Abstract
Addiction to prescribed opioids including oxycodone has reached tragic levels. Herein, we investigated the relevance of fibroblast growth factors (FGFs) and immediate early genes (IEGs) to withdrawal-induced incubation of drug craving following escalated oxycodone self-administration (SA). Rats were trained to self-administer oxycodone for 4 weeks. Seeking tests were performed at various intervals during one month of drug withdrawal. Rats were euthanized one day after the last test and nucleus accumbens and dorsal striata were dissected for use in PCR analyses. Rats given long access (LgA, 9 hours), but not short access (ShA, 3 hours), to drug escalated their oxycodone intake and exhibited incubation of oxycodone seeking during withdrawal. These rats exhibited dose-dependent increases in fgf2 expression in the dorsal striatum. Fgfr2 expression was also significantly increased in the striatum in LgA, but not ShA, groups. Similarly, striatal c-fos and junB mRNA levels showed greater increases in LgA rats. The observations that FGF mRNA levels were more altered in the dorsal striatum than in the NAc of LgA rats suggest that changes in striatal FGF expression may be more salient to incubation of oxycodone craving than alterations in the NAc. Targeting FGF signaling pathways might offer novel strategies against opioid addiction.
Keywords: Opioids, Addiction, Dorsal Striatum, Incubation, Relapse, glia
Introduction
Oxycodone is a semisynthetic agent that is prescribed to patients with chronic moderate to severe pain related to cancer and some neurological disorders (Gaskell et al., 2016; Riley et al., 2008; Schmidt-Hansen et al., 2017). Oxycodone behaves like morphine as an analgesic drug, but it exhibits better bioavailability and a longer half-life (Lugo and Kern, 2004; Olkkola et al., 2013; Poyhia et al., 1991; Poyhia et al.; Ruan et al., 2017), making it somewhat more clinically efficacious (Kalso et al., 1991). During the past few years, addiction to prescribed opioids has reached tragic proportions, with many deaths throughout the world attributed to accidental opioid overdoses (Dart et al., 2015; Roxburgh et al.). Many opioid abusers reach those high levels of use because of attempts to combat tolerance to the drug used by increasing the number of tablets that they consume (Ellis et al., 2018), by progressing to inhalation or intravenous routes (Gasior et al., 2016; McCabe et al., 2007), or by switching to more powerful opioids including heroin (Mars et al., 2014; Pouget et al., 2018).
Treatment of opioid-addicted populations can be effective (Ayanga et al., 2016; Stuart et al., 2018) but it has also been fraught with problems including repeated relapses (Lopez-Goni et al., 2014; Nunes et al., 2018; Stuart et al., 2018). To develop more efficacious therapeutic approaches, it is imperative to improve our understanding of the neurobiological underpinnings of addiction to opioid drugs. Therefore, we have begun to use the model of oxycodone self-administration in rats (Blackwood et al., 2018) to query potential molecular and biochemical consequences of opioid exposure. Our behavioral results are consistent with those of other investigators who have reported that rats will escalate their oxycodone intake (Bossert et al., 2018; Mavrikaki et al., 2017; Wade et al., 2015). In addition, we recently found that incubation of oxycodone seeking was related to decreased expression of mu opioid receptors in the dorsal striatum (Blackwood et al., 2018).
In the present study, we investigated the potential role of growth factors and immediate early genes in rats that had shown incubation of oxycodone craving following 4 weeks of forced abstinence from escalated drug self-administration (Blackwood et al., 2018). We focused our attention on the role of fibroblast growth factors (FGFs) (Guillemot and Zimmer, 2011; Ornitz and Itoh, 2015; Reuss and von Bohlen und Halbach, 2003) that, unlike BDNF (Li et al., 2017; Ornell et al., 2018), have not been extensively investigated in addiction models (Doncheck et al., 2018; Even-Chen and Barak, 2018; Flores et al., 1998; Flores and Stewart, 2000). FGFs constitute a family of peptides that influence diverse biological processes such as neuronal proliferation and differentiation in the central nervous system (CNS) (Guillemot and Zimmer, 2011; Ornitz and Ito, 2015; Reuss et al., 2003). Their actions are mediated by binding to transmembrane receptors whose activation occurs through autophosphorylation and activation of intracellular signaling pathways with secondary changes in gene expression (Ornitz and Ito, 2015). These FGF receptors are located in brain regions including the ventral and dorsal striata that are known to be involved in reward pathways (Asai et al., 1993; Fon Tacer et al., 2010; Itoh et al., 1994; Wanaka et al., 1990; Yazaki et al., 1994).
Herein we report that incubation of oxycodone craving during withdrawal from long access to oxycodone self-administration is associated with differential changes in the expression of some FGFs and their receptors in the ventral and dorsal striata of rats. The changes were also more prominent in the dorsal striatum, suggesting that the expression of FGFs in that brain structure may be more relevant to incubation of oxycodone seeking.
Materials and Methods
Subjects
Male Sprague-Dawley rats, (Charles River, Raleigh, NC, USA) weighing 350–400 g before surgery were maintained on a 12-h reversed light/dark cycle with food and water available ad libitum. All procedures were performed according to guidelines outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (eighth Edition, https://guide-for-the-care-and-use-of-laboratory-animals.pdf) and were approved by the local NIDA (National Institute of Drug Abuse) Intramural Research Program, Animal Care and Use Committee (ACUC).
Intravenous surgery and self-administration training
Animals underwent surgery for insertion of catheters in their jugular veins, essentially as previously described (Cadet et al, 2017; Blackwood et al., 2018). Subcutaneous injections of buprenorphine (0.1 mg/kg) after surgery to relieve pain and allowed the rats to recover for approximately one week before self-administration (SA) training. Rats were trained in self-administration chambers located inside sound-attenuated cabinets and controlled by a Med Associates System (Med Associates, St Albans, VT). Rats (n=38) were randomly assigned to either saline (Sal) (n=8) or oxycodone (n=30) conditions. Oxycodone-assigned rats were trained to self-administer oxycodone-HCL (NIDA Pharmacy, Baltimore, MD) using short and long access paradigms as described (Blackwood et al., 2018). Short access (ShA) rats (n=15) were trained for only one 3-h daily sessions throughout the experiment. Long access (LgA) rats (n=15) were trained to self-administer oxycodone for one 3-h daily session during the first 5 days, followed by two 3-h sessions during days 8–12, and then for three 3-h daily sessions during the last two weeks. Lever presses were reinforced using a fixed ratio-1 with a 20-s timeout accompanied by a 5-s compound tone-light cue. Rats self-administered oxycodone at a dose of 0.1mg/kg per infusion over 3.5-s (0.1 ml per infusion). At the end of each 3-hr session and at the end of the day, the tone-light cue was turned off and the levers retracted. After the last day of training rats were returned to the animal vivarium and individually housed with no access to oxycodone essentially as described previously (Cadet et al., 2017; Blackwood et al., 2018).
Oxycodone seeking tests
To perform cue-induced drug seeking tests, rats were brought back to their corresponding SA chambers on the morning of each test. Drug seeking was assessed at withdrawal day (WD) 5 and 31 under extinction conditions (no oxycodone was made available). The tests consisted of 3-h sessions (Blackwood et al., 2018).
mRNA extraction and quantitative RT-PCR
We euthanized rats 24-h after WD31 for removal of dorsal striata and nucleus accumbens. The dorsal striatum and nucleus accumbens (NAc) were then used for RNA extraction using RNeasy Mini Kit (Qiagen, Valencia, CA). Total RNA (0.5 μg) was reverse-transcribed (RT) with oligo dT primers using Advantage RT-for-PCR kit (Clontech, Mountain View, CA). RT-qPCR was performed as previously described (Cadet et al., 2017) with Roche LightCycler 480 II (Roche Diagnostics, Indianapolis, IN) using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). B2M or GAPDH were used as reference genes. The results are shown as fold changes calculated as the ratios of normalized gene expression data for oxycodone SA groups compared to the saline group. All quantitative data are presented as means ± SEM. Primer sequences are listed in Table 1. Primer sequences for fgfr2 recognized transcript variants 1 and 2.
Table 1.
List of Primer Sequences used in RT-PCR.
| Gene Name | Forward | Reverse |
|---|---|---|
| c-fos | GGGCAAAGTAGAGCAG | CTCTTTCAGTAGATTGGCA |
| fosB | GAAGCTGGAGTTCATGC | ATGGGCTTGATGACAGA |
| c-jun | TTGCCCCAACAGATCC | GCTGCGTTAGCATGAG |
| junB | TCTTTCTCTTCACGACTACA | CTAGCTTCAGAGATGCG |
| junD | CCTGGAGGAGAAAGTCAAGACC | GTGGCTGAGGACTTTCTGTTTG |
| fgf1 | AGAAAGCCATCTTGTTTCTC | GAAGCACTGCTTACAAATTCA |
| fgf2 | GATCCCAAGCGGCTCTA | ACACTCCCTTGATGGACA |
| fgf8 | TACATGGCCTTTACCCGCAAG | CGGGTAGTTGAGGAACTCGAAG |
| fgf9 | AAACTGAGTCTGGAAGGCGAAT | GCAAAGCTTGGGGATCATTTCA |
| fgfr1 | GGAGAAGAAACTGCACG | ACC ACA GAG TCC ATT ATG A |
| fgfr2 | TTTCAACTCTGCTGTCCGATGA | CATCTTGGGATGAGGACTCTGG |
| fgfr3 | CGGTGACATCAACTGACGAGT | GCTAGGGTCCGCGTAAACAT |
| fgfr4 | AGATGATGAAGCTAATCGG | TTTCTTATAGTAGTCGATGTGG |
| b2m | GATCTTTCTGGTGCTTGT | AGCTCAATTTCTATTTGAGGT |
| gadph | CCTTCTCTTGTGACAAAGTG | CCCATTTGATGTTAGCGG |
Statistical analyses
We used repeated-measures analysis of variance (ANOVA) to analyze behavioral data, with dependent variable being number of oxycodone infusions on training days and independent variables between subjects (saline, ShA, LgA-L, LgA-H), and within-subject SA day (training days 1–20) factors. Bonferroni post-hoc tests were used to compare reward types (SPSS version 24, IBM, Armonk, NY). Differences in lever pressing during WD3 and WD30 were analyzed by ANOVA. Fisher’s PLSD post-hoc tests were used to compare group differences. Biochemical data were analyzed by one-way ANOVA followed by the Fisher’s PLSD post hoc test (StatView version 4.0, SAS, to assess Cary, NC). Regression analyses were performed to detect potential correlations between the dose of oxycodone taken and mRNA expression. The null hypothesis was rejected at p <0.05.
Results
Rats exposed to LgA oxycodone self-administration escalate their drug intake over time
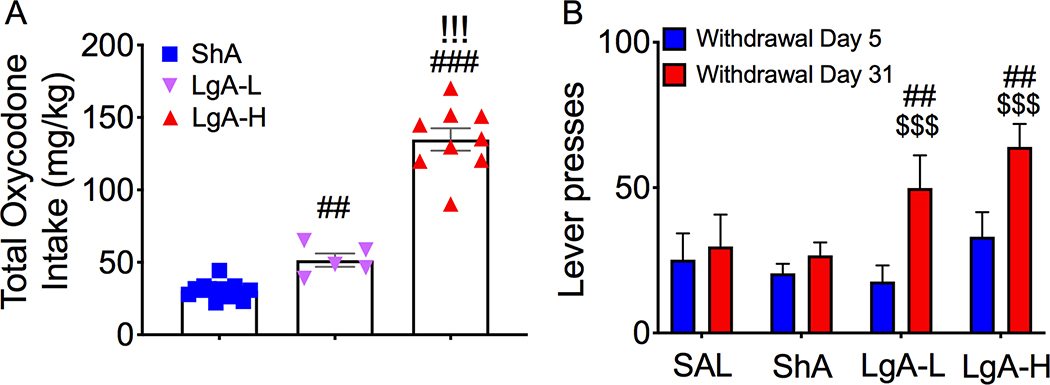
Figure 1 shows the results of the behavioral studies. As we previously reported, rats given long access to oxycodone, but not short access to the drug, escalated their intake over a period of 4 weeks (Blackwood et al., 2018). We also found that LgA rats could be divided into two phenotypes, LgA-L and LgA-H, based on the total amount of oxycodone that they self-administered during the 4 weeks of the experiment (Blackwood et al., 2018) (Fig. 1A). Both LgA-L and LgA-H phenotypes, but not the ShA rats, showed incubation of oxycodone seeking after a month of forced abstinence (Fig. 1B).
Figure 1. Differential total oxycodone intake and drug seeking behavior in rats given short or long access to the drug.
(A) LgA-L and LgA-H rats showed greater total oxycodone intake than ShA rats. (B) LgA-L and and LgA-H rats showed higher drug seeking behavior on WD31 than on WD5. The values represent means ± SEM. Key to statistics: ##, ### = p < 0.01, 0.001, respectively, in comparison to ShA rats; !!! = p < 0.001, in comparison to LgA-L rats. $ $ $ = p < 0.001, in comparison to withdrawal day 5.
Effects of oxycodone and withdrawal on FGF mRNA expression
Nucleus Accumbens
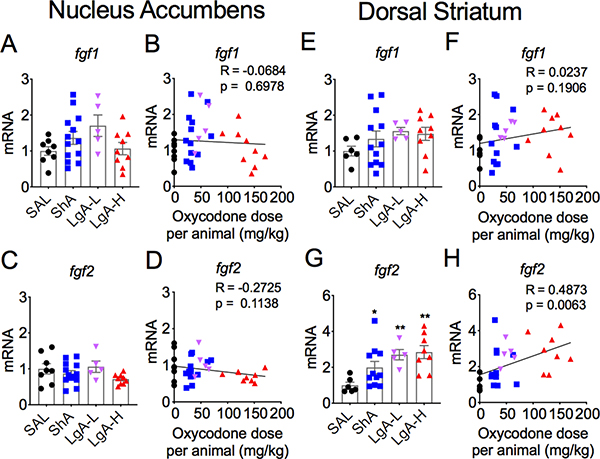
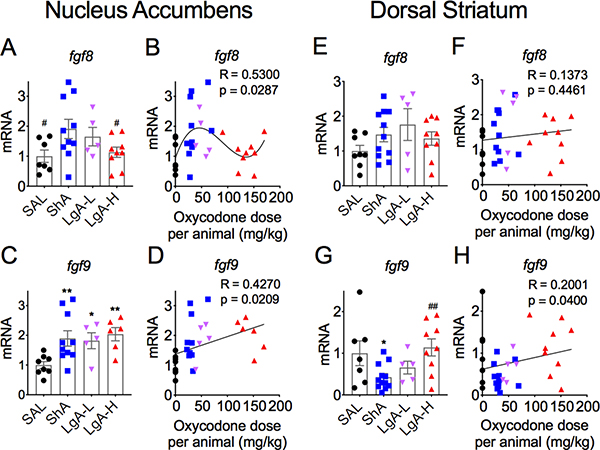
Figure 2 shows the effects of oxycodone SA on fgf1 (Figs. 2A and 2B) and fgf2 (Figs. 2B and 2D) in the nucleus accumbens (NAc). There were no significant changes in fgf1 expression [F (3,33) = 1.48, p = 0.238] (Figs. 2A and 2B). Fgf2 expression was also not significantly affected in any of the groups [F = 1.88, p = 0.154] (Figs. 2C and 2D). The results for fgf8 and fgf9 are shown in Figure 3. fgf8 mRNA expression showed a trend towards significance [F (3, 27) = 2.78, p = 0.06) (Fig. 3A), with post-hoc tests showing significant increases in the ShA oxycodone group in comparison to control and LgA-H groups. Regression analysis revealed a significant positive non-linear relationship of fgf8 mRNA levels to the amount of oxycodone self-administered (Fig. 3B). Fgf9 expression was significantly affected in all oxycodone rats [F (3, 25) = 4.30, p = 0.014] (Fig. 3C), with there being a significant positive correlation between fgf9 mRNA levels and doses of oxycodone (Fig. 3D).
Figure 2. Incubation of oxycodone seeking is accompanied by increased striatal fgf2 mRNA expression.
(A-B) Fgf1 mRNA expression showed no significant changes in the nucleus accumbens and (E-F) the dorsal striatum. (C-D) NAc fgf2 mRNA levels were not affected after incubation. (G-H) Striatal fgf2 mRNA expression was increased in an oxycodone dose-dependent fashion. The values in the bar graphs represent means ± SEM (n=5–14 animals per group). Note the differences in scales on the Y-axis. Key to statistics: *, ** = p < 0.05, 0.01 respectively, in comparison to saline rats.
Figure 3. Differential fgf8 and fgf9 mRNA levels in the nucleus accumbens and dorsal striatum.
(A-B) fgf8 mRNA expression is increased in the ShA rats in comparison to control and LgA-H rats. (C-D) NAc fgf9 mRNA levels are increased after exposure to oxycodone. (E-F) Striatal fgf8 mRNA expression showed no significant changes after oxycodone and withdrawal. (G-H) Striatal fgf9 mRNA levels are decreased in the ShA rats. The values in the bar graphs represent means ± SEM (n=5–15 animals per group). Note the differences in scales on the Y-axis. Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to saline rats; #, ## = p < 0.05, 0.01, respectively, in comparison to ShA rats.
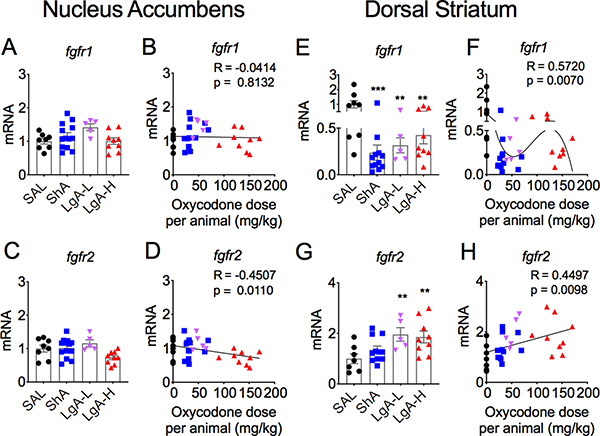
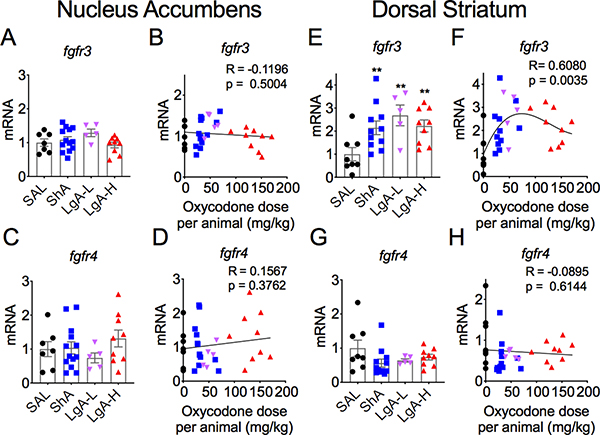
The effects of withdrawal from oxycodone SA on FGF receptors in the NAc are shown in Figures 4 and 5. Fgfr1 did not show any significant changes in oxycodone-exposed rats [F (3,31) = 2.46, p = 0.08] (Figs. 4A and 4B). Fgfr2 mRNA levels were also not significantly affected between the 3 groups of oxycodone [F (3,33) = 1.91, p = 0.15] (Fig. 4C). However, there was a significant negative correlation of fgfr2 mRNA levels to doses of oxycodone taken (Fig. 4D). The mRNA levels of fgfr3 [F (3,30) = 1. 70, p = 0.19] (Figs. 5A and 5B) and fgfr4 [F (3,31) = 0.94, p = 0.43] (Figs. 5C and 5D) in the NAc were not significantly impacted by withdrawal from oxycodone SA.
Figure 4. Differential mRNA expression of fgfr1 and fgfr2 in the NAc and dorsal striatum after withdrawal from oxycodone.
(A-B) fgfr1 and (C-D) fgfr2 mRNA levels showed no significant changes in the NAc. (E-F) Striatal fgfr1 mRNA levels are decreased in all oxycodone rats. (G-H) Striatal fgfr2 mRNA expression showed dose-dependent increases in LgA-L and LgA-H rats. The values in the bar graphs represent means ± SEM (n=5–15 animals per group). Note the differences in the scales in the Y-axis in G and H for fgfr2 expression in comparison to others. Key to statistics: **, *** = p < 0.01, 0.001, respectively, in comparison to saline rats.
Figure 5. Increased fgfr3 mRNA expression in the dorsal striatum.
(A-B) fgfr3 and (C-D) fgfr4 mRNA levels are not affected by oxycodone withdrawal in the NAc. In contrast, (E-F) there were significant increases in striatal fgfr3 mRNA expression in all oxycodone rats whereas (G-H) striatal fgfr4 mRNA levels were not significantly impacted. The values in the bar graphs represent means ± SEM (n=5–15 animals per group). Note the differences in scales on the Y-axis for fgfr3 in comparison to the others. Key to statistics: ** = p < 0.01, in comparison to saline rats.
Dorsal Striatum
Figure 2 also illustrates the effects of oxycodone SA on the mRNA expression of fgf1 (Figs 2E and 2F) and fgf2 (Figs 2G and 2H) in the dorsal striatum. Figure 2E shows no significant changes in striatal fgf1 [F (3, 28) = 1.074, p = 0.376]. There was also no significant correlation between doses of oxycodone and fgf1 expression (Fig. 2F). In contrast, fgf2 mRNA levels were increased after withdrawal from oxycodone SA [F (3,26) = 5.227, p = 0.006] (Fig. 2G), with changes in mRNA expression showing significant dose-dependent increases (Fig. 2H). There were no significant changes in striatal fgf8 expression [F (3,29) = 1.46, p = 0.25] (Figs. 3E and 3F). However, there were significant changes in the expression of striatal fgf9 [F (3, 29) = 3.63, p = 0.024]. These were due to 57% decreases in the ShA oxycodone group (Fig. 3G). There was also only a weak positive correlation between oxycodone doses and changes in mRNA expression (Fig. 3H).
We also tested the possibility that the expression of striatal FGF receptors might be impacted by oxycodone SA and withdrawal. Figures 4 and 5 illustrate the changes on four FGF receptors (fgfr1-fgfr4). There were significant decreases in striatal fgfr1 mRNA levels [F (3,30) = 5.89, p = 0.003] (Fig. 4E), with significant non-linear dose-dependent effects (Fig. 4F). In contrast, significant increases in fgfr2 expression [F (3,26) = 4.527, p = 0.011] were observed in both LgA-L and LgA-H groups (Fig. 4G) in a dose-dependent fashion (Fig. 4F). Fgfr3 mRNA levels were significantly increased in all oxycodone groups [F (3,29) = 4.611, p = 0.009] (Fig. 5F), with significant positive nonlinear relationship to drug doses (Fig. 4F). In contrast, there were no significant changes in fgfr4 mRNA levels [F (3,30) = 1.75, p = 0.18)] (Figs. 4G and 4H).
Effects of oxycodone and withdrawal on IEG mRNA levels
Nucleus Accumbens
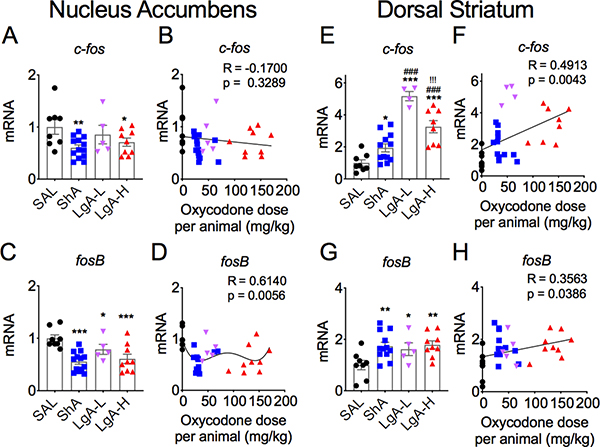
Figure 6 shows the effects of withdrawal from oxycodone SA on c-fos (Figs. 6A and 6B) and fosB (Figs. 6C and 6D) mRNA levels in the NAc. There were significant [F (3,31) = 3.35, p = 0.032] decreases in c-fos mRNA levels, with greater decreases (−40%) occurring in the ShA group (Fig. 6A). FosB mRNA levels were also significantly [F (3.31)= 8.30, p = 0.0003] decreased in all 3 oxycodone groups (Fig. 6C), with the ShA group (−45%) showing greater decreases than the LgA-L (−21%) and LgA-H (−39%) groups (Fig. 6C). The decreases in c-fos expression did not show any significant relationship to doses of oxycodone taken by the rats (Fig. 6B) while the changes in fosB expression showed non-linear relationships to doses (Fig. 6D).
Figure 6. Differential changes in c-fos and fosB mRNA levels in the NAc and dorsal striatum after oxycodone withdrawal.
(A-B) c-fos mRNA levels are significantly decreased in the NAc of ShA and LgA-H rats. (C-D) fosB mRNA levels are decreased in the NAc of all oxycodone rats. (E-F) There were greater increases in striatal c-fos mRNA in the LgA-L and LgA-L rats in comparison to the ShA group. (G-H) Striatal fosB mRNA expression is increased to the same degree in all oxycodone rats. The values in the bar graphs represent means ± SEM (n=5–15 animals per group). Note the differences in scales on the Y-axis for each separate sub-figure, with striatal c-fos and fosB showing greater magnitude of changes after oxycodone withdrawal. Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to saline rats; ##, ### = p < 0.01, 0.001, respectively, in comparison to ShA rats; !!! = p < 0.001, in comparison to LgA-L rats.
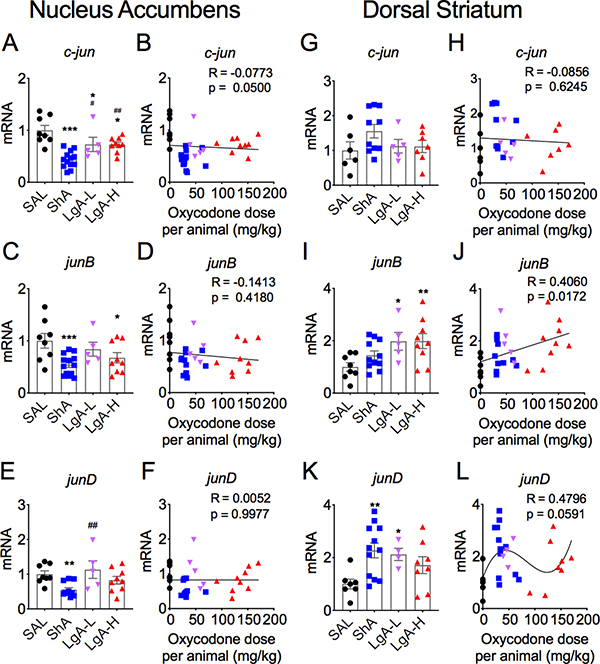
Figure 7 shows the observations for the jun family of IEGs. Figure 7A shows that c-jun mRNA levels were significantly [F (3,31) = 12.45, p < 0.0001] decreased in the 3 oxycodone groups after withdrawal from the drug. The ShA group showed greater decreases (−56%) than the LgA-L (−27%) and the LgA-H (−27%) groups (Fig. 7A). JunB mRNA expression was also significantly decreased [F (3,31) = 4.40, p = 0.01] (Fig. 7C), with the changes in ShA (−46%) and LgA-H (−33), but not LgA-L (−16%), being significant (Fig. 7C). JunD mRNA levels were also significantly influenced [F (3,29) = 4.81, p = 0.008] by oxycodone withdrawal (Fig. 7E). However, only the ShA group exhibited significant decreases (−45%) in junD expression (Fig. 7E). There was no correlation between expression of these 3 IEGs and doses of oxycodone taken (see Figs. 7B, 7D, and 7F).
Figure 7. Differential mRNA expression of jun family members in the NAc and dorsal striatum after oxycodone withdrawal.
(A-B) c-jun mRNA levels are decreased in all oxycodone rats. (C-D) jun-B mRNA levels are decreased in ShA and LgA-H rats. (E-F) NAc junD mRNA expression is decreased in ShA rats. (G-H) Striatal c-jun mRNA levels are not impacted by oxycodone withdrawal whereas (I-J) junB mRNA expression is increased in both LgA-L and LgA-H groups. (K-L) junD mRNA levels are increased in the striatum of ShA and LgA-L rats. The values in the bar graphs represent means ± SEM (n=5–15 animals per group). Note the differences in the scales on the Y-axis for the 3 mRNAs, with striatal junB and junD showing greater magnitude of changes in expression. Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to saline rats; #, ## = p < 0.05, 0.01, respectively, in comparison to ShA rats.
Dorsal striatum
Figure 6 also illustrates the results of oxycodone withdrawal on striatal expression of c-fos (6E) and fosB (Fig. 6G) mRNAs. There were significant increases [F (3, 28) = 26.08, p < 0.0001] in c-fos expression, with the largest changes being observed in the LgA-L (+5.1-fold) and LgA-H (+3.3-fold). There was also a significant positive correlation between oxycodone doses and c-fos expression (Fig. 6F). FosB expression was also increased [F (3,30) = 4.21, p = 0.013] in the oxycodone groups, with small significant positive correlation between oxycodone doses and gene expression (Fig. 6H).
In the dorsal striatum, c-jun expression was not significantly altered in any group [F (3,23) = 1.53, p = 0.23] (Figs 7G and 7H). In contrast, junB mRNA levels were significantly increased [F (3,29) = 3.95, p = 0.018] in both LgA-L (+98) and LgA-H (+99%) (Fig. 7G). There was also a positive correlation between oxycodone doses and junB mRNA levels (Fig. 7H). JunD expression was also increased [F (3,27) = 3.76, p = 0.022] (Fig. 7H), with no significant correlation between junD expression and oxycodone doses (Fig. 7I).
Discussion
The myriad clinical complications of opioid addiction have triggered renewed calls for better therapeutic approaches to pain syndromes and their varied consequences (Boscarino et al., 2010; Rudd et al., 2016). In order to reach these goals, elucidation of the neurobiological adaptions to exposure to these drugs is paramount. Therefore, our laboratory has recently focused its attention on identifying the biochemical and molecular effects of oxycodone exposure since this drug is prescribed by medical professionals for moderate to severe pain (Gaskell et al., 2016; Riley et al., 2008; Schmidt-Hansen et al., 2017). We recently reported that withdrawal from escalated oxycodone intake is accompanied by decreased expression of mu opioid receptor protein in the dorsal striatum of LgA-L and LgA-H rats that exhibited incubation of oxycodone seeking (Blackwood et al., 2018). In this follow-up study, we wanted to know to what extent changes observed in the expression of striatal mu opioid receptor protein might be accompanied by other molecular changes in the brains of these same rats. To do so, we compared the expression of FGFs and IEGs in the nucleus accumbens and dorsal striatum to test the possibility that there might be regional specificity in their responses to withdrawal from oxycodone self-administration. We found that: 1) fgf2 showed dose-related increases in the dorsal striatum; 2) striatal fgfr1 expression was decreased in the dorsal striatum; 3) fgfr2 was increased in the dorsal striatum of all LgA rats that showed incubation of oxycodone seeking; 4) similarly, c-fos mRNA levels were substantially increased in the dorsal striatum of LgA rats; and 5) striatal junB expression was also increased in the LgA rats. In what follows, we propose that these changes may potentially be secondary mechanisms that activate signaling pathways that drive incubation of oxycodone craving during abstinence from escalated oxycodone intake.
Previous studies have documented a potential role for BDNF in substance use disorders in humans and in animal models of addiction to drugs including opioids (Ornell et al., 2018). For example, BDNF protein expression is decreased in the NAc of heroin-injected rats (Li et al., 2017) whereas BDNF mRNA levels were increased in the locus coeruleus (LC) by repeated injections of morphine (Numan et al., 1998). Morphine treatment also increased BDNF protein levels in the frontal cortex (FC) and striatum of rats (Bachis et al., 2017). However, much less has been written about the members of the FGF family of trophic factors in relation to addiction and most studies have focused mainly on the effects of psychostimulants including cocaine and amphetamines on fgf2 gene expression (Even-Chen and Barak, 2018).
FGFs constitute a family of, at least, 23 growth factors that are involved in important biological processes including embryonic and neural development, tissue repair, regulation of endocrine functions, and adult neurogenesis, among others (Carter et al., 2015; Guillemot and Zimmer, 2011; Imamura, 2014; Itoh, 2010; Mudo et al., 2009) FGFs exert their functions by binding to five FGF receptors (FGFR1–5) that are very structurally similar (Porta et al., 2017; Sleeman et al., 2001). FGFR1–3 are widely expressed in the brain (Asai et al.; Yazaki et al., 1994), FGFR4 shows low levels of brain expression (Fon Tacer et al., 2010; Itoh et al., 1994) while FGFR5, a soluble protein, has not been reported in rodent brains. Importantly, FGFs and their receptors are located in brain regions such as the ventral and dorsal striatum, hippocampus, and the frontal cortices (Eckenstein et al., 1994; Itoh et al., 1994; Woodward et al., 1992; Yazaki et al., 1994), brain regions that represent nodal points in reward pathways that are involved in addiction processes (Koob and Volkow, 2016). The location of FGFs and FGFRs in these structures suggests that these signaling pathways and their downstream targets might be involved in brain health and diseases including substance use disorders (SUDs). Indeed, evidence in human and animal studies has accumulated to support the idea of FGF2 involvement in major depressive disorders and anxiety (Turner et al.). Of significant importance are the observations that fgf2, fgfr2, and fgfr3 mRNA levels are co-downregulated in the brains of human patients who suffered from major depressive disorders (MDD) (Evans et al., 2004), suggesting co-regulation of these genes in MDD patients.
As mentioned above, a role for the FGF signaling pathway in addiction has also been suggested in the case of psychostimulants focusing almost always on fgf2 expression (Even-Chen and Barak, 2018). Specifically, repeated cocaine injections increased fgf2 mRNA levels in the frontal cortex and striatum of adult (Fumagalli et al., 2006) and in the hippocampus of adolescent (Giannotti et al., 2013) rodents. Fgf2 mRNA expression is also increased in the NAc following cocaine conditioned place preference (CPP) (Doncheck et al., 2018). Animals bred for greater drug seeking behaviors showed increased FGF2 expression in the dentate gyrus of the hippocampus and neonatal injection of FGF2 increased cocaine self-administration by adult rats (Turner et al., 2009). Moreover, a single fgf2 injection on post-natal day 2 increased cocaine sensitization in rats bred for low responses for drugs (Clinton et al., 2012). The neonatal FGF2 injection was also reported to increase fgf2 mRNA expression in the core of the NAc (Clinton et al., 2012). Similar to cocaine, injections of amphetamine also impact FGF2 expression in the brain (Flores et al., 1998; Flores and Stewart, 2000), with significant increases in FGF2 protein levels having been reported in dopaminergic cell bodies and projection areas (Flores and Stewart, 2000). Other drugs including nicotine and alcohol can also influence fgf2 mRNA expression in the brain (Belluardo et al., 2008; Even-Chen and Barak, 2018) (Even-Chen and Barak, 2018). Those observations are consistent with our present observations that a month withdrawal from oxycodone SA is associated with changes in the expression of striatal fgf2 mRNA levels, with the long access groups showing greater increases (see Figure 2). These changes are of interest in view of the observations of increased expression of fgfr2 mRNA levels in the two LgA groups because FGF2, a mostly astrocytic protein, can interact with all the FGF receptors including FGFR2 (Porta et al., 2017), whose mRNA expression has been detected in the dorsal striatum (Asai et al., 1993). Importantly, because FGF2 and FGFR2 are mainly located in glial cells (Asai et al., 1993; Miyake et al., 1996; Woodward et al., 1992), these results also suggest the possibility that glial cells can play important roles in mediating incubation of oxycodone craving in rats and/or relapse to oxycodone abuse in humans. The observations of concomitant increased striatal fgf2 and fgfr2 mRNA levels also suggest potential secondary or tertiary feed-forward mechanisms wherein upregulation of both the ligand and its receptor might lead to potentiated behavioral responses (incubation) during withdrawal from oxycodone self-administration. Our observations are consistent with the report that fgf2 and fgfr2 mRNA levels are co-downregulated in the brains of MDD patients (Evans et, 2004). Co-regulation of another trophic factor, BDNF, and its receptor, TrkB, has also been reported after electroconvulsive treatment (ECT) (Nibuya et al., 1995), suggesting that trophic factor-induced neuroadaptions to exogenous stimuli may involve feed-forward mechanisms in some cases. These feed-forward mechanisms in the case of incubated oxycodone seeking might include plastic responses generated by fgf2 that is known induce neuroplastic changes in the CNS (Zechel et al., 2010). It is worthwhile noting that the rats that showed incubation also showed higher striatal c-fos and JunB mRNA levels than the ShA group that did not show incubation, thus implicating fgf2, fgfr2, c-fos, and junB in a mu opioid receptor-dependent pathway that is associated with augmented cue-induced drug seeking after prolonged abstinence. This idea will be tested in future experiments. It is also important to note, in relationship to potential effects of FGF2 in the brain, that the RNA expression of fgfr1, another FGF2 receptor which is located in neurons (Asai et al., 1993), was downregulated in all oxycodone groups, suggesting that activation of neuronal FGFR1 by FGF2 might have led to different adaptations than activation of glial FGFR2. The data also suggest that FGFR1 might not be directly involved in the manifestation of incubated behaviors since the ShA rats that did not show incubation of oxycodone seeking after prolonged abstinence also showed decreased fgfr1 mRNA expression (compare patterns of expression of the fgfrs in Figures 4G and 5G, respectively). Thus, our data also support the idea that regulation of FGFRs by their ligands may be cell type-specific (glial vs neurons) based on different intracellular mechanisms activated by these receptors.
As mentioned earlier, all rats given long access to oxycodone (LgA-L and LgA-H) showed similar increases in incubation of oxycodone seeking that was associated with decreases in striatal mu receptors (Blackwood et al., 2018). These observations had suggested that striatal opioid receptor mechanisms might, in part, be responsible for some of neurobiological drivers of incubation of oxycodone seeking in rodents and/or relapses in human addicted to opioid drugs. Our findings of increase striatal fgf2 mRNA levels in animals that showed incubation of oxycodone craving are consistent with reports of interactions of mu and FGF receptors (Belcheva et al., 2002; Di Liberto et al., 2014) and suggest that interactions of striatal Mu and FGF receptors might constitute links in the chain (s) of downstream molecular events that occur at various intervals in this model of oxycodone use disorder. This idea is supported that the observation that fgfr1-dominant negative glioma cells showed attenuated Mu receptor-induced ERK activation (Belcheva et al., 2002). This reasoning is also supported by the demonstration that FGF2 can influence the expression of Mu receptors in rats (Turner et al., 2019). Moreover, some of the proposed mechanisms for Mu/FGF receptor interactions may involve FGF signaling via Src kinases (Auciello et al., 2013; Cunningham et al., 2010) that appear to play significant roles in opioid withdrawal (Zhang et al., 2017). Furthermore, the proposal that activation of Mu receptors can lead to cleavage and shedding of FGF2 by matrix metalloproteinases is also of interest to this discussion (Di Liberto et al., 2014). The resulting increase in FGF2-induced stimulation of FGFRs after Mu receptor activation could potentially serve as a feed-forward mechanism to further potentiate activation of Mu receptors since these receptors are located at the site of convergence of Mu and FGF signaling pathways (Belcheva et al., 2002).
The differential increase in FGF2 expression is also interesting in relationship to data obtained from rats that were bred to show high responses to a novel environment (bHR) (Piazza et al., 1990). bHR animals self-administer amphetamine more than low response (bLR) rats (Piazza et al., 1990). The bHR rats also have higher levels of fgf2 mRNA in the hippocampus in comparison to bLR rats (Perez et al., 2009). Interestingly, administration of cocaine differentially impacted fgfr1 mRNA expression in the brains of these two phenotypes (Turner et al., 2008). Specifically, repeated injections of cocaine (15 mg/kg) for 7 days decreased fgfr1 mRNA expression in the hippocampus of bHR (not bLR) rats but increased fgfr1 mRNA levels in the prefrontal cortex of bLR (not bHR) rats (Turner et al., 2008). It is also noteworthy that bHR rats that have higher fgf2 mRNA expression also exhibited higher mu opioid receptor mRNA expression in their brain when compared to bLR rats (Turner et al., 2019). Importantly, injection of FGF2 early in life normalized mu opioid receptor expression in adult bLR rats (Turner et al., 2019). Taken together with our previous demonstration of altered Mu opioid expression in animals that showed incubation of oxycodone craving (Blackwood et al., 2018) and the papers discussed here, our present results provide further support for the idea that FGF signaling pathways might play a role in the behavioral manifestations of incubation during drug withdrawal.
In summary, we have found that incubation of oxycodone seeking after withdrawal from escalated drug self-administration is accompanied by increased expression of fgf2 and fgfr2 mRNAs in the rat striatum. These changes are also associated with greater increases in striatal c-fos and junB mRNA levels in incubated rats. In contrast, there were similar increases in fosB expression in all rats exposed to either escalated or non-escalated drug intake. Patterns of mRNA changes in the nucleus of these rats did not correspond to incubation of oxycodone seeking in LgA rats. In contrast to our results, fgf2 and fgfr2 mRNA levels have been reported to be decreased in the brains of MDD patients (Evans et al., 2004). Thus, it is intriguing to suggest that MDD and incubation of craving may represent behavioral manifestations of opposite FGF-mediated neurobiological phenomena. Finally, because changes in fgf2 expression had been observed in other models of drug abuse, the present observations suggest that manipulation of FGF2-dependent signaling pathways may offer potential novel strategies towards curbing addiction to psychostimulant and/or opioid drugs.
Acknowledgements and Disclosures
This work was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA. The authors also sincerely thank two reviewers and the editor whose comments substantially improved our paper.
Footnotes
Competing Interests: The authors declare no competing interests.
REFERENCES
- Asai T, Wanaka A, Kato H, Masana Y, Seo M, & Tohyama M. (1993). Differential expression of two members of FGF receptor gene family, FGFR-1 and FGFR-2 mRNA, in the adult rat central nervous system. Brain Res Mol Brain Res, 17(1–2), 174–178. [DOI] [PubMed] [Google Scholar]
- Auciello G, Cunningham DL, Tatar T, Heath JK, & Rappoport JZ. (2013). Regulation of fibroblast growth factor receptor signalling and trafficking by Src and Eps8. J Cell Sci, 126(Pt 2), 613–624. doi: 10.1242/jcs.116228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayanga D, Shorter D, & Kosten TR. (2016). Update on pharmacotherapy for treatment of opioid use disorder. Expert Opin Pharmacother, 17(17), 2307–2318. doi: 10.1080/14656566.2016.1244529 [DOI] [PubMed] [Google Scholar]
- Bachis A, Campbell LA, Jenkins K, Wenzel E, & Mocchetti I. (2017). Morphine Withdrawal Increases Brain-Derived Neurotrophic Factor Precursor. Neurotox Res, 32(3), 509–517. doi: 10.1007/s12640-017-9788-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcheva MM, Haas PD, Tan Y, Heaton VM, & Coscia CJ. (2002). The fibroblast growth factor receptor is at the site of convergence between mu-opioid receptor and growth factor signaling pathways in rat C6 glioma cells. J Pharmacol Exp Ther, 303(3), 909–918. doi: 10.1124/jpet.102.038554 [DOI] [PubMed] [Google Scholar]
- Belluardo N, Mudo G, Bonomo A, Di Liberto V, Frinchi M, & Fuxe K. (2008). Nicotine-induced fibroblast growth factor-2 restores the age-related decline of precursor cell proliferation in the subventricular zone of rat brain. Brain Res, 1193, 12–24. doi: 10.1016/j.brainres.2007.11.069 [DOI] [PubMed] [Google Scholar]
- Blackwood CA, Hoerle R, Leary M, Schroeder J, Job MO, McCoy MT, Ladenheim B, Jayanthi S, et al. (2018). Molecular Adaptations in the Rat Dorsal Striatum and Hippocampus Following Abstinence-Induced Incubation of Drug Seeking After Escalated Oxycodone Self-Administration. Mol Neurobiol. doi: 10.1007/s12035-018-1318-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino JA, Rukstalis M, Hoffman SN, Han JJ, Erlich PM, Gerhard GS, & Stewart WF. (2010). Risk factors for drug dependence among out-patients on opioid therapy in a large US health-care system. Addiction, 105(10), 1776–1782. doi: 10.1111/j.1360-0443.2010.03052.x [DOI] [PubMed] [Google Scholar]
- Bossert JM, Hoots JK, Fredriksson I, Adhikary S, Zhang M, Venniro M, & Shaham Y. (2018). Role of mu, but not delta or kappa, opioid receptors in context-induced reinstatement of oxycodone seeking. Eur J Neurosci. doi: 10.1111/ejn.13955 [DOI] [PubMed] [Google Scholar]
- Cadet JL, Brannock C, Krasnova IN, Jayanthi S, Ladenheim B, McCoy MT, Walther D, Godino A, et al. (2017). Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol Psychiatry, 22(8), 1196–1204. doi: 10.1038/mp.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EP, Fearon AE, & Grose RP. (2015). Careless talk costs lives: fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol, 25(4), 221–233. doi: 10.1016/j.tcb.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Clinton SM, Turner CA, Flagel SB, Simpson DN, Watson SJ, & Akil H. (2012). Neonatal fibroblast growth factor treatment enhances cocaine sensitization. Pharmacol Biochem Behav, 103(1), 6–17. doi: 10.1016/j.pbb.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham DL, Sweet SM, Cooper HJ, & Heath JK. (2010). Differential phosphoproteomics of fibroblast growth factor signaling: identification of Src family kinase-mediated phosphorylation events. J Proteome Res, 9(5), 2317–2328. doi: 10.1021/pr9010475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart RC, Severtson SG, & Bucher-Bartelson B. (2015). Trends in opioid analgesic abuse and mortality in the United States. N Engl J Med, 372(16), 1573–1574. doi: 10.1056/NEJMc1501822 [DOI] [PubMed] [Google Scholar]
- Di Liberto V, Mudo G, Fuxe K, & Belluardo N. (2014). Interactions between cholinergic and fibroblast growth factor receptors in brain trophism and plasticity. Curr Protein Pept Sci, 15(7), 691–702. [DOI] [PubMed] [Google Scholar]
- Doncheck EM, Hafenbreidel M, Ruder SA, Fitzgerald MK, Torres L, & Mueller D. (2018). bFGF expression is differentially regulated by cocaine seeking versus extinction in learning-related brain regions. Learn Mem, 25(8), 361–368. doi: 10.1101/lm.047530.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenstein FP, Kuzis K, Nishi R, Woodward WR, Meshul C, Sherman L, & Ciment G. (1994). Cellular distribution, subcellular localization and possible functions of basic and acidic fibroblast growth factors. Biochem Pharmacol, 47(1), 103–110. doi: 10.1016/0006-2952(94)90442-1 [DOI] [PubMed] [Google Scholar]
- Ellis MS, Cicero TJ, Dart RC, & Green JL. (2018). Understanding multi-pill ingestion of prescription opioids: Prevalence, characteristics, and motivation. Pharmacoepidemiol Drug Saf. doi: 10.1002/pds.4687 [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, et al. (2004). Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci U S A, 101(43), 15506–15511. doi: 10.1073/pnas.0406788101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Chen O, & Barak S. (2018). The role of fibroblast growth factor 2 in drug addiction. Eur J Neurosci. doi: 10.1111/ejn.14133 [DOI] [PubMed] [Google Scholar]
- Flores C, Rodaros D, & Stewart J. (1998). Long-lasting induction of astrocytic basic fibroblast growth factor by repeated injections of amphetamine: blockade by concurrent treatment with a glutamate antagonist. J Neurosci, 18(22), 9547–9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, & Stewart J. (2000). Changes in astrocytic basic fibroblast growth factor expression during and after prolonged exposure to escalating doses of amphetamine. Neuroscience, 98(2), 287–293. [DOI] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, Goetz R, Mohammadi M, et al. (2010). Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol, 24(10), 2050–2064. doi: 10.1210/me.2010-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Pasquale L, Racagni G, & Riva MA. (2006). Dynamic regulation of fibroblast growth factor 2 (FGF-2) gene expression in the rat brain following single and repeated cocaine administration. J Neurochem, 96(4), 996–1004. doi: 10.1111/j.1471-4159.2005.03627.x [DOI] [PubMed] [Google Scholar]
- Gasior M, Bond M, & Malamut R. (2016). Routes of abuse of prescription opioid analgesics: a review and assessment of the potential impact of abuse-deterrent formulations. Postgrad Med, 128(1), 85–96. doi: 10.1080/00325481.2016.1120642 [DOI] [PubMed] [Google Scholar]
- Gaskell H, Derry S, Stannard C, & Moore RA. (2016). Oxycodone for neuropathic pain in adults. Cochrane Database Syst Rev, 7, CD010692. doi: 10.1002/14651858.CD010692.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannotti G, Caffino L, Calabrese F, Racagni G, & Fumagalli F. (2013). Dynamic modulation of basic Fibroblast Growth Factor (FGF-2) expression in the rat brain following repeated exposure to cocaine during adolescence. Psychopharmacology (Berl), 225(3), 553–560. doi: 10.1007/s00213-012-2840-8 [DOI] [PubMed] [Google Scholar]
- Guillemot F, & Zimmer C. (2011). From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron, 71(4), 574–588. doi: 10.1016/j.neuron.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Imamura T (2014). Physiological functions and underlying mechanisms of fibroblast growth factor (FGF) family members: recent findings and implications for their pharmacological application. Biol Pharm Bull, 37(7), 1081–1089. [DOI] [PubMed] [Google Scholar]
- Itoh N (2010). Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res, 342(1), 1–11. doi: 10.1007/s00441-010-1024-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Yazaki N, Tagashira S, Miyake A, Ozaki K, Minami M, Satoh M, Ohta M, et al. (1994). Rat FGF receptor-4 mRNA in the brain is expressed preferentially in the medial habenular nucleus. Brain Res Mol Brain Res, 21(3–4), 344–348. [DOI] [PubMed] [Google Scholar]
- Kalso E, Poyhia R, Onnela P, Linko K, Tigerstedt I, & Tammisto T. (1991). Intravenous morphine and oxycodone for pain after abdominal surgery. Acta Anaesthesiol Scand, 35(7), 642–646. [DOI] [PubMed] [Google Scholar]
- Koob GF, & Volkow ND. (2016). Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry, 3(8), 760–773. doi: 10.1016/S2215-0366(16)00104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xia B, Li R, Yin D, Wang Y, & Liang W. (2017). Expression of brain-derived neurotrophic factors, neurotrophin-3, and neurotrophin-4 in the nucleus accumbens during heroin dependency and withdrawal. Neuroreport, 28(11), 654–660. doi: 10.1097/WNR.0000000000000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Goni JJ, Fernandez-Montalvo J, Cacho R, & Arteaga A. (2014). Profile of addicted patients who reenter treatment programs. Subst Abus, 35(2), 176–183. doi: 10.1080/08897077.2013.826614 [DOI] [PubMed] [Google Scholar]
- Lugo RA, & Kern SE. (2004). The pharmacokinetics of oxycodone. J Pain Palliat Care Pharmacother, 18(4), 17–30. [DOI] [PubMed] [Google Scholar]
- Mars SG, Bourgois P, Karandinos G, Montero F, & Ciccarone D. (2014). “Every ‘never’ I ever said came true”: transitions from opioid pills to heroin injecting. Int J Drug Policy, 25(2), 257–266. doi: 10.1016/j.drugpo.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikaki M, Pravetoni M, Page S, Potter D, & Chartoff E. (2017). Oxycodone self-administration in male and female rats. Psychopharmacology (Berl), 234(6), 977–987. doi: 10.1007/s00213-017-4536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe SE, Cranford JA, Boyd CJ, & Teter CJ. (2007). Motives, diversion and routes of administration associated with nonmedical use of prescription opioids. Addict Behav, 32(3), 562–575. doi: 10.1016/j.addbeh.2006.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A, Hattori Y, Ohta M, & Itoh N. (1996). Rat oligodendrocytes and astrocytes preferentially express fibroblast growth factor receptor-2 and −3 mRNAs. J Neurosci Res, 45(5), 534–541. doi: [DOI] [PubMed] [Google Scholar]
- Mudo G, Bonomo A, Di Liberto V, Frinchi M, Fuxe K, & Belluardo N. (2009). The FGF-2/FGFRs neurotrophic system promotes neurogenesis in the adult brain. J Neural Transm (Vienna), 116(8), 995–1005. doi: 10.1007/s00702-009-0207-z [DOI] [PubMed] [Google Scholar]
- Nibuya M, Morinobu S, & Duman RS. (1995). Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci, 15(11), 7539–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan S, Lane-Ladd SB, Zhang L, Lundgren KH, Russell DS, Seroogy KB, & Nestler EJ. (1998). Differential regulation of neurotrophin and trk receptor mRNAs in catecholaminergic nuclei during chronic opiate treatment and withdrawal. J Neurosci, 18(24), 10700–10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes EV, Gordon M, Friedmann PD, Fishman MJ, Lee JD, Chen DT, Hu MC, Boney TY, et al. (2018). Relapse to opioid use disorder after inpatient treatment: Protective effect of injection naltrexone. J Subst Abuse Treat, 85, 49–55. doi: 10.1016/j.jsat.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkola KT, Kontinen VK, Saari TI, & Kalso EA. (2013). Does the pharmacology of oxycodone justify its increasing use as an analgesic? Trends Pharmacol Sci, 34(4), 206–214. doi: 10.1016/j.tips.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Ornell F, Hansen F, Schuch FB, Pezzini Rebelatto F, Tavares AL, Scherer JN, Valerio AG, Pechansky F, et al. (2018). Brain-derived neurotrophic factor in substance use disorders: A systematic review and meta-analysis. Drug Alcohol Depend, 193, 91–103. doi: 10.1016/j.drugalcdep.2018.08.036 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, & Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol, 4(3), 215–266. doi: 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JA, Clinton SM, Turner CA, Watson SJ, & Akil H. (2009). A new role for FGF2 as an endogenous inhibitor of anxiety. J Neurosci, 29(19), 6379–6387. doi: 10.1523/JNEUROSCI.4829-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza PV, Deminiere JM, Maccari S, Mormede P, Le Moal M, & Simon H. (1990). Individual reactivity to novelty predicts probability of amphetamine self-administration. Behav Pharmacol, 1(4), 339–345. [DOI] [PubMed] [Google Scholar]
- Porta R, Borea R, Coelho A, Khan S, Araujo A, Reclusa P, Franchina T, Van Der Steen N, et al. (2017). FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol, 113, 256–267. doi: 10.1016/j.critrevonc.2017.02.018 [DOI] [PubMed] [Google Scholar]
- Pouget ER, Fong C, & Rosenblum A. (2018). Racial/Ethnic Differences in Prevalence Trends for Heroin use and Non-Medical use of Prescription Opioids Among Entrants to Opioid Treatment Programs, 2005–2016. Subst Use Misuse, 53(2), 290–300. doi: 10.1080/10826084.2017.1334070 [DOI] [PubMed] [Google Scholar]
- Poyhia R, Olkkola KT, Seppala T, & Kalso E. (1991). The pharmacokinetics of oxycodone after intravenous injection in adults. Br J Clin Pharmacol, 32(4), 516–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyhia R, Vainio A, & Kalso E. (1993). A review of oxycodone’s clinical pharmacokinetics and pharmacodynamics. J Pain Symptom Manage, 8(2), 63–67. [DOI] [PubMed] [Google Scholar]
- Reuss B, & von Bohlen und Halbach O. (2003). Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res, 313(2), 139–157. doi: 10.1007/s00441-003-0756-7 [DOI] [PubMed] [Google Scholar]
- Riley J, Eisenberg E, Muller-Schwefe G, Drewes AM, & Arendt-Nielsen L. (2008). Oxycodone: a review of its use in the management of pain. Curr Med Res Opin, 24(1), 175–192. doi: 10.1185/030079908X253708 [DOI] [PubMed] [Google Scholar]
- Roxburgh A, Hall WD, Dobbins T, Gisev N, Burns L, Pearson S, & Degenhardt L. (2017). Trends in heroin and pharmaceutical opioid overdose deaths in Australia. Drug Alcohol Depend, 179, 291–298. doi: 10.1016/j.drugalcdep.2017.07.018 [DOI] [PubMed] [Google Scholar]
- Ruan X, Mancuso KF, & Kaye AD. (2017). Revisiting Oxycodone Analgesia: A Review and Hypothesis. Anesthesiol Clin, 35(2), e163–e174. doi: 10.1016/j.anclin.2017.01.022 [DOI] [PubMed] [Google Scholar]
- Rudd RA, Aleshire N, Zibbell JE, & Gladden RM. (2016). Increases in Drug and Opioid Overdose Deaths--United States, 2000–2014. MMWR Morb Mortal Wkly Rep, 64(50–51), 1378–1382. doi: 10.15585/mmwr.mm6450a3 [DOI] [PubMed] [Google Scholar]
- Schmidt-Hansen M, Bennett MI, Arnold S, Bromham N, & Hilgart JS. (2017). Oxycodone for cancer-related pain. Cochrane Database Syst Rev, 8, CD003870. doi: 10.1002/14651858.CD003870.pub6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, et al. (2001). Identification of a new fibroblast growth factor receptor, FGFR5. Gene, 271(2), 171–182. [DOI] [PubMed] [Google Scholar]
- Stuart GL, Shorey RC, France CR, Macfie J, Bell K, Fortner KB, Towers CV, Schkolnik P, et al. (2018). Empirical Studies Addressing the Opioid Epidemic: An Urgent Call for Research. Subst Abuse, 12, 1178221818784294. doi: 10.1177/1178221818784294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Capriles N, Flagel SB, Perez JA, Clinton SM, Watson SJ, & Akil H. (2009). Neonatal FGF2 alters cocaine self-administration in the adult rat. Pharmacol Biochem Behav, 92(1), 100–104. doi: 10.1016/j.pbb.2008.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Eren-Kocak E, Inui EG, Watson SJ, & Akil H. (2016). Dysregulated fibroblast growth factor (FGF) signaling in neurological and psychiatric disorders. Semin Cell Dev Biol, 53, 136–143. doi: 10.1016/j.semcdb.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Flagel SB, Clinton SM, Akil H, & Watson SJ. (2008). Cocaine interacts with the novelty-seeking trait to modulate FGFR1 gene expression in the rat. Neurosci Lett, 446(2–3), 105–107. doi: 10.1016/j.neulet.2008.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Hagenauer MH, Aurbach EL, Maras PM, Fournier CL, Blandino P Jr., Chauhan RB, Panksepp J, et al. (2019). Effects of early-life FGF2 on ultrasonic vocalizations (USVs) and the mu-opioid receptor in male Sprague-Dawley rats selectively-bred for differences in their response to novelty. Brain Res, 1715, 106–114. doi: 10.1016/j.brainres.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CL, Vendruscolo LF, Schlosburg JE, Hernandez DO, & Koob GF. (2015). Compulsive-like responding for opioid analgesics in rats with extended access. Neuropsychopharmacology, 40(2), 421–428. doi: 10.1038/npp.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanaka A, Johnson EM Jr., & Milbrandt J (1990). Localization of FGF receptor mRNA in the adult rat central nervous system by in situ hybridization. Neuron, 5(3), 267–281. [DOI] [PubMed] [Google Scholar]
- Woodward WR, Nishi R, Meshul CK, Williams TE, Coulombe M, & Eckenstein FP. (1992). Nuclear and cytoplasmic localization of basic fibroblast growth factor in astrocytes and CA2 hippocampal neurons. J Neurosci, 12(1), 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki N, Hosoi Y, Kawabata K, Miyake A, Minami M, Satoh M, Ohta M, Kawasaki T, et al. (1994). Differential expression patterns of mRNAs for members of the fibroblast growth factor receptor family, FGFR-1-FGFR-4, in rat brain. J Neurosci Res, 37(4), 445–452. doi: 10.1002/jnr.490370403 [DOI] [PubMed] [Google Scholar]
- Zechel S, Werner S, Unsicker K, & von Bohlen und Halbach O. (2010). Expression and functions of fibroblast growth factor 2 (FGF-2) in hippocampal formation. Neuroscientist, 16(4), 357–373. doi: 10.1177/1073858410371513 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kibaly C, Wang YJ, Xu C, Song KY, McGarrah PW, Loh HH, Liu JG, et al. (2017). Src-dependent phosphorylation of mu-opioid receptor at Tyr(336) modulates opiate withdrawal. EMBO Mol Med, 9(11), 1521–1536. doi: 10.15252/emmm.201607324 [DOI] [PMC free article] [PubMed] [Google Scholar]