Rapid diagnosis of infections caused by carbapenem-resistant Enterobacteriaceae (CRE) is crucial for proper treatment and infection control. The Xpert Carba-R assay is a qualitative multiplex real-time PCR method that qualitatively detects and differentiates five common carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP) directly from rectal swabs or purified colonies within approximately 1 h.
KEYWORDS: Carba-R assay, sputum, blaKPC, blaNDM, carbapenem-resistant Enterobacteriaceae
ABSTRACT
Rapid diagnosis of infections caused by carbapenem-resistant Enterobacteriaceae (CRE) is crucial for proper treatment and infection control. The Xpert Carba-R assay is a qualitative multiplex real-time PCR method that qualitatively detects and differentiates five common carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP) directly from rectal swabs or purified colonies within approximately 1 h. We performed a multicenter evaluation of the investigational use of the Carba-R assay for detection and differentiation of carbapenemase genes from sputum specimens in patients with a clinical diagnosis of pneumonia. The intra- and interassay coefficients of variation values for the Carba-R assay were 0.2% to 2.0% and 1.4% to 2.3%, respectively. A total of 301 sputum specimens were collected and tested. Compared to bacterial culture followed by PCR identification of resistance genes from colonies, the Carba-R assay reduced turnaround time from 56 to 84 h to less than 2 h. Carbapenemase genes were detected by the Carba-R assay in Klebsiella pneumoniae (n = 236), Escherichia coli (n = 22), Enterobacter cloacae (n = 23), Klebsiella oxytoca (n = 8), Serratia marcescens (n = 6), Citrobacter freundii (n = 4), and Klebsiella aerogenes (n = 2). The Carba-R assay detected 112 blaKPC (33.5%), 70 blaNDM (21.0%), 8 blaIMP (2.4%), and 2 blaVIM (0.6%) genes, with positive percent agreement, negative percent agreement, and concordance rates of 92.9%, 86.7%, and 88.3%, respectively, for the dominant blaKPC and 85.0%, 87.8%, and 87.4%, respectively, for the blaNDM genes. Neither method detected the blaOXA-48 carbapenemase gene. The convenient, rapid, and simple characteristics of the Xpert Carba-R assay make it a potential tool for CRE detection and identification directly in sputum specimens.
INTRODUCTION
During the past several decades, the increase of multidrug-resistance (MDR) in Gram-negative bacteria (GNB) has become a major threat throughout the world. Carbapenem antimicrobial agents are currently considered the last line of defense to treat severe Gram-negative bacterial infections. Since the early 2000s, carbapenem-resistant Enterobacterales (CRE) isolates have emerged worldwide; these isolates are mainly due to acquired carbapenemases. CRE isolates now represent a significant threat in hospitals as well as an emerging public-health problem (1).
The most common mechanism of resistance to carbapenems in CRE strains is production of a carbapenemase. Carbapenemases are found in three classes of β-lactamases, class A or D serine β-lactamases and class B metallo-β-lactamases (MBLs). The most common carbapenemase genes are the Klebsiella pneumoniae carbapenemase gene (blaKPC, class A), the New Delhi metallo-β-lactamase gene (blaNDM, class B), the Verona integron-encoded metallo-β-lactamase gene (blaVIM, class B), the imipenemase metallo-β-lactamase gene (blaIMP, class B), and the oxacillinase-48 gene (blaOXA-48, class D). Because most of these carbapenemase genes are plasmid mediated, carbapenem resistance can spread quickly throughout different regions. Moreover, CRE strains frequently carry other resistance genes, which result in resistance to other classes of antimicrobial agents, such as fluoroquinolones and aminoglycosides. Consequently, these MDR CRE strains are very difficult to treat (2).
A rapid diagnostic method to detect CRE is required both to appropriately treat patients and to control the spread of CRE (3). Until recently, methods for detecting CRE resistance genes directly from clinical specimens have been limited. Previously, testing resistance genes of CRE strains required isolation of these strains from clinical specimens followed by PCR assay methods done on the purified colony as well as antibiotic susceptibility testing. With this previous approach, a diagnostic time of 54 to 86 h was typical and thus was not suitable for guiding antimicrobial therapy of these patients (4).
The Xpert Carba-R assay (Cepheid, Sunnyvale, CA) is a molecular diagnostic device specifically designed for the qualitative detection of the carbapenemase genes, including blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48, using real-time PCR with results available within less than an hour (5). The Carba-R assay has been cleared by the U.S. Food and Drug Administration for detection of carbapenemase genes directly from rectal and peri-rectal swabs and pure colonies. Published studies have shown that the Carba-R assay is able to detect carbapenemase genes from pure bacterial colonies (6, 7) as well as rectal swabs (8) with high sensitivities and specificities. Studies using the Carba-R assay for detecting carbapenemase genes from respiratory specimens rarely have been reported and are not included in the intended use of the assay (3, 9). Notably, unlike North America and Europe, where CRE were more frequently isolated from urinary samples, approximately two-thirds of CRE in China were isolated from lower respiratory tract infection specimens (10). It is thus essential to evaluate whether the Carba-R assay is able to directly detect CRE from sputum specimens in order to shorten the diagnostic turnaround time. In this study, we investigated the prevalence of carbapenemase genes in sputum specimens from patients admitted to six hospitals in different geographic areas in China by using the Carba-R assay in a study to detect resistance genes and compared these results with conventional methods (11).
MATERIALS AND METHODS
Specimen collection and preparation.
From March 2017 to February 2018, sputum specimens were collected from patients with a clinical diagnosis of pneumonia at six tertiary care Chinese hospitals. Gram stains were performed to assess the quality of the sputum prior to specimen processing. The specimens were collected and aliquoted into two portions. One aliquot was used for bacterial culture per routine microbiology practice. The other aliquot was stored in a –80°C freezer for Carba-R testing later. Information including patient’s gender, age, and accession number, as well as the bacterial species recovered and antimicrobial susceptibility profile, was acquired from each hospital.
Bacterial culture and isolate identification.
Sputum specimens were cultured on Columbia agar with 5% sheep blood and China blue lactose rosolic acid agar (Oxoide, Hampshire, England) for pathogen recovery (10). After 24 h of incubation at 35°C in 5% CO2, suspicious pathogen colonies (blue) were selected from the agar for identification using a Vitek 2 GN card and antimicrobial susceptibility testing using Vitek 2 AST-GN04 (bioMérieux, Marcy l’Etoile, France). Two ATCC prototype strains, Klebsiella pneumoniae BAA-1705 and BAA-1706, were included in each run as controls (12). CRE were identified according to the Clinical and Laboratory Standards Institute (CLSI) MIC breakpoints (13). All participating laboratories performed testing according to the same protocol to minimize the bias in sample processing and strain collection.
PCR characterization.
Nucleic acids were extracted from purified bacterial colonies and tested by PCR with primers specific to five carbapenemase target genes as previously described (3, 14). The appropriately sized PCR products were determined by DNA sequence analysis for confirmation (7, 14).
Xpert Carba-R assay.
All clinical sputum specimens were tested directly with the Carba-R assay. The sputum was preprocessed as previously described (3) before entering the Xpert cartridge. In brief, 300 μl of the specimen was mixed with 600 μl of 4% NaOH for 30 to 60 min. After the specimen was fully liquefied, 900 μl of sample reagent was added, and 1.7 ml of the mixture was transferred into the cartridge for testing. Quality control for the Xpert assay includes an internal probe check control (PCC) and a sample processing control (SPC). The GeneXpert instrument system (Cepheid, Sunnyvale, CA) is an automated real-time fluorescent quantitative PCR (qPCR) instrument designed for detecting the five carbapenemase genes blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP. The Carba-R assay was performed on all validation specimens according to the manufacturer’s package insert, and results were interpreted directly from the report generated by the GeneXpert instrument.
Ethics statement.
The six hospitals waived the need for written informed consent from patients, as included patients were not subject to extra procedures or questions. Samples were collected as part of standard care. This was purely a laboratory-based intervention without involvement of patients.
Data analysis and statistics analysis.
A positive result reported by the Carba-R assay means that at least one carbapenemase gene was detected in this specimen. A negative result means that there were no blaKPC, blaNDM, blaVIM, blaOXA-48, or blaIMP carbapenemase genes detected by the Carba-R assay. Test turnaround time (TAT), defined as hours between specimen processing and result reported, was calculated. For each carbapenemase gene, the first positive specimen with enough leftover volumes was chosen to determine the assay reproducibility by the coefficient of variation (CV) between three triplicate repeats within each run (intra-assay) and between three runs by different operators on different days (interassay). The positive percent agreement (PPA), negative percent agreement (NPA), and concordance rate of the Carba-R assay were calculated in comparison with culture followed by PCR assay using GraphPad Prism 8 statistical software analysis.
RESULTS
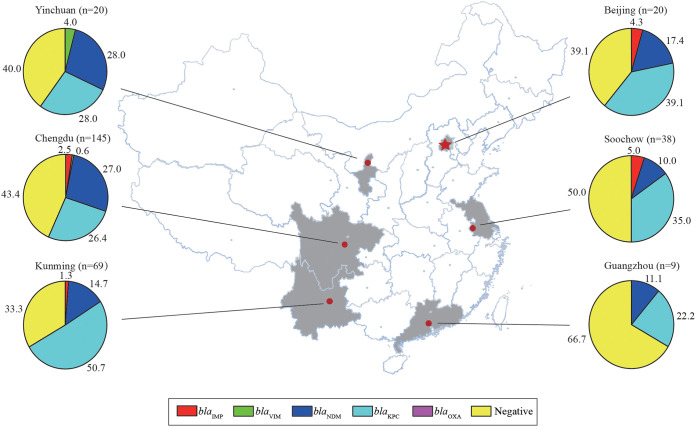
A total of 301 sputum specimens were included in this study. All specimens were collected from patients with a clinical diagnosis of pneumonia from six hospitals in China in the cities of Chengdu, Kunming, Soochow, Beijing, Yinchuan, and Guangzhou (Fig. 1). Among them, 221 (73.4%) patients were males and 80 (26.6%) were females, and the ages ranged from 3 months to 102 years.
FIG 1.
Distribution of five carbapenemase genes in six locations in China detected by the Xpert Carba-R assay.
The reproducibility of the Carba-R assay for detecting carbapenemase genes was assessed first. Three sputum specimens, which were positive for blaKPC, blaNDM, and blaIMP, respectively, were run in triplicate by three different operators. The intra-assay coefficient of variation (CV) values of the Carba-R assay ranged from 0.2 to 1.8% for blaKPC, 0.7 to 2.0% for blaNDM, and 0.9 to 1.3% for blaIMP (Table 1). The interassay CV values were 1.4%, 1.4%, and 2.3% for blaKPC, blaNDM, and blaIMP, respectively (Table 1).
TABLE 1.
Intra- and interassay variations of the Xpert Carba-R assay
| Carbapenemase gene | Operator | CT 1 | CT 2 | CT 3 | Mean CTa | SD CT | CV (%)b |
|---|---|---|---|---|---|---|---|
| Intra-assay | |||||||
| blaKPC | 1 | 24.7 | 24.4 | 25.0 | 24.7 | 0.3 | 1.2 |
| 2 | 24.6 | 24.5 | 24.6 | 24.6 | 0.1 | 0.2 | |
| 3 | 24.6 | 25.5 | 24.9 | 25.0 | 0.5 | 1.8 | |
| blaNDM | 1 | 22.0 | 21.6 | 22.0 | 21.9 | 0.2 | 1.1 |
| 2 | 21.6 | 21.5 | 21.8 | 21.6 | 0.2 | 0.7 | |
| 3 | 22.2 | 21.5 | 22.3 | 22.0 | 0.4 | 2.0 | |
| blaIMP | 1 | 35.4 | 34.8 | 35.2 | 35.1 | 0.3 | 0.9 |
| 2 | 36.7 | 37.4 | 36.5 | 36.9 | 0.5 | 1.3 | |
| 3 | 36.1 | 36.6 | 36.0 | 36.2 | 0.3 | 0.9 | |
| Interassay | |||||||
| blaKPC | 24.8 | 0.3 | 1.4 | ||||
| blaNDM | 21.8 | 0.3 | 1.4 | ||||
| blaIMP | 36.1 | 0.8 | 2.3 |
CT, threshold cycle.
CV, coefficient of variation.
The test turnaround time of the Carba-R assay was less than 2 h for detecting carbapenemase genes, which included specimen liquefication, testing, and result analysis. In contrast, the total turnaround time for culture was 56 to 84 h, followed by conventional culture (48 to 72 h), followed by PCR (8 to 12 h). The culture followed by PCR assay detected 122 positive results from 301 specimens, blaKPC (n = 79), blaNDM (n = 32), blaIMP (n = 2), blaVIM (n = 1), blaNDM + blaKPC (n = 6), and blaIMP + blaNDM (n = 2). The Carba-R assay detected 162 positive results, blaKPC (n = 86), blaNDM (n = 45), blaIMP (n = 2), blaVIM (n = 1), blaNDM + blaKPC (n = 22), blaIMP + blaKPC (n = 3), blaIMP + blaNDM (n = 1), blaIMP + blaNDM + blaVIM (n = 1), and blaIMP + blaNDM + blaKPC (n = 1). blaOXA48 was not detected by culture PCR or the Carba-R assay.
The distribution of carbapenemase genes detected by the Carba-R assay varied among the six locations, with blaKPC and blaNDM predominating across China (Fig. 1). The concordance rate of the two methods for detecting carbapenemase genes ranged from 87.4% to 99.1% with blaKPC (88.3%), blaNDM (87.4%), blaIMP (98.2%), and blaVIM (99.1%) (Table 2). PPA values ranged from 0% to 92.9%, with high values occurring for blaKPC and blaNDM and low values for the rare blaIMP and blaVIM genes (Table 2). The NPA value of the Carba-R assay ranged from 86.7% to 99.4%. Relatively low NPA values were noted for blaKPC and blaNDM (Table 2). It was noticed that the low values for blaIMP and blaVIM represented a single missed strain each, with the Carba-R assay detecting significantly more blaIMP- and blaVIM-positive specimens.
TABLE 2.
PPA, NPA, and concordance rate of the Xpert Carba-R assay (n = 334)a
| Carbapenemase gene | No. of specimens by result |
Test performance (95% CI) (%) |
|||||
|---|---|---|---|---|---|---|---|
| P + X+ | P − X+ | P + X− | P − X− | PPA (range) | NPA (range) | Concordance (range) | |
| blaNDM | 34 | 36 | 6 | 258 | 85.0 (70.9–92.9) | 87.8 (83.5–91.0) | 87.4 (83.4–90.6) |
| blaKPC | 79 | 33 | 6 | 216 | 92.9 (85.4–96.7) | 86.7 (82.0–90.4) | 88.3 (84.4–91.3) |
| blaIMP | 3 | 5 | 1 | 325 | 75.0 (30.1–98.7) | 98.5 (96.5–99.4) | 98.2 (96.1–99.2) |
| blaVIM | 0 | 2 | 1 | 331 | 0.0 (0.0–94.9) | 99.4 (97.8–99.9) | 99.1 (97.4–99.8) |
P, culture followed by PCR; X, Xpert Carba-R assay; +, detected; −, not detected; CI, confidence interval; PPA, positive percent agreement; NPA, negative percent agreement.
The ability of the Carba-R assay to detect carbapenemase genes from specimens containing different CRE species was evaluated. Each species carried different carbapenemase genotypes. Compared with culture followed by PCR, the Carba-R assay detected more genotypes from specimens with most of the bacterial species (see Table S1 in the supplemental material). Our data indicated that blaKPC and blaNDM are the two predominant genes detected in all bacterial species. The Carba-R assay demonstrated a high PPA for these genes in each species, with blaNDM ranging from 76.2% to 100.0% and blaKPC from 95.0% to 100.0% (except for Escherichia coli). However, the Carba-R assay had a relatively low NPA rate for blaKPC, with 80.0% in Citrobacter freundii, 66.7% in Klebsiella aerogenes, 84.9% in K. pneumoniae, and 83.3% in Serratia marcescens. In comparison to blaKPC and blaNDM, blaIMP and blaVIM were relatively rare genes which were only detected in Enterobacter cloacae, K. pneumoniae, and Klebsiella oxytoca. For the rarely detected blaVIM and blaIMP genes, the Carba-R assay performed poorly, detecting none of blaVIM genes in E. cloacae and only 66.7% of blaIMP in K. pneumoniae. In all three species, the Carba-R assay showed a high NPA rate for blaIMP and blaVIM genes, with blaIMP ranging from 98.1% to 100.0% and blaVIM from 99.2% to 100.0% (Table S1).
DISCUSSION
Currently, sputum specimens are the major specimen type submitted by clinicians for diagnosis of pneumonia in China. In this study, we evaluated the performance of the Xpert Carba-R assay for the detection of carbapenemase genes from sputum, a specimen type that has not been explored previously. The intra- and interassay CV values determined by the Carba-R assay ranged from 0.1 to 2.3%, demonstrating its excellent repeatability for detecting carbapenemase genes in sputum specimens. The assay had a high PPA for detecting the predominant blaKPC and blaNDM genes and a high NPA for detecting rare genes, including blaIMP and blaVIM. The convenient, rapid, and simple characteristics of the Carba-R assay make it a potential tool for CRE detection and identification directly in lower respiratory tract specimens.
CRE strains are among the major causes of pneumonia in hospitalized patients, and the choice of antimicrobial therapy for these types of pneumonia is difficult (15). Identifying CRE isolates from sputum specimens has been an important step for managing patients with the clinical diagnosis of pneumonia (16, 17). However, the methods for detection of CRE have required starting with a pure colony from a culture (18). Therefore, the development of a rapid and convenient method for detection and identification of CRE genes directly from sputum would improve the therapy of pneumonia. We found that the Carba-R assay showed a high PPA of 92.9% but a relatively low NPA of 86.7% in sputum specimens for the detection of blaKPC. One reason for the low NPA rate produced by the Carba-R assay for detecting blaKPC is that the Carba-R assay detects carbapenemase genes from specimens where no carbapenem-resistant organisms were detected by culture. The negative cultures could be due to testing patients who had already received antibiotics, resistant bacterial species that were fastidious and did not grow on the selective medium, or bacteria that contained resistance genes that were not expressed or only expressed at low levels. Importantly, the Carba-R assay can detect carbapenemase genes from other or non-Enterobacterales organisms (19). It has been reported that blaKPC genes that have been recovered from species other than Enterobacterales, including Pseudomonas aeruginosa, Pseudomonas putida, and Acinetobacter baumannii (20–23). Additionally, molecular tests detect all nucleic acids even if the bacteria are dead.
We noticed that the Carba-R assay resulted in a poor PPA in detecting blaIMP and blaVIM genes in comparison with culture followed by PCR assay, but this was only a single specimen for each resistance gene. blaIMP and blaVIM isolates are rare in China, and only limited positive samples were detected in this study. In addition, mutations in primer or probe binding regions may also affect detection of blaIMP- or blaVIM-containing variants (24). False-negative results were observed using the Carba-R assay when specimens contained a low concentration of the microorganisms harboring blaIMP or blaVIM genes (25). Limitations for blaIMP-7, blaIMP-13, and blaIMP-14 are listed in the package insert of the product. Larger numbers of sputum specimens with positive blaIMP and blaVIM genes are needed to further validate the performance of the Carba-R assay for detection and identification of rare carbapenemase genes.
Our study indicated that the Xpert Carba-R assay is a convenient and rapid method with high PPA and NPA rates compared with culture followed by PCR assay for detecting CRE genes from deep expectorated sputum, which remains a widely used specimen type in clinical practice in China. Our results suggest the possibility that the Carba-R assay can be used to screen for CRE genes directly from deep expectorated sputum specimens associated with high blaKPC and blaNDM prevalence.
Supplementary Material
ACKNOWLEDGMENTS
We thank the laboratory staff in the six participating hospitals for their excellent assistance.
The Xpert Carba-R cartridges were provided free of charge by Cepheid. The study was supported in part by grants from the China National Natural Science Foundation (81960386 to W.J.), from the Military Medical and Technology Twelfth Five-Year Science and Research Key Program (BWS11C073 to F.Q.), and from the U.S. National Institutes of Health (R01 AI090155 to L.C. and P30 CA008748 to Y.-W.T.).
Y.-W.T. is an employee of Cepheid, the commercial manufacturer of the Xpert Carba-R test. The rest of the authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lim CJ, Cheng AC, Kennon J, Spelman D, Hale D, Melican G, Sidjabat HE, Paterson DL, Kong DCM, Peleg AY. 2014. Prevalence of multidrug-resistant organisms and risk factors for carriage in long-term care facilities: a nested case-control study. J Antimicrob Chemother 69:1972–1980. doi: 10.1093/jac/dku077. [DOI] [PubMed] [Google Scholar]
- 2.Baran I, Aksu N. 2016. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microbiol Antimicrob 15:20–20. doi: 10.1186/s12941-016-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burillo A, Marin M, Cercenado E, Ruiz-Carrascoso G, Perez-Granda MJ, Oteo J, Bouza E. 2016. Evaluation of the Xpert Carba-R (Cepheid) assay using contrived bronchial specimens from patients with suspicion of ventilator-associated pneumonia for the detection of prevalent carbapenemases. PLoS One 11:e0168473. doi: 10.1371/journal.pone.0168473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich MP, Christensen DR, Coyne SR, Craw PD, Henchal EA, Sakai SH, Swenson D, Tholath J, Tsai J, Weir AF, Norwood DA. 2006. Evaluation of the Cepheid GeneXpert system for detecting Bacillus anthracis. J Appl Microbiol 100:1011–1016. doi: 10.1111/j.1365-2672.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim D-K, Kim HS, Pinto N, Jeon J, D’Souza R, Kim MS, Choi JY, Yong D, Jeong SH, Lee K. 2016. Xpert CARBA-R assay for the detection of carbapenemase-producing organisms in intensive care unit patients of a Korean tertiary care hospital. Ann Lab Med 36:162–165. doi: 10.3343/alm.2016.36.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mcmullen AR, Yarbrough ML, Wallace MA, Shupe A, Burnham CD. 2017. Evaluation of genotypic and phenotypic methods to detect carbapenemase production in Gram-negative bacilli. Clin Chem 63:723–730. doi: 10.1373/clinchem.2016.264804. [DOI] [PubMed] [Google Scholar]
- 7.Traczewski MM, Carretto E, Canton R, Moore NM, Brovarone F, Nardini P, Visiello R, García-Castillo M, Ruiz-Garbajosa P, Tato M. 2018. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in Gram-negative isolates. J Clin Microbiol 56. doi: 10.1128/JCM.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoyos-Mallecot Y, Ouzani S, Dortet L, Fortineau N, Naas T. 2017. Performance of the Xpert Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int J Antimicrobial Agents 49:774–777. doi: 10.1016/j.ijantimicag.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Vergara A, Moreno-Morales J, Roca I, Pitart C, Kostyanev T, Rodriguez-Baño J, Goossens H, Marco F, Vila J. 2020. A comparative study between real-time PCR and loop-mediated isothermal amplification to detect carbapenemase and/or ESBL genes in Enterobacteriaceae directly from bronchoalveolar lavage fluid samples. J Antimicrob Chemother 75:1453–1457. doi: 10.1093/jac/dkaa031. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, Xie L, Yang C, Ma X, Li H, Li W, Zhang X, Liao K, Man S, Wang S, Wen H, Li B, Guo Z, Tian J, Pei F, Liu L, Zhang L, Zou C, Hu T, Cai J, Yang H, Huang J, Jia X, Huang W, Cao B, Wang H. 2017. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother 62:e01882-17. doi: 10.1128/AAC.01882-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover FC, Canton R, Kop J, Chan R, Ryan J, Weir F, Ruiz-Garbajosa P, LaBombardi V, Persing DH. 2013. Detection of colonization by carbapenemase-producing Gram-negative bacilli in patients by use of the Xpert MDRO assay. J Clin Microbiol 51:3780–3787. doi: 10.1128/JCM.01092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S, Limbago B, Patel JB. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CLSI. 2016. Performance standards for antimicrobial susceptibility testing M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao M, Wen H, Xu P, Niu S, Lv J, Xie X, Mediavilla JR, Tang YW, Kreiswirth BN, Zhang X, Zhang H, Du H, Chen L. 2018. Genetic diversity of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in eastern China. Front Microbiol 9:3341. doi: 10.3389/fmicb.2018.03341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dwivedi M, Mishra A, Azim A, Singh R, Baronia A, Prasad K, Dhole T, Dwivedi U. 2009. Ventilator-associated pneumonia caused by carbapenem-resistant Enterobacteriaceae carrying multiple metallo-beta-lactamase genes. Indian J Pathol Microbiol 52:339–342. doi: 10.4103/0377-4929.54988. [DOI] [PubMed] [Google Scholar]
- 16.Chung DR, Song J-H, Kim SH, Thamlikitkul V, Huang S-G, Wang H, So TM-K, Yasin RMD, Hsueh P-R, Carlos CC, Hsu LY, Buntaran L, Lalitha MK, Kim MJ, Choi JY, Kim SI, Ko KS, Kang C-I, Peck KR, Asian Network for Surveillance of Resistant Pathogens Study Group. 2011. High prevalence of multidrug-resistant nonfermenters in hospital-acquired pneumonia in Asia. Am J Respir Crit Care Med 184:1409–1417. doi: 10.1164/rccm.201102-0349OC. [DOI] [PubMed] [Google Scholar]
- 17.Anevlavis S, Petroglou N, Tzavaras A, Maltezos E, Pneumatikos I, Froudarakis M, Anevlavis E, Bouros D. 2009. A prospective study of the diagnostic utility of sputum Gram stain in pneumonia. J Infect 59:83–89. doi: 10.1016/j.jinf.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z-G, Zheng X-Z, Guan J, Xiao S-N, Zhuo C. 2015. Direct detection of methicillin-resistant Staphylococcus aureus in sputum specimens from patients with hospital-associated pneumonia using a novel multilocus PCR assay. Pathogens 4:199–209. doi: 10.3390/pathogens4020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Chen C, Zhao M, Yu X, Lan K, Liao K, Guo P, Zhang W, Ma X, He Y, Zeng J, Chen L, Jia W, Tang YW, Huang B. 2019. High prevalence of metallo-beta-lactamase-producing Enterobacter cloacae from three tertiary hospitals in China. Front Microbiol 10:1610. doi: 10.3389/fmicb.2019.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 21.Martínez T, Vázquez GJ, Aquino EE, Ramírez-Ronda R, Robledo IE. 2012. First report of a Pseudomonas aeruginosa clinical isolate co-harbouring KPC-2 and IMP-18 carbapenemases. Int J Antimicrob Agents 39:542–543. doi: 10.1016/j.ijantimicag.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Caneiras C, Calisto F, Jorge da Silva G, Lito L, Melo-Cristino J, Duarte A. 2018. First description of colistin and tigecycline-resistant Acinetobacter baumannii producing KPC-3 carbapenemase in Portugal. Antibiotics (Basel) 7:96. doi: 10.3390/antibiotics7040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold RS, Thom KA, Sharma S, Phillips M, Johnson JK, Morgan DJ. 2011. Emergence of Klebsiella pneumoniae carbapenemase (KPC)-producing bacteria. South Med J 104:40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu Y-W, Afzal-Shah M, Houang ET, Palepou M-F, Lyon DJ, Woodford N, Livermore DM. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother 45:710–714. doi: 10.1128/AAC.45.3.710-714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goire N, Harnett GB, O’Reilly LC, Ingram PR, Leung MJ, Speers DJ, Healy PE, Inglis TJJ. 2016. The implications of endemic IMP-4 carbapenemase for clinical laboratory susceptibility testing. J Microbiol Methods 124:10–12. doi: 10.1016/j.mimet.2016.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.