisconsin strains or the hypermutator phenotype identified in one Wisconsin strain. Several pathogenicity islands were identified in both Australian and Wisconsin E. anophelis strains, suggesting they may play an important role in the species survival or pathogenesis.
Potential nosocomial transmission of E. anophelis in a transplant ward.
A recent case of hospital-acquired E. anophelis infection was suggested by the identification of a clonal cluster comprised of clinical and environmental isolates in this study. A pair of Australian E. anophelis clinical isolates, EkQ5 and EkQ17, collected almost a year apart in 2018 from two patients on the transplant ward, were characterized as differing by only eight SNPs and one deletion. Additionally, it was found that the hospital environmental sample collected from a hand-washing sink in the same transplant ward in late 2019 only differed from clinical sample EkQ5 by six of the same eight SNPs and the one deletion (Table 2). The combination of clinical and environmental genomic data with such low genetic diversity suggests that these strains were transmitted via the common reservoir of the hand-washing sink, given the extended time frame between patient infection and environmental collection. Nearly identical isolates have been described previously within E. anophelis, such as environmentally collected isolates OSUVM-1 and -2 (43), hospital outbreak strain NUHP (44), and Wisconsin CSID strains (6), suggesting that low genetic variation is not unusual among E. anophelis infections. The relatedness of sink or toilet environment hospital isolates EK4 and EK5 from the transplant ward to EK1 and EK3 from the oncology ward suggests that another transmission event may have also taken place, despite not identifying a related clinical isolate. Interestingly, these hospital environmental isolates (EK1, -3, -4, and -5) formed a clonal cluster and appear to share similarity to two 2012 Singaporean clinical isolates, NUH6 and NUH11, which were also isolated from hospital environments (11).
Additional studies have reported contaminated communal water sources as a reservoir for Elizabethkingia species infections within hospitals (1, 17), with handwashing stations in a pediatric intensive care unit the source of several Elizabethkingia species infections in Singapore, where staff transmitted the infection after handwashing (2). Although direct human-to-human transmission is seen in many other nosocomial infections (45, 46) and vertical transmission has been reported in E. anophelis (47), the role human-to-human transmission has in Elizabethkingia infections still remains unclear. However, given the severity of the infections, known patient risk factors, and the suggested longevity of the bacteria in the environment, the potential for horizontal transmission should not be overlooked.
MIC testing.
The MIC data generated in this study confirm that, like those in previous studies, the Australian clinical Elizabethkingia species isolates (with the exception of isolate EkQ6) (Tables S4 and S5 in the supplemental material) are resistant to many antimicrobial classes, including cephalosporins, carbapenems, and aminoglycosides (Tables 3 and 4) (12, 19, 20, 22, 48). From the literature, there is some variation in E. anophelis antimicrobial resistance profiles among isolates from the United States, Southeast Asia, and South Korea, while Australian isolates appear to phenotypically express some marked differences. For example, approximately 75 to 100% of E. anophelis isolates were reported to be resistant to trimethoprim-sulfamethoxazole (6, 19–22), while 75% of Australian strains remained sensitive. Additionally, 88 to 95% of isolates were susceptible to piperacillin-tazobactam (6, 19, 21, 22), while 68 to 70% of Australian and South Korean (20) isolates were resistant. Vancomycin has been suggested as a potential therapy for E. meningoseptica infections; therefore, we screened our E. anophelis strains against vancomycin and additional antimicrobials with Gram-positive activity, such as teicoplanin. Despite the recommendations for vancomycin use in Elizabethkingia infections (4, 20, 49, 50), our data show resistance among Australian clinical isolates, as the MICs were on the high end of the range tested and all isolates except E. bruuniana (EkQ11) carried the vanW gene. This is the first set of MIC data for teicoplanin, and with a modal MIC of 32 μg/ml, these strains appear to be resistant. Similar to the Wisconsin outbreak strains (6), Australian Elizabethkingia species strains may be susceptible to azithromycin, as the modal MIC of 4 μg/ml is on the lower end of the range tested. Although doxycycline testing against E. anophelis is not often reported in the literature, others have found their strains to be highly susceptible, unlike in our study (22). EUCAST breakpoints suggest that 6.25% and 43.75% of Australian E. anophelis isolates are resistant to levofloxacin and ciprofloxacin, respectively. Variability in fluoroquinolone susceptibility has also been observed in the majority of Southeast Asian and United States strains (6, 19–22). It is clear that numerous antimicrobials have been tested across E. anophelis isolates in previous studies, although susceptibility to multiple antimicrobial classes like that observed in EkQ6 has not been reported previously. Further testing of E. anophelis isolates from Australia and abroad would determine whether this type of sensitivity is unique to a subset of Australian strains or is present globally.
In silico AMR gene analysis.
Antimicrobial resistance (AMR) genes blaBlaB, blaGOB, and blaCME were identified within the genomes of all clinical Australian Elizabethkingia isolates, linking directly to their observed MIC profiles. All isolates except EkQ6 were resistant to cephalosporins and β-lactams, attributed to blaCME, and to carbapenemases and penicillin–β-lactamase combinations, attributed to blaBlaB and blaGOB (4, 51–53). Additionally, in conjunction with these three beta-lactamase genes, AmpC has been identified previously in a few Elizabethkingia genomes (E. anophelis and E. miricola) and was identified here in three E. miricola genomes and an E. bruuniana genome with high sequence similarity. Resistance attributed to AmpC and its exact role in Elizabethkingia species are still to be described, as there are no observed differences in susceptibility profiles of strains harboring AmpC to date (20). However, fluoroquinolone resistance varied, as described above, and is mediated by a single-step amino acid substitution (Ser83Ile or Ser83Arg) in GyrA (19, 21, 54) that was not identified in any of the Australian clinical Elizabethkingia isolates. The absence of the mutation has also been reported recently for a single isolate in Taiwan (22). Previous studies have linked DNA topoisomerase IV to an assistance-type role in fluoroquinolone resistance for Elizabethkingia spp. (22, 54), although this was not identified in our clinical isolate collection either. Phenicol resistance conferred by catB genes has been identified in Elizabethkingia previously, with all Australian clinical isolates harboring this gene (55, 56). Aminoglycoside and tetracycline resistance genes aadS and tetX were also present in all genomes, the former likely responsible for the observed aminoglycoside resistance (Tables 3 and 4), with similar genes shown to confer resistance in Elizabethkingia species (56, 57). Although tetX was present in all isolates, nearly ubiquitous susceptibility to tetracyclines was observed in MIC profiles, suggesting that tetX alone does not confer tetracycline resistance in some Elizabethkingia species or that it could be a silent gene, although it has been identified as a resistance mechanism in E. coli (56, 58).
In addition, clinical E. anophelis isolate EkQ6 carried several mutations not commonly described in blaBlaB and topA (21, 22, 50, 56) and yet remained susceptible to cephalosporins, carbapenems, tetracyclines, and fluoroquinolones. The substitutions and deletions, respectively, may or may not be linked to the susceptibility of this isolate. Comparative genomics, including more susceptible isolates like EkQ6, would provide great insight into the intrinsic antimicrobial resistance mechanisms of Elizabethkingia species (50, 59, 60).
Potential antimicrobial therapy for Elizabethkingia spp.
In this study, Australian isolates appear to be susceptible to fluoroquinolones, tetracyclines, and trimethoprim-sulfamethoxazole. Only levofloxacin and minocycline demonstrated 100% susceptibility using CLSI PK-PD breakpoints. Fluoroquinolone treatment alone has proven to be successful in Elizabethkingia species infections (61), but some recommend combination therapy (62) in order to mitigate high-level fluoroquinolone resistance for those susceptible to single-step mutations. From our and other studies, susceptibility is clearly strain dependent. Our findings suggest that rifampin (63) or azithromycin could also be effective antimicrobials, although this would require further testing. With this in mind and the recent success of newer antimicrobials against MDR Gram-negative bacteria (64–66), it would be of value to further test Elizabethkingia spp. against newer antimicrobials, such as cefiderocol (67). Due to the pathogenic nature of Elizabethkingia infections, therapy should always be guided by patient condition and MIC data.
Conclusions.
This study has characterized the diversity of Australian Elizabethkingia spp. genotypically and phenotypically using comparative genomics and antimicrobial resistance. We have revealed significant strain diversity in Australia and have shown that Vitek MS Knowledge Base version 3.2 can accurately identify E. anophelis, E. meningoseptica, and E. miricola species but is not yet able to correctly identify E. bruuniana. Furthermore, genomic exploration has provided insight into the breadth of the intrinsic MDR nature of Elizabethkingia species infections and revealed a potential reservoir of infection within a hospital setting, where two patients were infected with nearly identical strains. Antimicrobial resistance data suggest that clinical isolates are susceptible to fluoroquinolones, tetracyclines, and trimethoprim-sulfamethoxazole. Specifically, minocycline and levofloxacin showed suitable efficacy against Elizabethkingia isolates in vitro, although further clinical studies are required to define optimal therapy.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Study Education and Research Committee of Pathology Queensland (L.G.), University of the Sunshine Coast (D.B.), Advance Queensland (grant numbers AQRF13016-17RD2 for D.S.S. and AQIRF0362018 for E.P.P.), and the National Health and Medical Research Council (grant number GNT1157530 for P.N.A.H.) for funding this study. We would like to express our gratitude to Mater Pathology, Sullivan and Nicolaides Pathology, and Pathology Queensland for their involvement and support in this project. Finally, we thank the infection control nurses at participating hospitals for environmental sampling.
David L. Paterson reports nonfinancial support from Ecolab Pty. Ltd., Whiteley Corporation, and Kimberly-Clark Professional during the conduct of the study and personal fees from Merck, Shionogi, Achaogen, AstraZeneca, Leo Pharmaceuticals, Bayer, GlazoSmithKline, Cubist, Venatorx, Accelerate, and Pfizer and grants from Shionogi and Merck (MSD) outside the submitted work. Patrick N. A. Harris reports grants from Merck (MSD) and Shionogi and personal fees from Pfizer outside the submitted work. All other authors declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Moore LSP, Owens DS, Jepson A, Turton JF, Ashworth S, Donaldson H, Holmes AH. 2016. Waterborne Elizabethkingia meningoseptica in adult critical care. Emerg Infect Dis 22:9–17. doi: 10.3201/eid2201.150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yung C-F, Maiwald M, Loo LH, Soong HY, Tan CB, Lim PK, Li L, Tan NW, Chong C-Y, Tee N, Thoon KC, Chan YH. 2018. Elizabethkingia anophelis and association with tap water and handwashing, Singapore. Emerg Infect Dis 24:1730–1733. doi: 10.3201/eid2409.171843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, Emery B, Bell M, Loparev V, Juieng P, Gartin J, Bizet C, Clermont D, Criscuolo A, Brisse S, McQuiston JR. 2018. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 111:55–72. doi: 10.1007/s10482-017-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J-N, Lai C-H, Yang C-H, Huang Y-H. 2019. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms 7:295. doi: 10.3390/microorganisms7090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson AC, Humrighouse BW, Graziano JC, Emery B, McQuiston JR. 2016. Draft genome sequences of strains representing each of the Elizabethkingia genomospecies previously determined by DNA-DNA hybridization. Genome Announc 4:e00045-16. doi: 10.1128/genomeA.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin A, Larsonneur E, Nicholson AC, Edwards DJ, Gundlach KM, Whitney AM, Gulvik CA, Bell ME, Rendueles O, Cury J, Hugon P, Clermont D, Enouf V, Loparev V, Juieng P, Monson T, Warshauer D, Elbadawi LI, Walters MS, Crist MB, Noble-Wang J, Borlaug G, Rocha EPC, Criscuolo A, Touchon M, Davis JP, Holt KE, McQuiston JR, Brisse S. 2017. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun 8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew KL, Cheng B, Lin RTP, Teo JWP. 2018. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol 56:e01445-17. doi: 10.1128/JCM.01445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jean SS, Lee WS, Chen FL, Ou TY, Hsueh PR. 2014. Elizabethkingia meningoseptica: an important emerging pathogen causing healthcare-associated infections. J Hosp Infect 86:244–249. doi: 10.1016/j.jhin.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Frank T, Gody JC, Nguyen LBL, Berthet N, Le Fleche-Mateos A, Bata P, Rafaï C, Kazanji M, Breurec S. 2013. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet 381:1876. doi: 10.1016/S0140-6736(13)60318-9. [DOI] [PubMed] [Google Scholar]
- 10.Issack MI, Neetoo Y. 2011. An outbreak of Elizabethkingia meningoseptica neonatal meningitis in Mauritius. J Infect Dev Ctries 5:834–839. doi: 10.3855/jidc.1885. [DOI] [PubMed] [Google Scholar]
- 11.Teo J, Tan S-Y, Tay M, Ding Y, Kjelleberg S, Givskov M, Lin RT, Yang L. 2013. First case of E. anophelis outbreak in an intensive-care unit. Lancet 382:855–856. doi: 10.1016/S0140-6736(13)61858-9. [DOI] [PubMed] [Google Scholar]
- 12.Hsu M-S, Liao C-H, Huang Y-T, Liu C-Y, Yang C-J, Kao K-L, Hsueh P-R. 2011. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999–2006. Eur J Clin Microbiol Infect Dis 30:1271–1278. doi: 10.1007/s10096-011-1223-0. [DOI] [PubMed] [Google Scholar]
- 13.Choi MH, Kim M, Jeong SJ, Choi JY, Lee I-Y, Yong T-S, Yong D, Jeong SH, Lee K. 2019. Risk factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg Infect Dis 25:42–51. doi: 10.3201/eid2501.171985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janda JM, Lopez DL. 2017. New pathogen profiles: Elizabethkingia anophelis. Diagn Microbiol Infect Dis 88:201–205. doi: 10.1016/j.diagmicrobio.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Weaver KN, Jones RC, Albright R, Thomas Y, Zambrano CH, Costello M, Havel J, Price J, Gerber SI. 2010. Acute emergence of Elizabethkingia meningoseptica infection among mechanically ventilated patients in a long-term acute care facility. Infect Control Hosp Epidemiol 31:54–58. doi: 10.1086/649223. [DOI] [PubMed] [Google Scholar]
- 16.Chawla K, Gopinathan A, Varma M, Mukhopadhyay C. 2015. Elizabethkingia meningoseptica outbreak in intensive care unit. J Glob Infect Dis 7:43–44. doi: 10.4103/0974-777X.150890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyritsi MA, Mouchtouri VA, Pournaras S, Hadjichristodoulou C. 2018. First reported isolation of an emerging opportunistic pathogen (Elizabethkingia anophelis) from hospital water systems in Greece. J Water Health 16:164–170. doi: 10.2166/wh.2017.184. [DOI] [PubMed] [Google Scholar]
- 18.Lau SKP, Chow W-N, Foo C-H, Curreem SOT, Lo G-S, Teng JLL, Chen JHK, Ng RHY, Wu AKL, Cheung IYY, Chau SKY, Lung DC, Lee RA, Tse CWS, Fung KSC, Que T-L, Woo PCY. 2016. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep 6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J-N, Lai C-H, Yang C-H, Huang Y-H, Lin H-H. 2018. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother 73:2497–2502. doi: 10.1093/jac/dky197. [DOI] [PubMed] [Google Scholar]
- 20.Han M-S, Kim H, Lee Y, Kim M, Ku NS, Choi JY, Yong D, Jeong SH, Lee K, Chong Y. 2017. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol 55:274–280. doi: 10.1128/JCM.01637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J-N, Lai C-H, Yang C-H, Huang Y-H. 2018. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis isolated in Taiwan. J Clin Med 7:538. doi: 10.3390/jcm7120538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jian M-J, Cheng Y-H, Chung H-Y, Cheng Y-H, Yang H-Y, Hsu C-S, Perng C-L, Shang H-S. 2019. Fluoroquinolone resistance in carbapenem-resistant Elizabethkingia anophelis: phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J Antimicrob Chemother 74:1503–1510. doi: 10.1093/jac/dkz045. [DOI] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H. 2013. Seqtk: a fast and lightweight tool for processing FASTA or FASTQ sequences.
- 26.Huang W, Li L, Myers JR, Marth GT. 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, p 158–170. In Deng M, Jiang R, Sun F, Zhang X (ed), Research in computational molecular biology. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 29.Sarovich DS, Price EP. 2014. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 33.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahlmeter G, Brown DFJ, Goldstein FW, MacGowan AP, Mouton JW, Odenholt I, Rodloff A, Soussy C-J, Steinbakk M, Soriano F, Stetsiouk O. 2006. European Committee on Antimicrobial Susceptibility Testing (EUCAST) technical notes on antimicrobial susceptibility testing. Clin Microbiol Infect 12:501–503. doi: 10.1111/j.1469-0691.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; CLSI guideline M45, 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A10, 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 39.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J-N, Lai C-H, Yang C-H, Huang Y-H, Lin H-H. 2017. Genomic features, phylogenetic relationships, and comparative genomics of Elizabethkingia anophelis strain EM361-97 isolated in Taiwan. Sci Rep 7:14317. doi: 10.1038/s41598-017-14841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breurec S, Criscuolo A, Diancourt L, Rendueles O, Vandenbogaert M, Passet V, Caro V, Rocha EPC, Touchon M, Brisse S. 2016. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci Rep 6:30379. doi: 10.1038/srep30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson WL, Ramachandran A, Torres NJ, Nicholson AC, Whitney AM, Bell M, Villarma A, Humrighouse BW, Sheth M, Dowd SE, McQuiston JR, Gustafson JE. 2018. The draft genomes of Elizabethkingia anophelis of equine origin are genetically similar to three isolates from human clinical specimens. PLoS One 13:e0200731. doi: 10.1371/journal.pone.0200731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo J, Tan S-Y, Liu Y, Tay M, Ding Y, Li Y, Kjelleberg S, Givskov M, Lin RTP, Yang L. 2014. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol Evol 6:1158–1165. doi: 10.1093/gbe/evu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tagoe DNA, Baidoo S, Dadzie I, Tengey D, Agede C. 2011. Potential sources of transmission of hospital acquired infections in the Volta Regional Hospital in Ghana. Ghana Med J 45:22–26. doi: 10.4314/gmj.v45i1.68918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan HA, Baig FK, Mehboob R. 2017. Nosocomial infections: epidemiology, prevention, control and surveillance. Asian Pac J Trop Biomed 7:478–482. doi: 10.1016/j.apjtb.2017.01.019. [DOI] [Google Scholar]
- 47.Lau SKP, Wu AKL, Teng JLL, Tse H, Curreem SOT, Tsui SKW, Huang Y, Chen JHK, Lee RA, Yuen K-Y, Woo P. 2015. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg Infect Dis 21:232–241. doi: 10.3201/eid2102.140623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J-N, Lai C-H, Yang C-H, Huang Y-H, Lin H-H. 2019. Genomic features, comparative genomics, and antimicrobial susceptibility patterns of Elizabethkingia bruuniana. Sci Rep 9:2267. doi: 10.1038/s41598-019-38998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jean S-S, Hsieh T-C, Ning Y-Z, Hsueh P-R. 2017. Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int J Antimicrob Agents 50:507–511. doi: 10.1016/j.ijantimicag.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 50.Chang T-Y, Chen H-Y, Chou Y-C, Cheng Y-H, Sun J-R. 2019. In vitro activities of imipenem, vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. Eur J Clin Microbiol Infect Dis 38:2045–2052. doi: 10.1007/s10096-019-03639-3. [DOI] [PubMed] [Google Scholar]
- 51.Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. 2000. Genetic-biochemical analysis and distribution of the Ambler class A beta-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob Agents Chemother 44:1–9. doi: 10.1128/AAC.44.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossolini GM, Franceschini N, Lauretti L, Caravelli B, Riccio ML, Galleni M, Frère J-M, Amicosante G. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A beta-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER beta-lactamases. Antimicrob Agents Chemother 43:2193–2199. doi: 10.1128/AAC.43.9.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colapietro M, Endimiani A, Sabatini A, Marcoccia F, Celenza G, Segatore B, Amicosante G, Perilli M. 2016. BlaB-15, a new BlaB metallo-beta-lactamase variant found in an Elizabethkingia miricola clinical isolate. Diagn Microbiol Infect Dis 85:195–197. doi: 10.1016/j.diagmicrobio.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Jian M-J, Cheng Y-H, Perng C-L, Shang H-S. 2018. Molecular typing and profiling of topoisomerase mutations causing resistance to ciprofloxacin and levofloxacin in Elizabethkingia species. PeerJ 6:e5608. doi: 10.7717/peerj.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu S, Cao L, Wu Y, Zhou Y, Jiang T, Wang L, Wang Q, Ming D, Chen S, Wang M. 2019. Comparative genomic analysis of Myroides odoratimimus isolates. Microbiologyopen 8:e00634. doi: 10.1002/mbo3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M, Gao H, Lin N, Zhang Y, Huang N, Walker ED, Ming D, Chen S, Hu S. 2019. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. Microbiologyopen 8:e804. doi: 10.1002/mbo3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, Soehnlen M, Blom J, Terrapon N, Henrissat B, Walker ED. 2019. Comparative genomic analyses reveal diverse virulence factors and antimicrobial resistance mechanisms in clinical Elizabethkingia meningoseptica strains. PLoS One 14:e0222648. doi: 10.1371/journal.pone.0222648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang W, Moore IF, Koteva KP, Bareich DC, Hughes DW, Wright GD. 2004. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 279:52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- 59.Opota O, Diene SM, Bertelli C, Prod’hom G, Eckert P, Greub G. 2017. Genome of the carbapenemase-producing clinical isolate Elizabethkingia miricola EM_CHUV and comparative genomics with Elizabethkingia meningoseptica and Elizabethkingia anophelis: evidence for intrinsic multidrug resistance trait of emerging pathogens. Int J Antimicrob Agents 49:93–97. doi: 10.1016/j.ijantimicag.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 60.Balm MN, Salmon S, Jureen R, Teo C, Mahdi R, Seetoh T, Teo JT, Lin RT, Fisher DA. 2013. Bad design, bad practices, bad bugs: frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect 85:134–140. doi: 10.1016/j.jhin.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 61.Huang Y-C, Lin Y-T, Wang F-D. 2018. Comparison of the therapeutic efficacy of fluoroquinolone and non-fluoroquinolone treatment in patients with Elizabethkingia meningoseptica bacteraemia. Int J Antimicrob Agents 51:47–51. doi: 10.1016/j.ijantimicag.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 62.Chan JC, Chong CY, Thoon KC, Tee NWS, Maiwald M, Lam JCM, Bhattacharya R, Chandran S, Yung CF, Tan N. 2019. Invasive paediatric Elizabethkingia meningoseptica infections are best treated with a combination of piperacillin/tazobactam and trimethoprim/sulfamethoxazole or fluoroquinolone. J Med Microbiol 68:1167–1172. doi: 10.1099/jmm.0.001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassetti M, Peghin M, Vena A, Giacobbe DR. 2019. Treatment of infections due to MDR Gram-negative bacteria. Front Med 6:74. doi: 10.3389/fmed.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2017. In vitro activity of the siderophore cephalosporin, cefiderocol, against carbapenem-nonsusceptible and multidrug-resistant isolates of gram-negative bacilli collected worldwide in 2014 to 2016. Antimicrob Agents Chemother 62:e01968-17. doi: 10.1128/AAC.01968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, Doi Y, Hao B, Press EG, Cooper VS, Clancy CJ, Nguyen MH. 2017. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis 65:110–120. doi: 10.1093/cid/cix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shields RK, Clancy CJ, Pasculle AW, Press EG, Haidar G, Hao B, Chen L, Kreiswirth BN, Nguyen MH. 2017. Verification of ceftazidime-avibactam and ceftolozane-tazobactam susceptibility testing methods against carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa. J Clin Microbiol 56:e01093-17. doi: 10.1128/JCM.01093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhanel GG, Golden AR, Zelenitsky S, Wiebe K, Lawrence CK, Adam HJ, Idowu T, Domalaon R, Schweizer F, Zhanel MA, Lagacé-Wiens PRS, Walkty AJ, Noreddin A, Lynch JP III, Karlowsky JA. 2019. Cefiderocol: a siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant gram-negative bacilli. Drugs 79:271–289. doi: 10.1007/s40265-019-1055-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina sequence data for the 28 Elizabethkingia species genomes described in this study have been deposited in the NCBI in the SRA database under accession number SRP225137 and the BioProject database under accession number PRJNA576977 (BioSample accession numbers SAMN13016226 to SAMN13016247 and SAMN14081590 to SAMN14081595).