Pseudomonas aeruginosa is an opportunistic human pathogen that frequently causes health care-associated infections (HAIs). Due to its metabolic diversity and ability to form biofilms, this Gram-negative nonfermenting bacterium can persist in the health care environment, which can lead to prolonged HAI outbreaks. We describe the creation of a core genome multilocus sequence typing (cgMLST) scheme to provide a stable platform for the rapid comparison of P. aeruginosa isolates using whole-genome sequencing (WGS) data.
KEYWORDS: HAI, Pseudomonas aeruginosa, antibiotic resistance, cgMLST
ABSTRACT
Pseudomonas aeruginosa is an opportunistic human pathogen that frequently causes health care-associated infections (HAIs). Due to its metabolic diversity and ability to form biofilms, this Gram-negative nonfermenting bacterium can persist in the health care environment, which can lead to prolonged HAI outbreaks. We describe the creation of a core genome multilocus sequence typing (cgMLST) scheme to provide a stable platform for the rapid comparison of P. aeruginosa isolates using whole-genome sequencing (WGS) data. We used a diverse set of 58 complete P. aeruginosa genomes to curate a set of 4,440 core genes found in each isolate, representing ∼64% of the average genome size. We then expanded the alleles for each gene using 1,991 contig-level genome sequences. The scheme was used to analyze genomes from four historical HAI outbreaks to compare the phylogenies generated using cgMLST to those of other means (traditional MLST, pulsed-field gel electrophoresis [PFGE], and single-nucleotide variant [SNV] analysis). The cgMLST scheme provides sufficient resolution for analyzing individual outbreaks, as well as the stability for comparisons across a variety of isolates encountered in surveillance studies, making it a valuable tool for the rapid analysis of P. aeruginosa genomes.
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium responsible for 32,600 multidrug-resistant, health care-associated infections (HAIs) per year in the United States (1). P. aeruginosa can cause a variety of HAIs, including pneumonia and bloodstream and wound infections; it is also the main cause of mortality in cystic fibrosis patients (2).
P. aeruginosa has a large, complex genome with intrinsic, adaptive, and acquired resistance mechanisms (3) and multiple virulence factors (4), making infections both difficult to treat and highly morbid (5). P. aeruginosa can also form biofilms, allowing it to persist in the health care environment, including on surfaces (6), medical devices (7), and in water sources (8). Because of its ubiquity, outbreaks involving P. aeruginosa can be both clonal and involve multiple sequence types (STs) (9), as defined by the traditional seven-gene multilocus sequence typing (MLST) scheme (10, 11).
The advent of whole-genome sequencing (WGS) has changed HAI outbreak investigations by drastically increasing the resolution with which isolates can be genetically characterized (12, 13). Traditional MLST schemes can be expanded from seven genes to core genome MLST (cgMLST) schemes with WGS data by including the thousands of genes common to a particular species (14) or on an ad hoc basis (15). Herein, we describe the creation of a cgMLST scheme for P. aeruginosa and apply it to explore the diversity of the species in publicly available genomes. We also compare it to other phylogenetic methods using isolates from a convenient set of four HAI outbreaks.
MATERIALS AND METHODS
Development of the P. aeruginosa cgMLST scheme.
Genes from all 58 complete P. aeruginosa genomes available from RefSeq (16) in January 2017 were aligned and compared using BLAT v36x2 (17). All genes present as a single copy in each genome with ≥95% nucleotide similarity to the allele from the reference PAO1 genome were used to construct a core genome MLST (cgMLST) scheme. The cgMLST scheme was implemented using custom scripts (https://github.com/DHQP) on an additional 1,991 genomes downloaded from RefSeq (16) for evaluation, to refine the loci included in the scheme, and to expand the alleles for each gene. A Bonferroni correction threshold of P < 0.05 (P = 0.05/2,049 = 1.1 × 10−5) was used to determine whether any genes should be removed from the cgMLST scheme if they were missing, present as multiple copies, or varied by ≥10% in size at a statistically significant rate in the total set of 2,049 genomes (58 complete plus 1,991 assemblies). This was implemented by determining a Z-score associated with the absence of each gene, and genes with Z-scores higher than that of P were removed. For each gene in the scheme the number missing, present as multiple copies, and Z-score for all RefSeq genomes analyzed are included in Spreadsheet S2 in the supplemental material.
Identification of prophage and recombination genes.
The web server PHASTER (18) (phaster.ca) was used to identify genes included in the cgMLST scheme that originated form prophage sequences (Spreadsheet S2). ClonalFrameML (19) was run using default parameters on a dendrogram made with the cgMLST scheme for the 58 complete P. aeruginosa genomes to find genes that included regions of recombination (Spreadsheet S2).
Tree generation from cgMLST scheme.
DendroPy 4.2 (20) was used to create dendrograms using the unweighted pair group method with arithmetic mean (UPGMA) and neighbor joining (NJ) methods based on the number of allelic differences in the cgMLST scheme between genomes. The UPGMA method generated dendrograms more similar to the trees generated by the hqSNV pipeline (Table S2), so was used as the primary method in this work. Bootstrap support values were determined using the SumTrees package in DendroPy.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) patterns were determined using standard procedures for Gram-negative bacteria (https://www.cdc.gov/hai/pdfs/labsettings/Modified-PulsedNet-procedure-GNB.pdf) and analyzed through the BioNumerics software package from Applied Maths NV (Sint-Martens-Latem, Belgium). DendroPy 4.2 (20) was used to create dendrograms using UPGMA clustering based on the number of band differences in the PFGE patterns between isolates.
Whole-genome sequencing.
DNA was extracted using the Promega Maxwell 16 Cell Low Elution Volume (LEV) DNA purification kit and the automated Maxwell 16 MDx instrument (Madison, WI). High quality input genomic DNA (gDNA) was fragmented with the Covaris ME220 Focused-ultrasonicator (Woburn, MA). Sample libraries were prepared using the NuGEN Ovation Ultralow System V2 assay kit (San Carlos, CA). Sequencing was completed using the Illumina MiSeq platform (San Diego, CA) to produce 250-bp paired-end reads.
Genome assembly, annotation, and sequence typing.
Genomes were assembled de novo using high quality reads with SPAdes v3.9 (21). Genes were identified using Prokka v1.12 (22). Sequence types (STs) were determined using the multilocus sequence typing (MLST) definitions from PubMLST (https://pubmlst.org/paeruginosa/).
Generation of SNV trees.
The hqSNV pipeline SNVPhyl 1.3.0 (23) was used to generate phylogenetic trees using sequenced reads from the isolates associated with the four outbreaks, with the SNV abundance set to 0.75, minimum coverage set to 10, and the filter-density threshold and window set to 2 and 11, respectively. The median assembly based on Mash distance (24) was used as the reference. The k-mer-based trees were generated with the SNV pipeline kSNP v3.0 (25), using the assembled genomes from the outbreaks and a k-mer length of 13.
Tree comparisons.
Statistical comparisons of PFGE, cgMLST, and SNV tree topologies generated from the outbreak isolates were made using the unweighted Kendall-Colijn similarity metric implemented through the R package treespace (26). P values were calculated for the similarity of trees using a Z-test on a population standard deviation generated from 10,000 comparison scores from random trees (27). A threshold of P < 0.05 was used to determine similarity. Tree images were created and annotated using iTOL v3 (28).
Data availability.
Raw sequences have been deposited in NCBI BioProject ID PRJNA288601.
RESULTS
Development of the P. aeruginosa cgMLST scheme.
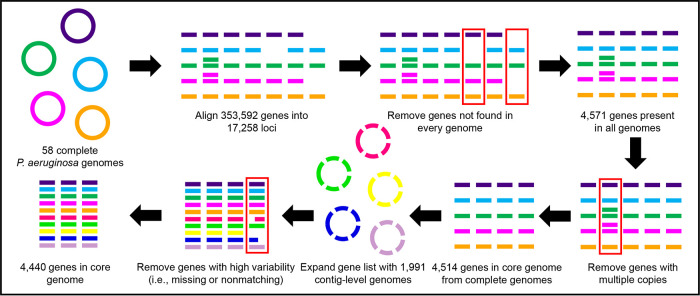
All 58 complete P. aeruginosa genomes available in NCBI’s RefSeq (16) in January of 2017 were used to construct the initial cgMLST gene set, as depicted in Fig. 1. There were 36 unique STs represented in the set, which also included eight genomes from high-risk clones (29), composed of three each of ST111 and ST253 and two from ST235 (Spreadsheet S1). The average genome size was 6.7 MB with 6,096 open reading frames. Three of the sequences included plasmids, which were excluded from the construction of the cgMLST scheme. There were 353,592 total gene alleles in the entire set, which when aligned represented 17,258 gene loci; 4,514 of the genes were present as a single copy in each genome and were included in the initial cgMLST gene list, which was later reduced to 4,440 genes after assessing an additional 1,991 contig-level genomes. These genes totaled 4.3 MB, representing ∼64% of the average P. aeruginosa genome.
FIG 1.
Creation of P. aeruginosa cgMLST gene set.
The core genome included 2,780 genes with known or probable functions, including the seven from the traditional MLST scheme. There were five virulence factors (exoT, lasB, lecA, toxA, and toxR) (4), as well as 23 genes involved in antimicrobial resistance, including one fosfomycin resistance enzyme, two beta-lactamases, and 20 components of multigene, multidrug efflux pump systems (30) (Spreadsheet S2).
The core gene list was also compared to representative genomes from the other 12 members of the P. aeruginosa species group (NCBI:txid136841) (31). They contained an average of 148 (3.4%) of the cgMLST genes, where Pseudomonas citronellolis had the most with 463 (10.5%), while Pseudomonas caeni had no overlapping genes (Table S1).
Exploration of P. aeruginosa diversity.
The 1,991 contig-level P. aeruginosa genome assemblies available in RefSeq (16) as of January of 2017 were assessed using the cgMLST scheme to explore the diversity of the species. These included 336 unique STs, 154 with more than one representative genome, as well as 190 genomes without defined STs.
Because assembled genomes may have artifacts of short-read sequencing, such as missing, incomplete, or duplicated genes (13), it was expected that some of the cgMLST loci would be absent or overrepresented from isolates in the expanded set. One hundred and fourteen genes were removed from the scheme because they were either missing from or present as multiple copies in too many genomes (see details in the Materials and Methods section), leaving a final set of 4,440 loci (Spreadsheet S2). On average there were 21 genes missing per sample, giving a coverage of 99.5% of the 4,440-gene cgMLST scheme, and in the average assembly there were 1.4 alleles with multiple copies (Spreadsheet S3).
We examined the core gene set for evidence of homologous recombination by applying ClonalFrameML (32) to a dendrogram made from the 58 complete genomes. There were 510 unique recombination sequences identified on 369 of the genes, representing 0.55% of the 92,837 unique cgMLST alleles in those genomes. Two-thirds (342/510) of the unique recombination sequences were found on multiple genomes, supporting the observation that homologous recombination helps shape the core genome of bacteria (33). Since recombination has been to shown to have greater effect on branch length rather than tree topology (34), which is of more concern when assessing relatedness in HAI outbreaks, and the alleles affected still met the statistical criteria for inclusion (Spreadsheet S2), they were not removed from the scheme.
Additionally, 20 (0.45%) of the 4,440 core genes were similar to prophage sequences, as determined by PHASTER (18). While prophage sequences can have higher rates of mutation, they can also accelerate evolution when stably incorporated into the genome (35), and thus contribute to phylogeny. Like the recombination alleles, there was no empirical evidence of increased variation in the prophage alleles in the cgMLST scheme (Spreadsheet S2), so they too were kept in the scheme.
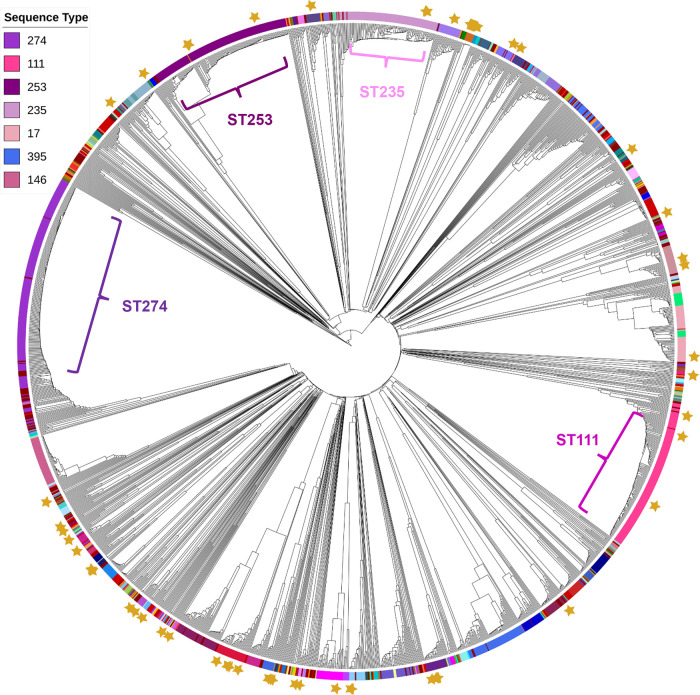
A pairwise comparison between the alleles found in each isolate was used to make a difference matrix for all isolates, from which the UPGMA was used to create a dendrogram (Fig. 2). The isolates generally clustered by ST, as different STs varied by an average of 3,889 alleles (median: 3,739; range: 8 to 4,300), while genomes from the same ST differed by an average of 331 alleles (median: 209; range: 0 to 2,903). Nineteen pairs of genomes differed by less than 10 alleles, including four pairs with zero allele differences. However, there were also instances of large diversity within STs, as 27 included isolate pairs that differed by more than 1,000 alleles. And the STs were not completely distinct in all cases, as 47 genomes had more common alleles with genomes from different STs rather than their own. Of course, some of the extreme observed differences within STs could be due to variations in sequence quality.
FIG 2.
Dendrogram from cgMLST scheme of 2,049 P. aeruginosa genomes. Stars indicate entries used to construct the core gene list. The most common MLST types are highlighted on the dendrogram.
Analysis of isolates from HAI outbreaks.
Isolates from four HAI outbreaks were used to compare the phylogeny created using the cgMLST scheme to those made from pulsed-field gel electrophoresis (PFGE) patterns and single nucleotide variant (SNV) pipelines. Outbreak 1 occurred in a single hospital unit (36) and included 12 isolates from three STs; outbreak 2 occurred in a skilled nursing facility with ventilated residents (9) and included 28 isolates from four different STs; outbreak 3 occurred in an intensive care unit and included nine isolates from three different STs; and outbreak 4 included 26 isolates from a single ST that were found across multiple facilities within the same geographic area. Only clinical isolates were included in this analysis.
PFGE, a means for determining relatedness between bacterial genomes using restriction digestion (37), generates dozens of fragments and offers more discriminatory power than traditional MLST for P. aeruginosa (38). Isolates from outbreaks 1, 2, and 3, for which both PFGE and WGS data were available, were selected for comparison. Only two isolates from outbreak 4 underwent both PFGE and WGS, so it was excluded from this analysis. Differences in PFGE patterns were used to construct UPGMA dendrograms. These were compared to trees created by the cgMLST scheme using the Kendall-Colijn metric (26), a quantitative measure of the difference in topology between two phylogenetic trees (27).
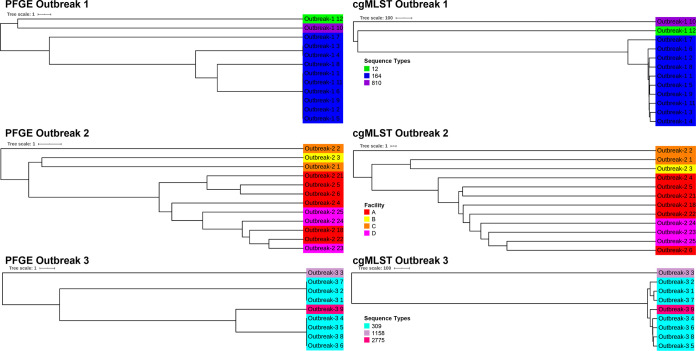
The topologies of the PFGE and cgMLST trees were statistically similar for outbreaks 1, 2, and 3 (Table S3). The tree topologies also showed similar clustering by ST for outbreaks 1 and 3, and by facility in outbreak 2 (as all the isolates were from the same ST) (Fig. 3), validating the results obtained from the statistical comparison. The isolates with identical PFGE patterns varied by a median value of 26 (0.59%) and 29 (0.66%) alleles for outbreaks 1 and 3, respectively. There were no isolates with identical PFGE patterns from outbreak 2.
FIG 3.
PFGE and cgMLST trees of isolates from three P. aeruginosa HAI outbreaks. Isolates from outbreaks 1 and 3 are colored by MLST sequence types (ST), while those from outbreak 2 (which were all from the same ST) are colored by the facility they were recovered from.
Given that P. aeruginosa genomes from the same ST vary by only a few hundred alleles on average, the cgMLST scheme was tested to see if it could differentially cluster outbreak isolates from genomes of the same ST from RefSeq. The median allele difference between the unrelated RefSeq and the outbreak isolates was more than four times greater than within the outbreak isolates themselves for all outbreaks (Table 1), and trees generated using the cgMLST scheme show distinct clusters separating the outbreak from RefSeq genomes (Fig. S1). There were no instances where an outbreak isolate was more closely related to a RefSeq genome of the same ST than to another outbreak isolate.
TABLE 1.
Differentiation of outbreak isolates versus unrelated RefSeq isolates of same sequence type
| Outbreak | MLST ST | No. of outbreak isolates | No. of RefSeq isolates | Median allele difference |
||
|---|---|---|---|---|---|---|
| Within outbreak | Within RefSeq | Outbreak versus RefSeq | ||||
| 1 | 164 | 10 | 2 | 51 | 118 | 916 |
| 2 | 233 | 25 | 10 | 51 | 134 | 323.5 |
| 3 | 309 | 8 | 6 | 69.5 | 103 | 299.5 |
| 4 | 308 | 26 | 23 | 64 | 196 | 376 |
SNV-based WGS comparison techniques are commonly used in HAI outbreak investigations to create phylogenetic trees to determine isolate relatedness and possible sources of infection (39). While cgMLST can measure thousands of allele-level differences, SNV-based approaches consider millions of possible nucleotide-level variations in both coding and noncoding sequences, providing much greater resolution, though analysis can be complicated by factors such as recombination (19) and convergent evolution (40). Trees made from the cgMLST scheme were compared to those from two SNV tools: SNVPhyl, an hqSNV pipeline (23), and kSNP, a k-mer based SNV program (25). SNV applications rely on large aligned core genomes for comparison (41), so only closely related isolates from the four outbreaks were included in the analysis. For outbreaks 1, 2 and 4, isolates from the predominant ST were included. For outbreak 3, the cgMLST tree showed that the isolate from ST2775 was similar to the ST309 isolates (Fig. 3), so it was also included in the SNV analysis.
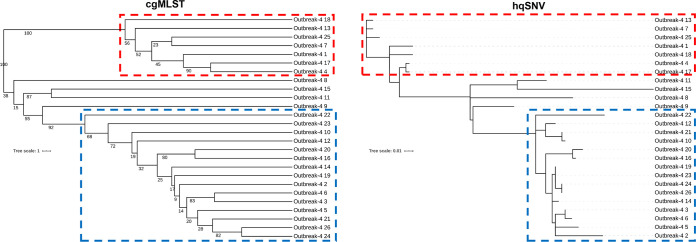
The dendrograms made from the cgMLST distances had quantitatively similar topologies to the trees from the k-mer based SNV pipeline for outbreaks 1, 3 and 4, and for outbreaks 3 and 4 for the hqSNV pipeline (Table 2). The trees from outbreak 4 showed similar clustering, as illustrated in the same two subgroups emerging in both the cgMLST and hqSNV trees (Fig. 4). The cgMLST allele differences and SNV counts correlated with each other as well, though there was much more variability for outbreaks 2 and 4, which both had more than twice as many isolates as the other two outbreaks (Fig. S2). Additionally, there were drastic differences in the slope of the correlations between SNVs and cgMLST allele differences for the k-mer and hqSNV pipelines, because areas of homologous recombination were filtered out by the hqSNV pipeline but included in the cgMLST scheme (Table S4) and the k-mer pipeline. While these regions affected only a fraction of the unique alleles in the outbreak isolates (0.76 to 3.8%), they are overrepresented in the k-mer pipeline SNV counts because of the higher density of SNVs in these alleles resulting from recombination.
TABLE 2.
Comparison of cgMLST to SNV Trees
| Outbreak | No. of isolates | MLST ST | Core genome size (hqSNV) |
P value of tree similarity scorea
|
|
|---|---|---|---|---|---|
| hqSNV | k-mer SNV | ||||
| 1 | 10 | 164 | 94.69% | 0.12 | 4.3 × 10-3,* |
| 2 | 25 | 233 | 94.86% | N/A (dissimilar) | N/A (dissimilar) |
| 3 | 8 | 309 (7), 2775 | 93.23% | 1.5 × 10-2,* | 1.1 × 10-2,* |
| 4 | 26 | 308 | 93.13% | 1.4 × 10-14,* | 1.2 × 10-3,* |
*, P value < 0.05; N/A, not applicable.
FIG 4.
Trees generated by cgMLST scheme and hqSNV pipeline (SNVPhyl) for outbreak 4 isolates. Both techniques revealed two clusters of the same genomes (outlined in red and blue) distinct from the larger set. Support values for 1,000 bootstraps are labeled on the cgMLST tree.
DISCUSSION
Because its pathogenicity is not limited to specific strains (42) and multiple STs are often found in the health care environment (9), investigations of outbreaks of P. aeruginosa HAIs require flexible classification tools that can rapidly assess relatedness among both diverse and genetically similar isolates. The P. aeruginosa cgMLST scheme presented herein provides a means for comparison of WGS data using a simple, static scheme that can easily accommodate diverse samples and, as well, rapidly add new isolates into the analyses.
As expected, the cgMLST scheme generated clusters consistent with traditional, seven-gene MLST typing, but the increased resolution from the thousands of genes included in the scheme revealed heterogeneity within STs when applied to thousands of publicly available P. aeruginosa genomes. The cgMLST scheme revealed 47 genomes in the public set with more similarity to members of different STs than their own. We also observed this same phenomenon in the isolates from outbreak 3, as a group of four ST309 isolates had more common alleles with an isolate from ST2775 than with the other ST309 isolates from the same facility. While ST2775 and ST309 are closely related (differing only by one of seven MLST alleles), an investigation of this outbreak using MLST as a filter of relatedness would exclude the ST2775 isolate.
PFGE, another tool traditionally used in outbreak investigations, incorporates dozens of fragments to provide more resolution than MLST, but still much less than cgMLST schemes, and simplifies phylogenetic relationships versus sequencing-based approaches. This was evident when comparing PFGE-derived dendrograms to those made using the cgMLST scheme for three P. aeruginosa HAI outbreaks. While both techniques generated similar clustering, the cgMLST trees revealed subtle relationships between the isolates, as expected from the increase in resolution.
While both approaches utilize WGS data, SNV-based techniques offer a more granular means of genomic comparison than cgMLST. But because they abstract thousands of gene sequences to simple allele names, cgMLST schemes are less computationally demanding than the higher resolution SNV-based techniques, and cgMLST analyses do not have to be rerun on the entire data set when new isolates are added because the core genome is static, making them ideal for longitudinal analysis of large collections (43–45). When applied to the outbreak isolates, the scheme produced topologically similar phylogenetic trees compared to an hqSNV pipeline for two of the four outbreaks. This variability is likely due to multiple factors in addition to the fundamental difference of comparing SNVs to cgMLST alleles, such as the larger aligned core genome size generated by SNV pipelines (>93% of the genome for the hqSNV pipeline, which includes noncoding and accessory genomic elements) compared to the cgMLST scheme (∼64%), which only considers the coding regions of species-wide core genes. The hqSNV pipeline also explicitly filtered regions of recombination, some of which were included in the cgMLST scheme. However, recombination events affected only a subset of core genes and, in general, would not have outsized influence on tree topology since the scheme considers any change to a single gene to be equivalent, whether it arises from one, ten, or hundreds of SNVs. The method of tree construction also differs between approaches (maximum likelihood for the hqSNV pipeline and UPGMA for the cgMLST scheme). These differences are largely a result of the cgMLST scheme’s simplicity, but this same feature allows for increased speed of execution, making it a useful first pass filter to identify outbreak clusters that may require higher resolution SNV analysis.
The cgMLST scheme for P. aeruginosa provides a standardized means of rapid typing and comparison for this diverse, ubiquitous pathogen. It can be applied to surveillance efforts to assess the population structure of the pathogen on larger scales and help uncover previously undetected clusters of related infections before outbreaks occur in a facility or region. In addition, because P. aeruginosa health care-associated outbreaks often involve unrelated and nonclonal samples, it can rapidly identify potential transmission events for further analysis. Given these applications, the P. aeruginosa cgMLST scheme is a useful addition to the public health toolkit for WGS analysis. The scheme has been adapted by Applied Maths NV (Sint-Martens-Latem, Belgium) for use on their BioNumerics software package.
Supplementary Material
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.2019. AR threats report. (Accessed Nov. 22, 2019). https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 2.Burdette SD, Herchline TE. 2006. Antimicrobe.org: an online reference for the practicing infectious diseases specialist. Clin Infect Dis 43:765–769. doi: 10.1086/507039. [DOI] [PubMed] [Google Scholar]
- 3.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Li X, Li W, Du X, He JQ, Tao C, Feng Y. 2015. Influence of carbapenem resistance on mortality of patients with Pseudomonas aeruginosa infection: a meta-analysis. Sci Rep 5:11715. doi: 10.1038/srep11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Abreu PM, Farias PG, Paiva GS, Almeida AM, Morais PV. 2014. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: a potential health hazard. BMC Microbiol 14:118. doi: 10.1186/1471-2180-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donlan RM. 2011. Biofilm elimination on intravascular catheters: important considerations for the infectious disease practitioner. Clin Infect Dis 52:1038–1045. doi: 10.1093/cid/cir077. [DOI] [PubMed] [Google Scholar]
- 8.Garvey MI, Bradley CW, Holden E. 2017. Waterborne Pseudomonas aeruginosa transmission in a hematology unit? Am J Infect Control 46:383–386. doi: 10.1016/j.ajic.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Clegg WJ, Pacilli M, Kemble SK, Kerins JL, Hassaballa A, Kallen AJ, Walters MS, Halpin AL, Stanton RA, Boyd S, Gable P, Daniels J, Lin MY, Hayden MK, Lolans K, Burdsall DP, Lavin MA, Black SR. 2018. Notes from the field: large cluster of Verona integron-encoded metallo-beta-lactamase-producing carbapenem-resistant Pseudomonas aeruginosa isolates colonizing residents at a skilled nursing facility—Chicago, Illinois, November 2016–March 2018. MMWR Morb Mortal Wkly Rep 67:1130–1131. doi: 10.15585/mmwr.mm6740a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2017. Carbapenemase-Producing Organisms: a Global Scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong JC, McCallum N, Sintchenko V, Howden BP. 2015. Whole genome sequencing in clinical and public health microbiology. Pathology 47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Royer G, Fourreau F, Boulanger B, Mercier-Darty M, Ducellier D, Cizeau F, Potron A, Podglajen I, Mongardon N, Decousser JW. 2020. Local outbreak of extended-spectrum beta-lactamase SHV2a-producing Pseudomonas aeruginosa reveals the emergence of a new specific sub-lineage of the international ST235 high-risk clone. J Hosp Infect 104:33–39. doi: 10.1016/j.jhin.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darch SE, McNally A, Harrison F, Corander J, Barr HL, Paszkiewicz K, Holden S, Fogarty A, Crusz SA, Diggle SP. 2015. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep 5:7649. doi: 10.1038/srep07649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Petkau A, Mabon P, Sieffert C, Knox NC, Cabral J, Iskander M, Iskander M, Weedmark K, Zaheer R, Katz LS, Nadon C, Reimer A, Taboada E, Beiko RG, Hsiao W, Brinkman F, Graham M, Van Domselaar G. 2017. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom 3:e000116. doi: 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 26.Jombart T, Kendall M, Almagro-Garcia J, Colijn C. 2017. treespace: statistical exploration of landscapes of phylogenetic trees. Mol Ecol Resour 17:1385–1392. doi: 10.1111/1755-0998.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA. 2017. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabrolier N, Sauget M, Bertrand X, Hocquet D. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identifies Pseudomonas aeruginosa high-risk clones. J Clin Microbiol 53:1395–1398. doi: 10.1128/JCM.00210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50 Pt 4:1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 32.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Torres P, Rodriguez-Mateos F, Anton J, Gabaldon T. 2019. Impact of homologous recombination on the evolution of prokaryotic core genomes. mBio 10:e02494-18. doi: 10.1128/mBio.02494-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedge J, Wilson DJ. 2014. Bacterial phylogenetic reconstruction from whole genomes is robust to recombination but demographic inference is not. mBio 5:e02158. doi: 10.1128/mBio.02158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramisetty BCM, Sudhakari PA. 2019. Bacterial ‘grounded’ prophages: hotspots for genetic renovation and innovation. Front Genet 10:65. doi: 10.3389/fgene.2019.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bicking Kinsey C, Koirala S, Solomon B, Rosenberg J, Robinson BF, Neri A, Laufer Halpin A, Arduino MJ, Moulton-Meissner H, Noble-Wang J, Chea N, Gould CV. 2017. Pseudomonas aeruginosa outbreak in a neonatal intensive care unit attributed to hospital tap water. Infect Control Hosp Epidemiol 38:801–808. doi: 10.1017/ice.2017.87. [DOI] [PubMed] [Google Scholar]
- 37.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol 45:3707–3712. doi: 10.1128/JCM.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasan NA, Epperson LE, Lawsin A, Rodger RR, Perkins KM, Halpin AL, Perry KA, Moulton-Meissner H, Diekema DJ, Crist MB, Perz JF, Salfinger M, Daley CL, Strong M. 2019. Genomic analysis of cardiac surgery-associated Mycobacterium chimaera infections, United States. Emerg Infect Dis 25:559–563. doi: 10.3201/eid2503.181282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen P, Papin JA. 2017. History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLoS Biol 15:e2001586. doi: 10.1371/journal.pbio.2001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. doi: 10.3389/fgene.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters MS, Grass JE, Bulens SN, Hancock EB, Phipps EC, Muleta D, Mounsey J, Kainer MA, Concannon C, Dumyati G, Bower C, Jacob J, Cassidy PM, Beldavs Z, Culbreath K, Phillips WE Jr, Hardy DJ, Vargas RL, Oethinger M, Ansari U, Stanton R, Albrecht V, Halpin AL, Karlsson M, Rasheed JK, Kallen A. 2019. Carbapenem-resistant Pseudomonas aeruginosa at US Emerging Infections Program sites, 2015. Emerg Infect Dis 25:1281–1288. doi: 10.3201/eid2507.181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Escalona N, Jolley KA, Reed E, Martinez-Urtaza J. 2017. Defining a core genome multilocus sequence typing scheme for the global epidemiology of Vibrio parahaemolyticus. J Clin Microbiol 55:1682–1697. doi: 10.1128/JCM.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. doi: 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou H, Liu W, Qin T, Liu C, Ren H. 2017. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae. Front Microbiol 8:371. doi: 10.3389/fmicb.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences have been deposited in NCBI BioProject ID PRJNA288601.