Carbapenem-nonsusceptible Citrobacter spp. (CNSC) are increasingly recognized as health care-associated pathogens. Information regarding their clinical epidemiology, genetic diversity, and mechanisms of carbapenem resistance is lacking. We examined microbiology records of adult patients at the University of Pittsburgh Medical Center (UMPC) Presbyterian Hospital (PUH) from 2000 to 2018 for CNSC, as defined by ertapenem nonsusceptibility. Over this time frame, the proportion of CNSC increased from 4% to 10% (P = 0.
KEYWORDS: carbapenem, carbapenemase, Citrobacter, multidrug resistance
ABSTRACT
Carbapenem-nonsusceptible Citrobacter spp. (CNSC) are increasingly recognized as health care-associated pathogens. Information regarding their clinical epidemiology, genetic diversity, and mechanisms of carbapenem resistance is lacking. We examined microbiology records of adult patients at the University of Pittsburgh Medical Center (UMPC) Presbyterian Hospital (PUH) from 2000 to 2018 for CNSC, as defined by ertapenem nonsusceptibility. Over this time frame, the proportion of CNSC increased from 4% to 10% (P = 0.03), as did daily defined carbapenem doses/1,000 patient days (6.52 to 34.5; R2 = 0.831; P < 0.001), which correlated with the observed increase in CNSC (lag = 0 years; R2 = 0.660). Twenty CNSC isolates from 19 patients at PUH and other UPMC hospitals were available for further analysis, including whole-genome short-read sequencing and additional antimicrobial susceptibility testing. Of the 19 patients, nearly all acquired CNSC in the health care setting and over half had polymicrobial cultures containing at least one other organism. Among the 20 CNSC isolates, Citrobacter freundii was the predominant species identified (60%). CNSC genomes were compared with genomes of carbapenem-susceptible Citrobacter spp. from UPMC and with other publicly available CNSC genomes. Isolates carrying genes encoding carbapenemases (blaKPC-2, blaKPC-3, and blaNDM-1) were also long-read sequenced, and their carbapenemase-encoding plasmid sequences were compared with one another and with publicly available sequences. Phylogenetic analysis of 102 UPMC Citrobacter genomes showed that CNSC from our setting did not cluster together. Similarly, a global phylogeny of 64 CNSC genomes showed a diverse population structure. Our findings suggest that both local and global CNSC populations are genetically diverse and that CNSC harbor carbapenemase-encoding plasmids found in other Enterobacterales.
INTRODUCTION
Carbapenem-resistant bacteria have become a major health concern worldwide (1). There are limited therapeutic options for treating infections caused by these multidrug-resistant organisms, resulting in greater morbidity and mortality than infections caused by susceptible organisms (2). Furthermore, multidrug-resistant infections place an additional economic burden on health care systems (3). In recognition of this threat, the treatment and control of carbapenem-resistant organisms have been prioritized by both the Centers for Disease Control and Prevention and the World Health Organization (4, 5).
The recent increase in infections caused by carbapenem-resistant organisms in the United States has been largely driven by the dissemination of plasmid-encoded carbapenemase genes, which are often carried by members of the Enterobacterales, particularly Klebsiella pneumoniae (6, 7). However, rates of carbapenem resistance in other bacterial species have also increased (8). Among them, carbapenem-nonsusceptible Citrobacter spp. (CNSC) have become increasingly recognized as health care-associated pathogens (9–13). CNSC isolates have been found to be both genotypically and phenotypically diverse (14, 15), and their resistance to carbapenems is frequently caused by plasmid-borne carbapenemase genes, which can be readily acquired through horizontal gene transfer (12, 16).
Information regarding the clinical epidemiology, genetic diversity, and mechanisms of carbapenem resistance among CNSC in the United States is currently limited to a small number of studies and very few isolates (9, 17–19). Here, we aimed to investigate the emergence of CNSC within our health care system using epidemiology and genomics approaches. We conducted a retrospective analysis of CNSC prevalence and carbapenem use over the last 2 decades and compared the genomes of CNSC isolates from our center with those of other local Citrobacter isolates, as well as with CNSC genomes sampled from around the globe. We found that while the CNSC sampled from our center are highly genetically diverse, their diversity is consistent with the local carbapenem-susceptible Citrobacter population, as well as with CNSC sampled elsewhere.
(Preliminary data included in this work were presented at the IDWeek 2019 conference [20].)
MATERIALS AND METHODS
Study design and isolate collection.
This study was conducted at the University of Pittsburgh Medical Center Presbyterian Hospital (UPMC-PUH), an adult medical and surgical tertiary-care hospital with 762 total beds, 150 critical care unit beds, more than 32,000 yearly inpatient admissions, and over 400 solid-organ transplants per year. CNSC isolates were collected from UPMC-PUH as well as other UPMC hospitals. Ethics approval for this study was obtained from the Institutional Review Board of the University of Pittsburgh.
To investigate CNSC epidemiology at UPMC-PUH, microbiology records of adult patients with a positive clinical culture for CNSC from 1 January 2000 to 31 December 2018 were evaluated. Cases were excluded from reinclusion within 90 days of any CNSC culture. Carbapenem nonsusceptibility was defined as nonsusceptibility to any carbapenem according to the 2017 Clinical and Laboratory Standards Institute (CLSI) interpretative criteria (21). Antibiotic consumption was measured by daily defined doses (DDDs) of any carbapenem (22). To further phenotype and genotype CNSC at UPMC, 20 available CNSC isolates from 19 patients collected between 2013 and 2019 were included. Isolates were considered community associated if the organism was isolated from a specimen collected within 72 h following hospital admission; isolates collected after 72 h were considered health care-associated (23). Clinical characteristics and outcomes of patients with CNSC isolates that underwent further characterization were collected through retrospective chart review. The primary clinical outcome was in-hospital mortality and/or transfer to hospice.
CNSC isolate characterization.
Initial species assignment was performed using standard clinical microbiology laboratory methods and was confirmed or modified after whole-genome sequencing. Carbapenem nonsusceptibility was initially determined by standard clinical microbiology laboratory methods and was confirmed by the Kirby-Bauer disk diffusion method as per the 2017 CLSI interpretative criteria (21). Susceptibility to additional agents was determined by the broth microdilution method (21). The presence of carbapenemase enzyme activity was assessed by modified carbapenem inactivation method (mCIM) (24).
Genome sequencing and analysis.
Genomic DNA was extracted from pure overnight cultures of single bacterial colonies using a Qiagen DNeasy blood and tissue kit according to the manufacturer’s instructions (Qiagen, Germantown, MD). Library construction and sequencing were conducted using the Illumina Nextera NGS library prep kit or the Illumina Nextera XT DNA library prep kit (Illumina, San Diego, CA). Libraries were sequenced on an Illumina NextSeq system with 150-bp paired-end reads or an Illumina MiSeq system with 300-bp paired-end reads. Isolates with suspected plasmid-encoded carbapenemases were sequenced with long-read technology on a MinION device (Oxford Nanopore Technologies, Oxford, United Kingdom). Long-read sequencing libraries were prepared and multiplexed using a rapid multiplex barcoding kit (Oxford Nanopore Technologies catalog no. SQK-RBK004) and sequenced on R9.4.1 flow cells. Base-calling on raw reads was performed using Guppy v2.3.1 (Oxford Nanopore Technologies, Oxford, United Kingdom), and hybrid assembly was performed with both short Illumina reads and long Oxford Nanopore reads using Unicycler v0.4.8beta (25).
Illumina reads were quality filtered and assembled de novo using SPAdes v3.11. Species were identified by Kraken and by performing pairwise comparisons of average nucleotide identity on the assembled genomes using fastANI using the many-to-many method (26). Assemblies were clustered using the hierarchy module of the python package SciPy by single linkage method and a distance criterion of 5% difference in average nucleotide identity. Multilocus sequence typing (MLST) was performed with the mlst tool at https://github.com/tseemann/mlst. Genomes were annotated using Prokka v1.13 (27). Core genes were defined using Roary v3.12.0 with a 90% sequence identity cutoff (28). A phylogenetic tree based on a core gene alignment containing 1,606 genes identified by Roary was generated using RAxML v8.2.11 (29) by running 1,000 bootstrap replicates under the generalized time-reversible model of evolution, a categorical model of rate heterogeneity (GTR-CAT), with Lewis correction for ascertainment bias. The tree was visualized and annotated using Interactive Tree of Life (iTOL) v4 (30). The genomes of closely related isolates were compared with one another with breseq (31). Antimicrobial resistance gene and plasmid content were assessed by BLASTn of assembled contigs against downloaded ResFinder and PlasmidFinder databases, with 80% sequence identity and an 80% sequence coverage cutoff (32). Virulence gene content was assessed using the VirulenceFinder web interface with default settings and the Escherichia coli database (33). Carbapenemase-encoding contigs resolved from hybrid assembly of CNSC genomes were annotated using Prokka v1.13 (27), and resistance genes were identified using the ResFinder web interface with default settings (32). Carbapenemase-encoding contigs were compared to one another and to plasmid sequences downloaded from the RefSeq database (n = 18,364) using BLASTn of assembled contigs (34, 35). RefSeq and CNSC contigs whose sequences yielded at least 90% coverage of one another in either search direction were aligned to one another using EasyFig (36). Sequences were mapped to the ompC and ompF porin gene sequences from Citrobacter freundii ATCC 8090 (GenBank accession no. CP049015.1) using Geneious v11.1.5 to assess putative loss-of-function mutations, such as those resulting in premature stop codons, frameshift mutations, or large deletions. Available CNSC genomes were downloaded from the GenBank or Sequence Read Archive repositories maintained by the National Center for Biotechnology Information (NCBI).
Statistics.
The proportion of CNSC was measured by dividing the number of CNSC isolates by the total number of Citrobacter species isolates tested for carbapenem susceptibility each year. Carbapenem (ertapenem, doripenem, meropenem, and imipenem) DDDs were measured per year at UPMC-PUH (22). Changes in the rate of carbapenem-nonsusceptible pathogen isolation over time were measured by linear regression, and comparison with the rate of antibiotic DDDs per year was conducted with time series cross-correlation analysis. Categorical data were compared using the χ2 test. Statistical analyses were performed using Stata V15 (StataCorp, College Station, TX) and R V3.5.1 (37).
Data availability.
Genome sequence data generated in this study have been deposited in SRA or GenBank with accession numbers listed in Tables S1, S2, and S3 in the supplemental material. Accession numbers for genomes newly sequenced for this study are SAMN14007636 to SAMN14007655, SRR11038037 to SRR11038052, and SAMN14082844 to SAMN14082856.
RESULTS
Clinical epidemiology of CNSC.
During the study period from 2000 through 2018, 78 unique patients with CNSC were identified from 2,817 Citrobacter isolates tested. Citrobacter spp. were the seventh most common carbapenem-nonsusceptible Gram-negative bacteria and fifth most common carbapenem-nonsusceptible Enterobacterales at our center during this time period. The proportion of Citrobacter species isolates that were CNSC increased significantly over time (R2 = 0.257; P = 0.03), from 4% in 2000 to 10% in 2018 (Fig. 1). Daily defined doses (DDDs) of carbapenems per 1,000 patient days also increased during the same time period, from 6.52 in 2000 to 34.5 in 2018 (R2 = 0.831; P < 0.001). We found that the increase in DDDs correlated with the increase in CNSC over the same period (lag = 0 years; R2 = 0.660) (Fig. 1).
FIG 1.
Carbapenem consumption and proportion of carbapenem-nonsusceptible Citrobacter spp. (CNSC), 2000 to 2018. Carbapenem daily defined doses (DDDs) per 1,000 patient days (yellow bars) and the proportion of Citrobacter species isolates that were carbapenem nonsusceptible (solid blue line) were quantified for each year between 2000 and 2018 at UPMC-PUH. Testing of 2,817 total Citrobacter species isolates revealed that 78 unique patients had CNSC (defined as ertapenem nonsusceptible). The dotted blue line shows a linear regression for increased CNSC proportion over time (R2 = 0.257; P = 0.03). Carbapenem DDDs per 1,000 patient days also increased over time (R2 = 0.831; P < 0.001) and correlated with the increase in CNSC (lag = 0 years; R2 = 0.660).
Isolation and characterization of CNSC.
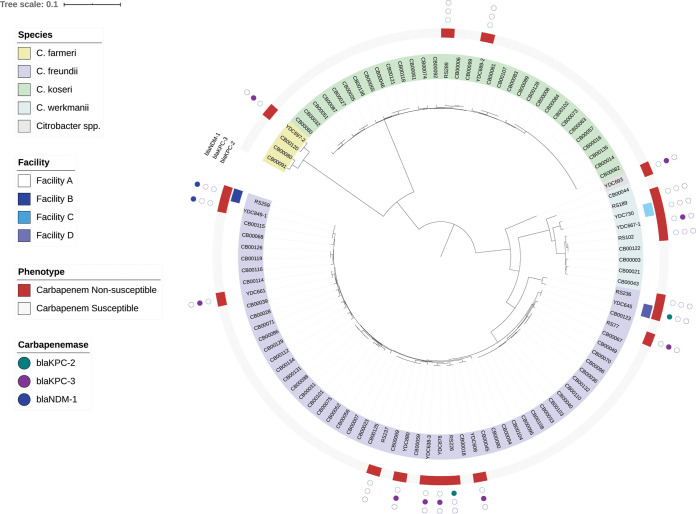
Twenty CNSC isolates from 19 patients from UPMC-PUH and three additional UPMC hospitals were available for further analysis (Table 1). Among these patients, the median age was 65 (range, 26 to 92), and 37% were female (7/19). The majority of patients had multiple comorbidities, frequently acquired CNSC in the health care setting (84% [16/19]), had polymicrobial cultures (57% [11/19]), and had high rates of in-hospital mortality and discharge to hospice (47% [9/19]) (Table 1). We sequenced the genomes of all 20 CNSC isolates on the Illumina platform (Table S1) and constructed a phylogenetic tree that also included an additional 82 carbapenem-susceptible Citrobacter isolates collected by the Enhanced Detection System for Hospital-Associated Transmission (EDS-HAT) project (Fig. 2; Table S4) (38, 39). Among the 20 CNSC isolates, C. freundii was the predominant species (60% [12/20]), followed by Citrobacter werkmanii (20% [4/20]), C. koseri (10% [2/20]), and C. farmeri (5% [1/20]). One CNSC isolate, YDC693, was originally identified as C. freundii but showed only 90 to 92% average nucleotide identity to other C. freundii genomes (Table S4). This isolate appears to belong to a new, unnamed Citrobacter species. YDC693 (Citrobacter sp.) and YDC697-2 (C. farmeri) were cultured from the same patient and were from samples taken approximately 2 weeks apart. Their distribution throughout the genome phylogeny suggested that the CNSC isolates were largely genetically distinct from one another (Fig. 2; Table S4). The one exception was RS259 and YDC849-1, which were found to have fewer than 20 genetic variants (single nucleotide polymorphisms and insertion/deletion variants) that distinguished them from one another, despite being isolated from patients at two different facilities (facility A versus facility B).
TABLE 1.
Clinical characteristics of patients with CNSCa
| Isolate ID | Patient age | Gender | Yr | Source | Facility | CNSC | Culture site | Clinical syndrome | Polymicrobial culture | Other organism(s) | Comorbid condition(s) | Outcome of hospitalization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS77 | 49 | M | 2018 | Hospital | A | C. freundii | Rectal swab | Colonization | N | None | ESLD, liver transplant | Discharge to home |
| RS102 | 74 | F | 2018 | Hospital | A | C. werkmanii | Rectal swab | Colonization | N | None | ESLD, liver transplant | In-hospital death |
| RS189 | 66 | F | 2017 | Hospital | A | C. werkmanii | BAL fluid | Pneumonia | N | None | Diverticulosis | Discharge to facility |
| RS226 | 56 | F | 2018 | Hospital | A | C. freundii | Urine | Colonization | Y | E. coli | Opiate dependency, CHF, HTN | In-hospital death |
| RS236 | 70 | M | 2018 | Hospital | A | C. freundii | Peritoneal fluid | Intra-abdominal | Y | C. freundii (ESBL), Enterococcus faecium (VRE), Candida tropicalis, Candida parapsilosis | Pancreatic cancer, COPD, DM, HTN | Transfer to hospice |
| RS237 | 80 | F | 2018 | Community | A | C. freundii | Urine | Colonization | N | None | Sinus OM | Discharge to home |
| RS259 | 61 | F | 2018 | Hospital | B | C. freundii | Peritoneal fluid | Intra-abdominal | Y | E. coli, Pseudomonas aeruginosa | Metastatic lung cancer, COPD, DM | In-hospital death |
| RS289 | 92 | M | 2018 | Community | A | C. koseri | Urine | UTI | Y | Enterococcus faecalis | Bladder cancer, dementia, CKD, COPD, CHF | Discharge to home |
| YDC608 | 57 | M | 2013 | Hospital | A | C. freundii | BAL fluid | Colonization | N | None | HLD | Transfer to hospice |
| YDC638-2 | 27 | M | 2013 | Hospital | A | C. freundii | Biliary drainage | Intra-abdominal | Y | E. faecium (VRE) | ESLD, Crohn’s diseases, PSC, liver transplant | Discharge to home |
| YDC645 | 67 | F | 2013 | Hospital | C | C. freundii | Blood | SSTI/Endocarditis | Y | Bacteroides sp., Enterococcus raffinosus | CAD, CHF, DM, ESRD, dementia | Transfer to hospice |
| YDC661 | 64 | M | 2014 | Hospital | A | C. freundii | BAL fluid | Pneumonia | Y | Stenotrophomonas maltophilia | Heart transplant | Discharge to facility |
| YDC667-1 | 73 | M | 2014 | Hospital | A | C. werkmanii | BAL fluid | Pneumonia | Y | K. pneumoniae (ESBL) | CHF, DM, CAD, CKD | In-hospital death |
| YDC689-2 | 61 | M | 2015 | Hospital | A | C. koseri | BAL fluid | Pneumonia | N | None | ESLD, liver transplant | Discharge to facility |
| YDC693b | 65 | M | 2015 | Hospital | A | Citrobacter spp. | BAL fluid | Pneumonia | Y | E. coli (ESBL) | SBT, adrenal insufficiency | In-hospital death |
| YDC697-2b | 65 | M | 2015 | Hospital | A | C. farmeri | Tracheostomy site drainage | SSTI | Y | Klebsiella oxytoca (KPC) | SBT, adrenal insufficiency | In-hospital death |
| YDC730 | 71 | M | 2015 | Hospital | D | C. werkmanii | Pelvic abscess | Intra-abdominal | Y | E. faecium (VRE) | Multiple myeloma | Discharge home |
| YDC849-1 | 26 | F | 2018 | Hospital | A | C. freundii | Urine | UTI | Y | C. freundii (NDM) | CVID | In-hospital death |
| YDC876 | 53 | M | 2019 | Hospital | A | C. freundii | BAL fluid | Pneumonia | N | None | CAD, CHF | Discharge home |
| YDC880 | 73 | M | 2019 | Hospital | A | C. freundii | Rectal swab | Colonization | N | None | Lung transplant | In-hospital death |
Abbreviations: BAL, bronchoalveolar lavage; CAD, coronary artery disease; CHF, congestive heart failure; CKD, chronic kidney disease; CNSC, carbapenem nonsusceptible Citrobacter spp.; COPD, chronic pulmonary disease; CVID, common variable immunodeficiency; DM, diabetes mellitus; ESBL, extended-spectrum beta-lactamase; ESLD, end-stage liver disease; ESRD, end-stage renal disease; F, female; HAP, hospital-acquired pneumonia; HCC, hepatocellular carcinoma; HLD, hyperlipidemia; HTN, hypertension; KPC, Klebsiella pneumoniae carbapenemase; M, male; NDM, New Delhi metallo-beta-lactamase; OM, osteomyelitis; PSC, primary sclerosing cholangitis; SBT, small bowel transplant; SSTI, skin and soft tissue infection; UTI, urinary tract infection; VRE, vancomycin-resistant Enterococcus.
Same-patient isolates.
FIG 2.
Local phylogeny of carbapenem-susceptible and -nonsusceptible Citrobacter spp. from UPMC. A phylogenetic tree of 102 local Citrobacter sp. genomes (82 carbapenem-susceptible and 20 carbapenem-nonsusceptible isolates) was generated based on an alignment of 1,606 core genes using RAxML (28). The tree was visualized and annotated using Interactive Tree of Life (iTOL) (29). The tree is annotated based on species, facility source, carbapenem susceptibility phenotype, and carbapenemase genes identified, if any, in the genome of each isolate.
Antimicrobial susceptibility and identification of antibiotic resistance and virulence genes.
While the CNSC isolates we collected were originally defined as nonsusceptible to ertapenem, they displayed variable patterns of susceptibility to other carbapenem antibiotics. Only about half of the isolates (55% [11/20]) were nonsusceptible to meropenem, with three isolates showing intermediate resistance and eight being resistant (Table 2). We tested all isolates for the presence of a carbapenemase using a modified carbapenem inactivation method (mCIM) (24), in which 13 isolates (65%) tested positive. Carbapenemase genes were present in the genomes of all 13 isolates (Table 2). Among the carbapenemases identified, blaKPC-3 was predominant (9/13), followed by blaNDM-1 (2/13) and blaKPC-2 (2/13). Analysis of the major porin-encoding genes revealed that ompC was intact in all CNSC isolates, while ompF was disrupted in at least six CNSC genomes (Table 2; Table S8). ompF disruption was not associated with increased meropenem MICs; however, we did not find evidence of ompC or ompF disruptions in any of the carbapenem-susceptible isolates. Next, we tested all 20 CNSC isolates against novel β-lactam–β-lactamase inhibitor agents and found that they were frequently susceptible to ceftazidime-avibactam (17/20 [85%]) and meropenem-vaborbactam (18/20 [90%]). As expected, both isolates with blaNDM-1 exhibited phenotypic resistance to both agents (Table 2). Isolate RS237 was also found to be resistant to ceftazidime-avibactam, even though its genome did not contain a carbapenemase sequence or evidence of porin mutations. We compared RS237 with the most closely related carbapenem-susceptible isolate, CB00023, and found a large number of mutations separating them from one another (Table S5). One of these was a missense mutation (S219I) in acrE, which is predicted to encode a multidrug export protein and could be a candidate resistance-associated gene.
TABLE 2.
Carbapenemase genes, porin genotypes, antimicrobial susceptibilities, and mCIM results for 20 CNSC isolatesa
| Isolate | Organism | Carbapenemase gene | Porin genotypeb
|
MIC (μg/ml) ofc
: |
mCIM result | |||
|---|---|---|---|---|---|---|---|---|
| ompC | ompF | Meropenem | Ceftazidime-avibactam | Meropenem-vaborbactam | ||||
| RS77 | C. freundii | blaKPC-3 | Intact | Intact | 1 | 0.5 | 0.015 | Positive |
| RS102 | C. werkmanii | Intact | Intact | 0.25 | 0.5 | 0.06 | Negative | |
| RS189 | C. werkmanii | Intact | Intact | ≤0.06 | 0.5 | 0.015 | Negative | |
| RS226 | C. freundii | blaKPC-2 | Intact | Disrupted | 2 (I) | <0.25 | 0.015 | Positive |
| RS236 | C. freundii | Intact | Intact | 2 (I) | 2 | 0.5 | Negative | |
| RS237 | C. freundii | Intact | Intact | 4 (R) | 64 (R) | 0.5 | Negative | |
| RS259 | C. freundii | blaNDM-1 | Intact | Intact | 16 (R) | >256 (R) | >8 (R) | Positive |
| RS289 | C. koseri | Intact | Unknown | 0.12 | 2 | 0.12 | Negative | |
| YDC608 | C. freundii | blaKPC-3 | Intact | Intact | 16 (R) | 4 | 0.06 | Positive |
| YDC638-3 | C. freundii | blaKPC-3 | Intact | Disrupted | 2 (I) | 2 | 0.06 | Positive |
| YDC645 | C. freundii | blaKPC-2 | Intact | Intact | ≤0.06 | <0.25 | 0.03 | Positive |
| YDC661 | C. freundii | blaKPC-3 | Intact | Disrupted | 1 | 4 | 0.06 | Positive |
| YDC667-1 | C. werkmanii | blaKPC-3 | Intact | Disrupted | 1 | 0.5 | 0.03 | Positive |
| YDC689-2 | C. koseri | Intact | Intact | 0.5 | 4 | 0.12 | Negative | |
| YDC693d | Citrobacter sp. | blaKPC-3 | Intact | Unknown | 16 (R) | 1 | 0.03 | Positive |
| YDC697-2d | C. farmeri | blaKPC-3 | Intact | Intact | 32 (R) | 4 | 0.12 | Positive |
| YDC730 | C. werkmanii | Intact | Intact | 0.12 | 0.5 | 0.12 | Negative | |
| YDC849-1 | C. freundii | blaNDM-1 | Intact | Intact | 16 (R) | >256 (R) | 16 (R) | Positive |
| YDC876 | C. freundii | blaKPC-3 | Intact | Disrupted | 8 (R) | 1 | 0.06 | Positive |
| YDC880 | C. freundii | blaKPC-3 | Intact | Disrupted | 4 (R) | 1 | 0.03 | Positive |
All isolates were determined to be carbapenem nonsusceptible based on ertapenem nonsusceptibility.
OmpC and OmpF porin genotypes were evaluated by comparison to C. freundii ATCC 8090. “Intact” indicates a protein sequence of expected length; “disrupted” indicates a premature stop codon, frameshift, or large deletion; “unknown” indicates that we were unable to assess the genotype due to a contig break (RS289) or highly divergent sequence (YDC693).
Intermediate (I) and resistance (R) designations are based on CLSI breakpoints.
Same-patient isolates.
In addition to carbapenemase genes, we also compared the non-β-lactam acquired antibiotic resistance gene content between CNSC and carbapenem-susceptible EDS-HAT isolates (Table S6). CNSC isolate genomes often carried genes encoding resistance to aminoglycoside, fluoroquinolone, and tetracycline antibiotic classes. Excluding β-lactam resistance genes, the average number of resistance genes was significantly higher among CNSC isolates than carbapenem-susceptible EDS-HAT isolates (mean [standard deviation], 6 [4] versus 2 [3]; P < 0.001) (Table S6). Finally, we identified three CNSC isolates with virulence genes previously identified in E. coli (33). RS289 and YDC689-2 both carried a senB gene (GenBank accession no. AAZ89288), which encodes an enterotoxin (40). Additionally, YDC667-1 carried an astA gene (GenBank accession no. AF411067), which encodes an EAST-1 heat-stable toxin (41). These genes were also found among carbapenem-susceptible isolates (Table S7).
Global phylogeny of CNSC.
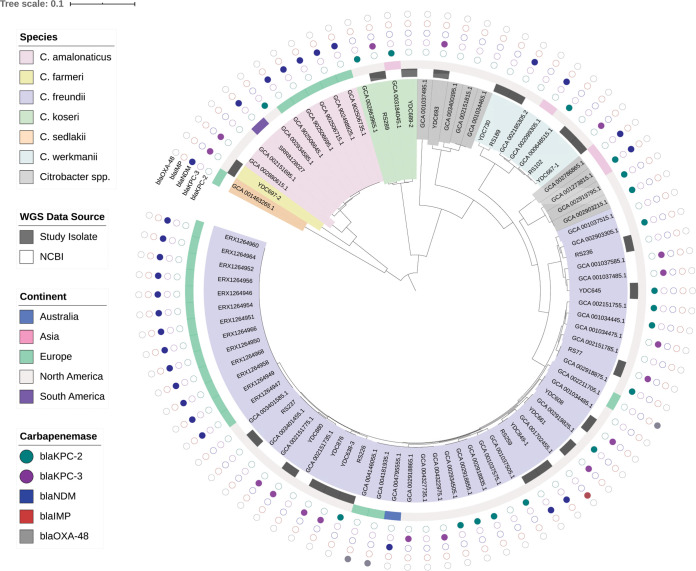
To understand how the genomic diversity of the study isolates compared to that of CNSC isolates from other locations, we searched the NCBI databases for additional publicly available CNSC genomes. Using search terms “Citrobacter” and “carbapenem,” we identified 64 additional CNSC genomes (Table S3). A global phylogeny of these 64 CNSC genomes combined with the 20 from this study showed abundant genetic heterogeneity (Fig. 3). We investigated the species distribution of the global CNSC population using fastANI and found that, similar to our UPMC isolates, the global CNSC population was dominated by C. freundii (41/64 [64%]), followed by Citrobacter amalonaticus (9/64 [14%]), C. werkmanii (3/64, 5%), and C. koseri (2/64, 3%). Carbapenem-nonsusceptible C. amalonaticus was not found among our UPMC isolates, but it has been isolated in the United States, South America, and Europe (Fig. 3; Table S3). Citrobacter sp. YDC693 was found to cluster with an additional three isolates from the United States (Table S3). Three other global CNSC (two from the United States and one from China) appeared to belong to another distinct Citrobacter species with 90 to 93% average nucleotide identity to C. freundii. The proportion of global CNSC isolate genomes that encoded carbapenemases (49/64 [77%]) was similar to that of our UPMC isolate set (Fig. 3); however, the diversity of enzyme types was greater and included blaNDM-5, blaIMP-38, and blaOXA-48-like enzymes.
FIG 3.
Global phylogeny of available CNSC genomes. A phylogenetic tree of 84 CNSC genomes (20 from this study and 64 from the NCBI) was generated based on an alignment of 1,842 core genes using RAxML (28). The tree was visualized and annotated using Interactive Tree of Life (iTOL) (29). The tree is annotated based on species, whole-genome sequencing data source, continent of isolation, and carbapenemase genes identified, if any, in the genome of each isolate.
Diversity of carbapenemase-encoding plasmids.
To better understand the genetic context of the carbapenemase enzymes encoded by our UPMC CNSC isolates, we conducted Oxford Nanopore long-read sequencing and hybrid assembly of the 13 carbapenemase-carrying isolate genomes (Table 3). A total of 11 complete, circular contigs were resolved from 10 isolates and nine patients, and all but one of these contigs contained replicons belonging to the IncA/C2, IncL/M, IncN, and unnamed RepA families. The genome assembly of RS259 contained a 21.4-kb circular contig encoding the blaNDM-1 carbapenemase on a class 1 integron, but it lacked readily identifiable plasmid replication machinery. This circular contig was highly similar to the blaNDM-1-encoding region of a 161-kb IncA/C2 plasmid resolved from the YDC849-1 genome. The RS259 genome appeared to also contain the remaining regions of the 161-kb plasmid from YDC849-1; however, the coverage was split across multiple contigs, suggesting excision of the class 1 integron into an unstable intermediate structure and/or issues with mobile element sequence assembly. Additionally, the YDC876 genome contained two plasmids of different sizes with distinct replicons that both harbored blaKPC-3 carbapenemase genes. Direct comparison of these two plasmids confirmed that they were distinct, even though they encoded the same carbapenemase and carried other acquired antimicrobial resistance genes (Table 3). Finally, three carbapenemase-encoding contigs were not completely resolved by hybrid assembly; in all cases, the contigs were short (less than 20 kb), and additional experiments would be needed to completely resolve their structures.
TABLE 3.
Carbapenemase-encoding contigs identified in CNSC genomes
| Isolate_contig | Length (bp) | Circulara | Replicon(s) | Carbapenemase gene | Carbapenemase-carrying element | Additional acquired antimicrobial resistance gene(s)b |
|---|---|---|---|---|---|---|
| RS77_21 | 10,011 | No | None | blaKPC-3 | Tn4401b-like | None |
| RS226_4 | 43,621 | Yes | RepA | blaKPC-2 | Tn4401-like | blaTEM-1B |
| RS259_9 | 21,420 | Yes | None | blaNDM-1 | Class 1 integron | aac(3)-IId, aac(6′)-Ib-cr, aadA16, arr-3, catB3, dfrA27, mph(A), sul1 |
| YDC608_5 | 172,511 | Yes | IncA/C2 | blaKPC-3 | Tn4401b-like | aac(6′)-Ib, aadA1, ant(2′')-Ia, blaOXA-9, blaSHV-7, blaTEM-1A |
| YDC638-3_3 | 213,257 | Yes | IncA/C2 | blaKPC-3 | Tn4401b-like | aac(6′)-Ib, aac(6′)-Ib-cr, aadA1, ant(2”)-Ia, blaOXA-9, blaSHV-7, blaTEM-1A, qnrA1, sul1 |
| YDC645_3 | 44,364 | Yes | RepA | blaKPC-2 | Tn4401-like | None |
| YDC661_9 | 15,031 | No | None | blaKPC-3 | Tn4401b-like | None |
| YDC667-1_41 | 4,894 | No | None | blaKPC-3 | Incomplete Tn4401 | None |
| YDC693_4c | 258,721 | Yes | IncA/C2, IncN | blaKPC-3 | Tn4401-like | aac(6′)-Ib, aadA1, blaOXA-9, blaSHV-7, blaTEM-1A, dfrA14 |
| YDC697-2_6c | 62,530 | Yes | IncN | blaKPC-3 | Tn4401-like | dfrA14 |
| YDC849-1_2 | 160,983 | Yes | IncA/C2 | blaNDM-1 | Class 1 integron | aac(3)-Iid, aac(6′)-Ib-cr, aadA16, aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, catB3, dfrA27, floR, mph(A), qnrS1, sul1, sul2, tet(A) |
| YDC876_2d | 176,497 | Yes | IncA/C2 | blaKPC-3 | Tn4401b-like | aac(6′)-Ib, aac(6′)-Ib-cr, aadA1, ant(2′')-Ia, blaOXA-9, blaSHV-7, blaTEM-1A, qnrA1, sul1 |
| YDC876_4d | 78,220 | Yes | IncL/M | blaKPC-3 | Tn4401b-like | aac(6′)-Ib, aac(6′)-Ib-cr, aadA1, blaOXA-9, blaTEM-1A |
| YDC880_4 | 88,095 | Yes | IncL/M | blaKPC-3 | Tn4401-like | aadA2, blaSHV-30, dfrA12, sul1 |
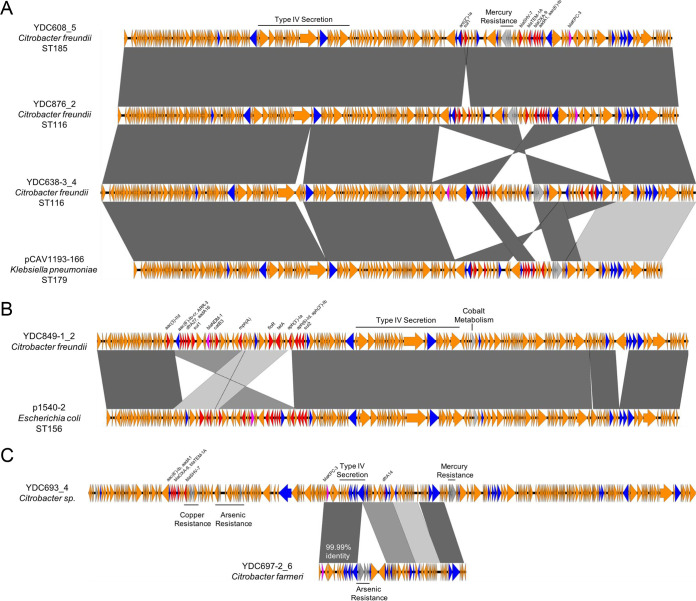
To determine how the carbapenemase-encoding plasmids in this study compared to one another and whether they were unique to our study, we conducted pairwise comparisons of the resolved plasmids. In addition, we searched the RefSeq database (35) for plasmids that showed substantial homology and high sequence identity to one or more of the CNSC plasmids from our UPMC isolates. One of the plasmids we resolved (RS226_4) matched a blaKPC-2-carrying plasmid from a publicly available C. freundii isolate genome (GenBank accession CP037739.1) with 100% coverage and 100% nucleotide identity, despite the isolates themselves being genetically distinct from one another. Three other plasmids that we identified (YDC608_5, YDC876_2, and YDC638-3) were highly similar to one another and were found in C. freundii isolates belonging to two different sequence types (ST185 and ST116) (Fig. 4A). While YDC608 and YDC876 belonged to two different C. freundii sequence types, their plasmids were more similar to one another than the plasmid from YDC638-3, which belonged to the same sequence type as YDC876. These plasmids were also highly similar to the pCAV1193-166 plasmid found in a blaKPC-carrying K. pneumoniae isolate from Virginia (Fig. 4A) (42). Separately, the YDC849-1_2 plasmid carrying blaNDM-1 had high similarity with plasmid p1540-2, which was found in a carbapenem-resistant E. coli isolate from Hong Kong (GenBank accession no. CP019053.1) (Fig. 4B). In addition, the blaKPC-3-carrying plasmids YDC693_4 and YDC697-2_6 were from isolates of different species that came from the same patient. Despite being different sizes (259 kb versus 63 kb), the plasmids showed some similarity to one another, and in particular, the Tn4401-like elements carrying blaKPC-3 on each plasmid contained only 1 mutation in more than 17 kb of sequence (Fig. 4C). These data suggest possible transfer of a blaKPC-3-encoding mobile element between the isolates from this patient; however, independent acquisition or independent transfer from another species cannot be ruled out.
FIG 4.
Carbapenemase-encoding plasmid diversity among and between CNSC genomes. CNSC genome contigs were compared to each other and to sequences deposited in the National Center for Biotechnology Information (NCBI). Sequences were aligned to one another with EasyFig. Sequence names are in the format “isolate_contig” based on hybrid assembly or correspond to the sequence name from NCBI. Bacterial species and sequence type are listed, where available. Open reading frames are colored by function (blue, mobilization; pink, carbapenemase; red, other antibiotic resistance; gray, metal interacting; orange, other/hypothetical). Antibiotic resistance genes, metal-interacting operons, and type IV secretion system components are labeled. Gray blocks between sequences indicate regions of >5 kb with >98% nucleotide identity, with darker shading indicating higher identity. Nucleotide identity between the blaKPC-3-encoding Tn4401-like regions of YDC693_4 and YDC697-2_6 (from two isolates of different Citrobacter species from the same patient) is noted with white text in panel C.
DISCUSSION
In this study, we conducted a retrospective review of the clinical and genomic epidemiology of CNSC over the past 2 decades at a large health care center in the United States. We analyzed the genomes of 20 CNSC and 82 carbapenem-susceptible Citrobacter spp. sampled locally, as well as 64 publicly available genomes sampled from around the globe. We found that the rates of CSNC had increased significantly over the last 2 decades at our center, that CSNC were frequently acquired in the health care setting along with other health care-associated organisms, and that patients from whom CNSC were isolated often had poor clinical outcomes. Our phylogenetic analyses revealed genetically diverse CNSC populations both locally and globally, suggesting that CNSC most often arise independently from one another. We also found that carbapenem nonsusceptibility was often mediated by acquisition of carbapenemase genes, with blaKPC-3 being the predominant carbapenemase identified among CNSC isolates in our setting.
Citrobacter spp. have become increasing recognized as a cause of multidrug-resistant health care-associated infections around the world (10, 43–45), and prior reports identified CNSC predominantly from health care sources and often in association with nosocomial outbreaks (9, 12, 46). We detected a significant increase in the proportion of Citrobacter isolates that were carbapenem nonsusceptible over the last 2 decades, which correlated with increased use of carbapenems at our center. While the incidence of carbapenem-resistant organisms has increased worldwide over recent years (7, 47), attention has been largely focused on other carbapenem-resistant members of the Enterobacterales, such as Enterobacter spp., E. coli, and Klebsiella spp. (7). As with other Gram-negative species, increasing antibiotic resistance among Citrobacter spp. is of significant concern, as our findings show that many CNSC appear to have acquired resistance genes from other bacteria by horizontal gene transfer.
Our analysis revealed extensive genomic diversity among both CNSC and carbapenem-susceptible Citrobacter isolates in samples from our center. This is similar to previous analyses of CNSC from non-U.S. centers that used more classical molecular typing methods (14, 48, 49). Among Citrobacter species, C. freundii is most commonly associated with both clinical disease (45) and multidrug-resistant phenotypes (14, 15, 50). Our findings were consistent with these prior reports; C. freundii was the most frequent CNSC species we observed, though we also found CNSC belonging to four additional species. The global CNSC population was similarly diverse, but C. freundii was again the most prevalent species observed, which may be due to its higher rate of antibiotic resistance compared to other Citrobacter spp. (44, 45).
The term “carbapenem nonsusceptibility” encompasses a wide range of phenotypic susceptibilities, which can be caused by different mechanisms. Among the CNSC isolates we collected, roughly two-thirds were found to produce carbapenemases, a rate that is similar to those in prior reports (14) and to that in the global CNSC population. CSNC have been found to encode a diverse array of carbapenemases. For example, a study by Arana et al. found five different carbapenemase types among Citrobacter species isolates collected from Spain (14), and similar results were also demonstrated in a study of carbapenemase-producing Enterobacterales in China (15). It has been suggested that carbapenemase diversity depends on local geography (47), and future studies of larger populations may confirm or refute this notion. The high diversity of carbapenemase-encoding plasmids we found, all from isolates of a single genus at a single hospital, highlights the complexity of antibiotic resistance gene transfer between pathogens in the hospital setting (51). Even with a relatively small number of isolates, we observed identical and closely related plasmids in genetically distinct bacteria, identical blaKPC-3-encoding mobile elements on different plasmids carried by the same bacterial isolate, and similar carbapenemase-encoding plasmids in CNSC of different species that were isolated from the same patient. These findings underscore the highly dynamic and variable transfer of carbapenemase-encoding mobile genetic elements into and among CNSC isolates.
There were several limitations to this study. While our study presents the largest genomic analysis of CNSC from the United States, the number of isolates we included was still rather limited. Moreover, the isolates we collected represent a convenience sample of available isolates, which may introduce bias. Furthermore, the correlation between carbapenem consumption and proportion of CNSC is a strictly ecologic analysis. Additionally, our genomic analysis of resistance determinants was limited to acquired carbapenemase genes, and we did not investigate other resistance mechanisms, such as chromosomal ampC genes and efflux pumps, or perform functional testing of outer membrane protein mutations, which are known to be associated with carbapenem resistance. Furthermore, many of the global isolate genomes we analyzed were from the United States and/or were part of outbreak investigations; thus, they may not be representative of the true global diversity of CNSC. Additionally, we made only in silico, sequence-based comparisons of the plasmids we resolved; as such, we cannot comment on their capacity for conjugative transfer. Finally, we were unable to determine whether the poor clinical outcomes among the patients from whom CNSC were isolated were indeed attributable to CNSC infection.
As CNSC become more prevalent in the health care system, further studies will be needed to increase our understanding of their genomic diversity and resistance mechanisms. In particular, examining the local and global epidemiology of horizontal transfer of drug resistance elements among Citrobacter spp. and between Citrobacter and other Enterobacterales species would provide valuable insights into risk factors and other trends that could be targeted to limit the occurrence and spread of CNSC.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Hayley Nordstrom, Daniel Snyder, and Vaughn Cooper for generating genome sequence data.
This study was funded by the National Institute of Allergy and Infectious Diseases (R21AI109459 and R01AI127472 to L.H.H.) and by the University of Pittsburgh Department of Medicine. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no commercial or other associations that might pose a conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lozano R, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadri SS, Lai YL, Ricotta EE, Strich JR, Babiker A, Rhee C, Klompas M, Dekker JP, Powers JH III, Danner RL, Adjemian J. 2019. External validation of difficult-to-treat resistance prevalence and mortality risk in Gram-negative bloodstream infection using electronic health record data from 140 US hospitals. Open Forum Infect Dis 6:ofz110. doi: 10.1093/ofid/ofz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch SM, McKinnell JA, Mueller LE, Miller LG, Gohil SK, Huang SS, Lee BY. 2017. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect 23:48.E9–48.E16. doi: 10.1016/j.cmi.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discover, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.CDC. 2019. Antibiotic resistance threats in the United States. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 6.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM, Vagnone PMS, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z, Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D, Rasheed JK, Limbago BM, Kallen AJ. 2015. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iovleva A, Doi Y. 2017. Carbapenem-resistant Enterobacteriaceae. Clin Lab Med 37:303–315. doi: 10.1016/j.cll.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson BM, El Chakhtoura NG, Patel S, Saade E, Donskey CJ, Bonomo RA, Perez F. 2017. Carbapenem-resistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006–2015. Emerg Infect Dis 23:878–880. doi: 10.3201/eid2305.162034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiménez A, Castro JG, Munoz-Price LS, de Pascale D, Shimose L, Mustapha MM, Spychala CN, Mettus RT, Cooper VS, Doi Y. 2017. Outbreak of Klebsiella pneumoniae carbapenemase-producing Citrobacter freundii at a tertiary acute care facility in Miami, Florida. Infect Control Hosp Epidemiol 38:320–326. doi: 10.1017/ice.2016.273. [DOI] [PubMed] [Google Scholar]
- 10.Gaibani P, Ambretti S, Farruggia P, Bua G, Berlingeri A, Tamburini MV, Cordovana M, Guerra L, Mazzetti M, Roncarati G, Tenace C, Moro ML, Gagliotti C, Landini MP, Sambri V. 2013. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian hospital, identified during the carbapenemases screening actions, June 2012. Int J Infectious Diseases 17:e714–e717. doi: 10.1016/j.ijid.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Cunha CB, Kassakian SZ, Chan R, Tenover FC, Ziakas P, Chapin KC, Mermel LA. 2016. Screening of nursing home residents for colonization with carbapenem-resistant Enterobacteriaceae admitted to acute care hospitals: incidence and risk factors. Am J Infect Control 44:126–130. doi: 10.1016/j.ajic.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Venditti C, Fortini D, Villa L, Vulcano A, D'Arezzo S, Capone A, Petrosillo N, Nisii C, Carattoli A, Di Caro A. 2017. Circulation of bla(KPC-3)-carrying IncX3 plasmids among Citrobacter freundii isolates in an Italian hospital. Antimicrob Agents Chemother 61:e00505-17. doi: 10.1128/AAC.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, Program NCS, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arana DM, Ortega A, González-Barberá E, Lara N, Bautista V, Gómez-Ruíz D, Sáez D, Fernández-Romero S, Aracil B, Pérez-Vázquez M, Campos J, Oteo J, Group SARSPC. 2017. Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J Antimicrob Chemother 72:3283–3287. doi: 10.1093/jac/dkx325. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, Zhang F, Cao B, Xie L, Liao K, Gu B, Yang C, Liu Z, Ma X, Jin L, Zhang X, Man S, Li W, Pei F, Xu X, Jin Y, Ji P, Wang H. 2018. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis 67:S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 16.Yoon E-J, Kang DY, Yang JW, Kim D, Lee H, Lee KJ, Jeong SH. 2018. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front Microbiol 9:571–571. doi: 10.3389/fmicb.2018.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerqueira GC, Earl AM, Ernst CM, Grad YH, Dekker JP, Feldgarden M, Chapman SB, Reis-Cunha JL, Shea TP, Young S, Zeng Q, Delaney ML, Kim D, Peterson EM, O'Brien TF, Ferraro MJ, Hooper DC, Huang SS, Kirby JE, Onderdonk AB, Birren BW, Hung DT, Cosimi LA, Wortman JR, Murphy CI, Hanage WP. 2017. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conlan S, Lau AF, Deming C, Spalding CD, Lee-Lin S, Thomas PJ, Park M, Dekker JP, Frank KM, Palmore TN, Segre JA. 2019. Plasmid dissemination and selection of a multidrug-resistant Klebsiella pneumoniae strain during transplant-associated antibiotic therapy. mBio 10:e00652-19. doi: 10.1128/mBio.00652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasheed JK, Biddle JW, Anderson KF, Washer L, Chenoweth C, Perrin J, Newton DW, Patel JB. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 darbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J Clin Microbiol 46:2066–2069. doi: 10.1128/JCM.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babiker A, Evans DR, Griffifth [sic] MP, Mettus RT, McElheny CL, Clarke L, Harrison L, Doi Y, Shields RK, Van Tyne D. 2019. Clinical and moleular epidemiology of carbapenem non-susceptible Citrobacter sp. Open Forum Infect Dis 6(Suppl 2):S237–S238. doi: 10.1093/ofid/ofz360.558. [DOI] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; M100-S27, 27th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.WHO Collaborating Centre for Drug Statistics Methodology. 2018. Definition and general considerations. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 23.CDC. 2019. CDC/NHSN surveillance definitions for specific types of infections. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 24.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB, Jenkins SG, Limbago BM, Das S. 2017. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 28.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcillan-Barcia MP, Redondo-Salvo S, Vielva L, de la Cruz F. 2020. MOBscan: automated annotation of MOB relaxases. Methods Mol Biol 2075:295–308. doi: 10.1007/978-1-4939-9877-7_21. [DOI] [PubMed] [Google Scholar]
- 35.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. 2019. R: a language and environment for statistical computing. https://www.r-project.org/.
- 38.Sundermann AJ, Babiker A, Marsh JW, Shutt KA, Mustapha MM, Pasculle AW, Ezeonwuka C, Saul MI, Pacey MP, Van Tyne D, Ayres AM, Cooper VS, Snyder GM, Harrison LH. 2020. Outbreak of vancomycin-resistant Enterococcus faecium in interventional radiology: detection through whole genome sequencing-based surveillance. Clin Infect Dis 70:2336–2343. doi: 10.1093/cid/ciz666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundermann AJ, Miller JK, Marsh JW, Saul MI, Shutt KA, Pacey M, Mustapha MM, Ayres A, Pasculle AW, Chen J, Snyder GM, Dubrawski AW, Harrison LH. 2019. Automated data mining of the electronic health record for investigation of healthcare-associated outbreaks. Infect Control Hosp Epidemiol 40:314–319. doi: 10.1017/ice.2018.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao B-H, Chang Y-F, Scaria J, Chang C-C, Chou L-W, Tien N, Wu J-J, Tseng C-C, Wang M-C, Chang C-C, Hsu Y-M, Teng C-H. 2012. Identification of Escherichia coli genes associated with urinary tract infections. J Clin Microbiol 50:449–456. doi: 10.1128/JCM.00640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez S, Llorente MT, Herrera-León L, Ramiro R, Nebreda S, Remacha MA, Herrera-León S. 2017. Mucus-activatable Shiga toxin genotype stx2d in Escherichia coli O157:H7. Emerg Infect Dis 23:1431–1433. doi: 10.3201/eid2308.170570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheppard AE, Stoesser N, Sebra R, Kasarskis A, Deikus G, Anson L, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Complete genome sequence of KPC-producing Klebsiella pneumoniae strain CAV1193. Genome Announc 4:e01649-15. doi: 10.1128/genomeA.01649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, Hu F, Liu Y, Zhu D, Wang H, Zhang Y. 2011. Detection and spread of carbapenem-resistant Citrobacter freundii in a teaching hospital in China. Am J Infect Control 39:e55–e60. doi: 10.1016/j.ajic.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Maraki S, Vardakas KZ, Mavromanolaki VE, Kyriakidou M, Spais G, Kofteridis DP, Samonis G, Falagas ME. 2017. In vitro susceptibility and resistance phenotypes in contemporary Citrobacter isolates in a university hospital in Crete, Greece. Infect Dis (Lond) 49:532–539. doi: 10.1080/23744235.2017.1297896. [DOI] [PubMed] [Google Scholar]
- 45.Samonis G, Karageorgopoulos DE, Kofteridis DP, Matthaiou DK, Sidiropoulou V, Maraki S, Falagas ME. 2009. Citrobacter infections in a general hospital: characteristics and outcomes. Eur J Clin Microbiol Infect Dis 28:61–68. doi: 10.1007/s10096-008-0598-z. [DOI] [PubMed] [Google Scholar]
- 46.Hammerum AM, Hansen F, Nielsen HL, Jakobsen L, Stegger M, Andersen PS, Jensen P, Nielsen TK, Hansen LH, Hasman H, Fuglsang-Damgaard D. 2016. Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J Antimicrob Chemother 71:3117–3124. doi: 10.1093/jac/dkw289. [DOI] [PubMed] [Google Scholar]
- 47.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villa J, Arana DM, Viedma E, Perez-Montarelo D, Chaves F. 2017. Characterization of mobile genetic elements carrying VIM-1 and KPC-2 carbapenemases in Citrobacter freundii isolates in Madrid. Int J Med Microbiol 307:340–345. doi: 10.1016/j.ijmm.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Liu L, Lan R, Liu L, Wang Y, Zhang Y, Wang Y, Xu J. 2017. Antimicrobial resistance and cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front Microbiol 8:1357. doi: 10.3389/fmicb.2017.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Praharaj AK, Khajuria A, Kumar M, Grover N. 2016. Phenotypic detection and molecular characterization of beta-lactamase genes among Citrobacter species in a tertiary care hospital. Avicenna J Med 6:17–27. doi: 10.4103/2231-0770.173578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lerminiaux NA, Cameron A. 2019. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol 65:34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequence data generated in this study have been deposited in SRA or GenBank with accession numbers listed in Tables S1, S2, and S3 in the supplemental material. Accession numbers for genomes newly sequenced for this study are SAMN14007636 to SAMN14007655, SRR11038037 to SRR11038052, and SAMN14082844 to SAMN14082856.