Abstract
Background
Colorectal cancer (CRC) cell-derived extracellular vesicles (EVs) contribute to tumor progression. Differentially expressed long non-coding (lnc)RNAs may serve as biomarkers for CRC diagnosis. This study aimed to discuss the diagnostic value of serum EV-derived lncRNA X inactive-specific transcript (XIST) in CRC.
Material/Methods
Serum EVs were extracted and identified. Microarray analysis was performed to screen out the differentially expressed lncRNAs in serum EVs. The expression and diagnostic efficacy of the most differentially expressed lncRNA were measured. Kaplan-Meier survival analysis was performed to evaluate the association between survival time and XIST expression in EVs. The expression profile of serum EV-carried XIST in 94 CRC patients with different tumor-node-metastasis stages, lymph node metastasis, and differentiation was assessed. The serum contents of CEA, CA242, CA199, and CA153 were measured.
Results
XIST in serum EVs in CRC patients was upregulated, with greatest diagnostic value. CRC patients with higher expression of XIST in serum EVs had worse 5-year survival rates and shorter life cycles, lower differentiation, higher lymph node metastasis, and tumor-node-metastasis than patients with lower XIST expression. XIST expression in serum EVs was positively correlated with CRC marker contents.
Conclusions
XIST upregulation in serum EVs is related to CRC progression, which may be helpful to the clinical diagnosis and prognosis of CRC.
MeSH Keywords: Colorectal Neoplasms; Diagnosis; Kaplan-Meier Estimate; Prognosis; RNA, Long Noncoding
Background
Colorectal cancer (CRC) is the third most prevalent cancer and the fourth most common cause of cancer-related deaths worldwide, with approximately 1.4 million new diagnoses and nearly 700 000 deaths every year [1,2]. In the typical CRC formation model, most cancers evolve from a polyp with abnormal crypt to early adenoma, then late adenoma, and finally to CRC [3]. Metastatic tumors that spread through the blood or lymphatic vessels might occur in most forms of human cancers. Regional lymph node metastasis (LNM), tumor-node-metastasis (TNM) stage, primary location, and molecular markers of the tumors are also regarded as important prognostic factors for CRC [4,5]. Actually, most CRC develops through a series of histological, morphological, and genetic changes, which, accumulating over time, make it extremely important to screen and detect precancerous polyps before they become cancerous in individuals with an average risk of CRC [6]. The 5-year survival rate of CRC patients is 90% at stage I-II, 71% at stage III, and 14% at stage IV [7]. In addition, 40 to 50% of CRC patients develop metastatic disease, which hinders effective treatment and makes 5-year survival rate extremely low [8]. The dismal outcome of CRC has increased attention to the critical importance of early detection and novel therapeutic strategies.
As small membranous vesicles, extracellular vesicles (EVs) are found in blood, urine, and saliva, and can facilitate intercellular communication and affect the homeostasis of blood vessels by transporting microRNAs, long non-coding RNAs (lncRNAs), and mRNAs [9,10]. Additionally, circulating EVs are abundant in serum, one of the most accessible sources [11]. EVs exert versatile functions in vascular biology, embryonic development, tissue repair, liver homeostasis, cancer progression, and immune and nervous system functioning [12]. Tumor-derived EVs, as new tumor biomarkers and functional regulators of tumor development, play pivotal roles in the interactions between tumor cells and immune cells in the tumor microenvironment, particularly in CRC [13]. In recent years, it has been recognized that lncRNAs stably exist in the blood and may mirror the physiological and pathological changes of cancer patients, which has aroused great interest in the possibility of using circulating lncRNAs as minimally invasive biomarkers for cancer detection [14]. Importantly, some circulating lncRNAs in serum EVs have been reported to differentially express in CRC patients and are therefore potential biomarkers for CRC diagnosis [15]. In light of this, we extracted serum EVs from CRC patients and healthy subjects. We then carried out microarray analysis to screen out the differentially expressed lncRNAs in serum EVs to determine the relationship between serum EV-derived lncRNA and their clinical significance in CRC.
Material and Methods
Ethics statement
This study was approved and supervised by the ethics committee of Yantaishan Hospital. All procedures were strictly conducted as per the Code of Ethics. All patients signed informed consent.
Clinical sample collection
From January 2012 to June 2013, 94 CRC patients diagnosed and treated in Yantaishan Hospital were enrolled in this study, including 54 males and 40 females, with an average age of 59.22±9.64 years. All patients were followed up every 3 months for 5 years. The inclusion criteria were diagnosis with CRC by postoperative pathological examination; no treatment with radiotherapy or chemotherapy before surgery; and complete clinical data. Patients with chronic system diseases or other malignant tumors were excluded. During the same time, 41 healthy people who came to our hospital for physical examinations were selected as normal controls, including 27 males and 14 females, with an average age of 54.70±10.83 years. The diastolic and systolic blood pressures of the selected subjects were measured by sphygmomanometers, and 5 mL fasting venous blood was collected in the morning from each subject.
Isolation and identification of HMCs
Blood samples from all subjects were coagulated at room temperature and centrifuged at 3000 g for 10 min. The serum was centrifuged at 4°C at 3000 g for 10 min and then at 4°C at 10 000 g for 30 min to remove cell debris. Then 1 mL of supernatant was centrifuged (Class H, R, and S preparatory ultracentrifuges; type 50.4 Ti Rotor; Beckman Coulter, Brea, CA, USA) at 4°C for 2 h to obtain the precipitate. The precipitates were washed with phosphate buffer saline (PBS), then filtered through a 0.22-μm millipore filter, and centrifuged at 4°C at 100 000 g for 2 h to precipitate the EVs. The EV precipitates were resuspended in 100 μL PBS to analyze EVs or in 100 μL lysate buffer (Beyotime Biotechnology Co., Ltd, Shanghai, China) for protein quantification. A DC protein assay kit (5000116, Bio-Rad, Inc., Hercules, CA, USA) was used for EV quantification. Western blot analysis was used to detect the EV marker proteins CD9, CD81, and CD63, and cis-Golgi matrix protein (GM130).
Microarray analysis
Microarray analysis was performed following the steps described previously in the literature [16]. In brief, total RNA was extracted from the serum EVs from 6 CRC patients and 6 healthy subjects. Subsequently, total RNA from the EVs was extracted and 0.5 μg RNA was used for cDNA synthesis using a GeneChip 3′. In vitro transcription express kit (902789, Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, the cDNA was segmented and hybridized with a human lncRNA expression array V3.0 (AS-LNC-H-V4.0, Arraystar Inc. Rockville, MD, USA). After hybridization, the microarray was washed and scanned with a GeneChip Scanner 3000 7G System (000213, Thermo Fisher Scientific).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA in serum was acquired using the RNAiso Plus and Trizol LS Reagent (both from Takara, Otsu, Shiga, Japan). Then, formaldehyde denaturation electrophoresis was used to verify high-quality RNA for subsequent experiments. RT-PCR was conducted as per the manufacturer’s protocol using a PrimeScript RT reagent kit (Takara). Finally, the mRNA expression was quantified by standard real-time qPCR protocol via SYBR Premix Ex Taq (Takara) with glyceraldehyde-3-phosphate dehydrogenase as an internal reference. The primer sequences are shown in Table 1.
Table 1.
Primer sequences of RT-qPCR.
| Sequence | Forward | Reverse |
|---|---|---|
| XIST | CGCGGCGCCTGTGCGTGTGACA | AACUGGCCCUCAAAGUCCCGCU |
| LINC02037 | TCTATGAGGGCTTAGCTGCTTG | UAUGGCUUUCAUUCCUAUGUGA |
| LINC01987 | CCGCCGATCAGTGCACTACAGAA | CUGUGCGUGUGACAGCGGCUGA |
| LINC02041 | UCAGUGCACUACAGAACUUUGU | CCGCCGATCAGTGCACTACAGAA |
| TET-AS1 | CCGCCGAAACTGGCCCTCAAAGT | AACUGGCCCUCAAAGUCCCGCU |
| LINC00174 | CAUCAUCGUCUCAAAUGAGUCU | CGCGGCGCTATGGCTTTTCATTCC |
| GAPDH | AUCUGGAGGUAAGAAGCACUUU | TCTATGATCTGGAGGTAAGAAG |
RT-qPCR – reverse transcription quantitative polymerase chain reaction; XIST – X-inactive specific transcript; GAPDH – glyceraldehyde-3-phosphate dehydrogenase.
Measurement of CRC markers in serum
The levels of carcinoembryonic antigen (CEA), cancer antigen (CA)242, CA199 and CA153 in serum were determined by an immunoassay analyzer (Cobas E602, Roche Diagnostics, Indianapolis, Indiana, USA) in accordance with previous literature [17].
Statistical analysis
Statistical analysis was conducted with SPSS21.0 (IBM Corp. Armonk, NY, USA). Kolmogorov-Smirnov test was used to determine whether the data were normally distributed. The measurement data were exhibited in mean±standard deviation. The t test was applied for comparisons between 2 groups, while one-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test were used for multi-groups. Pearson’s correlation coefficient was used to test the correlation, and a receiver operating characteristic (ROC) curve was drawn to evaluate the predictive effects of serum EV-carried lncRNAs on CRC. A survival curve was drawn by the Kaplan-Meier method and analyzed by log rank test. The P value was obtained by a two-tailed test and P<0.05 indicated statistical differences.
Results
Clinical baseline characteristics of CRC patients
In our study, serum was harvested from 41 healthy subjects participating in physical examinations, including 27 males and 14 females, with an average age of 54.70±10.83 years. During the same period, 94 CRC patients were recruited, including 54 males and 40 females, with an average age of 59.22±9.64 years. There were no significant differences between the 2 groups in body mass index, age, and gender (Table 2). More detailed demographic data of the clinical samples are shown in Supplementary Table 1.
Table 2.
Clinical baseline characteristics of CRC patients.
| Healthy participants | CRC patients | P value | |
|---|---|---|---|
| Age (years) | 54.70±10.83 | 59.22±9.64 | 0.48 |
| Gender | 0.20 | ||
| Male | 27 | 54 | |
| Female | 14 | 40 | |
| BMI (kg/m2) | 23.65±3.36 | 21.86±2.88 | 0.22 |
CRC – colorectal cancer; BMI – body mass index.
Identification of serum EVs
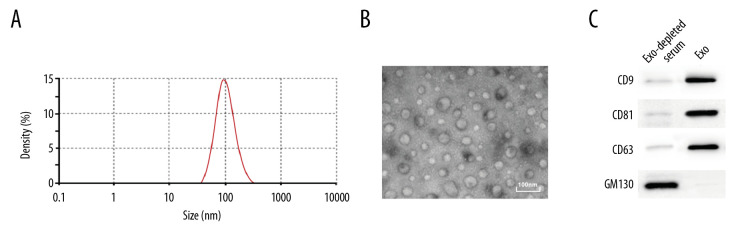
EVs were extracted from the serum of the subjects by transmission electron microscope, nanoparticle tracking, and western blot analysis. The results showed that the size of extracted EVs was 104.3±6.94 nm, which accorded with the definition of small EVs in the revised edition of MISEV 2018 [18] (Figure 1A–1C). Then, the protein content in the EVs was quantified using a DC protein assay kit, and the resulting content was 314.67±23.58 μg.
Figure 1.
Identification of serum EVs. (A) Nanoparticle tracking analysis of serum EVs, represented as size vs. concentration. (B) Transmission electron microscope observed serum EVs. Scale bar=100 nm. (C) Representative immunoblots indicated that EVs isolated from blood serum of present enrichment of EVs markers CD9, CD81, and CD63 but do not contain Golgi matrix protein GM130, as detected by western blot analysis.
LncRNA XIST is highly expressed in serum EVs in CRC patients
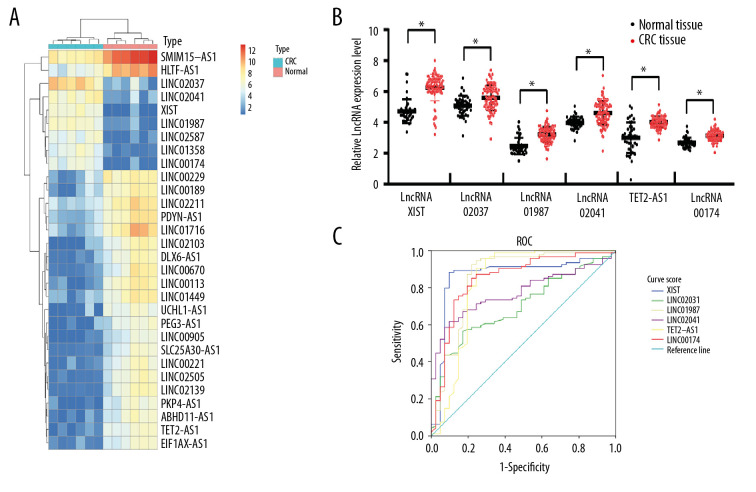
First, serum EVs from 6 CRC patients and 6 healthy subjects were analyzed by lncRNA microarray. With |LogFC >2| and P<0.05 as the screening criteria, 96 differentially expressed lncRNAs were screened out, 68 of which were upregulated, while 32 were downregulated. The heat map showed the first 30 differentially expressed lncRNAs (Figure 2A). Then the first 6 significantly upregulated lncRNAs were detected by RT-qPCR, which was consistent with the microarray analysis (Figure 2B). Subsequently, the diagnostic efficacy of lncRNA XIST, linc02037, linc01987, linc02041, TET2-AS1, and linc00174 to CRC was analyzed by an ROC curve, and the results showed that lncRNA XIST had the largest area under the curve (AUC) (Figure 2C) (all P<0.05), indicating lncRNA XIST had the best diagnostic efficacy.
Figure 2.
Upregulation of lncRNA XIST in serum EVs in CRC patients. Microarray analysis of different expressed lncRNAs in serum EVs in 6 healthy participants and 6 CRC patients. (A) Heat map of 30 differentially expressed lncRNAs. (B) RT-qPCR was performed to determine lncRNA XIST, linc02037, linc01987, linc02041, TET2-AS1, and linc00174 expression in serum EVs in 41 healthy participants and 94 CRC patients. (C) ROC analysis for expression of lncRNA XIST, linc02037, linc01987, linc02041, TET2-AS1, and linc00174. For lncRNA XIST, areas under the ROC curve: 0.864; sensitivity: 88.3%; and specificity: 90.2%; for linc02037, areas under the ROC curve: 0.698; sensitivity: 56.4%; and specificity: 82.9%; for linc01987, areas under the ROC curve: 0.856; sensitivity: 92.6%; and specificity: 78.0%; for linc02041, areas under the ROC curve: 0.774; sensitivity: 61.7%; and specificity: 90.2%; for TET2-AS1, areas under the ROC curve: 0.828; sensitivity: 95.7%; and specificity: 70.7%; for linc174, areas under the ROC curve: 0.849; sensitivity: 85.1%; and specificity: 78.0%.
High lncRNA XIST expression predicts worse prognosis
According to the lncRNA XIST expression in serum EVs, patients were assigned into a high expression group (>6.4) or a low expression group (<6.4), and Kaplan-Meier survival analysis was performed to evaluate the association between survival time and lncRNA XIST expression in serum EVs. The 5-year follow-up survey identified that patients with higher expression of lncRNA XIST (average life expectancy=34.26 months) in serum EVs had a worse 5-year survival rate and a shorter life cycle than those with lower expression of lncRNA XIST (average life expectancy=47.96 months) (Figure 3) (P<0.05).
Figure 3.
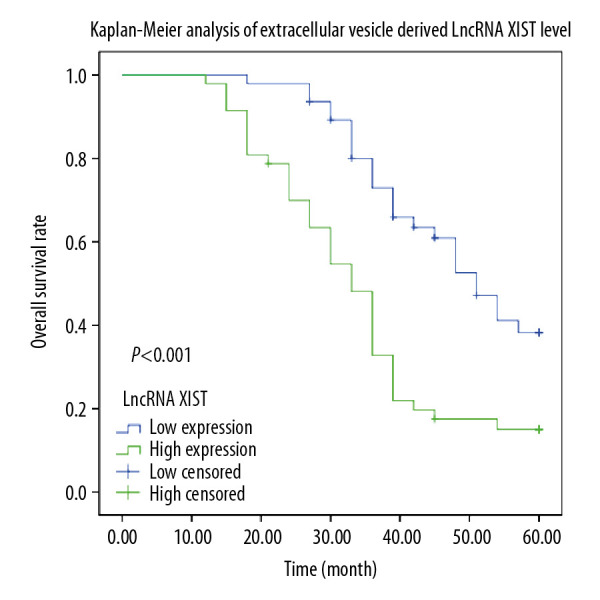
Kaplan-Meier analysis was performed to overview the OS rate and median survival time among CRC patients allocated by median expression of serum EV-carried lncRNA XIST.
LncRNA XIST expression is related to the malignant degree of CRC patients
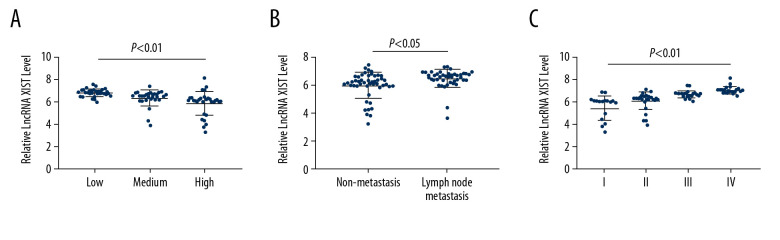
To further analyze the relationship between lncRNA XIST expression in serum EVs and CRC, the expression profile of serum EV-carried lncRNA XIST in the 94 CRC patients with different TNM stages, LNM, and differentiation degree was assessed. It was revealed that CRC patients with higher expression of serum EV-carried lncRNA XIST had lower differentiation, higher LNM status, and TNM stage than those with lower lncRNA XIST expression (all P<0.05) (Figure 4A–4C).
Figure 4.
Serum EV-carried lncRNA XIST is associated with CRC malignance. We analyzed the correlation between CRC patients’ tumor differentiation level (A), LNM status (B) and TNM stage (C) with serum EV-carried lncRNA XIST expression. Each spot represents an individual’s data. One-way ANOVA and Tukey’s multiple comparison test were used for significance determination.
Positive correlation between lncRNA XIST expression and serum indicators in CRC patients
To determine the diagnostic efficacy of serum EV-carried lncRNA XIST to CRC, CEA, CA242, CA199, and CA153, the marker contents in the serum of the 94 CRC patients were measured. Results of a Pearson correlation analysis showed that lncRNA XIST expression in serum EVs was positively correlated with CRC marker contents (all P<0.05) (Figure 5A–5D).
Figure 5.
Serum EV-carried lncRNA XIST level is associated with CRC serum biomarker. We analyzed the correlation between CRC patients’ serum CEA content (r=0.806) (A), CA242 content (r=0.627) (B), CA199 content (r=0.254) (C), and CA242 content (r=0.706) (D) with serum EV-carried lncRNA XIST expression. Each spot represents an individual’s data. Pearson correlation analysis was used for significance and co-relation determination.
Discussion
Although fecal occult blood testing and colonoscopy examination in the past decades has moderately increased the detection rate of early stage tumors, occult blood testing has relatively poor sensitivity, and colonoscopy is invasive and costly [14]. Meanwhile, many functional molecules are stably encapsulated in EVs, which are increasingly considered as promising biomarkers for cancer detection [19]. Also, biomarker candidates presented in EVs have been reported in pancreatic cancer and CRC [20,21]. Based on this background, we aimed to search for novel CRC biomarkers in EVs from the serum of patients.
Many studies have revealed that XIST is highly expressed in CRC. For example, Chen et al. noticed that XIST was overexpressed in CRC cell lines and tissues, and high XIST expression was correlated with adverse overall survival (OS) in CRC patients [22]. Zhang et al. found high XIST expression was correlated with larger tumor size and higher TNM stages and predicted poor progression-free survival and OS of CRC patients [23]. To summarize, these researchers reached a consensus that lncRNA XIST is an oncogene in CRC and serves as a potential biomarker to predict prognosis in CRC patients. However, to the best of our knowledge, no studies have investigated the significant effects of serum EV-carried XIST in CRC until now. The present study showed that XIST in serum EVs in CRC patients was upregulated. Circulating EVs expressing tumor-specific proteins are potential biomarkers for various cancers [24]. Many clinical studies indicate that tumor-derived EVs and their contents have great potential as diagnostic tools for cancers [25,26]. In addition, serum EVs from CRC patients were shown to promote cellular mobility and epithelial-mesenchymal transition [27]. CRC cell-derived EVs suppressed the proliferation of T cells and contributed to CRC progression [28]. Interestingly, a prior study demonstrated that circulating lncRNAs in serum/plasma EVs could serve as biomarkers for CRC diagnosis [15,29]. Further, METTL14 inhibits proliferation and metastasis of CRC by downregulating oncogenic lncRNA XIST [30]. But there is little research investigating serum EV-carried XIST in CRC, which indicates, to some degree, the innovation of our study.
In any organ system, TNM staging, LNM, and distant metastasis detection are tricky problems for pathologists and clinicians [31]. By analyzing the relationship between XIST expression and CRC clinicopathological features, we found that CRC patients with higher XIST expression in serum EVs had a worse 5-year survival rate and shorter life cycle, lower differentiation, higher LNM status, and TNM stage than patients having lower XIST expression in serum EVs. From the results of their Kaplan-Meier survival analysis, Xiao et al. found that CRC patients with high serum XIST expression exhibited poor OS rates, chemoresponse, and recurrence-free survival rates, and thus identified XIST as an independent prognostic factor for CRC patients receiving 5-fluorouracil treatment [32]. Additionally, cancer cell-derived EVs activate fibroblasts, which degrade the extracellular matrix and induce oncogenic cytokines, contributing to further tumor metastasis and ultimately promoting cancer malignancy [9]. Several lncRNAs enriched in CRC cell-derived EVs are of prime significance in cancer progression and diagnosis [33]. Similarly, exosomal colorectal neoplasia differentially expressed-h in the serum of CRC patients was strongly correlated with LNM, distant metastasis, and OS rates, thus acting as a novel noninvasive serum-based biomarker for diagnosis and prognosis of CRC [34].
In addition, we assessed the correlation between XIST expression and the specific markers of CRC patients to explore their potential for prognostic prediction. Data supported that serum EV-carried XIST in CRC patients was positively correlated with CEA, CA242, CA199, and CA153 contents. Serum EVs of right-sided CRC may promote metastasis by upregulating extracellular matrix-related proteins, and thus potentially serve as diagnosis and prognosis biomarkers in CRC [27]. CEA is one of the most widely used and reliable blood CRC markers today [19]. CEA levels are correlated with the metastasis and recurrence of CRC after surgery [35,36]. CA242 has high specificity and sensitivity in CRC patients, and the combination of CEA and CA242 is even more sensitive in CRC diagnosis [37]. The increased preoperative CA199 level indicates a low survival rate of CRC [38]. Increased CEA, CA199, and CA242 contents are associated with TNM stages and differentiation, and direct impact on metastasis, recurrence status, and survival time of CRC patients at stages II and III [37]. This is a preliminary finding and larger studies with proper testing and validation cohorts will allow for better understanding of the utility of this biomarker in CRC.
Conclusions
Serum EV-transported lncRNA XIST has diagnostic and predictive effects on CRC. Preventing EV secretion by tumor cells or removing tumor cell-derived EVs is a potential therapeutic strategy worthy of further study. In addition, an in-depth understanding of the physiology of EVs during cancer development may pave the way for the design of diagnostic tools and therapeutic strategies for CRC. Further studies including large clinical samples and diverse ethnic populations are required to confirm the usefulness of lncRNAs as noninvasive markers in CRC patients.
Supplementary Data
Supplementary Table 1.
Demographic characteristics of the CRC patients.
| No. | Type | Stage | M | D | TS | Gender | Age | OS (months) | Status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CRC | I | Y | High | <3 cm | Male | 66 | 18 | Dead |
| 2 | CRC | II | N | Medium | <3 cm | Female | 51 | 24 | Dead |
| 3 | CRC | III | Y | Low | >3 cm | Male | 63 | 36 | Dead |
| 4 | CRC | IV | Y | Low | <3 cm | Male | 38 | 60 | Live |
| 5 | CRC | II | N | High | <3 cm | Male | 60 | 60 | Live |
| 6 | CRC | IV | Y | Low | <3 cm | Female | 53 | 30 | Dead |
| 7 | CRC | IV | N | Low | <3 cm | Female | 68 | 42 | Dead |
| 8 | CRC | II | Y | Low | <3 cm | Male | 52 | 39 | Dead |
| 9 | CRC | IV | N | Low | <3 cm | Male | 56 | 18 | Dead |
| 10 | CRC | IV | Y | High | <3 cm | Female | 62 | 12 | Dead |
| 11 | CRC | IV | Y | Low | >3 cm | Female | 66 | 18 | Dead |
| 12 | CRC | IV | Y | Low | <3 cm | Male | 71 | 15 | Dead |
| 13 | CRC | IV | Y | High | >3 cm | Male | 61 | 36 | Dead |
| 14 | CRC | IV | N | Low | >3 cm | Male | 71 | 42 | Live |
| 15 | CRC | IV | Y | Low | <3 cm | Female | 67 | 30 | Dead |
| 16 | CRC | III | Y | Low | <3 cm | Female | 66 | 39 | Dead |
| 17 | CRC | IV | N | Medium | <3 cm | Male | 66 | 33 | Dead |
| 18 | CRC | IV | Y | Medium | <3 cm | Male | 71 | 36 | Dead |
| 19 | CRC | IV | Y | Low | >3 cm | Male | 58 | 45 | Live |
| 20 | CRC | III | Y | Low | <3 cm | Female | 64 | 30 | Dead |
| 21 | CRC | IV | Y | Medium | >3 cm | Female | 58 | 15 | Dead |
| 22 | CRC | IV | Y | Medium | <3 cm | Female | 63 | 18 | Dead |
| 23 | CRC | IV | N | Low | >3 cm | Female | 72 | 33 | Dead |
| 24 | CRC | I | Y | Low | >3 cm | Female | 65 | 36 | Dead |
| 25 | CRC | IV | N | Low | <3 cm | Male | 60 | 60 | Live |
| 26 | CRC | IV | Y | Medium | >3 cm | Male | 60 | 24 | Dead |
| 27 | CRC | IV | N | Medium | <3 cm | Male | 61 | 24 | Dead |
| 28 | CRC | III | Y | Medium | >3 cm | Female | 66 | 33 | Dead |
| 29 | CRC | III | N | Medium | >3 cm | Male | 71 | 39 | Dead |
| 30 | CRC | II | Y | Medium | <3 cm | Female | 58 | 24 | Dead |
| 31 | CRC | III | Y | Low | >3 cm | Male | 66 | 60 | Live |
| 32 | CRC | III | N | Low | <3 cm | Female | 66 | 27 | Dead |
| 33 | CRC | IV | Y | Low | >3 cm | Female | 65 | 39 | Dead |
| 34 | CRC | IV | Y | Low | >3 cm | Female | 64 | 36 | Dead |
| 35 | CRC | III | N | Medium | <3 cm | Male | 73 | 39 | Dead |
| 36 | CRC | III | Y | Medium | >3 cm | Female | 73 | 27 | Dead |
| 37 | CRC | III | Y | Medium | <3 cm | Female | 66 | 45 | Dead |
| 38 | CRC | III | Y | Low | >3 cm | Male | 68 | 54 | Dead |
| 39 | CRC | III | N | Medium | >3 cm | Female | 67 | 30 | Dead |
| 40 | CRC | III | Y | Medium | <3 cm | Male | 63 | 60 | Live |
| 41 | CRC | III | Y | Medium | <3 cm | Female | 61 | 36 | Dead |
| 42 | CRC | III | Y | Low | >3 cm | Male | 67 | 18 | Dead |
| 43 | CRC | III | N | Medium | >3 cm | Female | 68 | 15 | Dead |
| 44 | CRC | II | Y | Medium | <3 cm | Female | 47 | 27 | Dead |
| 45 | CRC | III | Y | Medium | <3 cm | Male | 70 | 21 | Dead |
| 46 | CRC | II | Y | Medium | >3 cm | Female | 67 | 36 | Dead |
| 47 | CRC | III | Y | Medium | >3 cm | Male | 73 | 21 | Dead |
| 48 | CRC | III | Y | Low | <3 cm | Male | 40 | 39 | Dead |
| 49 | CRC | III | Y | Medium | <3 cm | Female | 55 | 54 | Dead |
| 50 | CRC | III | N | Medium | >3 cm | Female | 53 | 51 | Live |
| 51 | CRC | II | N | Low | >3 cm | Male | 45 | 30 | Dead |
| 52 | CRC | II | N | Medium | >3 cm | Male | 65 | 33 | Dead |
| 53 | CRC | II | N | Medium | >3 cm | Male | 39 | 18 | Dead |
| 54 | CRC | II | N | High | >3 cm | Male | 47 | 27 | Dead |
| 55 | CRC | II | Y | Medium | >3 cm | Female | 72 | 60 | Live |
| 56 | CRC | II | N | High | >3 cm | Male | 60 | 27 | Live |
| 57 | CRC | III | N | High | <3 cm | Male | 49 | 36 | Dead |
| 58 | CRC | II | N | High | <3 cm | Male | 54 | 33 | Dead |
| 59 | CRC | II | N | High | <3 cm | Male | 41 | 60 | Live |
| 60 | CRC | II | N | High | >3 cm | Male | 70 | 51 | Dead |
| 61 | CRC | II | Y | High | >3 cm | Female | 45 | 48 | Dead |
| 62 | CRC | II | N | Low | <3 cm | Male | 44 | 45 | Dead |
| 63 | CRC | II | N | Low | <3 cm | Male | 56 | 45 | Dead |
| 64 | CRC | II | N | Low | <3 cm | Male | 68 | 33 | Dead |
| 65 | CRC | II | Y | Low | >3 cm | Female | 62 | 39 | Dead |
| 66 | CRC | IV | N | High | <3 cm | Female | 47 | 27 | Dead |
| 67 | CRC | IV | N | High | <3 cm | Female | 43 | 60 | Live |
| 68 | CRC | II | N | High | >3 cm | Male | 54 | 60 | Live |
| 69 | CRC | III | N | Medium | >3 cm | Male | 62 | 60 | Live |
| 70 | CRC | II | N | Medium | <3 cm | Male | 65 | 33 | Dead |
| 71 | CRC | II | Y | High | >3 cm | Male | 43 | 45 | Dead |
| 72 | CRC | II | Y | High | <3 cm | Male | 50 | 48 | Dead |
| 73 | CRC | I | N | High | >3 cm | Male | 65 | 51 | Live |
| 74 | CRC | I | N | Low | <3 cm | Male | 67 | 54 | Dead |
| 75 | CRC | I | Y | High | <3 cm | Male | 45 | 57 | Dead |
| 76 | CRC | IV | N | High | >3 cm | Female | 60 | 51 | Dead |
| 77 | CRC | IV | N | High | >3 cm | Male | 51 | 60 | Live |
| 78 | CRC | I | N | Low | >3 cm | Male | 53 | 48 | Dead |
| 79 | CRC | I | N | High | <3 cm | Male | 58 | 60 | Live |
| 80 | CRC | I | Y | High | >3 cm | Male | 50 | 39 | Dead |
| 81 | CRC | III | N | High | >3 cm | Female | 68 | 27 | Live |
| 82 | CRC | I | Y | Low | >3 cm | Female | 53 | 36 | Dead |
| 83 | CRC | I | N | High | <3 cm | Male | 46 | 30 | Live |
| 84 | CRC | III | N | Low | <3 cm | Male | 68 | 42 | Dead |
| 85 | CRC | I | N | Low | <3 cm | Male | 41 | 30 | Dead |
| 86 | CRC | III | N | High | <3 cm | Male | 45 | 57 | Dead |
| 87 | CRC | III | N | Medium | >3 cm | Female | 64 | 42 | Dead |
| 88 | CRC | IV | N | Medium | <3 cm | Male | 73 | 42 | Live |
| 89 | CRC | I | N | High | >3 cm | Male | 43 | 39 | Dead |
| 90 | CRC | IV | Y | High | <3 cm | Female | 58 | 45 | Dead |
| 91 | CRC | I | N | Low | >3 cm | Female | 62 | 42 | Dead |
| 92 | CRC | III | N | High | >3 cm | Male | 47 | 33 | Live |
| 93 | CRC | I | Y | High | >3 cm | Male | 65 | 36 | Dead |
| 94 | CRC | I | N | High | >3 cm | Female | 68 | 30 | Dead |
M – stands for metastasis; D – stands for differentiation; TS – stands for tumor size; CRC – colorectal cancer.
Abbreviations
- ANOVA
analysis of variance
- AUC
area under the curve
- BMI
body mass index
- CA
cancer antigen
- CEA
carcinoembryonic antigen
- CRC
colorectal cancer
- EVs
extracellular vesicles
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- lncRNAs
long non-coding RNAs
- LNM
lymph node metastasis
- OS
overall survival
- PBS
phosphate buffer saline
- ROC
receiver operating characteristic
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- TNM
tumor-node-metastasis
- XIST
X inactive-specific transcript
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Emambux S, Tachon G, Junca A, Tougeron D. Results and challenges of immune checkpoint inhibitors in colorectal cancer. Expert Opin Biol Ther. 2018;18(5):561–73. doi: 10.1080/14712598.2018.1445222. [DOI] [PubMed] [Google Scholar]
- 2.Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23(28):5086–96. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107(3):dju427. doi: 10.1093/jnci/dju427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Hao L, Wang L, et al. Elevated IGFIR expression regulating VEGF and VEGF-C predicts lymph node metastasis in human colorectal cancer. BMC Cancer. 2010;10:184. doi: 10.1186/1471-2407-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–76. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worm Orntoft M-B. Review of blood-based colorectal cancer screening: How far are circulating cell-free DNA methylation markers from clinical implementation? Clin Colorectal Cancer. 2018;17(2):e415–33. doi: 10.1016/j.clcc.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Miao F, Huang R, et al. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J Exp Clin Cancer Res. 2018;37(1):22. doi: 10.1186/s13046-018-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126(4):1163–72. doi: 10.1172/JCI81130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomura S. Extracellular vesicles and blood diseases. Int J Hematol. 2017;105(4):392–405. doi: 10.1007/s12185-017-2180-x. [DOI] [PubMed] [Google Scholar]
- 11.Cavallari C, Ranghino A, Tapparo M, et al. Serum-derived extracellular vesicles (EVs) impact on vascular remodeling and prevent muscle damage in acute hind limb ischemia. Sci Rep. 2017;7(1):8180. doi: 10.1038/s41598-017-08250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanez-Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning S, Danielson KM. The immunomodulatory role of tumor-derived extracellular vesicles in colorectal cancer. Immunol Cell Biol. 2018 doi: 10.1111/imcb.12038. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Du L, Yang X, et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142(11):2291–301. doi: 10.1007/s00432-016-2238-9. [DOI] [PubMed] [Google Scholar]
- 15.Dong L, Lin W, Qi P, et al. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1158–66. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Song Y, Liu W, et al. The noncoding RNA linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral disc degeneration through inhibiting ADAMTS5 expression. Clin Sci (Lond) 2017;131(10):965–79. doi: 10.1042/CS20160918. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Zhang M, Zhang H, et al. Exosomal long noncoding RNA lnc-GNAQ-6: 1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252–57. doi: 10.1016/j.cca.2019.10.047. [DOI] [PubMed] [Google Scholar]
- 18.Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1535750. 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiromizu T, Kume H, Ishida M, et al. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci Rep. 2017;7(1):12782. doi: 10.1038/s41598-017-13092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–81. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen DL, Chen LZ, Lu YX, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8(8):e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XT, Pan SX, Wang AH, et al. Long non-coding RNA (lncRNA) X-inactive specific transcript (XIST) plays a critical role in predicting clinical prognosis and progression of colorectal cancer. Med Sci Monit. 2019;25:6429–35. doi: 10.12659/MSM.915329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, Xu K, Zhou B, et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J Extracell Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1750202. 1750202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32(4):983–97. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–90. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 27.Zhong ME, Chen Y, Xiao Y, et al. Serum extracellular vesicles contain SPARC and LRG1 as biomarkers of colon cancer and differ by tumour primary location. EBioMedicine. 2019;50:211–23. doi: 10.1016/j.ebiom.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada N, Kuranaga Y, Kumazaki M, et al. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-beta1-mediated suppression. Oncotarget. 2016;7(19):27033–43. doi: 10.18632/oncotarget.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu D, Zhan Y, Zhu K, et al. Plasma exosomal long non-coding RNAs serve as biomarkers for early detection of colorectal cancer. Cell Physiol Biochem. 2018;51(6):2704–15. doi: 10.1159/000495961. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin M, Frankel WL. Lymph node metastasis in colorectal cancer. Surg Oncol Clin N Am. 2018;27(2):401–12. doi: 10.1016/j.soc.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Y, Yurievich UA, Yosypovych SV. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget. 2017;8(47):83171–82. doi: 10.18632/oncotarget.20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, Xu R, Ji H, et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep. 2016;6:38397. doi: 10.1038/srep38397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–63. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal C, Meropol NJ, Punt CJ, et al. Relationship among circulating tumor cells, CEA and overall survival in patients with metastatic colorectal cancer. Ann Oncol. 2013;24(2):420–28. doi: 10.1093/annonc/mds336. [DOI] [PubMed] [Google Scholar]
- 36.Primrose JN, Perera R, Gray A, et al. Effect of 3 to 5 years of scheduled CEA and CT follow-up to detect recurrence of colorectal cancer: The FACS randomized clinical trial. JAMA. 2014;311(3):263–70. doi: 10.1001/jama.2013.285718. [DOI] [PubMed] [Google Scholar]
- 37.You W, Sheng N, Yan L, et al. The difference in prognosis of stage II and III colorectal cancer based on preoperative serum tumor markers. J Cancer. 2019;10(16):3757–66. doi: 10.7150/jca.31660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stiksma J, Grootendorst DC, van der Linden PW. CA 19-9 as a marker in addition to CEA to monitor colorectal cancer. Clin Colorectal Cancer. 2014;13(4):239–44. doi: 10.1016/j.clcc.2014.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Demographic characteristics of the CRC patients.
| No. | Type | Stage | M | D | TS | Gender | Age | OS (months) | Status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CRC | I | Y | High | <3 cm | Male | 66 | 18 | Dead |
| 2 | CRC | II | N | Medium | <3 cm | Female | 51 | 24 | Dead |
| 3 | CRC | III | Y | Low | >3 cm | Male | 63 | 36 | Dead |
| 4 | CRC | IV | Y | Low | <3 cm | Male | 38 | 60 | Live |
| 5 | CRC | II | N | High | <3 cm | Male | 60 | 60 | Live |
| 6 | CRC | IV | Y | Low | <3 cm | Female | 53 | 30 | Dead |
| 7 | CRC | IV | N | Low | <3 cm | Female | 68 | 42 | Dead |
| 8 | CRC | II | Y | Low | <3 cm | Male | 52 | 39 | Dead |
| 9 | CRC | IV | N | Low | <3 cm | Male | 56 | 18 | Dead |
| 10 | CRC | IV | Y | High | <3 cm | Female | 62 | 12 | Dead |
| 11 | CRC | IV | Y | Low | >3 cm | Female | 66 | 18 | Dead |
| 12 | CRC | IV | Y | Low | <3 cm | Male | 71 | 15 | Dead |
| 13 | CRC | IV | Y | High | >3 cm | Male | 61 | 36 | Dead |
| 14 | CRC | IV | N | Low | >3 cm | Male | 71 | 42 | Live |
| 15 | CRC | IV | Y | Low | <3 cm | Female | 67 | 30 | Dead |
| 16 | CRC | III | Y | Low | <3 cm | Female | 66 | 39 | Dead |
| 17 | CRC | IV | N | Medium | <3 cm | Male | 66 | 33 | Dead |
| 18 | CRC | IV | Y | Medium | <3 cm | Male | 71 | 36 | Dead |
| 19 | CRC | IV | Y | Low | >3 cm | Male | 58 | 45 | Live |
| 20 | CRC | III | Y | Low | <3 cm | Female | 64 | 30 | Dead |
| 21 | CRC | IV | Y | Medium | >3 cm | Female | 58 | 15 | Dead |
| 22 | CRC | IV | Y | Medium | <3 cm | Female | 63 | 18 | Dead |
| 23 | CRC | IV | N | Low | >3 cm | Female | 72 | 33 | Dead |
| 24 | CRC | I | Y | Low | >3 cm | Female | 65 | 36 | Dead |
| 25 | CRC | IV | N | Low | <3 cm | Male | 60 | 60 | Live |
| 26 | CRC | IV | Y | Medium | >3 cm | Male | 60 | 24 | Dead |
| 27 | CRC | IV | N | Medium | <3 cm | Male | 61 | 24 | Dead |
| 28 | CRC | III | Y | Medium | >3 cm | Female | 66 | 33 | Dead |
| 29 | CRC | III | N | Medium | >3 cm | Male | 71 | 39 | Dead |
| 30 | CRC | II | Y | Medium | <3 cm | Female | 58 | 24 | Dead |
| 31 | CRC | III | Y | Low | >3 cm | Male | 66 | 60 | Live |
| 32 | CRC | III | N | Low | <3 cm | Female | 66 | 27 | Dead |
| 33 | CRC | IV | Y | Low | >3 cm | Female | 65 | 39 | Dead |
| 34 | CRC | IV | Y | Low | >3 cm | Female | 64 | 36 | Dead |
| 35 | CRC | III | N | Medium | <3 cm | Male | 73 | 39 | Dead |
| 36 | CRC | III | Y | Medium | >3 cm | Female | 73 | 27 | Dead |
| 37 | CRC | III | Y | Medium | <3 cm | Female | 66 | 45 | Dead |
| 38 | CRC | III | Y | Low | >3 cm | Male | 68 | 54 | Dead |
| 39 | CRC | III | N | Medium | >3 cm | Female | 67 | 30 | Dead |
| 40 | CRC | III | Y | Medium | <3 cm | Male | 63 | 60 | Live |
| 41 | CRC | III | Y | Medium | <3 cm | Female | 61 | 36 | Dead |
| 42 | CRC | III | Y | Low | >3 cm | Male | 67 | 18 | Dead |
| 43 | CRC | III | N | Medium | >3 cm | Female | 68 | 15 | Dead |
| 44 | CRC | II | Y | Medium | <3 cm | Female | 47 | 27 | Dead |
| 45 | CRC | III | Y | Medium | <3 cm | Male | 70 | 21 | Dead |
| 46 | CRC | II | Y | Medium | >3 cm | Female | 67 | 36 | Dead |
| 47 | CRC | III | Y | Medium | >3 cm | Male | 73 | 21 | Dead |
| 48 | CRC | III | Y | Low | <3 cm | Male | 40 | 39 | Dead |
| 49 | CRC | III | Y | Medium | <3 cm | Female | 55 | 54 | Dead |
| 50 | CRC | III | N | Medium | >3 cm | Female | 53 | 51 | Live |
| 51 | CRC | II | N | Low | >3 cm | Male | 45 | 30 | Dead |
| 52 | CRC | II | N | Medium | >3 cm | Male | 65 | 33 | Dead |
| 53 | CRC | II | N | Medium | >3 cm | Male | 39 | 18 | Dead |
| 54 | CRC | II | N | High | >3 cm | Male | 47 | 27 | Dead |
| 55 | CRC | II | Y | Medium | >3 cm | Female | 72 | 60 | Live |
| 56 | CRC | II | N | High | >3 cm | Male | 60 | 27 | Live |
| 57 | CRC | III | N | High | <3 cm | Male | 49 | 36 | Dead |
| 58 | CRC | II | N | High | <3 cm | Male | 54 | 33 | Dead |
| 59 | CRC | II | N | High | <3 cm | Male | 41 | 60 | Live |
| 60 | CRC | II | N | High | >3 cm | Male | 70 | 51 | Dead |
| 61 | CRC | II | Y | High | >3 cm | Female | 45 | 48 | Dead |
| 62 | CRC | II | N | Low | <3 cm | Male | 44 | 45 | Dead |
| 63 | CRC | II | N | Low | <3 cm | Male | 56 | 45 | Dead |
| 64 | CRC | II | N | Low | <3 cm | Male | 68 | 33 | Dead |
| 65 | CRC | II | Y | Low | >3 cm | Female | 62 | 39 | Dead |
| 66 | CRC | IV | N | High | <3 cm | Female | 47 | 27 | Dead |
| 67 | CRC | IV | N | High | <3 cm | Female | 43 | 60 | Live |
| 68 | CRC | II | N | High | >3 cm | Male | 54 | 60 | Live |
| 69 | CRC | III | N | Medium | >3 cm | Male | 62 | 60 | Live |
| 70 | CRC | II | N | Medium | <3 cm | Male | 65 | 33 | Dead |
| 71 | CRC | II | Y | High | >3 cm | Male | 43 | 45 | Dead |
| 72 | CRC | II | Y | High | <3 cm | Male | 50 | 48 | Dead |
| 73 | CRC | I | N | High | >3 cm | Male | 65 | 51 | Live |
| 74 | CRC | I | N | Low | <3 cm | Male | 67 | 54 | Dead |
| 75 | CRC | I | Y | High | <3 cm | Male | 45 | 57 | Dead |
| 76 | CRC | IV | N | High | >3 cm | Female | 60 | 51 | Dead |
| 77 | CRC | IV | N | High | >3 cm | Male | 51 | 60 | Live |
| 78 | CRC | I | N | Low | >3 cm | Male | 53 | 48 | Dead |
| 79 | CRC | I | N | High | <3 cm | Male | 58 | 60 | Live |
| 80 | CRC | I | Y | High | >3 cm | Male | 50 | 39 | Dead |
| 81 | CRC | III | N | High | >3 cm | Female | 68 | 27 | Live |
| 82 | CRC | I | Y | Low | >3 cm | Female | 53 | 36 | Dead |
| 83 | CRC | I | N | High | <3 cm | Male | 46 | 30 | Live |
| 84 | CRC | III | N | Low | <3 cm | Male | 68 | 42 | Dead |
| 85 | CRC | I | N | Low | <3 cm | Male | 41 | 30 | Dead |
| 86 | CRC | III | N | High | <3 cm | Male | 45 | 57 | Dead |
| 87 | CRC | III | N | Medium | >3 cm | Female | 64 | 42 | Dead |
| 88 | CRC | IV | N | Medium | <3 cm | Male | 73 | 42 | Live |
| 89 | CRC | I | N | High | >3 cm | Male | 43 | 39 | Dead |
| 90 | CRC | IV | Y | High | <3 cm | Female | 58 | 45 | Dead |
| 91 | CRC | I | N | Low | >3 cm | Female | 62 | 42 | Dead |
| 92 | CRC | III | N | High | >3 cm | Male | 47 | 33 | Live |
| 93 | CRC | I | Y | High | >3 cm | Male | 65 | 36 | Dead |
| 94 | CRC | I | N | High | >3 cm | Female | 68 | 30 | Dead |
M – stands for metastasis; D – stands for differentiation; TS – stands for tumor size; CRC – colorectal cancer.