Abstract
The similarity between folding and binding led us to posit the concept that the number of protein–protein interface motifs in nature is limited, and interacting protein pairs can use similar interface architectures repeatedly, even if their global folds completely vary. Thus, known protein–protein interface architectures can be used to model the complexes between two target proteins on the proteome scale, even if their global structures differ. This powerful concept is combined with a flexible refinement and global energy assessment tool. The accuracy of the method is highly dependent on the structural diversity of the interface architectures in the template dataset. Here, we validate this knowledge-based combinatorial method on the Docking Benchmark and show that it efficiently finds high-quality models for benchmark complexes and their binding regions even in the absence of template interfaces having sequence similarity to the targets. Compared to “classical” docking, it is computationally faster; as the number of target proteins increases, the difference becomes more dramatic. Further, it is able to distinguish binders from nonbinders. These features allow performing large-scale network modeling. The results on an independent target set (proteins in the p53 molecular interaction map) show that current method can be used to predict whether a given protein pair interacts. Overall, while constrained by the diversity of the template set, this approach efficiently produces high-quality models of protein–protein complexes. We expect that with the growing number of known interface architectures, this type of knowledge-based methods will be increasingly used by the broad proteomics community.
Keywords: template-based docking, flexible refinement, 3D modeling, protein interaction prediction
INTRODUCTION
Identification of protein–protein interactions at the structural level is pivotal to the understanding of protein function. Experimentally, X-ray crystallography and NMR techniques are used to determine structures at atomic resolution. Despite the fast growth in the number of experimentally solved structures of protein complexes, they are vastly fewer when compared to the available pair-wise interactions coming from high-throughput techniques. For this reason, computationally fast and accurate algorithms to model protein complexes are increasingly needed.1,2 The details of protein interactions provide crucial information, which allows fuller representation of the cellular network and the design of therapeutic molecules.
Computationally, structural modeling of protein interactions is performed predominantly in two ways: docking and knowledge-based prediction (reviewed in Ref. 3). Docking is the most widely used approach to predict the interactions of a given pair of unbound protein structures.4–9 Despite the improvements in blind docking tests,10,11 scoring functions are not yet optimized and still constitute a major hurdle.12,13 Docking is challenging on large scale because of two main reasons. First and most important reason is that docking will practically always find “reasonable” solutions with apparent favorable interactions and shape complementarity. For this reason, for reliable docking, it is desirable to not only have experimental information that the pair of proteins interact but also some biochemical data related to the location of the binding site. Second, large-scale docking is computationally very demanding. Currently, a main advantage of docking is the refinement of the modeled complexes and assessment of their energies.
Knowledge-based modeling of protein interactions uses information related to known structures of protein–protein complexes to target protein structures whose interactions are unknown. Such a strategy is based on the concept that binding and folding are similar events, and protein–protein interactions resemble protein cores.14–19 Similar to protein cores, the number of interface motifs is also limited in nature.14,15,19–21 This type of modeling started with the pioneering work of Aloy and Russell where global structural homology of the templates was considered in the modeling.22 Several works that use global sequence or structural similarities have also appeared (reviewed in Ref. 3). Later, the concept that binding sites of the proteins are structurally more conserved than the remaining surface regions23–25 has also been introduced. Further, it has been shown that unique interface architectures can be used repeatedly by different protein pairs independent from their global folds14,26 (reviewed in Ref. 27). These theoretical considerations and observations motivated the idea that interface architectures can be utilized instead of global similarity to infer structural models of protein complexes. The first approach, based on interface templates, was released by Aytuna et al.28 (and its webserver by Ogmen et al.29); there, two complementary parts of the template-interface architectures served as templates in search for structurally similar target protein surfaces. This approach handles both spatially local and implicitly global similarities. Later, additional similar approaches appeared30,31 with some differences, such as the size and type of the template dataset, and the structural alignment method. A recent study showed that an interface architecture can also be used to perform multiple functions,32 which supports the template-based modeling approaches. Further, these approaches have been found to be sufficiently accurate for proteome scale studies.33 Another advantage as compared to docking is that template-based approaches can predict whether two given proteins interact (or not). Nonetheless, in all cases, the predicted interactions need refinements, because all template-based methods consider only rigid-body structural alignment for docking of the target proteins. Proteins are not rigid molecules; the orientation of the side chains and the movements of the backbone need to be taken into account during or following the rigid-body alignment.
A multiscale strategy, which combines knowledge-based structural alignments and docking, can lead to a powerful method to model the structural proteome.34 The predicted complexes in which the interacting proteins have surfaces matching experimental interfaces in a structurally nonredundant template dataset undergo flexible refinement using a new efficient docking method.35 The energy assessments make the prediction more physical and provide a way to score the modeled complexes. This approach is being released as a protocol to be used freely community wide.34
In this work, we present the validation of this template-based docking method integrated with flexible refinement (PRISM—prediction of protein interactions by structural matching) on the Docking Benchmark. As long as structurally similar template interfaces are available in the dataset, it efficiently produces the near native or acceptable protein complex models independent from the global folds. Further, unlike docking, it can also be used for prediction of pair-wise protein interactions, besides modeling binding pose and prediction of binding site. To show this feature, verification of the predicted pair-wise interactions on large scale is performed on the proteins compiled and organized in the p53 molecular interaction map (MIM).36 This template-based method is computationally very fast compared to classical docking on large scale. The running time analysis shows that its computational effectiveness is getting more obvious with an increasing number of target proteins.
METHODS
Target dataset
Eighty-eight rigid-body test cases (from 165 protein chains) in Docking Benchmark 3.037 are used for validation of the method. The benchmark contains 28 enzyme/inhibitor, 21 antibody/antigen, and 39 other type of complexes. All possible pairs of the 165 target protein chains are searched on the templates, and a 165 × 165 interaction matrix is constructed to see, if the method distinguishes binders from nonbinders.
To show the pathway-scale performance of the method, the protein structures available in the MIM36 is used. Several proteins do not have complete structures, only fragments. For example, the full-length human DNA excision protein ERCC1 has 297 residues; however, the available structures are for residues 96–227 (PDB: 2a1i, chain A) and 220–297 (1z00, chain A). Both fragments are considered in the target set. In this pathway, 77 proteins have structural information but when considering all protein fragments, the number of chains increases to 112.
Template dataset
Three template sets are used throughout the work. Only the first and second are used for benchmarking: (i) An optimal template set for target proteins in the Docking Benchmark: this set is extracted from the bound states of the proteins resulting in 88 interfaces, which contain discontinuous residue subsets of the target protein chains. Using benchmark templates, we first check if the method can find all “true” solutions and no “false” ones with the optimal template set. (ii) A more diverse template dataset, which is utilized for the validation of the method on the benchmark proteins: this second template set is constructed from a nonredundant interface dataset composed of 49,512 interfaces structurally clustered into 8205 different interface architectures. Eliminating nonprotein complexes from these 8205 protein interfaces resulted in 7922 protein–protein interfaces. We aim to see how many interactions the method predicts with this unbiased template set for our targets. Also, we selected heterodimeric protein interfaces (1036 interfaces) among this dataset to be used for the MIM analysis. (iii) A template set, composed of the interfaces already available in the MIM (59 interfaces): this set is used for the verification of other interactions in MIM.
Here, the interface definition is as follows: the contacts between two complementary chains are calculated from the distance between any two atoms each from one chain. If this distance is less than a threshold, which is defined as the sum of van der Waals radii of the corresponding atoms plus 0.5 Å, they are considered as contacting residues. The neighbors of the contacting residues are searched within the same chain and the threshold for the distance between the Cα atoms is 6 Å. These residues, called “nearby residues,” are important for a correct structural alignment of the interface architectures.14,19,26 Hot spots in the template interfaces, that is, residues contributing more to the binding energy, are identified using the Hotpoint web server.38
The prediction algorithm
As noted above, this algorithm combines template-based prediction with flexible refinement. Proteins interact using their surfaces. Different from other approaches, instead of considering the overall structure, we extract the surface residues of the target proteins. This prevents an inaccurate matching of the templates to the core of the proteins, especially in proteins with large sizes. Here, we define the surface as a shell around the protein. The surface residues are found by calculating the relative accessibilities of the residues using NACCESS.39 If the relative accessibility, that is, the ratio of the accessible surface to the maximum accessibility of that residue in an extended peptide conformation, is more than 15%, these residues are defined as surface residues. As in the template interfaces, structural scaffolds on the surfaces are very important for an accurate matching; thus, nearby residues of surface residues are also calculated, as described earlier.
Rigid-body alignment
Structural aligment of surface regions requires a method that can compare discontinuous fragments in a sequence-order-independent fashion.40,41 MultiProt41 is appropriate for structural alignment of target surfaces to template partners. Geometry and residue type (hydrophobic, hydrophilic, aromatic, or glycine) are considered in the structural alignment. Forty percent of the residues of template chains should geometrically match the target surfaces to pass to the next step. This threshold is 60% for template chains containing less than 50 residues. To get rid of interfaces, which have small contact areas that could reflect crystal packing effects or spurious matchings, at least 15 residues should be matched in both cases. If there are computational hot spots in the template interface, at least one hot spot in each template partner should correctly match with the target surface. Hot spot filtering incorporates evolutionary similarity between the target surface and template interface in addition to structural similarity.38 The maximal RMSD allowed in the structural alignment is 2 Å. Thus, while the alignment is rigid, it can handle movements within 2 Å. Candidate target proteins passing the alignment threshold are transformed onto the corresponding template interface. If the two partners present (more than five) spatially colliding residues after tranformation, the match is eliminated. Side-chain clashes are not considered at this stage.
Flexible refinement
Flexible refinement of the rigid docking solutions involves resolving steric clashes, especially of side chains, followed by ranking putative complexes by the global energy. FiberDock35 is used for flexible refinement, which considers both side-chain and backbone flexibility. The side-chain orientations are optimized using a rotamer library, and the combination of rotamers having lowest total energy is selected. Side-chain optimization is restricted to only the clashing residues in the predicted interface. Up to 20% clashes between side chains are allowed. Backbone flexibility is modeled using the first 50 normal modes of the corresponding protein. The quality of the predicted models is assessed using the calculated global energy value, which is the single score to rank the models. The lowest global energy value implies the highest ranking prediction. Other thresholds for residue match ratio, hot spots matching, and geometrical clashes in the previous steps are used only for filtering the possible interactions.
RESULTS AND DISCUSSIONS
PRISM effectively finds near native models and distinguishes the nonbinders from binders on an optimal template set
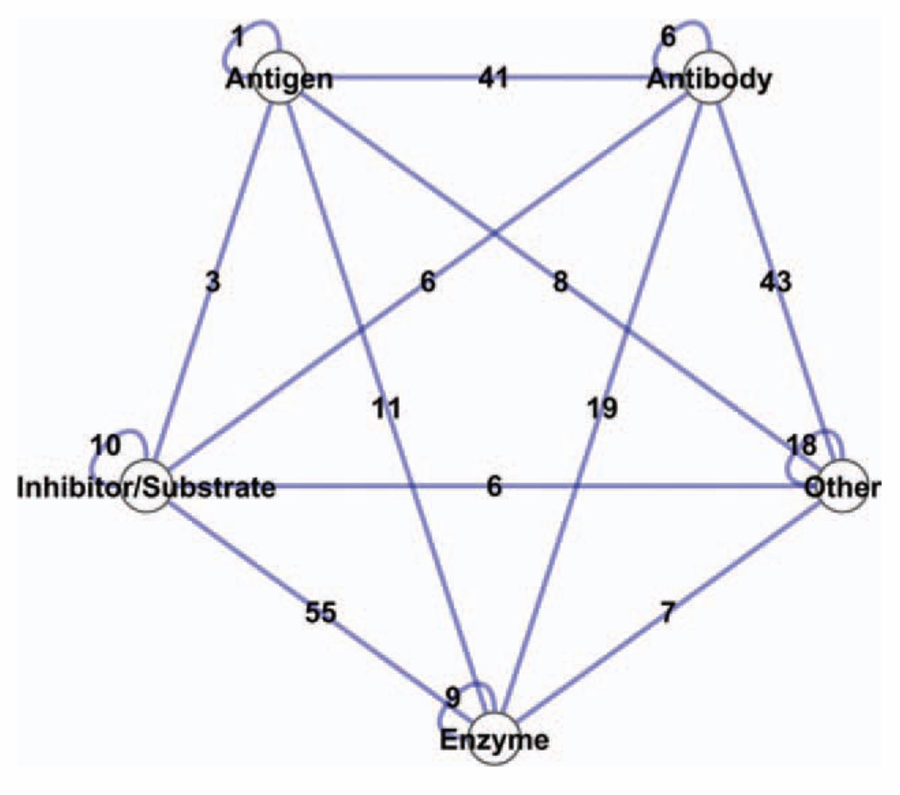
At the validation stage, our first aim is to examine how the method performs on an optimal template set. As expected, this is the best scenario that the method can face, because all the templates are coming from the native complexes of the target proteins. Although running the method on templates coming from “self-hits” seems redundant, it provides important information about the performance, such as, learning how the structural alignment is performed and selecting the optimum parameters for alignment, because both target surfaces and template partners are discontinuous sets of residues. This analysis also provides information on how the method distinguishes binders from nonbinders. Here, the aim is to verify the matching parameters and to show that given a template set containing similar interfaces, the method finds near native modes of the protein complexes with relatively less false positives. Each of the target protein surfaces is aligned with the partner chains of those 88 interfaces. The method is applied to all possible pairs (165 × 165), and a matrix of interacting pairs is generated. At the matching phase, correct binding regions are found for all 88 protein complexes, except one case, which is an antibody/antigen complex (Fab N10/Staphylococcal nuclease complex). These correct protein complex models are highly ranked by FiberDock. Besides the 87 complex models, 243 protein complexes are also modeled at the end of this run. If all 165 nodes would interact with each other, there would be 13,530 edges in the network. Our algorithm gives only 243 extra interactions; 41 of them are modeled as antibody/antigen, 55 as enzyme and inhibitor/substrate complexes, 74 as one-side antibody, and the remaining are between other types of complexes. The network representation of these extra interactions is illustrated in Figure 1. Most of these arise from antibodies or antigens. Prediction of antibody/antigen complexes is challenging; many efforts in library construction aimed to understand the binding specificity of antibodies to antibody.42
Figure 1.
Illustration of the extra interactions predicted by our method on the benchmark templates (88 interfaces). The 165 protein chains are categorized into five classes in line with Docking Benchmark classification. Each node represents one class, and each edge represents the number of predicted interactions between two classes. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
As an example for the extra interactions, the modeled complexes of bovine trypsin are illustrated. In addition to the soybean trypsin inhibitor (1ba7), our algorithm predicts that bovine trypsin (1qqu) can interact with Bowman–Birk inhibitor (1k9b:A), pancreatic secretory trypsin inhibitor (1hpt), bovine pancreatic trypsin inhibitor (9pti), CMTI-1 squash (1lu0:B) inhibitor, and TDPI from tick (2uux), which are all trypsin inhibitors. Although the overall structures and sequences of the partner proteins are dissimilar, they can bind to the bovine trypsin on the same surface, and the energy-based rankings of these interactions are high. In Figure 2, the partners of the bovine trypsin are superimposed onto each other to illustrate clearly the structural similarity in the interface region.
Figure 2.
Bovine trypsin (colored white) can interact with several trypsin inhibitors using the same region and three of these partners are superimposed to show the structural similarity in their binding sites only. Although the overall structures of Bowman–Birk inhibitor (1k9b:A, yellow), bovine pancreatic trypsin inhibitor (9pti, pink), and TDPI from tick (2uux, cyan) are dissimilar, the binding region to bovine trypsin is structurally very conserved. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
PRISM finds high-quality models for the proteins in the Docking Benchmark independent from the global folds of the template interfaces
We next use an unbiased and more diverse template dataset containing 7922 protein interfaces, which are structurally nonredundant. We eliminated template interfaces, if at least one side of these interfaces is similar in sequence to one of the target proteins.
The comparison with the native complex and the scoring of the quality of the binding mode are performed by the classical docking scoring parameters, such as lRMSD (the RMSD between the native and modeled ligands after superimposition of the receptors), iRMSD (the RMSD between the native and modeled interfaces), and fnat (fraction of the native contacts). I-Score, a new metric to score the quality of docking predictions,43 is also used to compare the predicted model with the native complex. If the I-Score is greater than 0.17, the predicted binding mode is a near native model, and if it is between 0.12 and 0.17, it is an acceptable model. If I-Score is less than 0.12, the predicted model is incorrect. We checked the distribution of the I-Score versus fnat and I-Score versus iRMSD values for the modeled complexes by PRISM (Fig. 3). Here, we performed two distinct PRISM runs. The former is performed using default thresholds; the latter is performed by relaxing the hot spot matching threshold. In the default case, PRISM produces seven binding modes on average for each target protein pair, while in the relaxed case, it produces 136 binding modes on average for each target pair. Therefore, the distribution in Figure 3(a) is less populated when compared to Figure 3(b). This distribution shows that fnat and I-Score are linearly correlated with each other. Further, predicted complexes having high I-Score have low iRMSD values. As a result, measuring the quality of the predicted models using I-Score performs well.
Figure 3.

Distribution of I-Score versus fnat and iRMSD values based on (a) default parameters and (b) hot spot threshold relaxed parameters. Higher I-Score implies higher fraction of native contacts. The iRMSD values of high I-Scores fluctuate between 0 and 3. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The template dataset is not biased to the native complexes in the Docking Benchmark, but it includes several self-hits in it. We remove these from the template dataset. In the default case, for 25 out of 88 targets pairs, at least one near native conformation is found (I-Score ≥0.17); also two more complexes are predicted as acceptable models (0.12 < I-Score < 0.17). In the relaxed version, 41 target pairs are modeled as near native (I-Score ≥0.17) after removal of self-hits. In addition, at least one acceptable model is produced for 25 target pairs among the remaining ones (0.12 < I-Score <0.17). To see how our method performs on target proteins, if we eliminate homologous template chains, we tried different sequence similarity thresholds between target proteins and template-interface partners. In Figure 4(a), the total number of near native and acceptable models predicted by PRISM are illustrated versus the sequence similarity threshold. In Figure 4(b), the distributions of the I-Score versus calculated global energies (∆Gcalc) and I-Score versus combined matching score (total of 0.6 × fhotspot + 0.4 × fmatch for each matching side of the template interface) values for the modeled complexes by PRISM are shown, where fhotspot is the fraction of identically matched hot spot residues, and fmatch is the fraction of the matched residues. Also, predicted protein complexes having negative energy values are mostly near native complexes. In addition, predicted protein complexes having a combined matching score greater than one are mostly near native. The matching scores and calculated global energies are correlated with each other. Besides providing a ranking metric as global energy, flexible refinement solves side-chain clashes in the interface region, minimizes the overall protein complex; in this way, produces physically meaningful models. Later, we will show several examples among the correctly modeled protein complexes.
Figure 4.

(a) The change in the total number of near native and acceptable models predicted by PRISM with the default parameters and the hot spot threshold relaxed parameters versus the sequence similarity elimination thresholds between the target proteins and template-interface partners. At 100% sequence similarity, template interfaces from native complexes in the Docking Benchmark are excluded. (b) The distribution of the I-Score versus calculated global energies (∆Gcalc) and I-Score versus combined matching score (total of 0.6 × fhotspot + 0.4 × fmatch for each matching side of the template interface) values for predicted complexes. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
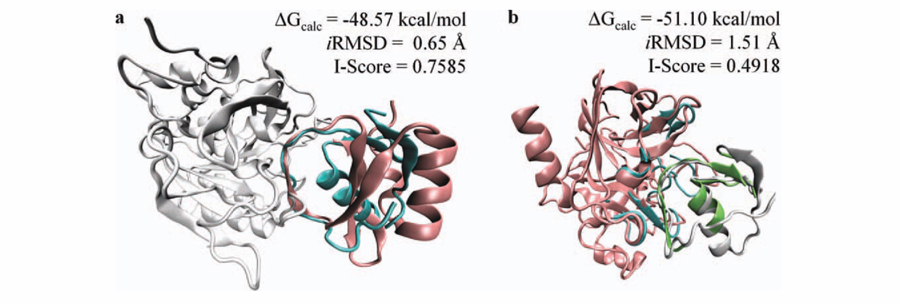
The subtilisin (2gkr)/ovomucoid (1scn) complex is modeled using the template interface between subtilisin/chymotrypsin inhibitor 2 (2sniEI). The sequence similarity between ovomucoid and chymotrypsin inhibitor is low (only 7%). Although their global folds are dissimilar, the structural similarity between the binding regions is very high. The global energy is calculated as −48.57 kcal/mol for this interaction. To show the similarity between the binding regions and dissimilarity in the global folds, chymotrypsin inhibitor 2 is superimposed on ovomucoid, and the predicted model is illustrated in Figure 5(a). The iRMSD for this binding mode is 0.65 Å, and the I-Score is 0.7585. Our method successfully identifies the binding region on ovomucoid and correctly models the subtilisin/ovomucoid complex.
Figure 5.
(a) The subtilisin (2gkr, white)/ovomucoid (1scn, cyan) complex is modeled on the template subtilisin/chymotrypsin inhibitor 2 (2sniEI). Chymotrypsin inhibitor 2 (pink) is superimposed on ovomucoid to show the structural similarity in the interface region between target and template chains. (b) The interaction between bovine chymotrypsinogen (2cga, pink) and pancreatic secretory trypsin inhibitor (1hpt, white) is modeled on the interface region of human leukocyte elastase/the turkey ovomucoid inhibitor complex. Template interface (1ppfEI) is colored cyan and green to show the structural matching between target surfaces and template partners.
The interaction between bovine chymotrypsinogen (2cga) and pancreatic secretory trypsin inhibitor (1hpt) is found using the interface region in human leukocyte elastase/the turkey ovomucoid inhibitor complex (1ppfEI). The sequence similarity between elastase and chymotrypsinogen is 32% and between trypsin inhibitor and ovomucoid inhibitor is 28%. As illustrated in Figure 5(b), the template-interface matches well with target surfaces and the calculated global energy for this interaction is −51.10 kcal/mol. The iRMSD value for this binding mode is 1.51 Å, and the I-Score is 0.4918. This complex is also predicted on the template interface in rhodniin in complex with thrombin (1tbrHR). The sequence similarity between thrombin (1tbrH) and chymotrypsinogen (2cga) is 35% and between rhodniin and trypsin inhibitor is 25%. I-Score is 0.304 and shows that this predicted model is near native. The calculated global energy is −32.96 kcal/mol. The lRMSD is 2.73 Å, and the iRMSD is 1.82 Å for this model.
The template interface within idiotope–anti-idiotope complex (1cicBC) produced a near native model for the human CD40 ligand (1aly) and the immunoglobulin Fab fragment (1i9rH) complex. The sequence similarity between 1cicB and 1i9rH is 67% and between 1cicC and 1aly is 8%. The lRMSD measure is 7.6 Å, and iRMSD is 2.13 Å for this model. The I-Score is 0.329 and 58% of the native contacts are correctly predicted (fnat = 0.58). The calculated global energy is 13.18 kcal/mol.
The matripase (1eax)/trypsin inhibitor (9pti) complex is modeled on the template interface extracted from thrombin/trypsin inhibitor (1bthKQ). The metrics for this model are as follows: I-Score = 0.745, iRMSD = 0.58 Å, and fnat = 0.79. The sequence similarity between thrombin and matripase is 35%. This complex is also correctly modeled on the template interfaces within trypsinogen and trypsin inhibitor (2tpiZI, I-Score = 0.71, iRMSD = 0.63 Å, and fnat = 0.79) and chymotrypsin and trypsin inhibitor (1t8oAB, I-Score = 0.64, iRMSD = 0.85 Å, and fnat = 0.72). The sequence similarity between matripase (1eax) and trypsin (2tpiZ) is 38% and between matripase (1eax) and chymotrypsin (1t8oA) is 33%.
Beta-trypsin and tryptase inhibitor complex is also modeled on the template from thrombin–trypsin inhibitor complex (1bthKQ). The sequence similarity between thrombin and beta-trypsin is 36% and between tryptase inhibitor and trypsin inhibitor is 25%. This model is identified as near native according to the scoring measures (lRMSD = 2.66 Å, iRMSD = 0.62 Å, I-Score = 0.62, and fnat = 0.75). The calculated global energy for this model is −37.67 kcal/mol.
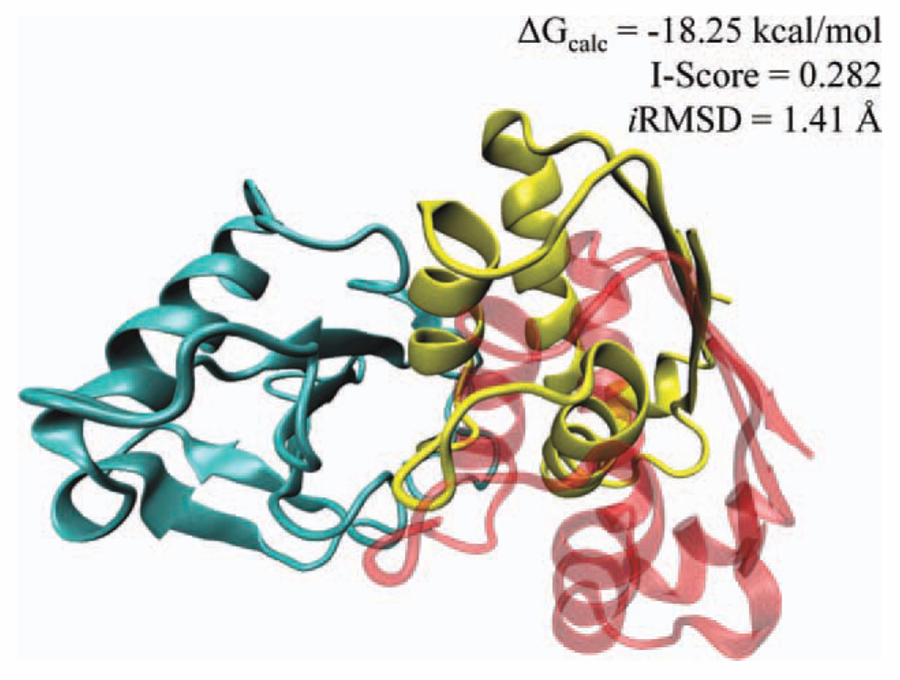
Although there is no sequence similarity, a high-quality model of the ribonuclease (1rghA)/barstar (1a19) complex is obtained on the template interface of catalytic antibody 4B2 complex (1f3dHJ), where the I-Score is 0.282, and the iRMSD is 1.41 Å. In Figure 6, the predicted complex is superimposed onto the native complex. The fraction of the native contacts in the modeled complex is found to be 0.50. The calculated global energy for this model is −18.25 kcal/mol. As shown in the figure, while the binding region is highly overlapping with the native complex, the predicted orientation is shifted 10.27 Å, when we compare overall structures of the ligand proteins.
Figure 6.
The barnase–barstar complex predicted from the template interface between catalytic antibody4B2 complex. The cyan colored structure is barnase. The yellow-colored structure is predicted orientation of the barstar and transparent red-colored one is the native orientation of the barstar. The lRSMD value shows that the predicted partner is 10 Å shifted from the native structure. Fifty percent of the contacts between barnase/barstar are correctly found and the RMSD between the interface of predicted and native barstar/barnase complex is 1.41 Å. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Overall, the validation results show that even if we eliminate interfaces having more than 50% sequence similarity from the template set, the method still provides both near native and acceptable models (17 out of 88 with default thresholds and 61 out of 88 with hot spot threshold relaxed case) using this method independent from the sequence similarity and global folds of the corresponding template interfaces. The flexible refinement of these modeled protein complexes makes these models physically more meaningful. We considered only rigid-body cases in Docking Benchmark 3.0 during validation, because the first stage of this template-based method is rigid-body alignment. Therefore, if a target monomer changes its conformation substantially in the binding region, when it binds to its partner proteins (called difficult cases), it is hard to handle this conformational change in the rigid-body alignment stage. The PRISM results on difficult cases are consistent with this statement, with only two near native and two acceptable complexes that are modeled out of the 17 difficult cases in Docking Benchmark 3.0. As a future direction, we are working on integrating flexible aligment into the first stage instead of rigid-body alignment. Another option is collecting all the conformations of the templates available in the PDB and using them to handle conformational changes between the unbound and bound states of the target proteins.
Template-based modeling of protein complexes is computationally faster than “classical” docking
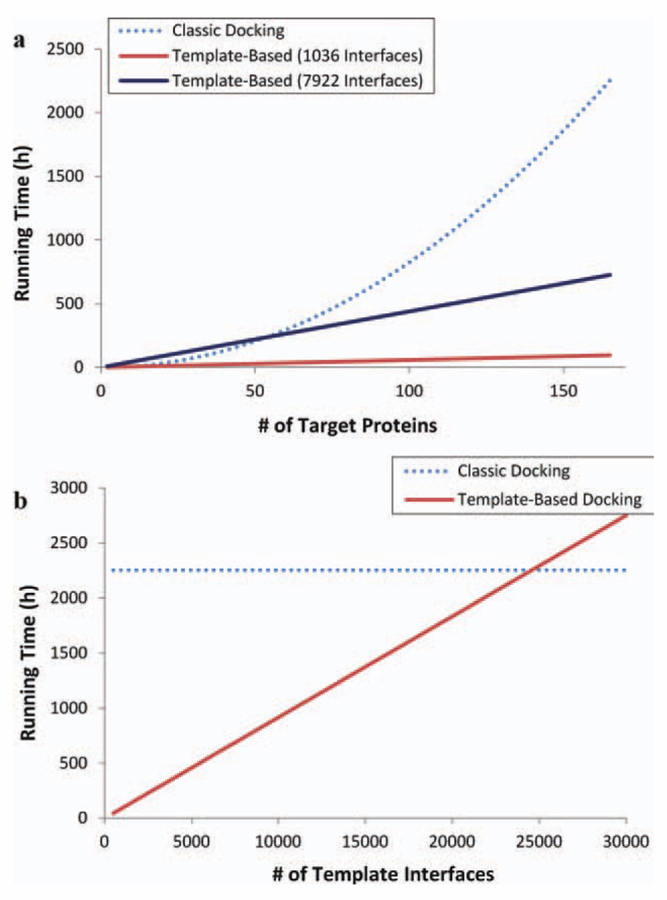
When modeling the interactions between target proteins, most of the time is spent on rigid-body structural alignment and flexible refinement of the filtered complexes. Here, we show the running time differences between rigid-body alignment and rigid-body docking as a function of the number of target proteins in the target dataset (N). Because the surface of the target protein and the interface of the template partners are only subsets of their corresponding protein structures, their alignment is faster than the alignment of two complete proteins. On average, rigid-body structural alignment of a target surface and a template-interface chain takes 1 s. To compare the first part of this approach with rigid-body docking algorithms, we selected PatchDock,44 which performs docking in less than 10 min on average on a single processor. In the running time analysis, we assume that the template set is composed of 7922 interfaces. Also, each target to template partner alignment is assumed to be performed in 1 s as measured earlier, and each docking run for a protein pair is assumed to be performed in 10 min.
Docking must be performed for all pairs of proteins separately; so for N target proteins, N × (N − 1)/2 docking runs are needed. In total, docking for N targets takes 10 min × N × (N − 1)/2, which is O(N2). On the other hand, during structural matching, one target surface is compared to one side of a template interface only once. In total, structural matching takes 1s × 2 × 1036 × N for 1036 templates and 1s × 2 × 7922 × N for 7922 templates; that is, it increases linearly with increasing number of targets, O(N). The rigid-body alignment part already defeats the docking in computational time. Also, the total running time of the flexible refinement part is dependent on the number of output solutions, and the increase is again linear. Thus, even if we add the time spent for flexible refinement, the difference is still very large. Figure 7(a) illustrates a comparison of running times as a function of target dataset size. As shown in this figure, for a small set of target proteins, both methods perform the docking in a rather equal time frame. However, at large scale, it changes, and the knowledge-based method dramatically decreases the solution space as a result the running times. As the size of the target dataset enlarges, the difference between running times gets larger. Here, we show the time comparison for up to 165 target proteins in the Docking Benchmark. For a larger set, the difference gets more dramatic. Also, the running time of our method is a function of the template dataset size in addition to that of the target dataset. In Figure 7(b), the running time of the docking for 165 targets is compared to our template-based method. The figure shows that if the template dataset size would be composed of 25,000 interfaces, the running times of both methods would be the same, which is larger than the current template sets.14,19,21,26–28
Figure 7.
Comparison of running times of our template-based method with classical docking (using PatchDock) on the Docking Benchmark. (a) Running times are plotted as a function of the number of target chains in Docking Benchmark. The analysis is performed on two template datasets (composed of 1036 and 7922 interfaces). (b) Running time of our template-based method is plotted as a function of the number of template interfaces for 165 target chains in Docking Benchmark. If there were around 25,000 template interfaces, two methods would have the same running times for 165 target proteins. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Our method is restricted by the diversity of the interface architectures. Although the number of distinct interface architectures increases exponentially, not all interface types have been discovered till date. As in all template-based methods (such as homology modeling, motif finding-based predictions, and threading), the performance of the method mainly depends on the quality of the template dataset. The contribution of the parameters used during prediction loses its importance, when an optimal template set is used. Overall, the running time analysis shows the clear advantage of this method in docking on a large scale. With the continuous growth of the PDB, knowledge-based methods will get more attractive for the community, especially because of their fast running times; further, the reliability of using known motifs from nature will permit their application to proteins, which a priori are not known to interact.
PRISM is a possible tool for prediction of binding partners of proteins: a pathway-scale case study on p53 MIM
As above mentioned, we proposed the predominance of this method over classical docking making it a possible tool for the prediction of whether any two proteins interact. Therefore, following validation, the next step is verification of the pair-wise interactions. For this purpose, we apply our multiscale combinatorial docking algorithm to the proteins available in the human MIM.36 Additional interactions from other databases such as DIP,45 MINT,46 BIND,47 and IntAct48 enrich this map. Overall, 328 interactions between 104 proteins are found from MIM and interaction databases. Among them, only for 25 interactions the structures of the complexes are available in the PDB. At this point, our method intervenes to complete the lacking network components. Using the template interfaces in MIM with a default matching threshold, 108 interactions between 49 proteins are obtained, and 30 of these 108 are known experimentally (without complex structures). We also searched the STRING49 database, which gives known and predicted interactions along with a confidence score. Here, all active prediction methods (neighborhood, coexpression, gene fusion, co-occurence, coexpression, experiments, databases, homology, and text mining) are used to obtain the confidence score. In this way, 34 interactions are found in STRING, besides those 30 interactions (Table I). In this network, transcription factors such as E2F1–2-3, Max, Myc, Jun, and Fos interconnect via multiple interactions. As expected, there is also a large number of interactions between cyclins and kinases. The template set containing 1036 interfaces gives just 53 interactions between 38 proteins with default thresholds, of which 18 interactions have experimental evidence and 17 more are verified in STRING49 (35 verified interactions in total). When the matching thresholds are relaxed by 10% (with new thresholds being 30 and 50%, respectively), the template set in the MIM gives 396 putative protein complexes between 68 protein chains of which a total of 157 interactions are verified by interaction databases. Using the relaxed matching thresholds, 721 interactions between 71 proteins are found from the 1036 template interfaces of which a total of 244 interactions are verified (Table I). The results show that using strict matching thresholds not only give more reliable predictions but also miss true positives. When thresholds are relaxed, the true positive rate increases; however, false positives also increase.
Table I.
The Number of Predicted Interactions and Verification on the Experimental Data
| Template dataset | No. of predicted interactionsa | No. of verified interactionsb | No. of further interactionsc | Total no. of verified interactions |
|---|---|---|---|---|
| Defaultd | ||||
| P53 templates (default) | 108 (49) | 30 | 34 | 64 |
| 1036 templates | 53 (38) | 18 | 17 | 35 |
| Total | 114 (50) | 31 | 37 | 68 |
| Relaxede | ||||
| p53 templates | 396 (68) | 52 | 105 | 157 |
| 1036 templates | 721 (71) | 67 | 177 | 244 |
| Total | 870 (71) | 83 | 222 | 305 |
Numbers in parantheses represent the number of proteins; that is, 108 interactions between 49 proteins.
Experimental interaction data for verification are obtained from the human molecular interaction map, DIP, MINT, BIND, and IntAct.
For further evidence for the predicted interactions, we used the STRİNG49 search tool where we considered all active prediction methods (neighborhood, coexpression, gene fusion, co-occurence, experiments, databases, homology, and text mining) with medium confidence threshold (0.4).
The default structural matching thresholds are as follows: 40% of the residues of template chains should geometrically match the target surfaces to pass to the next step. This threshold is 60% for template chains containing less than 50 residues.
In the relaxed case, default matching thresholds are reduced by 10% where new thresholds are 30 and 50%.
CONCLUSIONS
Here, we presented the validation of a combinatorial approach to effectively model the 3D structures of protein complexes and the verification of the predicted pair-wise interactions on a pathway scale. This approach relies on the expectation that the number of protein–protein interface architectures in nature is limited; thus, extrapolation of the known architecture space to target protein surfaces may help to identify protein interactions. This knowledge-based approach is made more physical by combining it with flexible refinement of the solutions and energy assessments to rank them. The docking performance of the method is examined on the Docking Benchmark proteins. The validation results show that if a structurally similar interface is available in the template dataset, the method can find the binding surface accurately and efficiently. After self hits are eliminated from the template dataset, the method produces near native models for 25 complexes and acceptable models for two complexes out of 88 with default matching thresholds. When the hot spot matching threshold is relaxed, this number increases to 41 near native models and 25 acceptable models; however, the number of false positive models increases. Even if the sequence similarity between template interfaces and target proteins are decreased dramatically, the method still finds near native and acceptable models (17 out of 88 with default thresholds, 61 out of 88 with the hot spot threshold relaxed case). The case studies provide a detailed view how the method predicts accurate models. For example, the ribonuclease/barstar complex is modeled on a catalytic antibody 4B2 homodimer. Although the overall folds and sequences of these two complexes are dissimilar, both contain structurally similar interface architectures. Also, an accurate model for a protein complex can be found from multiple template interfaces. For instance, the matripase/trypsin inhibitor is modeled on the template interfaces extracted from the thrombin/trypsin inhibitor complex, trypsinogen/trypsin inhibitor complex, and chymotrypsin/trypsin inhibitor complex.
In the validation, we used only rigid cases in the benchmark. As a limitation of our current method, it does not perform well on difficult cases. Because the first phase of this algorithm is rigid-body alignment, it is hard to handle conformational changes in the protein from the bound to the unbound states, if the conformational change is in the binding region. As a future direction, we are working on integrating flexible aligment into the first phase of the method instead of rigid-body alignment and also integrating experimentally known multiple conformations of template interfaces in the PDB. An additional key feature of this strategy is that the method effectively distinguishes the nonbinders from binders. The verification of the predicted interactions on an independent protein set (obtained from MIM) shows that this method can be used to predict whether two proteins interact, in addition to the 3D modeling of the protein complexes. Comparison of the running times with docking illustrates that the template-based approach is dramatically faster than docking. The limitation of this template-based flexible docking approach is the diversity of the template dataset. Currently, most of the available high-resolution structures are of monomers, and the number of experimentally determined different interface architectures is still limited. However, the projected fast growth in the number of experimentally determined protein complexes in the near future will lead to an increasing number of different interface architectures, which we expect to result in an increased use of such fast and reliable approaches by the community.
ACKNOWLEDGMENTS
The authors thank Dr. Dina Duhovny-Schneidman for suggestions. The content of this publication necessarily neither does reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Grant sponsor: TUBITAK; Grant numbers: 109T343 and 109E207; Grant sponsor: Turkish Academy of Sciences; Grant sponsors: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, Federal Funds from National Cancer Institute, National Institutes of Health; Grant number: HHSN261200800001E.
REFERENCES
- 1.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB. Structure-based assembly of protein complexes in yeast. Science 2004;303:2026–2029. [DOI] [PubMed] [Google Scholar]
- 2.Kiel C, Beltrao P, Serrano L. Analyzing protein interaction networks using structural information. Annu Rev Biochem 2008;77:415–441. [DOI] [PubMed] [Google Scholar]
- 3.Tuncbag N, Gursoy A, Keskin O. Prediction of protein–protein interactions: unifying evolution and structure at protein interfaces. Phys Biol 2011;8:035006. [DOI] [PubMed] [Google Scholar]
- 4.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein–protein docking. Proteins 2008;73:271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray JJ. High-resolution protein–protein docking. Curr Opin Struct Biol 2006;16:183–193. [DOI] [PubMed] [Google Scholar]
- 6.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 2002;47:409–443. [DOI] [PubMed] [Google Scholar]
- 7.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 2010;5:883–897. [DOI] [PubMed] [Google Scholar]
- 8.Lesk VI, Sternberg MJ. 3D-Garden: a system for modelling protein–protein complexes based on conformational refinement of ensembles generated with the marching cubes algorithm. Bioinformatics 2008;24:1137–1144. [DOI] [PubMed] [Google Scholar]
- 9.Cheng TM, Blundell TL, Fernandez-Recio J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein–protein docking. Proteins 2007;68:503–515. [DOI] [PubMed] [Google Scholar]
- 10.Janin J Protein–protein docking tested in blind predictions: the CAPRI experiment. Mol Biosyst 2010;6:2351–2362. [DOI] [PubMed] [Google Scholar]
- 11.Wodak SJ, Mendez R. Prediction of protein–protein interactions: the CAPRI experiment, its evaluation and implications. Curr Opin Struct Biol 2004;14:242–249. [DOI] [PubMed] [Google Scholar]
- 12.Kastritis PL, Bonvin AM. Are scoring functions in protein–protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J Proteome Res 2010;9:2216–2225. [DOI] [PubMed] [Google Scholar]
- 13.Feliu E, Aloy P, Oliva B. On the analysis of protein–protein interactions via knowledge-based potentials for the prediction of protein–protein docking. Protein Sci 2011;20:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. A dataset of protein–protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol 1996;260:604–620. [DOI] [PubMed] [Google Scholar]
- 15.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Protein–protein interfaces: architectures and interactions in protein–protein interfaces and in protein cores. Their similarities and differences. Crit Rev Biochem Mol Biol 1996;31:127–152. [DOI] [PubMed] [Google Scholar]
- 16.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Studies of protein–protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci 1997;6:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CJ, Xu D, Nussinov R. Structural motifs at protein–protein interfaces: protein cores versus two-state and three-state model complexes. Protein Sci 1997;6:1793–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai CJ, Xu D, Nussinov R. Protein folding via binding and vice versa. Fold Des 1998;3:R71–R80. [DOI] [PubMed] [Google Scholar]
- 19.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein–protein interfaces. J Mol Biol 2008;381:785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keskin O, Ma B, Rogale K, Gunasekaran K, Nussinov R. Protein–protein interactions: organization, cooperativity and mapping in a bottom-up systems biology approach. Phys Biol 2005;2: S24–S35. [DOI] [PubMed] [Google Scholar]
- 21.Keskin O, Nussinov R. Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng Des Sel 2005;18:11–24. [DOI] [PubMed] [Google Scholar]
- 22.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA 2002;99:5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell RE, Ben-Tal N. In silico identification of functional protein interfaces. Comp Funct Genomics 2003;4:420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 2003; 19:163–164. [DOI] [PubMed] [Google Scholar]
- 25.Keskin O, Ma B, Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol 2005;345:1281–1294. [DOI] [PubMed] [Google Scholar]
- 26.Keskin O, Tsai CJ, Wolfson H, Nussinov R. A new, structurally nonredundant, diverse data set of protein–protein interfaces and its implications. Protein Sci 2004;13:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem Rev 2008;108:1225–1244. [DOI] [PubMed] [Google Scholar]
- 28.Aytuna AS, Gursoy A, Keskin O. Prediction of protein–protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics 2005;21):2850–2855. [DOI] [PubMed] [Google Scholar]
- 29.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res 2005;33:W331–W336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH—prediction of interactions using known interfaces. Proteins 2007;69:839–844. [DOI] [PubMed] [Google Scholar]
- 31.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein–protein interfaces. Proteins 2010;78:3235–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, Xiao R, Krug RM, Montelione GT. Dimer interface of the effector domain of nonstructural protein 1 from influenza A virus: an interface with multiple functions. J Biol Chem 2011;286:26050–26060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundrotas PJ, Vakser IA. Accuracy of protein–protein binding sites in high-throughput template-based modeling. PLoS Comput Biol 2010;6:e1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein–protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 2011;6:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashiach E, Nussinov R, Wolfson HJ. FiberDock: flexible inducedfit backbone refinement in molecular docking. Proteins 2010;78: 1503–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohn KW. Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell 1999;10:2703–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein–protein docking benchmark version 3.0. Proteins 2008;73:705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuncbag N, Keskin O, Gursoy A. HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res 2010;38 (suppl):W402–W406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard SJ, Thornton JM. NACCESS. University College, London: Department of Biochemistry and Molecular Biology; 1993. [Google Scholar]
- 40.Nussinov R, Wolfson HJ. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc Natl Acad Sci USA 1991;88:10495–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins 2004;56:143–156. [DOI] [PubMed] [Google Scholar]
- 42.Chailyan A, Marcatili P, Tramontano A. The association of heavy and light chain variable domains in antibodies: implications for antigen specificity. FEBS J 2011;278:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao M, Skolnick J. New benchmark metrics for protein–protein docking methods. Proteins 2011;79:1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Patch-Dock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005;33:W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res 2004;32:D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res 2007;35:D572–D574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bader GD, Betel D, Hogue CW. BIND: the biomolecular interaction network database. Nucleic Acids Res 2003;31:248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. IntAct—open source resource for molecular interaction data. Nucleic Acids Res 2007;35:D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]