Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an emergent RNA virus that spread around the planet in about 4 months. The consequences of this rapid dispersion are under investigation. In this work, we analyzed thousands of genomes and protein sequences from Africa, America, Asia, Europe, and Oceania. We provide statistically significant evidence that SARS-CoV-2 phylogeny is spatially structured. Remarkably, the virus phylogeographic patterns were correlated with ancestral amino acidic substitutions, suggesting that such mutations emerged along colonization events. We hypothesize that geographic structuring is the result of founder effects occurring as a consequence of, and local evolution occurring after, long-distance dispersion. Based on previous studies, the possibility that this could significantly affect the virus biology is not remote.
Keywords: SARS-CoV-2, COVID19, Phylogeny, Phylogenetic structure
Highlights
-
•
SARS-CoV-2 spatial variation is significant.
-
•
Variation affects both RNA and protein sequences.
-
•
Vicariant events can explain spatial variation.
-
•
Long-distance dispersion could boost viral diversification.
1. Introduction
A cluster of acute atypical pneumonia syndrome cases of unknown etiology was reported late December 2019 in China. Rapidly, metagenomic RNA sequencing of bronchoalveolar lavage fluid showed that the etiological agent was a new human coronavirus (Wu et al., 2020; Zhou et al., 2020), now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; Gorbalenya et al., 2020). Human coronaviruses are well known to produce mild to moderate upper-respiratory tract illnesses. However, severe acute respiratory syndrome coronavirus (SARS-CoV) responsible for a 2002–2003 epidemic, Middle East respiratory syndrome coronavirus (MERS-CoV) responsible for a 2012 outbreak and the novel SARS-CoV-2, cause severe syndromes with high fatality rates (10%, 36% and 4%, respectively) (Cui et al., 2019; WHO, 2020). SARS-CoV-2 is closely related to SARS-CoV, the prototype of the species Severe acute respiratory syndrome-related coronavirus. Thus, it has been classified in the said species, which belongs to the subgenus Sarbecovirus of the genus Betacoronavirus (Gorbalenya et al., 2020). SARS-CoV-2 and SARS-CoV are ~79% identical at the genetic level and share important features as the same cell receptor and immunopathogenic mechanism.
SARS-CoV-2 has dispersed to hundreds of countries, causing millions of infections. This spread consequences on the virus evolution are still unclear. Recent studies suggested that SARS-CoV-2 genotypes are unevenly distributed (Forster et al., 2020; Rambaut et al., 2020). However, quantitative, and phenotypic assessments are still lacking. When relatedness between spatially coexisting lineages is greater than expected, their distribution is said to be phylogenetically structured (Webb et al., 2002). One of the consequences of structuring is spatially close sequences being more similar to each other than expected by chance (Fig. S1). The phenomenon can be assessed by comparing measured phylogenetic distances with those expected under no structuring, which can be accomplished by Monte Carlo simulations. Here, we describe an analysis of SARS-CoV-2 genomes collected from the beginning of the pandemic to April 25, 2020. We observed an uneven distribution of genetic and amino acidic variants, and a statistically significant spatial phylogenetic structure. Notably, ancestral amino acidic substitutions were highly fitted to the virus phylogeny and geographic patterns, strongly suggesting that long-distance dispersion can facilitate the establishment of otherwise rare, and/or the emergence of new, viral phenotypes.
2. Materials and methods
Dataset and Operational Taxonomic Units delimitation. Structure analyses require to determine the abundance patterns of a set of operational taxonomic units (Webb et al., 2002). We studied 8612 genomes obtained from GISAID’s EpiCoV™ presenting less than 1% non-DNA characters (high coverage only option) (Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017). The corresponding sequences and metadata are described in Table S1. As a first step previous to sequence analyses, we performed extra quality controls. The sequences were first inspected for the presence of ambiguous and undetermined positions by the base. freq() function from the Ape package (Paradis and Schliep, 2018). The selected data (genomes with no non-DNA characters) were aligned with Mafft (Katoh and Toh, 2008) (details below) and the diversity along the obtained alignment was assessed by inspecting the entropy at each aligned position using the HDMD package (https://CRAN.R-project.org/package=HDMD). The alignment positions homologous to nucleotides 1–77 and 29,804–29,903 of the isolate Wuhan-Hu-1 (NC_045512.2) presented too high diversities compared to the rest of the genome (Fig. S2). Thus, these regions were masked-out in posterior analyses. Furthermore, the sequences presenting missing data after and before positions 77 and 28,804, respectively, and rare internal gaps, visually identified using Jalview (Waterhouse et al., 2009), were dismissed. The final, high-quality dataset had 4,333 genomes. Pairwise distances were obtained from the aligned sequences using R string comparison tools, and used to identify sequence variants (SV), which were treated as the operational taxonomic units. Shortly, we made an R script that takes a sequence (the query) from the dataset, determines which sequences are identical to it, put these together in an identity cluster, and record the size (number of sequences, interpreted as SV abundance) of the cluster. After this, the script starts a new iteration. The process was repeated until all the sequences in the dataset were processed.
SVs distribution assessment. If SV abundances were the same everywhere, the global frequencies would be good approximations of the SV abundances expected in any sample, regardless of where it comes from. This is to say that the probability of detecting a given SV i, in some area A j, P(V i |A j ), should be the same for all j: P(V i |A 1 ) = P(V i |A 2 ) = … = P(V i |A N ) = P(V i ), where N is the number of sampled areas. Thus, we modeled the probability of observing a given SV, P(V i ), by its frequency in the data. Then, the expected number of SV i sequences in region A j can be obtained as P(i) × M j, where M j is the number of sequences from A j. Standardized deviations from expectation can be obtained by dividing the squared deviations (O ij – E ij ) 2 by E ij, where O ij and E ij are the observed and expected numbers of SV i sequences in region j, respectively. This statistic is not affected by the order of magnitude of the data, which allows to straightforwardly compare very abundant and rare SVs, and shallowly and much sampled regions.
Phylogenetic analysis. Sequence alignments were obtained with Mafft’s FFT-NS-2 algorithm. Under this strategy, the program first generates a rough alignment (namely FFT-NS-1 alignment) from a guide tree created from a distance matrix made by computing the number of hexamers shared between all sequence pairs. Then the FFT-NS-1 alignment is used to generate a new tree, which is used to guide a second progressive alignment. Once aligned, our high-quality dataset presented 2713 polymorphic sites of which 885 were parsimony informative. The dataset was phylogenetically analyzed with IQTREE (Minh et al., 2020) using the GTR + I + G model of nucleotide substitution (selected by the IQTREE’s BIC routine) with base frequencies inferred from the data and the default tree search algorithm. Branch supports were calculated by ultrafast bootstrap (Hoang et al., 2017) (n = 1000) in IQTREE. Trees were inspected and plotted by Dendroscope (Huson and Scornavacca, 2012), Ape and ggtree (Yu, 2020). Ancestral character states were inferred with the ace() function from Ape using the SVs’ sequences and phylogeny.
Spatial phylogenetic structure analysis. Structure analyses were performed by the picante package (Kembel et al., 2010). Null distributions were inferred by shuffling SVs across tree tips 10,000 times. Using the obtained permutations and the actual data, the software calculates the following weighted metric for each region:
| SESMPD = (MPDOBS– mean(MPDH0))/sd(MPDH0) |
where MPD OBS is the mean phylogenetic distance (MPD) between all sequences from the region, mean(MPD H0 ) is the average of the MPD between the sequences from the region in the randomized data and sd(MPD H0 ) is the corresponding standard deviation. Negative and low SES MPD values support structured distributions. Significance levels are calculated as the MPD OBS rank divided by the number of permutations minus one.
3. Results
SVs distribution. The 4333 high quality genomes studied here corresponded to 2336 operational taxonomic units or SVs. These SVs’ distributions were very uneven (detailed in Table S2). Here we highlight the most relevant cases. Of the SVs represented by at least 10 sequences, only SVs 17 and 37 were distributed more or less homogeneously. SV 1 abundance was smaller than expected in Asia, and larger than expected in North America. SV 2 was represented only among the North American sequences and SV 3 was over-abundant in Europe and scarcely represented in North America. SVs 4, 9, 12, 23 and 29 were over-represented (SVs 4, 9 and 12) or exclusively observed (SVs 12 and 23) in Asia. SVs 5 and 6 were too abundant in Europe and South America and were under-represented in Asia and North America. SVs 7, 8, 10, 14, 16, 19, 20, 21, 24, 26, 30, 31, 33, 34, 35 and 38 were over-represented or exclusively detected in Europe. SVs 11, 13, 15, 18, 22, 27 and 36 displayed more sequences than expected by chance in North America. The number of SV 28 genomes observed among the Oceania samples was about 20 times greater than expected. Likewise, the SVs 25 and 32 counts in South America and Africa, respectively, were approximately 22 and 21 times larger than the corresponding expected numbers.
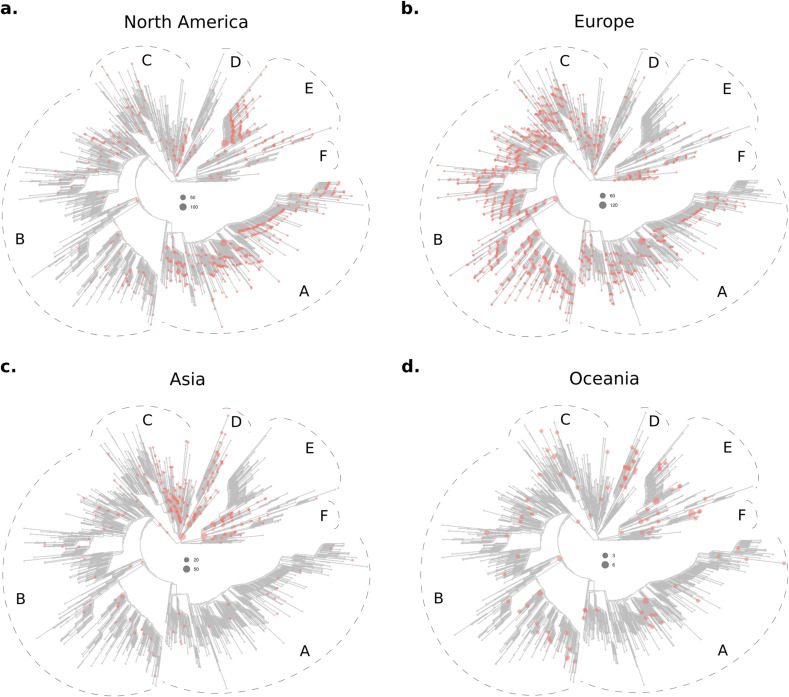
Phylogenetic structure. The above results show that SARS-CoV-2 spatial diversity is substantial. As explained in the Introduction, a possible explanation is spatially coexisting lineages being more related to each other than expected by chance. To check this hypothesis, we performed a phylogenetic structure analysis. The SVs phylogeny and distributions are detailed in Fig. S3. A thorough inspection of these results revealed six major sections of the tree in which the sequences were clumped according to their origins, as shown in a simplified manner in Fig. 1 . For practical reasons, we refer to these six tree sections as A, B, C, D, E, and F. The North American sequences were more abundant in sections A and E while the European ones were prevalent in sections B, C and F, and moderately represented in section A. The Asian sequences clustered preferentially in section D, and in sections C, E and F but in proximal positions relative to the positions occupied by the North American (region E) and European (regions C and F) sequences. Limitation in space was significant (p < 0.01) for Asia, Europe, and North America (Table 1 ).
Fig. 1.
Phylogeny and distribution of 2336 SARS-CoV-2 SVs representative of 4333 genomes from around the world. The SVs present in North America (a), Europe (b), Asia (c) and Oceania (d) are indicated by colored dots. Six sections of the tree are indicated (A, B, C, D, E, and F) to facilitate results interpretation (please see main text). Dots diameters are proportional to the number of genomes accrued in each SV, as indicated in each panel separately.
Table 1.
Spatial phylogenetic structure analysis.
| ntaxaa | obsb | rand.meanc | rand.sdd | obs.ranke | SESMPDf | p-valueg | |
|---|---|---|---|---|---|---|---|
| Africa | 17 | 0.00029 | 0.00036 | 0.00004 | 182 | -2.05848 | 0.01820 |
| Asia | 304 | 0.00026 | 0.00038 | 0.00002 | 1 | -5.84756 | 0.00010 |
| Europe | 1293 | 0.00032 | 0.00039 | 0.00001 | 1 | -5.78452 | 0.00010 |
| North America | 688 | 0.00034 | 0.00038 | 0.00002 | 29 | -2.20543 | 0.00290 |
| Oceania | 96 | 0.00043 | 0.00038 | 0.00002 | 9970 | 2.81459 | 0.99690 |
| South America | 18 | 0.00028 | 0.00036 | 0.00004 | 185 | -1.99065 | 0.01850 |
Number of sequence variants from each region (rows).
Observed Mean Phylogenetic Distances (MPD).
Average MPD in Monte Carlo (M-C) randomizations.
Standard deviation of M-C MPDs.
Observed MPDs ranks.
Standardized effect of geographical structure.
H0: evenly distributed sequence variants.
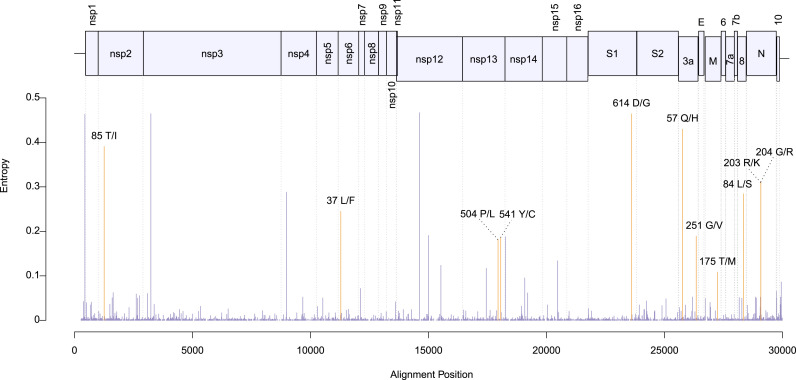
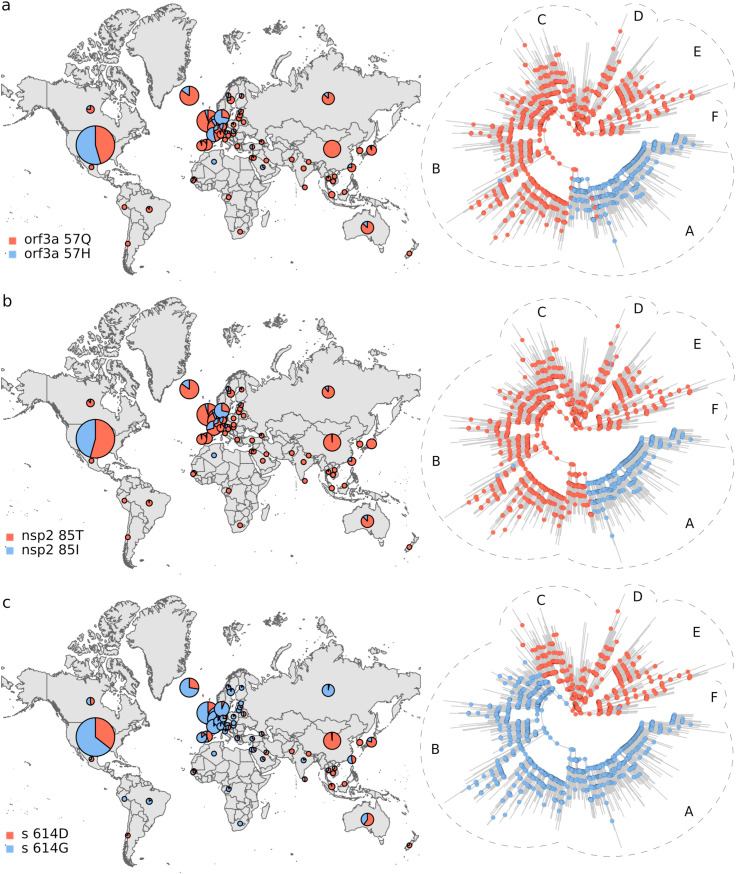
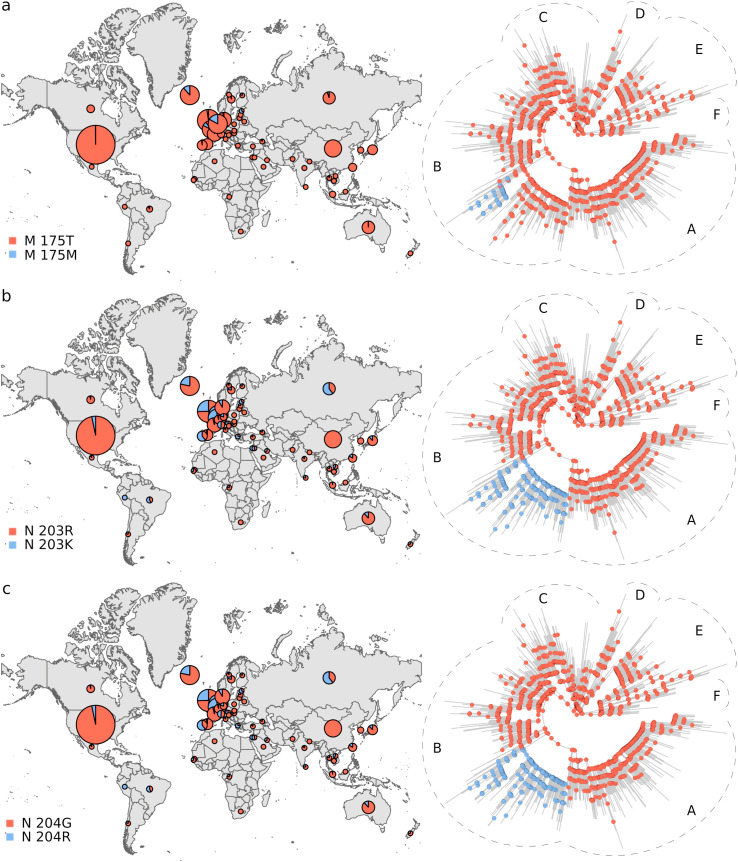
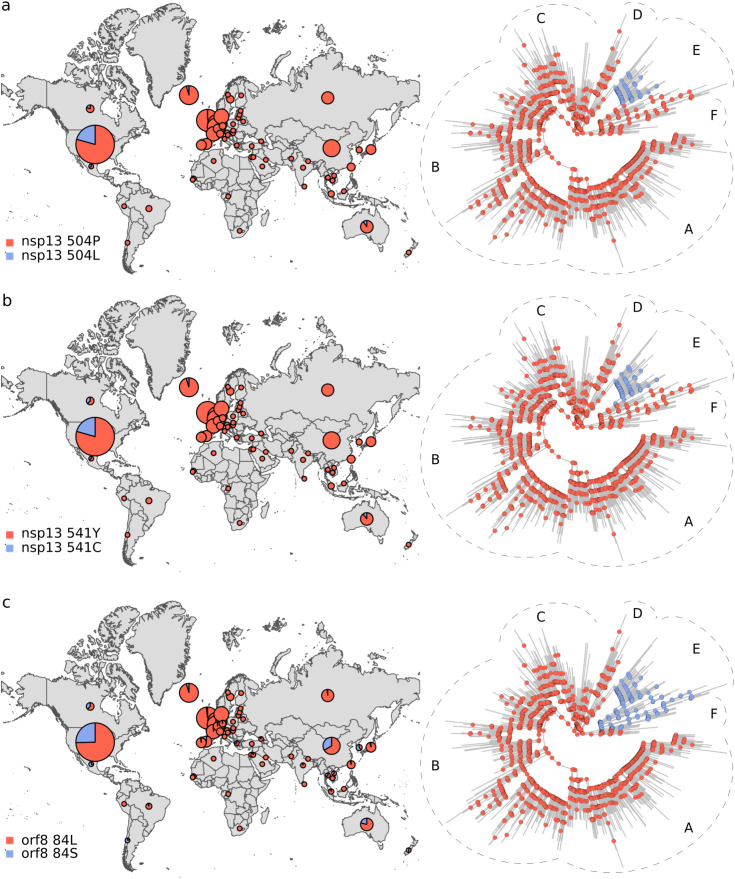
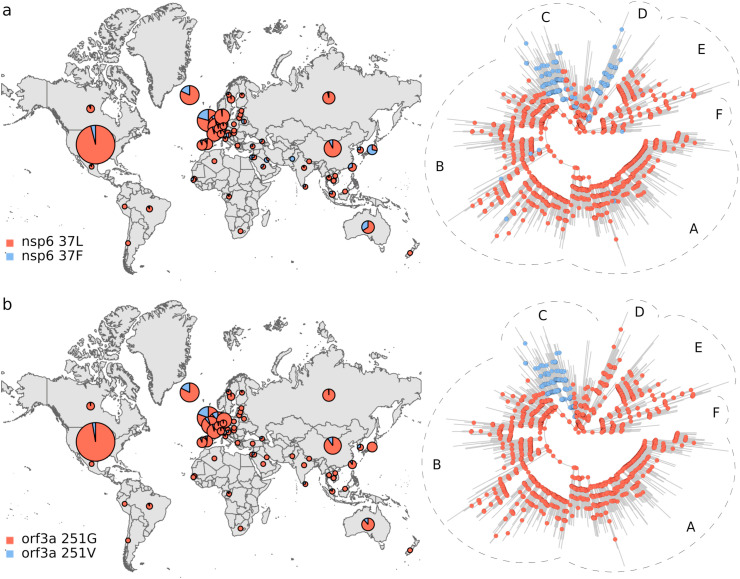
Phylogeographic analysis. The biological insight gained from the results above is that the viral genetic repertoire can vary depending on world region. However, the analyses do not show if spatial variation affects the virus phenotype. Thus, we complemented the above results with a phylogeographic analysis of amino acid variants. We only considered mutations that were observed in at least 100 genomes. By inferring these polymorphisms evolutionary trajectories, it was also possible to evaluate if the present amino acidic diversity could be explained by ancestral mutations occurred in a coordinated way with dispersion events, in which case ancestral changes should resemble the observed association between the spatial distribution and the phylogeny (Fig. 1; Tables 1 and S2). We identified 11 amino acidic polymorphisms endorsed by at least 100 sequences (Fig. 2 ). The less frequent amino acidic variant, protein M 175M, was observed in 166 genomes. The amino acidic variants distributions were very uneven. Furthermore, ancestral amino acidic transitions were highly fitted to the virus phylogeny and occurred at branches dividing the main tree sections described above, or separating large tree chunks inside these sections. These results are summarized in Fig. 3, Fig. 4, Fig. 5, Fig. 6 . The distributions of the 57Q/H, 85T/I and 614 D/G polymorphisms of the orf3, nsp2 and S proteins, respectively, were very contrasting between the east and west (Fig. 3). The amino acidic variants 57H, 85I, and 614G were frequent in Europe and North America, whereas 57Q, 85T, and 514D were relatively abundant in Asia. Unlike the majority of countries, China presented neither the 175M variant of the M protein nor amino acids K and R at positions 203 and 204, respectively, of the N protein (Fig. 4). The nsp13 protein amino acids 504L and 541C were almost exclusively observed among the sequences from North America and Australia (Fig. 5 a and b). The 84L/S polymorphism (orf8 protein) displayed a similar geographic pattern. However, the 84S variant turned out to be abundant in China and South Korea as well as in North America and Australia (Fig. 5 c). The 37 and 251 positions of the nsp6 and orf3a proteins, respectively, experienced mutations inside the C tree section (Fig. 6). However, position 37 also experienced a change in tree section D. In agreement with this, the 251V variant was prevalent in Europe, China, and Australia, whereas 37F was also abundant in Japan and relatively more abundant than 251V in Australia.
Fig. 2.
Location in the virus genome of 11 amino-acidic polymorphism used in phylogeographic analyses. The positional entropies (y-axis) were derived from the genomic sequences. The orange-highlighted bars correspond to the positions that displayed non-synonymous mutations in at least 100 sequences. The substitutions underlying polymorphisms 57Q/H, 203R/K and 204G/R also affected the protein sequences of two hypothetical ORFs that overlap the 3a and N proteins’ ORFs (Pavesi, 2020).
Fig. 3.
Geographic distribution and evolutionary trajectories of polymorphisms 57Q/H (a), 85T/I (b) and 614D/G (c).
Fig. 4.
Distribution and evolutionary trajectories of polymorphisms 175T/M (a), 203R/K (b), and 204G/R.
Fig. 5.
Distribution and evolutionary trajectories of polymorphisms 504P/L (a), 541Y/C (b), and 84L/S (c).
Fig. 6.
Distribution and evolutionary trajectories of polymorphisms 37L/F (a) and 251G/V (b).
4. Discussion
Here, we show that SARS-CoV-2 spatial variation is significant, which, as discussed below, is likely due to vicariant events occurred along the pandemic. As expected, since SARS-CoV-2 is a variant within the species Severe acute respiratory syndrome-related coronavirus (Gorbalenya et al., 2020), sequence diversity was low, as has been observed in previous studies. However, the here performed pairwise comparisons revealed an extra level of complexity: Out of 9,385,278 genome pairs analyzed, only 50,839 (0.5%) consisted of identical sequences and 5,056,965 (53.8%) presented between 6 (Q1) and 12 (Q2) reciprocal mutations (Fig. S4). A raw extrapolation of these results suggests that, only among the ~70,000 SARS-CoV-2 genomes sequenced so far, there could be some 35,000 SVs. The database used in this study is the major SARS-CoV-2 sequence repository and can be considered reliable. Additionally, the here studied sequences were subjected to a strict quality control. However, a few sequences might have been recalcitrant to control procedures of the contributing laboratories and our own ones. Thus, the above estimate of the number of circulating viral genotypes requires further assessment.
The results in Fig. 1 and Table 1 constitute quantitative, significant evidence that SARS-CoV-2 phylogeny is spatially structured. Structuring has a remarkable correlate at the protein level (Fig. 2; pie graphs on maps in Fig. 3, Fig. 4, Fig. 5, Fig. 6). In addition, the studied amino acidic mutations likely occurred in combination with dispersion events (ancestral trajectories in Fig. 3, Fig. 4, Fig. 5, Fig. 6). Many ancestral nodes inside tree section A presented the 57H and 85I amino acidic variants (Fig. 3 a and b). This indicates that the divergence event leading to the split between tree section A and the rest of the phylogeny was accompanied by protein mutations in the orf3a and nsp2 proteins. The fact that the strains from tree section A were abundant in North America and Europe (Fig. 1 a and b), strongly suggests that viral dispersion to, or from, these regions was associated with polymorphisms 57Q/H and 85T/I emergence. In a similar way, the 614D/G, 504P/L, 541Y/C and 84L/S polymorphism may have resulted from the spread of the virus between the east and west (Figs. 3 c and 5 a-c, respectively). The 175T/M, 203R/K and 204G/R transitions occurred inside tree section B (Fig. 4), which was highly represented among the European sequences (Fig. 1 b). This, together with the presence of 203K and 204R in Russia, suggests that these polymorphisms may have emerged in Europe or Russia. Likewise, the 37L/F and 251G/V polymorphisms probably emerged in association with dispersion to, or from, Europe (Fig. 6). Our ancestral states inferences indicated that the amino acidic position 37 of the nsp6 protein experienced several independent L/F transitions. Two such transitions occurred close to the phylogeny branches separating tree sections C and D from the rest of the virus phylogeny (Fig. 6 a). These two tree sections were well represented in Europe and Japan, respectively, strongly suggesting that 37L/F emergence may has occurred twice as a consequence of independent dispersion events.
Biogeographic patterns have been previously observed in other coronaviruses (Chu et al., 2018) and very different viruses as phages and retroviruses (Díaz-Muñoz et al., 2013; Rodriguez et al., 2009). That SARS-CoV-2 has developed a biogeography despite its high propagation rate may seem contradictory. However, spatial diversification depends not only on dispersion constraints but also on evolutionary rates. In particular, spatially structured phylogenies can be the consequence of speciation rates being very high relative to dispersion rates (Webb et al., 2002). As far as we know, travels between remote places constitute the only SARS-CoV-2 dispersion mechanism. This implies that founder viruses usually carry very small fractions of the total genetic variation of the source populations. Therefore, each time the virus spreads, substantial losses of diversity can occur, possibly combined with rare mutations settlement, due to founder effects. In addition, it stands to reason that, after dispersion, mutations accumulate quickly as newly established populations enlarge, due to viral polymerases high error rates (Gago et al., 2009; Moya et al., 2004). Some of these mutations can lead to novel phenotypes, as shown in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6. Based on these considerations, it is reasonable to hypothesize that long-distance dispersion constitutes an opportunity for the virus to fix otherwise rare, and/or develop new, mutations.
The 2002–2003 SARS-CoV epidemic was subdivided into three genetically different phases, indicating that coronaviruses can mutate recurrently along relatively short periods of time (The Chinese SARS Molecular Epidemiology Consortium, 2004). By the other side, it has been shown that slight mutations can produce significant phenotypic effects in MERS-CoV and other coronaviruses (Chu et al., 2018; Rasschaert et al., 1990; Zhang et al., 2007). Furthermore, recent experimental studies suggest that the 614G spike protein variant increases SARS-CoV-2 infectivity (Korber et al., 2020; Zhang et al., 2020). Here, we showed that at least 10 further protein polymorphisms may have emerged along the recent SARS-CoV-2 evolutionary history. This must be taken as a call for attention. The virus evolution should continue to be monitored and relationships should be sought between viral diversity and pathogenesis.
CRediT authorship contribution statement
Leandro R. Jones: Conceptualization, Data curation, Methodology, Software, Formal analysis, Investigation, Writing. Julieta M. Manrique: Conceptualization, Data curation, Methodology, Investigation, Writing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiCoV™ Database used in this study. This work was partially supported by Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 11220130100255CO), Agencia Nacional de Promoción Científica y Tecnológica (PICT 2016-2795) and Asociación Civil Argentina Genetics. The authors would like to acknowledge the helpful comments raised throughout the review process.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.08.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Chu D.K.W., Hui K.P.Y., Perera R.A.P.M., Miguel E., Niemeyer D., Zhao J., Channappanavar R., Dudas G., Oladipo J.O., Traoré A., Fassi-Fihri O., Ali A., Demissié G.F., Muth D., Chan M.C.W., Nicholls J.M., Meyerholz D.K., Kuranga S.A., Mamo G., Zhou Z., So R.T.Y., Hemida M.G., Webby R.J., Roger F., Rambaut A., Poon L.L.M., Perlman S., Drosten C., Chevalier V., Peiris M. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Muñoz S.L., Tenaillon O., Goldhill D., Brao K., Turner P.E., Chao L. Electrophoretic mobility confirms reassortment bias among geographic isolates of segmented RNA phages. BMC Evol. Biol. 2013;13:206. doi: 10.1186/1471-2148-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago S., Elena S.F., Flores R., Sanjuan R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. 1308. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon1 L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Briefings Bioinf. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kembel S.W., Cowan P.D., Helmus M.R., Cornwell W.K., Morlon H., Ackerly D.D., Blomberg S.P., Webb C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E., Bhattacharya T., Foley B., Hastie K., Parker M., Partridge D., Evans C., Freeman T., de Silva T., McDanal C., Perez L., Tang H., Moon-Walker A., Whelan S., LaBranche C., Saphire E., Montefiori D., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D. Cell; 2020. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic Inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Holmes E.C., González-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Pavesi A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology. 2020;546:51–56. doi: 10.1016/j.virol.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0770-5. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasschaert D., Duarte M., Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71:2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- Rodriguez S.M., Golemba M.D., Campos R.H., Trono K., Jones L.R. Bovine leukemia virus can be classified into seven genotypes: evidence for the existence of two novel clades. J. Gen. Virol. 2009;90:2788–2797. doi: 10.1099/vir.0.011791-0. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.O., Ackerly D.D., McPeek M.A., Donoghue M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Systemat. 2002;33:475–505. [Google Scholar]
- WHO Emergencies. 2020. https://www.who.int/topics/emergencies/en/.2020
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. Using ggtree to visualize data on tree-like structures. Current Protocols in Bioinformatics. 2020;69 doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Vlasova A., Janies D., Jones L.R., Ghedin E., Saif L.J. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology. 2007;363:1–10. doi: 10.1016/j.virol.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. BioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.