The increasing prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli is worrisome. Coordinated efforts to better understand global prevalence and risk factors are needed. Developing lower- and middle-income countries need reliable, readily available, and cost-effective solutions for detecting ESBL E. coli to contribute to global surveillance. We evaluated MacConkey agar supplemented with ceftriaxone or cefotaxime as a screening method for accurately detecting and quantifying potential ESBL E. coli.
KEYWORDS: ESBL, Escherichia coli, MacConkey agar, global surveillance
ABSTRACT
The increasing prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli is worrisome. Coordinated efforts to better understand global prevalence and risk factors are needed. Developing lower- and middle-income countries need reliable, readily available, and cost-effective solutions for detecting ESBL E. coli to contribute to global surveillance. We evaluated MacConkey agar supplemented with ceftriaxone or cefotaxime as a screening method for accurately detecting and quantifying potential ESBL E. coli. MacConkey agar from eight manufacturers, representing seven countries, was prepared with 2 or 4 μg/ml ceftriaxone or cefotaxime. Four E. coli strains (NC11, ATCC 25922, CM-13457, and CM-10455) and one Klebsiella pneumoniae strain (CM-11073) were grown overnight, serially diluted, and plated in triplicate for enumeration on all medium combinations. After recovery was assessed, US-1 MacConkey agar with cefotaxime was used to further evaluate the reproducibility and detection of potential ESBL E. coli from poultry cecal (n = 30) and water (n = 30) samples. Results indicated the recovery of E. coli 13457 from four MacConkey agar manufacturers was reduced by up to 4 log CFU/ml, and phenotypic differences in colony size and color were apparent for each manufacturer for control E. coli strains. A true ESBL, NC11, was not reduced with 4 μg/ml cefotaxime. From ceca and water, potential ESBL E. coli isolates were only confirmed from MacConkey agar with 4 μg/ml cefotaxime, where 45% and 16.6% of E. coli isolates phenotypically expressed ESBL production. The quality and reproducibility of MacConkey agar varied by manufacturer, which suggests that a single manufacturer and medium type should be selected for global monitoring efforts so that training and interpretation can be standardized.
INTRODUCTION
In 2015, the World Health Organization released its Global Action Plan (GAP) on Antimicrobial Resistance, with five objectives aimed at ensuring our future ability to treat and prevent infectious diseases with safe and effective antimicrobials (1). The second strategic objective in the GAP is to strengthen the knowledge and evidence base through surveillance and research. Surveillance systems are critical to predict the magnitude and to map the emergence of antibiotic resistance (2). In the United States, the Centers for Disease Control and Prevention (CDC) estimated the burden of antimicrobial resistance and acknowledged the report underestimates the burden because of the lack of availability of accurate data (3). In other developed or developing countries, the problem is no different or potentially worse. Challenges in methodology and criteria for defining antimicrobial resistance are complex, yet for global, harmonized monitoring systems, the quality and reproducibility of methods is critical for data comparison.
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae are a subset of antimicrobial-resistant bacteria that the CDC has declared a serious threat (3). Escherichia coli, a member of the Enterobacteriaceae family, has repeatedly demonstrated ESBL activity from both clinical and environmental biological matrices (4, 5). Production of ESBLs by E. coli is challenging for the treatment of both human and veterinary infections, because the resistance mechanisms are generally nonspecific to many beta-lactam antimicrobials (6). Adding to the challenge, resistance to multiple different classes of antimicrobials is common among ESBL-producing E. coli isolates (7).
In many European countries and the United States, detecting and characterizing ESBL-producing E. coli from human or veterinary clinical cases is common; automated equipment, molecular inference, and specialized chromogenic media have been evaluated for routine use (8–10). For a global surveillance application in lower- and middle-income countries (LMIC) without consistent inventory and fewer resources, other solutions are needed. Developing LMIC need reliable, readily available, and cost-effective solutions for detecting these organisms. Additionally, for projects supported by GAP initiatives, detection techniques should span sample matrices and be useful in both clinical and environmental settings.
The objective of our work was to evaluate a widely available microbiological medium and simple methodology to detect potential ESBL-producing E. coli from different sample types. If effective, this medium could be used as a screening methodology in a global surveillance program, particularly in LMIC, with biological matrices from human clinical specimens, human and animal feces, and environmental water samples (11). We specifically targeted MacConkey agar because of its reasonable cost and availability, familiarity in human and veterinary clinical settings, and relatively simple selectivity and interpretation. Because this medium is globally manufactured and used in the microbiological assessment of many different sample matrices, it is a good candidate for use in global surveillance systems. Additionally, we evaluated two third-generation cephalosporins for use as a selective component. Both cefotaxime and ceftriaxone have previously been used in studies of ESBL-producing Enterobacteriaceae; cefotaxime has previously demonstrated slightly higher selectivity than other agents in the detection of ESBL-producing E. coli (12). Optimizing the concentration of this selective agent is critical for achieving the highest sensitivity and specificity of detection methods; optimization can also be complicated by different breakpoints used to determine resistance in human clinical testing.
MATERIALS AND METHODS
Medium acquisition and sources.
MacConkey agar was purchased from eight manufacturers, representing seven countries (the United States [two manufacturers, US-1 and US-2], China, India, Italy, France, Canada, and the United Kingdom). The manufacturers and compositions of MacConkey agar are provided in Table 1; composition was not identical across manufacturers. For all experiments, MacConkey agar was prepared per the manufacturer’s instructions. Cefotaxime (Sigma-Aldrich, Merck, St. Louis, MO) and ceftriaxone (Sigma-Aldrich) were added to MacConkey agar at 2 μg/ml or 4 μg/ml after the media had cooled below 50°C. Each MacConkey agar plate (with antimicrobials) was poured to a volume of 20 ml. Plates were properly labeled and stored at 4°C until further use (within 48 h).
TABLE 1.
Composition of MacConkey agar from seven countries and eight different manufacturers
| Countries | Manufacturer | Composition (g/liter) |
|---|---|---|
| United States-1 | BD | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 13.5; neutral red, 0.03; crystal violet, 0.001 |
| United States-2 | Microbiology Intl. | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 13.5; neutral red, 0.03; crystal violet, 0.001 |
| United Kingdom | Mast | Peptone, 14.6; lactose, 10; bile salts, 0.004; sodium chloride, 5; agar, 14; neutral red, 0.03; crystal violet, 0.001 |
| Canada | Oxoid | Peptone, 20; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 15; neutral red, 0.03; crystal violet, 0.001 |
| France | Biokar Diagnostics | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 13.5; neutral red, 0.03; crystal violet, 0.001 |
| India | Hi medium | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 13.5; neutral red, 0.03; crystal violet, 0.001 |
| Italy | Liofilchem | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 15; neutral red, 0.03; crystal violet, 0.001 |
| China | HKM | Pancreatic digest of gelatin, 17; peptone, 3; lactose, 10; bile salts, 1.5; sodium chloride, 5; agar, 13.5; neutral red, 0.03; crystal violet, 0.001 |
Bacterial strains.
Five strains of bacteria were used in the initial evaluation of MacConkey agar supplemented with 2 or 4 μg/ml cefotaxime or ceftriaxone. Four E. coli strains, including ATCC 25922 (negative control), 13457 (veterinary clinical isolate), 10455 (veterinary clinical isolate), NC11 (cattle feces; nonclinical isolate), and K. pneumoniae 11073 (veterinary clinical isolate) were streaked onto blood agar, checked for purity, and evaluated for their antimicrobial susceptibility profile using broth microdilution (Vitek2 compact; Biomeriux, Marcy-lEtoile, France). E. coli 13457 and NC11 and K. pneumoniae 11073 demonstrated phenotypic ESBL profiles (indicated as ESBL positive on Vitek2), while E. coli 10455 did not demonstrate an ESBL phenotypic profile. Whole-genome sequencing (WGS) was carried out for the three reference E. coli strains (NC11, 10455, and 13457) using an Illumina MiSeq instrument at NC State University (FDA-GenomeTrakr Program laboratory) as previously described (13). The raw sequence (FASTA) files were annotated and assembled using CLC genomics work bench (CLC Genomics, Qiagen). The assembled files then were uploaded to an open-source resistance finder tool at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). WGS analysis of isolates confirmed the presence of blaCMY-2 in both E. coli 13457 and E. coli 10455; TEM-1B was detected in E. coli 13457. The NC11 strain contained the blaCTX-M gene, confirming it as a true ESBL producer.
A combination disk diffusion test was performed to further confirm reference strains as true ESBL or AmpC producers according to CLSI and EUCAST guidelines (14, 15). Briefly, overnight cultures of potential ESBL E. coli (NC11), AmpC E. coli (10455), and a negative control (E. coli ATCC 25922) were adjusted to 0.5 McFarland standard, and 100 μl was streaked onto Mueller-Hinton agar plates. The discs, containing cefotaxime (30 μg), cefotaxime (30 μg) plus clavulanic acid (10 μg), ceftazidime (30 μg), and ceftazidime (30 μg) plus clavulanic acid (10 μg), were placed on a plate. All of the plates were incubated at 37°C for 24 h. A ≥5-mm increase in the zone diameter for either antimicrobial agent tested in combination with clavulanic acid versus the zone diameter of the agent tested alone is considered to indicate true ESBL producers; otherwise, they are AmpC producers.
For the evaluation of media, an isolated colony of each strain was grown overnight in 10 ml of tryptic soy broth (TSB; Fisher Scientific, Waltham, MA) at 37°C. Overnight cultures were serially diluted 10-fold using 9 ml of phosphate-buffered saline (PBS). Dilutions were plated (0.1 ml) in triplicate onto all media using a cell spreader, and the plates were incubated at 37°C for 24 h. In addition to MacConkey agar, all cultures were plated onto tryptic soy agar (TSA; Fisher Scientific) as a nonselective control. After incubation, the number of colonies for the dilution with between 30 and 300 colonies was counted and recorded for all medium types. The recovered concentration of each isolate was calculated and compared to the starting concentration recovered on TSA.
Assessment of medium quality and reproducibility.
Based on outcomes from the evaluation of all MacConkey agars (n = 8) supplemented with 2 or 4 μg/ml cefotaxime or ceftriaxone (Fig. 1) and the timely availability of media, we selected US-1 (BD-Becton, Dickinson, USA) MacConkey agar with cefotaxime for further evaluation of reproducibility. Again, each strain (ESBL-positive E. coli [NC11], AmpC-positive E. coli 10455, E. coli 13457, and negative-control ATCC 25922) was grown overnight in 10 ml of TSB at 37°C. Overnight cultures were serially diluted 10-fold using 9 ml PBS. Appropriate dilutions were plated (0.1 ml) in triplicate onto US-1 MacConkey agar with cefotaxime at 1, 2, and 4 μg/ml, and TSA without drug, using a cell spreader. All of the plates were incubated at 37°C for 18 to 24 h and the concentration assessed as described above.
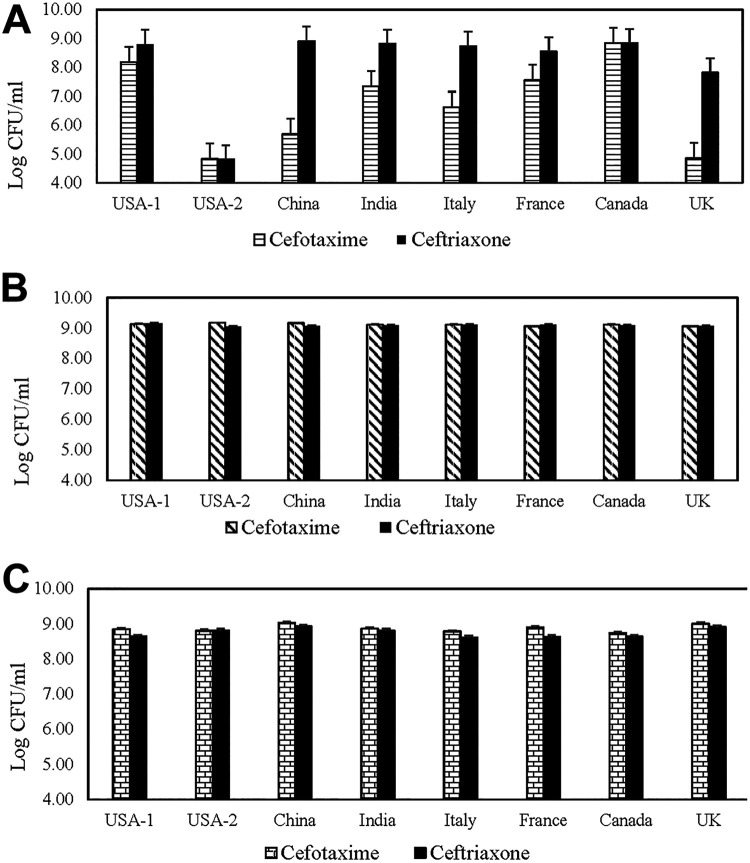
FIG 1.
Concentration recovered and standard errors for E. coli 13457 (A), E. coli 10455 (B), and Klebsiella pneumoniae 11073 (C) on MacConkey agar from various manufacturers, representing seven countries, supplemented with 4 μg/ml cefotaxime or 4 μg/ml ceftriaxone.
Additionally, four different combinations or cocktails of strains were created to evaluate the recovery of different beta-lactamase phenotypes in a mixed population. Each strain was grown independently in 20 ml of TSB for 18 h at 37°C. After incubation, 1-ml volumes of each strain were combined and serially diluted 10-fold in 9 ml of PBS. The cocktail groups included (i) E. coli 13457 and E. coli 10455; (ii) E. coli 13457, E. coli 10455, and K. pneumoniae 11073; (iii) E. coli 10455 and K. pneumoniae 11073; and (iv) E. coli 10455 and E. coli NC11. Appropriate dilutions were plated and assessed, as described above, onto US-1 MacConkey agar with 1, 2, and 4 μg/ml cefotaxime.
Recovery of ESBL E. coli from chicken ceca and water.
The sample size needed to detect an estimated 90% prevalence (as determined after expert input for water samples) with 95% confidence and a precision of 0.2 was 25 samples. Therefore, ceca (n = 30) from adult chickens were collected from necropsy specimens at NC State University (n = 15) and independent poultry farms across North Carolina (n = 15). A loopful of cecal content from each sample was plated directly onto US-1 MacConkey agar supplemented with cefotaxime at 2 μg/ml and 4 μg/ml for the recovery of ESBL-producing E. coli. To assess environmental application, water samples from a local river/creek (n = 30) were collected over 4 weeks. For the recovery of ESBL E. coli from water, 90 ml of buffered peptone water (BPW) was added to 10-ml water samples for preenrichment. Samples were incubated at 37°C for 18 to 24 h. After 24 h, 10 μl of enriched water was plated onto US-1 MacConkey agar supplemented with cefotaxime at 4 μg/ml. All MacConkey plates were incubated at 37°C for 18 to 24 h. Up to eight presumptive E. coli colonies (morphologically consistent lactose-fermenting colonies) were picked from each plate and confirmed to be E. coli by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) (Vitek MS; Biomeriux). Confirmed E. coli isolates were streaked for isolation onto 5% Columbia blood agar plates for overnight incubation at 37°C. Isolated colonies were evaluated for antimicrobial susceptibility testing (Vitek2 Compact; bioMérieux) to determine potential ESBL production. Additionally, a subset of 28 cecal isolates was randomly selected for multiplex PCR detection of TEM, SHV, OXA1-like, and various CTX-M genes using a previously described protocol (16).
RESULTS
In the evaluation of eight different MacConkey manufacturers with either 2 or 4 μg/ml cefotaxime or ceftriaxone, the starting concentrations of E. coli 13457, E. coli 10455, E. coli NC11, and E. coli 25922 inocula on TSA plates were 8.99, 8.96, 9.02, and 8.96 log10 CFU/ml, respectively. Additionally, no E. coli ATCC 25922 growth was observed on any MacConkey plate supplemented with cefotaxime or ceftriaxone (2 μg/ml or 4 μg/ml).
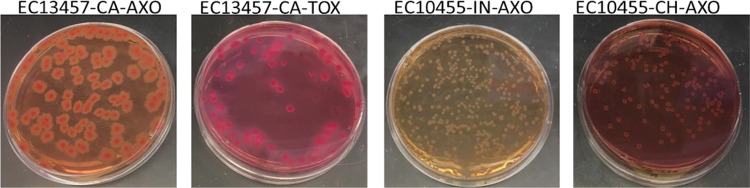
The recovered concentration of E. coli 10455 and K. pneumoniae 11073 on MacConkey agar from each manufacturer with either cefotaxime or ceftriaxone was the same as recovery on the TSA control plates. The recovery of E. coli 13457 on MacConkey from China, US-2, Italy, and the United Kingdom was reduced by between 3 and 4 log10 CFU/ml from the initial count (8.96 log10 CFU/ml) on plates with 4 μg/ml cefotaxime (Fig. 1A). Recovery of E. coli 13457 from MacConkey agar with 4 μg/ml ceftriaxone was similar to that of the control plate, with the exception of reductions in US-2 (4.8 log10 CFU/ml) and UK (7.8 log10 CFU/ml) manufacturers (Fig. 1A). Additionally, phenotypic differences, including size, shape, and color, were apparent in this strain (Fig. 2). Although there were no differences in the recovery of E. coli 10455 or K. pneumoniae 11073 among manufacturers and antimicrobials, similar differences in phenotypic appearance, including colony size and color, were apparent for each manufacturer (Fig. 2 and 3).
FIG 2.
Phenotypic appearance of pure cultures of Escherichia coli (EC) 13457 and E. coli 10455 on MacConkey agar manufactured in Canada (CA), India (IN), and China (CH) supplemented with 4 μg/ml either cefotaxime (TOX) or ceftriaxone (AXO).
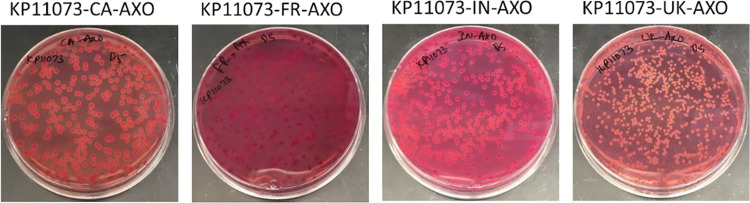
FIG 3.
Phenotypic appearance of pure cultures of Klebsiella pneumoniae (KP) on MacConkey agar manufactured in Canada (CA), India (IN), France (FR), and the United Kingdom (UK) supplemented with 4 μg/ml ceftriaxone (AXO).
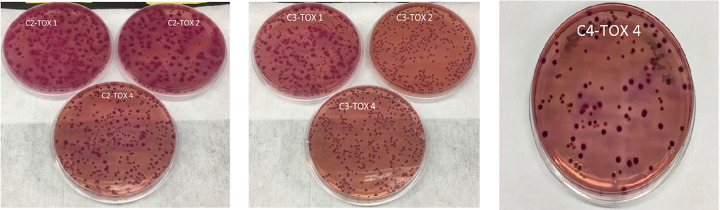
Upon further evaluation of US-1, similar concentrations were obtained from all of the bacterial strains on MacConkey agar with cefotaxime at 1 or 2 μg/ml. At 4 μg/ml, there was also no difference in the recovered concentration of E. coli 10455, E. coli NC11, or K. pneumoniae 11073; however, the level of E. coli 13457 was reduced by 1.26 log10 CFU/ml (Table 2). The recovery of control E. coli strains on MacConkey agar with 1 μg/ml cefotaxime and inoculated with a cocktail of strains was similar to the initial count on TSA (9.0 to 9.1 log10 CFU/ml) (Table 3). We observed a reduction of approximately 0.25 log10 CFU/ml in recovered beta-lactamase-producing E. coli control strains on MacConkey agar with 2 and 4 μg/ml cefotaxime, especially in the cocktails with K. pneumoniae (Table 3). Based on the appearance or lack of pale pink capsulated colonies, it was evident that there was no growth of K. pneumoniae on cocktail two or three at 2 and 4 μg/ml cefotaxime (Fig. 4). Colonies selected from MacConkey agar inoculated with cocktail group 2 or 3 inocula confirmed that there was no growth of K. pneumoniae, as all isolates were confirmed to be E. coli. Cocktail group 4 (AmpC E. coli 10455 and ESBL E. coli NC11) resulted in the recovery of equal concentrations of 10455 and the ESBL producer NC11, which was consistent with the original concentration on MacConkey agar. There was a phenotypic difference in the colonies on the MacConkey agar (Fig. 4). We picked these two different colonies and confirmed their respective phenotypes using a combination disk method (Fig. 5). In addition, we counted the number of colonies on a 106 dilution for the recovery of AmpC and ESBL producers based on phenotypic appearance (colony characteristics), and we found a larger number of colonies of true ESBL producers than AmpC producers (Fig. 4).
TABLE 2.
Recovery of E. coli and K. pneumoniae strains inoculated after overnight incubation onto TSA and MacConkey agar with cefotaxime
| Strain tested | Growth (log CFU/ml) ona
: |
||||
|---|---|---|---|---|---|
| TSA | Mac-no AB | Mac-TOX1 | Mac-TOX2 | Mac-TOX4 | |
| E. coli 13457 | 8.99 | 8.89 | 8.94 | 8.98 | 7.73 |
| E. coli 10455 | 8.96 | 8.91 | 8.92 | 8.97 | 8.99 |
| E. coli NC11 | 9.02 | 8.99 | 8.97 | 8.99 | 9.01 |
| K. pneumoniae 11073 | 8.94 | 8.87 | 8.83 | 8.86 | 8.82 |
| E. coli 25922 | 8.96 | 8.95 | 0 | 0 | 0 |
TOX1, 1 μg/ml cefotaxime; TOX2, 2 μg/ml cefotaxime; TOX4, 4 μg/ml cefotaxime; no AB, no cefotaxime in agar.
TABLE 3.
Recovery of Escherichia coli and Klebsiella pneumoniae in cocktail combinations inoculated after overnight incubation onto TSA or MacConkey agar
| Cocktail groupa | Growth (log10 CFU/ml) onb
: |
|||
|---|---|---|---|---|
| TSA | Mac-TOX1 | Mac-TOX2 | Mac-TOX4 | |
| C1 (EC13457, EC10455) | 9.03 | 9.03 | 8.91 | 8.93 |
| C2 (EC13457, EC10455, KP700603) | 9.06 | 9.06 | 8.83 | 8.69 |
| C3 (EC10455, KP700603) | 9.1 | 9.07 | 8.86 | 8.86 |
| C4 (EC10455, NC11) | 8.99 | 8.93 | 8.95 | 8.97 |
EC, E. coli; KP, K. pneumoniae.
TOX1, 1 μg/ml cefotaxime; TOX2, 2 μg/ml cefotaxime; TOX4, 4 μg/ml cefotaxime; no AB, no cefotaxime in agar.
FIG 4.
Phenotypic appearance of Escherichia coli and Klebsiella pneumoniae strains plated in cocktail combinations (C1, C2, C3, and C4) onto US-1 MacConkey agar supplemented with 1, 2, or 4 μg/ml cefotaxime.
FIG 5.
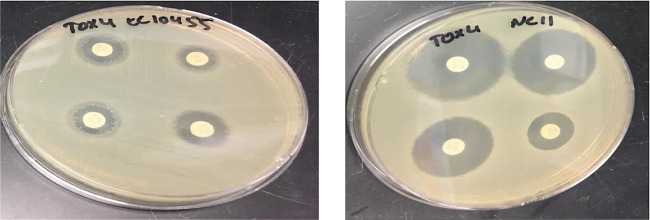
ESBL confirmation using combination disk diffusion test of AmpC-positive E. coli 10455 and true ESBL producer E. coli NC11.
The US-1 medium with 2 or 4 μg/ml was used to screen chicken cecal contents for potential ESBL-producing E. coli. A total of 21 (out of 30; 70%) and 20 (67%) ceca were positive for presumptive AmpC/ESBL E. coli on MacConkey agar with 2 and 4 μg/ml cefotaxime, respectively. Presumptive E. coli organisms were considered lactose-fermenting colonies with size and morphology consistent with those of E. coli. A total of 170 presumptive E. coli (n = 85 each from MacConkey with 2 μg/ml and 4 μg/ml of cefotaxime) isolates from direct plating were confirmed as E. coli after analysis by MALDI-TOF mass spectrometry; only one isolate from MacConkey with 2 μg/ml cefotaxime was not E. coli.
One isolate from each positive ceca (n = 21 from US-1 with 2 μg/ml cefotaxime; n = 20 from US-1 with 4 μg/ml cefotaxime) was tested for antimicrobial susceptibility and for the phenotypic appearance of ESBL production. None of the E. coli (0/21) strains isolated from US-1 with 2 μg/ml cefotaxime were considered positive for ESBL production phenotypically (e.g., demonstrating resistance to cefpodoxime and defined as ESBL phenotype by Vitek2), whereas 9 out of 20 (45%) potential ESBL E. coli strains isolated from MacConkey agar with 4 μg/ml cefotaxime demonstrated an ESBL phenotype per interpretation from Vitek2. Twenty-eight randomly selected, presumptive ESBL isolates from the 20 presumptive positive samples were screened by PCR for ESBL-associated genes; 71% of isolates were positive for CTX-M-9/14 (17 of 28; 60.7%) or CTX-M-1/3/15 (4 of 28; 14.2%). One isolate contained both CTX-M-9/14 and CTX-M-1/3/15. Additionally, 2 of 28 isolates (7.1%) were positive for TEM. In water samples, the prevalence of ESBL E. coli on US-1 MacConkey agar with 4 μg/ml cefotaxime was lower than that of chicken ceca (5/30; 16.6%). All of the presumptive E. coli ESBL-producing isolates (5/5; 100%) were confirmed to be E. coli and had phenotypically expressed ESBL production. Due to the results from chicken ceca, water was not evaluated using 2 μg/ml cefotaxime.
DISCUSSION
The global dissemination and ecology of beta-lactamase-producing E. coli, including ESBLs, is understudied and complex, although the occurrence of these organisms has been frequently reported from both clinical specimens and environmental sources, including wastewater (4, 5). Our improved understanding of the transmission and fate of these organisms within global health care, agricultural, and environmental settings is critical for the management and implementation of interventions to control their spread. The importance of ESBL-producing Enterobacteriaceae, including E. coli, have been noted by the CDC and others as critical to public health (3, 17). The reported prevalence of ESBL E. coli in human clinical infections has ranged from 7.8% to 18.3% (18). Still, despite frequent reports of these organisms, their full burden on human or animal health has not been realized.
The specific objective of our work was to identify a simple, cost-effective, and reliable methodology to accurately screen for the detection of ESBL-producing E. coli for use in a global monitoring program of different sample types, particularly for use in LMIC. This study was not intended to estimate the prevalence of ESBL-producing E. coli in any sample type or to characterize the specific beta-lactamase- or ESBL-associated genetic mechanisms of recovered bacteria. There have been numerous studies evaluating chromogenic media and molecular detection techniques for ESBL-producing E. coli (9). This work is specialized for the detection of ESBL-producing E. coli from clinical settings. We chose MacConkey agar from a variety of other media used in environmental (water), agricultural, veterinary, and human laboratory testing to specifically address the one-health approach outlined in the WHO GAP for combatting antimicrobial resistance. Especially in environmental sampling protocols, media such as tryptone bile X-glucuronide and chromogenic agars are much more commonly reported (19). Broad application of standard medium types is a newly recognized challenge as more integrated one-health initiatives are undertaken in LMIC, and standardization should be evaluated. For our study, the accessibility of MacConkey agar appeared to be broad, and we were able to obtain at least one product from eight of ten requested manufacturers originating in seven different countries (two manufacturers in the United States were selected, for a total of 8 agar formulations [Table 1]). Additionally, MacConkey agar is routinely used in both human and veterinary clinical sectors, and familiarity with interpretation should be widespread. As expected, lactose-fermenting colonies recovered on MacConkey agar was highly sensitive for E. coli in this study, where 169 of 170 selected colonies were correctly identified from chicken ceca and 5 of 5 from water samples. However, of critical importance is the fact that there was a wide variation in phenotypic appearance of E. coli across medium manufacturers. Because of this observation, we recommend the use of a common source of media for surveillance programs. If standardized protocols are deployed with descriptive characteristics and phenotypic differences are realized, an under- or overestimation of E. coli will occur.
In addition to examining medium type, the selection of E. coli isolates capable of ESBL production was done in this study by evaluating two different third-generation cephalosporins, cefotaxime and ceftriaxone. The chemical structures of these two antibiotics are highly similar (20). Previous work has shown that environmental bacteria have slightly greater resistance activity against cefotaxime than other third-generation cephalosporins (19); additionally, resistance to this antibiotic is commonly present on mobile genetic elements (7). In our study, clinically confirmed AmpC beta-lactamase-producing E. coli (10455) recovery was slightly higher with cefotaxime than ceftriaxone; however, 13457, which has blaCMY-2 and blaTEM-1B mechanisms, was slightly more variable (Fig. 1a). The recovery difference was considered minimal, and we chose to move forward with cefotaxime because of its prior use in European surveillance efforts (21). More importantly, the number of confirmed phenotypical ESBL-resistant E. coli isolates recovered from cecal samples was greatly improved (45% versus 0%) when we used 4 μg/ml instead of 2 μg/ml cefotaxime, the selective agent. In a subset of 28 isolates recovered from chicken ceca, 71% had the presence of at least one CTX-M-associated gene. It is important to note that there are different mechanisms of resistance to third-generation cephalosporins, and not all bacteria capable of growing in the presence of third-generation cephalosporins will be confirmed at the molecular level as ESBL producers. The concentration of 4 μg/ml resulted in more true putative ESBL-positive isolates from chicken ceca in this study, and previous work has shown that AmpC-producing E. coli may be resistant to 2 μg/ml third-generation cephalosporin but inhibited by 4 μg/ml. Although this was not demonstrated in our pure-culture studies, it was evident in a mixed cecal content matrix. Additionally, a simplistic screening methodology is unlikely to have both high sensitivity and specificity for ESBL-producing E. coli detection in all sample types; recovered isolates should be stored and evaluated with more sensitive methods, including whole-genome sequencing, to fully elucidate the underlying resistance mechanism. Perhaps a more readily available technique is evaluating the susceptibility of recovered isolates to third-generation cephalosporins alone and in the presence of clavulanic acid, which would aid in differentiating ESBL- and AmpC-producing E. coli, both of which are expected under the proposed conditions.
Numerous calls have been issued for improvements in monitoring antimicrobial resistance, including ESBL-producing E. coli, from different sample types (4). A one-health approach accounting for human, veterinary, and environmental sectors is critical to our improved understanding of risk factors and potential interventions of antimicrobial resistance; however, the challenges for harmonized and global engagement are complex (11). Results from our study suggest MacConkey agar with a third-generation cephalosporin used as selective pressure, particularly 4 μ/ml cefotaxime, is a reliable screening method for the recovery of ESBL-producing E. coli from fecal and water sources and could be an inexpensive, reliable method for use in LMIC, particularly if the product was standardized. Continued work to optimize methods acceptable to the environmental sector are warranted, and diagnostic sensitivity and specificity should be evaluated. Further characterization of isolates with more robust molecular methods would aid in better epidemiological assessment of transmission and should be considered when available.
ACKNOWLEDGMENTS
We thank the staff at the Clinical Microbiology and Molecular Diagnostics Laboratory at the NC State University College of Veterinary Medicine for their expertise and assistance in the isolation and evaluation of bacterial strains.
REFERENCES
- 1.WHO, FAO, OIE. 2016. Antimicrobial resistance: a manual for developing national action plans. WHO, Geneva, Switzerland: http://www.who.int/antimicrobial-resistance/national-action-plans/manual/en/. [Google Scholar]
- 2.Grundmann H, Klugman KP, Walsh T, Ramon-Pardo P, Sigauque B, Khan W, Laxminarayan R, Heddini A, Stelling J. 2011. A framework for global surveillance of antibiotic resistance. Drug Resist Updat 14:79–87. doi: 10.1016/j.drup.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 4.Bréchet C, Plantin J, Sauget M, Thouverez M, Talon D, Cholley P, Guyeux C, Hocquet D, Bertrand X. 2014. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 58:1658–1665. doi: 10.1093/cid/ciu190. [DOI] [PubMed] [Google Scholar]
- 5.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 6.Gniadkowski M. 2001. Evolution and epidemiology of extended-spectrum-β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect 7:597–608. doi: 10.1046/j.1198-743x.2001.00330.x. [DOI] [PubMed] [Google Scholar]
- 7.Pitout JDD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 8.Grohs P, Tillecovidin P, Caumont-Prim A, Carbonnelle E, Day N, Podglajen I, Gutmann L. 2013. Comparison of five media for detection of extended-spectrum-β-lactamase by use of the wasp instrument for automated specimen processing. J Clin Microbiol 51:2713–2716. doi: 10.1128/JCM.00077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazin M, MOSAR WP2 and SATURN WP1 Study Teams, Paasch F, Goossens H, Malhotra-Kumar S. 2012. Current trends in culture-based and molecular detection of extended-spectrum-β-lactamase-harboring and carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 50:1140–1146. doi: 10.1128/JCM.06852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P, Gniadkowski M, Giske CG, Poire L, Woodford N, Miriagou V, European Network on Carbapenemases. 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 11.Matheu J, Aidara-Kane A, Andremont A. 2017. The ESBL Tricycle AMR surveillance project: a simple, one health approach to global surveillance. Global Health Dynamics, Suffolk, United Kingdom: http://resistancecontrol.info/2017/the-esbl-tricycle-amr-surveillance-project-a-simple-one-health-approach-to-global-surveillance/. [Google Scholar]
- 12.Wasyl D, Hasman H, Cavaco LM, Aarestrup FM. 2012. Prevalence and characterization of cephalosporin resistance in nonpathogenic Escherichia coli from food producing animals slaughtered in Poland. Microb Drug Resist 18:79–82. doi: 10.1089/mdr.2011.0033. [DOI] [PubMed] [Google Scholar]
- 13.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical Laboratory Standard Institute (CLSI). 2019. Performance standards for antimicrobial susceptibility testing. CLSI, Wayne, PA: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED29:2019&format=SPDF. [Google Scholar]
- 15.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2013. Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf.
- 16.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 17.Walsh TR, Toleman MA. 2012. The emergence of pan-resistant Gram-negative pathogens merits a rapid global political response. J Antimicrob Chemother 67:1–3. doi: 10.1093/jac/dkr378. [DOI] [PubMed] [Google Scholar]
- 18.Doi Y, Iovleva A, Bonomo RA. 2017. The ecology of extended-spectrum-β-lactamases (ESBLs) in the developed world. J Travel Med 24:S44–S51. doi: 10.1093/jtm/taw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franz E, Veenman C, Van Hoek A, Husman ADR, Blaak H. 2015. Pathogenic Escherichia coli producing extended-spectrum-β-lactamases isolated from surface water and wastewater. Sci Rep 5:14372. doi: 10.1038/srep14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Shaboury SR, Saleh GA, Mohamed FA, Rageh AH. 2007. Analysis of cephalosporin anitibiotics. J Pharm Biomed Anal 45:1–19. doi: 10.1016/j.jpba.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Hasman H, Agerso Y, Hendriksen R, Cavaco LM, Guerra-Roman B. 2019. Laboratory protocol: Isolation of ESBL-, AmpC- and carbapenemase-producing E. coli from fresh meat. National Food Institute, Lyngby, Denmark: https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/529_esbl-ampc-cpeprotocol-version-meat-v7-09-12-19.pdf. [Google Scholar]