Abstract
The –1 ribosomal frameshifting is vital for the translation of the open reading frame (ORF)1b in SARS-CoV-2. The products of ORF1b participate in viral replication. Therefore, changing the frameshift frequency reduces the survival of the virus. This study aimed to successfully develop a toolkit for screening antiviral drugs. Finally, the FDA-approved drug library was screened, revealing that ivacaftor and (–)-Huperzine A worked well in changing the –1 ribosomal frameshifting of SARS-CoV-2 in vitro.
Keywords: Biomedical engineering, Microbial biotechnology, Protein engineering, Peptides, Virology, Molecular biology, Drug screen, SARS-Cov-2, -1 ribosomal frameshifting
Biomedical engineering; Microbial biotechnology; Protein engineering; Peptides; Virology; Molecular biology; Drug screen; SARS-Cov-2; -1 ribosomal frameshifting.
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by a coronavirus called SARS-CoV-2. Options to prevent or control SARS-CoV-2 include vaccines, neutralizing antibodies, interferon therapies, oligonucleotide-based therapies, and small-molecule drugs. The screening system based on a living virus is important for the development of small-molecule drugs. However, the use of the drug screening system based on a living virus is limited because of the high risk of infection. The coronavirus SARS-CoV-2 is a positive single-stranded RNA with a 30-kb genome. Like other coronaviruses, two thirds of the genomic RNA is occupied by two long open reading frames (ORF1a and ORF1b) [1]. After entry into the cell, ORF1a is translated and cleaved into 11 nonstructural proteins to disrupt the host innate immune response, and ORF1b is translated and cleaved into 15 replicase enzymes involved in the transcription and replication of the virus genome. When the translation of ORF1a is complete, ribosomes bypass the stop codon through a -1 programmed ribosomal frameshifting (–1 PRF) strategy to synthesize ORF1b [2].
Absolute values for –1 PRF frequency in the wild-type SARS-CoV in vitro are 15 ± 3% [3]. Alterations in –1 PRF efficiencies, rather than turning −1 PRF completely on or off, disturbs the replication of the virus by changing the abundance of viral particles, leading to the rapid disappearance of the virus [4, 5, 6]. The –1 PRF apparatus of SARS-CoV comprises three cis-acting mRNA elements: the slippery site (U_UUA_AAC), three stem-looped mRNA pseudoknots, and a short spacer sequence. Besides, recent studies have found an attenuator hairpin upstream of the sliding sequence [2, 3, 5, 7]. Several attempts have been made to reduce the –1 PRF frequency of SARS-CoV, such as using interference RNAs [8] or small-molecule compounds [9]. The sequence alignment of SARS-CoV-2 with SARS-CoV shows that the slippery site and the spacer sequence are identical, with one nucleotide difference in the mRNA pseudoknot. However, only 60% similarity is found in upstream attenuator hairpin between these two viruses. Therefore, effective –1 PRF-inhibiting drugs in SARS may not be effective in SARS-CoV-2.
Two tools were developed in this study to select effective antiviral compounds based on luciferase and fluorescent protein, respectively. The FDA-approved drug library was screened, revealing six candidate drugs to change –1 PRF in SARS-CoV-2 in vitro.
2. Results
2.1. Construction of luciferase-based drug screening tool
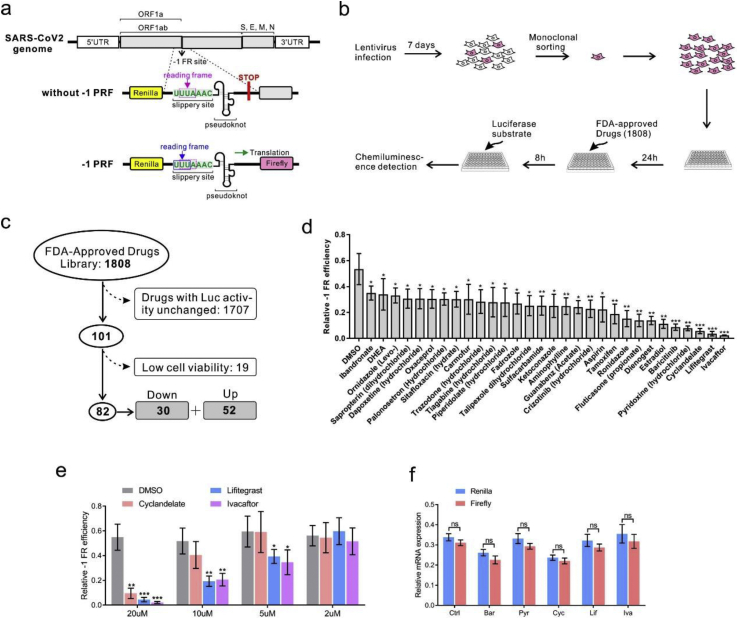
This study was performed to develop tools used in drug screening. A reporter system was built by putting the –1 PRF apparatus of SARS-CoV-2 between Renilla and firefly luciferase genes (Figure 1a). These reporter elements were then introduced into the lentivirus backbone. The lentivirus particles were collected and used to infect the human lung epithelial cell line 16HBE and human embryonic kidney cell HEK293. One week after the lentivirus infection, the cells were resuspended and sorted into single-cell clones by fluorescence-activated cell sorting (FACS). The cells were cultured for weeks with puromycin, and the firefly luciferase–expressing cell clone was used for the following tests. Under normal conditions, the firefly luciferase mRNA was translated because of –1 PRF.
Figure 1.
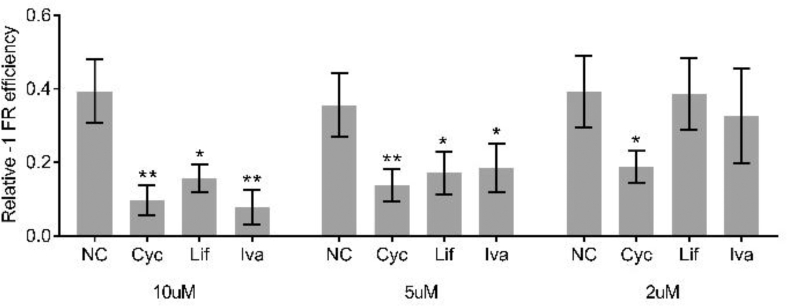
Screening of FDA-approved drugs using –1 PRF luciferase assay. (a) Design of the report system. The slippery site was inserted into a dual luciferase reporter gene plasmid. A stop codon was put in front of the firefly luciferase coding sequence; if –1 PRF occurred, the stop codon was read through because the reading frame changed. (b) Workflow of drug screening. 16HBE cell lines were infected and plated in 96-well plates. After treatment with FDA-approved drugs at a concentration of 20μM for 8 h, the luminescence signal was detected. (c) In the 1808 drugs tested, ineffective drugs or drugs that caused low cell viability were excluded. (d) Relative quantification of –1 PRF frequency in 16HBE by comparing the expression of firefly luciferase with Renilla luciferase. (e) Decreases in –1 PRF frequency by drugs at different concentrations. (f) Relative mRNA expression of Renilla and firefly luciferase in 16HBE cells (mean ± SD; ns: no significant difference; n = 5 batches; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by the two-tailed Student t test). Abbreviations: Cyc, Cyclandelate; Ery, erythromycin; Hup, (–)-huperzine A; IVA, ivacaftor; Lif, lifitegrast; Tri, trimethobenzamide.
2.2. High-throughput drug screening of FDA-approved drugs
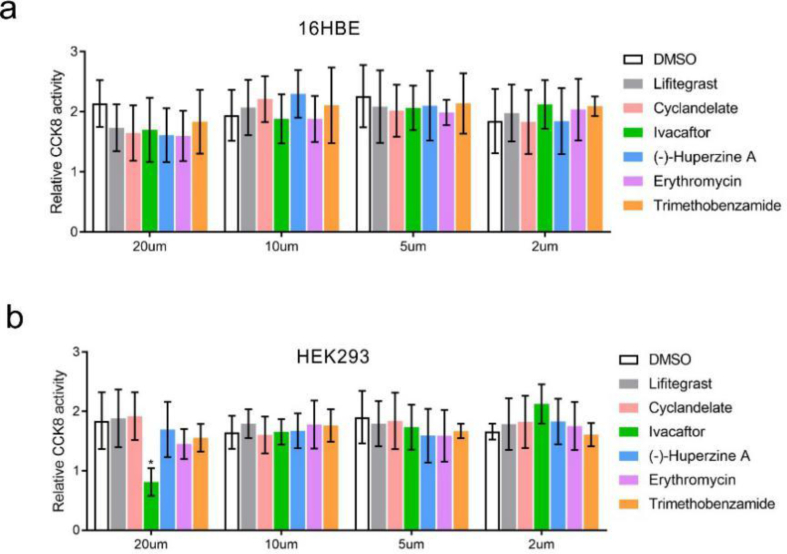
Next, FDA-approved drugs (1808 in total) were tested using this system; if –1 PRF was blocked, the ratio of firefly/Renilla declined (Figure 1b). The residual firefly luciferase, both mRNA and protein, could interfere with the experimental results. Half-life of the luciferase mRNA could be as long as 5 h in vitro [10], and the decay rate of the luciferase protein by proteasome has been estimated to be 3 h in mammalian cells [11]. Thus, we treated cells with FDA-approved drugs for 8 h to ensure the synthesized firefly luciferase being completely degraded. In the first step, 1707 drugs were ruled out because the firefly/Renilla ratio remained unchanged. Next, 19 drugs were ruled out for low cell viability. Finally, 82 drugs were chosen (Figure 1c). Of these drugs, 30 resulted in a decrease in the proportion of –1 PRF. The most effective inhibitory drugs were ivacaftor, lifitegrast, and cyclandelate. The –1 PRF levels were reduced to 4%, 7%, and 11% of the mock group, respectively (Figure 1d). Further, 20μM, but not 10μM, of ivacaftor, caused a decrease in cell viability in HEK293 (Figure S1a and S1b). The drug was diluted with a concentration gradient of 20μM, 10μM, 5μM, and 2μM to eliminate the influence of cell vitality on –1 PRF. Lifitegrast and ivacaftor remained effective in reducing –1 PRF in 16HBE at a concentration of 2μM (Figure 1e). When the aforementioned experiments were repeated in the HEK293 cell line, cyclandelate displayed an inhibitory effect even at the concentration of 2μM (Figure S2). The viral −1 PRF signal acted as a cis-acting mRNA-destabilizing element [12]. To further analyze the effect of these drugs on RNA stability, the mRNA levels of firefly luciferase and Renilla luciferase were compared. The transcription of these genes was not affected by drug treatment (Figure 1f).
2.3. Construction of drug screening tool based on fluorescent protein
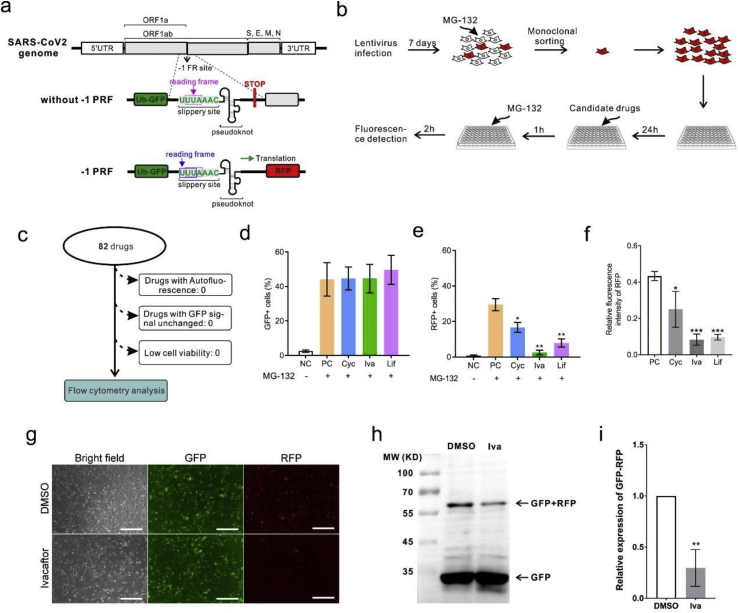
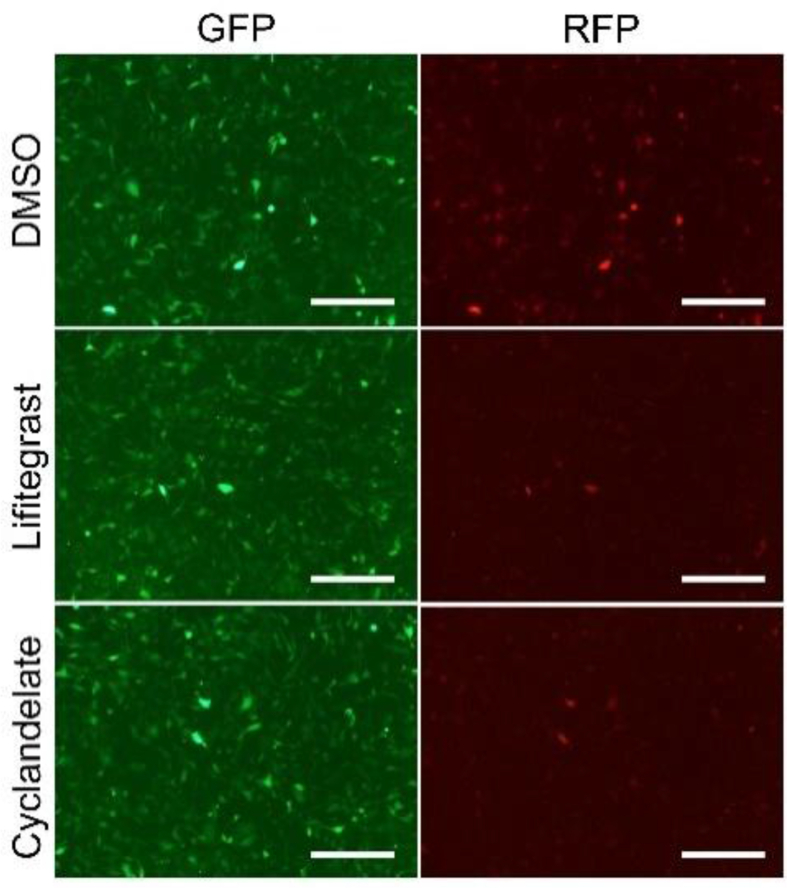
In the luciferase reporter system, the levels of luciferases are also affected by the activity of the protease. A dual fluorescent protein reporter assay system was built to exclude the probability of protease activity instability caused by different drugs. In this system, luciferase was changed with ubiquitin fluorescent proteins (Figure 2a). Without a protease inhibitor, the fluorescent proteins were degraded immediately after synthesis. When treated with the protease inhibitor MG-132, the fluorescent proteins accumulated and were detected using a fluorescence microscope or flow cytometry. However, if small-molecule compounds inhibited the –1 PRF activity, the RFP downstream of the slippery site was undetectable. Like the luciferase system, 16HBE cells were infected with a fluorescent protein–expressing lentivirus for 1 week. The cells were treated with MG-132 and sorted into monoclonal cells. Since fluorescent protein is fused with ubiquitin, it is rapidly degraded by proteasome after synthesis. Therefore, there is no fluorescent protein in the cells before the drug treatment. For drug screening, the cells were treated with the 82 candidate drugs for 1 h, followed by treatment with MG-132 for 2 h to inhibit the proteasome activity. Expression of the fluorescent protein could be detected by fluorescence microscope 2 h after MG-132 treatment (Figure 2b). All drugs were tested with no autofluorescence or cell viability decline. Next, the expression of GFP and RFP was measured using flow cytometry. The proportion of RFP-expressing cells significantly decreased, consistent with luciferase assay results (Figures 2d–2f). The expression of GFP and RFP was observed under a fluorescence microscope. Before adding MG-132, most of the fluorescent proteins were degraded because of ubiquitin fusion. After MG132 treatment for 1 h, the fluorescence intensity increased significantly (Figures 2d and 2e). The imaging analysis showed that ivacaftor, lifitegrast, and cyclandelate treatment significantly inhibited the expression of RFP but not that of GFP (Figure 2f, g and S3). Since RFP was integrated into GFP, the molecular weights of RFP and GFP were all close to 25 kDa. Therefore, the positive band of RFP should be more than 50 KDa in Western blot analysis. The Ub–GFP–slippery site–RFP fusion protein band was weakened in ivacaftor-treated 16-HBE cells (Figures 2h and 2i), consistent with previous findings. In conclusion, the –1 PRF-decreasing activity of ivacaftor, lifitegrast, and cyclandelate was protease independent.
Figure 2.
Reconfirmation of the effective drugs using a dual fluorescent protein reporter assay system. (a) Design of the reporter system. Renilla and firefly luciferase were changed with ubiquitin fluorescent proteins GFP and RFP, respectively. (b) Workflow of drug screening. 16HBE cells were infected and plated in 96-well plates after treatment with mock, ivacaftor, lifitegrast, and cyclandelate at a concentration of 20μM for 1 h. Then, the cells were treated with MG132 for 2 h, followed by fluorescence microscopy and flow cytometry. (c) Eighty-two candidate drugs were tested for autofluorescence, unchanged RFP signal, or low cell viability. (d–f) Functions of the three inhibitory drugs were detected by flow cytometry (mean ± SD; n = 5 tests; ∗P < 0.05; ∗∗P < 0.01; by the two-tailed Student t test). (g) Microscopic image of fluorescent protein expression (scale bar = 200 μm). (h) Western blot analysis of GFP and GFP–RFP fusion protein. (i) Densitometric quantification of GFP–RFP fusion protein levels in western blot normalized against GFP in 16HBE cells (mean ± SD; n = 3 tests; ∗P < 0.05 by the two-tailed Student t test). Abbreviations: Cyc, cyclandelate; Ery, erythromycin; Hup, (–)-huperzine A; IVA, ivacaftor; Lif, lifitegrast; Tri, trimethobenzamide.
2.4. Drugs that increased the –1 PRF frequency found by the luciferase screening system
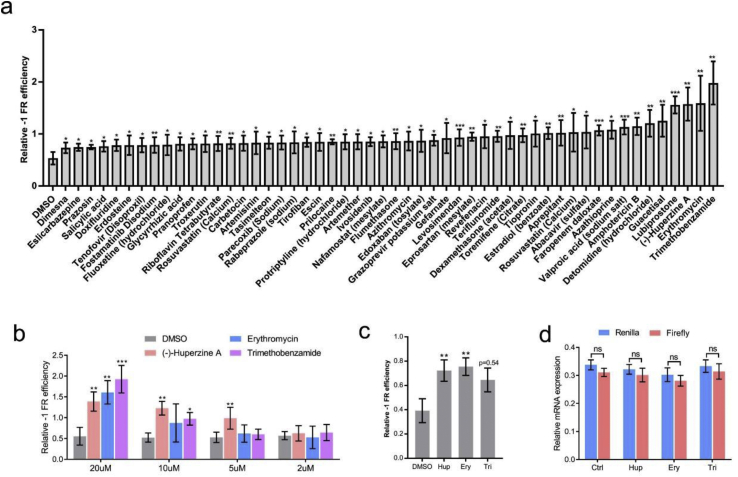
A total of 52 drugs increased the –1 PRF frequency (Figure 3a). Of these, (–)-huperzine A, erythromycin, and trimethobenzamide were the three most effective drugs (Figure 3a). The gradient drug concentration treatment showed that (–)-huperzine A was effective in increasing the –1 PRF activity even at a concentration of 5 μM (Figure 3b). Previous studies found that the cytopathic effects of SARS-CoV-2 were cell type dependent [13]. The –1 PRF efficiency in HEK293 cells was tested to further determine the effects of (–)-huperzine A, erythromycin, and trimethobenzamide among different cell lines. The –1 PRF activity increased in (–)-huperzine A– and erythromycin-treated cells, but not in trimethobenzamide-treated cells (Figure 3c). The mRNA expression was not changed by drug treatment (Figure 3d). These results suggested that the –1 PRF frequency of SARS-CoV-2 was sensitive to given chemical reagents; also, the effect of these drugs was cell type dependent.
Figure 3.
Drugs that increased the –1 PRF frequencies. (a) List of drugs that increased the –1 PRF activity in 16HBE cells. (b) Increase in the –1 PRF frequency by drugs at different concentrations. (c) Frequencies of –1 PRF were tested in HEK293 cells. (d) The mRNA level of firefly and Renilla luciferase was detected by RT-PCR (mean ± SD; n = 3 tests; ns: no significant difference; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by two-tailed Student t test). Abbreviations: Ctrl, control; Ery, erythromycin; Hup, (–)-huperzine A; Tri, trimethobenzamide.
2.5. Reconfirmation of the –1 PRF enhancing activity using the dual fluorescent protein screening tool
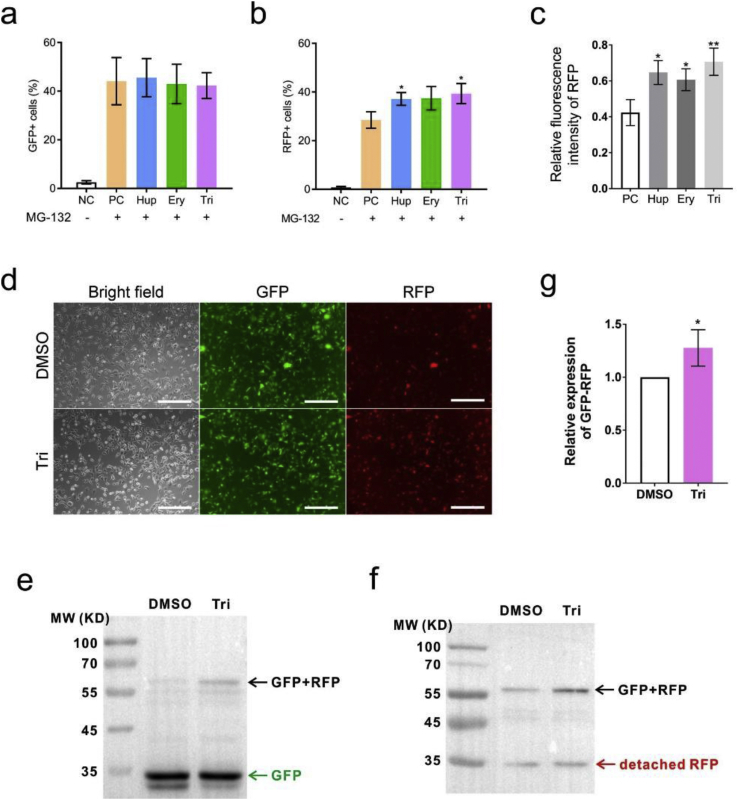
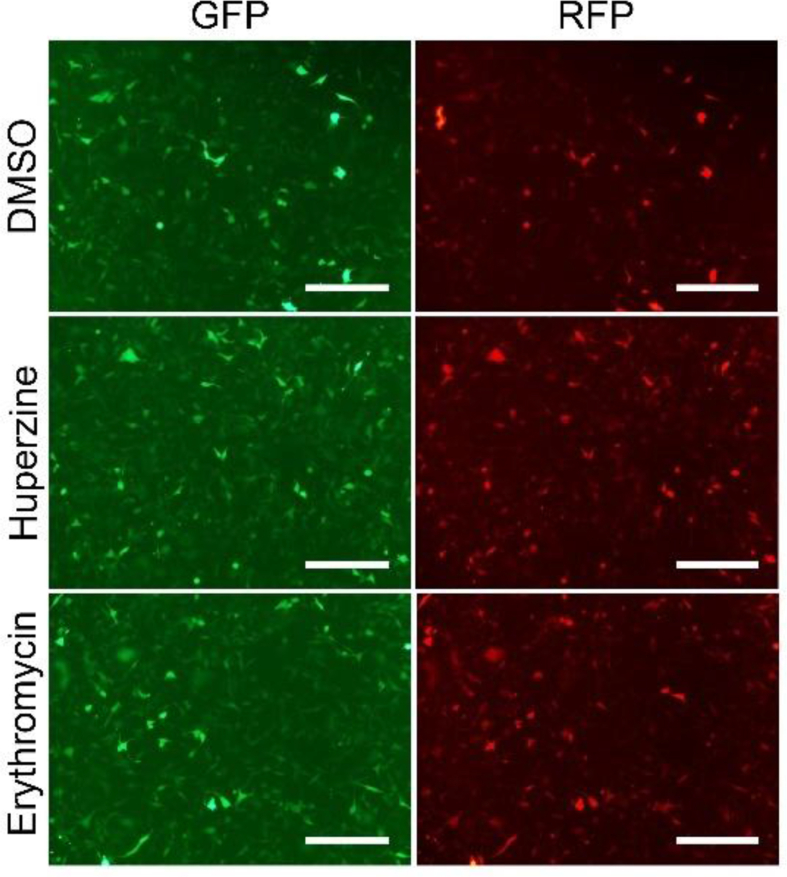
The –1 PRF frequency was further assayed using the protease-free system. The number of GFP-positive cells was not changed. (–)-Huperzine A and trimethobenzamide, but not erythromycin, increased the number of RFP-expressing cells (Figures 4a and 4b). The –1 PRF activity was measured using the ratio of RFP- and GFP-positive cells. These drugs increased the –1 PRF activity, from 60% in mock-treated cells to about 80% in drug-treated cells (Figures 4a and 4b). The detection of the fluorescence intensity using flow cytometry showed that all three drugs significantly increased the expression of RFP (Figure 4c). The effect of trimethobenzamide treatment on the expression of red fluorescent protein was analyzed using a fluorescence microscope to further testify the validity of these results. The RFP signal increased significantly, while the signal of GFP did not change (Figure 4d and S4). The detection of the expression of GFP–RFP fusion protein using Western blot analysis showed that the expression of RFP increased in trimethobenzamide-treated 16-HBE cells (Figures 4e–4g).
Figure 4.
Drugs that increased the –1 PRF frequencies. (a–c) Functions of the the inhibitory drugs were detected by flow cytometry (mean ± SD; n = 5 tests; ∗P < 0.05; ∗∗P < 0.01; by two-tailed Student t test). (d) Microscopic image of fluorescent protein expression (scale bar = 200 um). (e) Western blot analysis of GFP and GFP–RFP fusion protein. (f) Western blot analysis of RFP and GFP–RFP fusion protein. (g) Densitometric quantification of GFP–RFP fusion protein levels in western blot normalized against GFP in 16HBE cells (mean ± SD; n = 3 tests; ∗P < 0.05 by the two-tailed Student t test). Abbreviations: Ery, Erythromycin; Hup, (–)-huperzine A; NC, negative control; PC, positive control; Tri, trimethobenzamide.
3. Discussion
This study demonstrated the feasibility of in vitro drug screening using an anti-SARS-CoV-2 drug screening toolkit. The monoclonal cell line expressing firefly luciferase or red fluorescent protein under the control of –1 PRF of SARS-CoV-2 element could be used in the high-throughput drug screening. A number of drug screening efforts based on live viruses were made [14, 15, 16, 17]. Toremifene (citrate) and tamoxifen, two drugs used to change –1 PRF efficacy in this study, were reported to inhibit the replication of living SARS-CoV-2 virus [18]. In the previous work of H. J. Park group, compound 43 has been identified as SARS-CoV inhibitor through a virtual screening. 2-methylthiazole and anilin were introduced to each side of 1,4-diazepane scaffold to give the compound 43 [9]. There was no structural similarity between these drugs and compound 43, suggests that the targets of these drugs may be different. SARS-CoV-2 is highly infectious and can spread through droplets or even aerosols [19]. A high risk of infection in high-throughput drug screening limited the development of drug screening. The toolkit developed in this study recruited only a small part of the virus genome as the “sensor,” which had no infection risk at all and hence could be used in the laboratory of biosafety class II. After screening, the candidate drugs were then further confirmed using a live virus, thus greatly reducing the probability of infection.
The –1 PRF activity was different among cell types. Besides, the antiviral effect of the drugs varied a lot among different cell line–based screening tools. Therefore, it is necessary to verify the candidate drugs in different systems. In addition, the luciferase–substrate reaction is a protease activity-dependent process. If the protease activity is interfered by some drugs, the screening results are biased. In addition, the screening results based on fluorescent proteins were interfered by disturbed pH or protein folding. In this study, luciferase and fluorescent protein detections were combined to improve the reliability of drug screening.
The activity of –1 PRF is affected by many factors. Specific RNA structure stimulates or inhibits the –1 PRF frequency. Small-molecule inhibitors against SARS-CoV have been designed by destroying the pseudoknot structure [9]. The candidate drugs used in this study might alter the pseudoknot structure by disrupting the base-pairing interactions, thus changing the frequency of –1 PRF. Therefore, drugs such as ivacaftor, lifitegrast, and trimethobenzamide, but not eythromycin, was effective in changing the –1 PRF both in 16HBE and HEK293 cells. Besides, mutations in frameshift factors, translation factors, and ribosomal proteins could alter frameshift efficiency [20]. A recent study found that shiftless was a cellar inhibitor of programmed-1 PRF of HIV [21]. Therefore, these drugs might also affect the activity of these proteins or directly affect the expression, translation, post-translational modification, and stability of these proteins. It is worth noting that in the luciferase based assay, cyclandelate only inhibited the - 1 PRF in HEK293 cells, while ivacaftor and lifegrade were both effective in HEK293 and 16-HBE cells. This may be due to the different repressive mechanisms between these compounds.
In conclusion, a mutually confirming antiviral screening toolkit was successfully constructed in this study. Using this kit, the FDA-approved drugs were screened, revealing that ivacaftor, lifitegrast, and cyclandelate are the three most promising candidates in blocking the –1 PRF, (–)-huperzine A, erythromycin, and trimethobenzamide were three candidates in increasing the –1 PRF activity in vitro. The results demonstrated that the –1 PRF activity of SARS-CoV-2 was changed by some drugs. However, further studies are required to confirm the effectiveness of drugs on live SARS-CoV-2 virus or use these drugs as a backbone to develop more effective agents.
4. Materials and methods
4.1. Reagents
The FDA-approved drug library was purchased from MedChemExpress (HY-L022). MG132 was purchased from Selleck (S2619). Polyethylenimine, branched (PEI) was purchased from Sigma (408727). Anti-GFP antibody (66002-1-Ig) was purchased from Proteintech Group, and anti-RFP antibody (T0055) was purchased from Affinity Biosciences.
4.2. Plasmids
pMD2.G was a gift from Didier Trono (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID: Addgene_12259), and psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID: Addgene_12260). pLVX-IRES-Puro was purchased from Clontech Laboratories (632183). Ubiquitin, AcGFP, and mCherry coding sequence were synthesized by Beijing TsingKe Biotechnology and cloned into pLVX-IRES-Puro between EcoRI and BamHI. The -299 and +296 sequence around “–1 frameshift site,” firefly luciferase, and Renilla luciferase coding sequence were synthesized by Beijing TsingKe Biotechnology and cloned into pLVX-IRES-Puro between EcoRI and BamHI.
4.3. Cell culture
16HBE, HEK293 and HEK293T cells were from ATCC. All cells were maintained in complete medium containing Dulbecco's modified Eagle's medium (DMEM, Biological Industries 01-052-1A), 10% fetal bovine serum (FBS, Biological Industries 04-007-1A), and antibiotic–antimycotic (1×, Gibco 15240112). All cell lines were ruled out of mycoplasma contamination using PCR. The cells were plated at a density of 1 × 104 per well of a 96-well plate 24 h before drug screening.
4.4. Lentivirus packaging and enriching
All cells were maintained in the incubator containing 5% CO2. HEK293T cells were plated at a density of 1 × 106 per well of a 10-cm culture dish 16 h before transfection. Then, 20 μg backbone plasmid, 10 μg Plpax2, and 5 μg pMD2.G were dissolved in 1 mL of DMEM containing 70 μg PEI without FBS and antibiotic. Four hours after transfection, DMEM medium was changed with complete medium. The cell culture supernatant was collected 48 and 72 h after transfection. Lentivirus particles were enriched using 10-kDa ultrafiltration centrifuge tube (Millipore, UFC905096). The culture medium containing lentivirus particles was aliquoted into 200 μL and stored at -80 °C.
4.5. Flow cytometry cell analysis and sorting
16HBE and HEK293 cells were infected with lentivirus for 7 days. The cells were digested with trypsin (0.25 mg/mL, Gibco 25300054) and centrifuged in complete medium (500 g, 3 min, room temperature). For flow cytometry cell analysis, 16HBE cells with stable expression of ub-GFP-SARS-COV-2 slippery site –RFP were treated with FDA-approved drugs for 1 h and MG-132 for 2 h. The expression of fluorescent protein was determined using a fluorescence microscope (Olympus, IX73) at room temperature. For cell sorting, all cells were filtered using a 70-μm filter, analyzed, and sorted with FACSAria SORP (BD Biosciences) and FlowJo program.
4.6. Western blot analysis
The cells were collected and lysed in cell lysis buffer for Western blot analysis and IP (Beyotime Biotechnology, P0013) with the addition of protease inhibitor PMSF (MedChemExpress, HY-B0496). Antibodies against GFP (Proteintech Group, 50430-2-AP) and RFP (Affinity Biosciences, T0055) were used at a 1:1000 dilution in 5% BSA at 4 °C overnight. HRP-linked anti-mouse IgG (Cell Signaling Technology, 7076S) and HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074S) were used as secondary antibodies at a 1:5000 dilution at 37 °C for 1 h. The membranes were explored using chemiluminescence instrument and quantified using Image J software.
4.7. Luciferase assays
16HBE (1 × 104 cells/well) cells with stable expression of luciferase and frameshift sequence of SARS-COV-2 in 96-well plates were treated with FDA-approved drug library (2–20μM). Three hours after drug treatment, the culture supernatant was discarded and the cells were lysed using 100 μL of lysis buffer for 20 min at room temperature. The activities of firefly and Renilla luciferase were measured using dual luciferase reporter gene assay lit (Beyotime Biotechnology, RG027). Data were normalized by calculating the ratio between firefly and Renilla luciferase activities.
4.8. Statistical analysis
All statistical tests were performed using GraphPad Prism software v. 8.4.2. (GraphPad Software). Data were represented as mean ± standard deviation. All statistical analyses were two-sided, and P values ≤ 0.05 (95% confidence interval) were considered statistically significant (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Declarations
Author contribution statement
X. Bai: Conceived and designed the experiments; Wrote the paper.
X. Fu: Conceived and designed the experiments.
Y. Chen and H. Tao: Performed the experiments.
S. Shen, L. Li and Z. Miao: Analyzed and interpreted the data.
H. Zhang and Y. Jia: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Ministry of Science and Technology of the People's Republic of China (2018YFC1003502) and West China Hospital, Sichuan University (Z20192008).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank Xuemei Chen, Li Chai, Huifang Li, Yi Zhong, Tao Su, Yan Wang, Shasha Wu, Dachao Mou, Jingjing Ran, and Pingxian Liu (West China Hospital) for experimental assistance, and Ting Yang (West China Second Hospital) for writing assistance, professor Tao Yang from West China Hospital for discussion on the structure of compounds.
Appendix.
Figure S1.
Cell activities at different drug concentrations. 16HBE (a) and HEK293 (b) cells were treated with different drugs (2–20μM) for 8 h. Cell activity was tested using CCK8 (Mean ± SD; n = 9 tests; ∗P < 0.05 by the two-tailed Student t test).
Figure S2.
Detection of drug efficiency in HEK293 cells. HEK293 cells were treated with ivacaftor, lifitegrast, and cyclandelate in 2–10 μM for 8 h. Drug efficiency was detected by adding the luciferase and substrate to the cell lysate (mean ± SD; n = 3 tests; ∗P < 0.05, ∗∗P < 0.01 by the two-tailed Student t test). Abbreviations: Cyc, cyclandelate; Ery, erythromycin; Hup, (–)-huperzine A; IVA, ivacaftor; Lif, lifitegrast; Tri, trimethobenzamide.2
Figure S3.
The -1 PRF inhibitory effect of drugs in 16-HBE cells. 16-HBE cells were treated with Lifitegrast and Cyclandelate in 2μM for 1 h, followed by treated with MG-132 for 2 h. Drug efficiency was detected by fluorescent microscope (bars = 200 μm).3
Figure S4.
The -1 PRF promoting effect of drugs in 16-HBE cells. 16-HBE cells were treated with Huperzine A and Erythromycin in 2μM for 1 h, followed by treated with MG-132 for 2 h. Drug efficiency was detected by fluorescent microscope (bars = 200 μm).4
References
- 1.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly J.A., Olson A.N., Neupane K., Munshi S., Emeterio J.S., Pollack L., Woodside M.T., Dinman J.D. bioRxiv; 2020. Structural and Functional Conservation of the Programmed -1 Ribosomal Frameshift Signal of SARS-CoV-2. 2020.2003.2013.991083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinman J.D. Mechanisms and implications of programmed translational frameshifting. Wiley interdisciplinary reviews. RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su M.-C., Chang C.-T., Chu C.-H., Tsai C.-H., Chang K.-Y. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for −1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 2005;33:4265–4275. doi: 10.1093/nar/gki731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tommy R.T. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infect. Disord. - Drug Targets. 2009;9:223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- 7.Plant E.P., Rakauskaitė R., Taylor D.R., Dinman J.D. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn D.-G., Lee W., Choi J.-K., Kim S.-J., Plant E.P. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 2011;91:1–10. doi: 10.1016/j.antiviral.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S.-J., Kim Y.-G., Park H.-J. Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 10.Gurdon J.B., Javed K., Vodnala M., Garrett N. Long-term association of a transcription factor with its chromatin binding site can stabilize gene expression and cell fate commitment. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:15075–15084. doi: 10.1073/pnas.2000467117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson J.F., Hayes L.S., Lloyd D.B. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 12.Plant E.P., Wang P., Jacobs J.L., Dinman J.D. A programmed –1 ribosomal frameshift signal can function as a cis-acting mRNA destabilizing element. Nucleic Acids Res. 2004;32:784–790. doi: 10.1093/nar/gkh256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N., Zhang D., Wang W., Li X., Yang B. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmar M., Lee J.S., Whig K., Segrist E., Li M. bioRxiv; 2020. Drug Repurposing Screens Reveal FDA Approved Drugs Active against SARS-Cov-2. 2020. 2006.2019.161042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N. bioRxiv; 2020. A Large-Scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. 2020.2004.2016.044016. [Google Scholar]
- 16.Yuan S., Chan J.F.W., Chik K.K.H., Chan C.C.Y., Tsang J.O.L. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol. Res. 2020;159:104960. doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia G., Sharma A., Ramaiah A., Sen C., Kohn D. bioRxiv; 2020. Antiviral Drug Screen of Kinase Inhibitors Identifies Cellular Signaling Pathways Critical for SARS-CoV-2 Replication. 2020.2006.2024.150326. [Google Scholar]
- 18.Weston S., Coleman C.M., Haupt R., Logue J., Matthews K. bioRxiv; 2020. Broad Anti-coronaviral Activity of FDA Approved Drugs against SARS-CoV-2 in Vitro and SARS-CoV in Vivo. 2020.2003.2025.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-CoV-2. Science (New York, N.Y.) 2020;368:1422–1424. doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- 20.Barry J.K., Miller W.A. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Xuan Y., Han Y., Ding X., Ye K. Regulation of HIV-1 gag-pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell. 2019;176:625–635. doi: 10.1016/j.cell.2018.12.030. e614. [DOI] [PMC free article] [PubMed] [Google Scholar]