Abstract
Receptor Interacting Protein Kinases (RIPKs) are cellular signaling molecules that are critical for homeostatic signaling in both communicable and non-communicable disease processes. In particular, RIPK1, RIPK2, RIPK3 and RIPK7 have emerged as key mediators of intracellular signal transduction including inflammation, autophagy and programmed cell death, and are thus essential for the early control of many diverse pathogenic organisms. In this review, we discuss the role of each RIPK in host responses to bacterial and viral pathogens, with a focus on studies that have used pathogen infection models rather than artificial stimulation with purified pathogen associated molecular patterns. We also discuss the intricate mechanisms of host evasion by pathogens that specifically target RIPKs for inactivation, and finally, we will touch on the controversial issue of drug development for kinase inhibitors to treat chronic inflammatory and neurological disorders, and the implications this may have on the outcome of pathogen infections.
Abbreviations: 3Cpro, viral non-structural protein 3C; AEC, airway epithelial cell; AIM2, absent in melanoma 2; ALRs, AIM2-like receptors; AP-1, activator protein 1; ATG16L1, autophagy-related 16-like 1; ASC, apoptosis-associated speck-like protein; BAV, BeAn 58058 poxvirus; Bcl-2, B-cell lymphoma 2; BclxL, Bcl-extra large; BMDMs, bone marrow-derived macrophages; BVDV, bovine viral diarrhoea virus; CA6, coxsackievirus A6; CARD, caspase activation and recruitment domain; CD, Crohn’s disease; c-FLIP, cellular FLICE inhibitory protein; cIAPs, cellular inhibitor of apoptosis proteins; Cif, cycle inhibiting factor; CMV, cytomegalovirus; CNF1, cytotoxic necrotising factor 1; CNS, central nervous system; COR, C-terminal of Roc; COTV, Cotia poxvirus; COVID-19, coronavirus disease 2019; CVB, coxsackievirus B3; DAI, DNA-dependent activator of IRFs; DAMPs, danger-associated molecular patterns; DAP, diaminopimelic acid; DENV, dengue virus; DISCs, death-inducing signaling complexes; DR, death receptor; DRP1, dynamin-related protein 1; dsRNA, double stranded RNA; E3L, vaccinia virus protein E3 long; EBV, Epstein-Barr virus; ER, endoplasmic reticulum; ERK, extracellular signal-regulated kinases; GEF, guanidine exchange factor; GSDMD, gasdermin D; HAEC, human aortic endothelial cells; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; HCMV, human cytomegalovirus; HCV, hepatitis C virus; HFMD, hand foot and mouth disease; HIV-1, human immunodeficiency virus type 1; HMGB1, high mobility group box 1; HPV, human papilloma virus; HSV, Herpes simplex virus; IAV, influenza A virus; ID, intermediate domain; iE-DAP, Ɣ-d-glutamyl-mesodiaminopimelic acid; IFI44L, IFN-induced protein 44-like; IFN, interferon; IL, interleukin; IĸB, inhibitor of ĸB; IKK, IĸB kinase; IRFs, IFN regulatory factors; IRG1, immunoresponsive gene 1; ISGF3, IFN-stimulated gene factor 3; ISGs, interferon stimulated genes; JEV, Japanese encephalitis virus; JNK, c-Jun N-terminal kinase; KD, kinase domain; Kgp, lysine-specific protease gingipain; KO, knockout; LMP1, latent membrane protein; LPS, lipopolysaccharide; LRR, leucine-rich repeat; LUBAC, linear ubiquitin chain assembly complex; M45, murine CMV virion associated protein M45; MAPKs, mitogen-activated protein kinases; MAVS, mitochondrial antiviral signaling proteins; MCMV, murine cytomegalovirus; MDA5, melanoma differentiation-associated protein 5; MDP, muramyl dipeptide; MLKL, mixed-lineage kinase-domain like protein; MKKs, MAPK kinases; Mtb, Mycobacterium tuberculosis; MyD88, myeloid differentiation factor 88; NAIPs, NLR family apoptosis inhibitory proteins; Nec-1, necrostatin-1; NET, neutrophil extracellular traps; NF-ĸB, nuclear factor-ĸB; NK, natural killer; NleB, non-LEE encoded effector protein B; NLRs, nucleotide-binding domain–like receptors; NLRC4, NLR family CARD domain containing 4; NLRP3, NLR family pyrin domain containing 3; NO, nitric oxide; NOD, nucleotide-binding oligomerization domain; NoV, norovirus; NP, nucleoprotein; NSA, necrosulfonamide; NSP, non-structural protein; OMV, outer membrane vesicle; ORF, open reading frame; PAMPs, pathogen associated molecular patterns; PD, Parkinson disease; PFT, pore-forming toxins; PGN, bacterial peptidoglycan; PKR, protein kinase R; PI3P, phosphatidylinositol 3-phosphate; PI3K, PI3 kinase; PnCW, pneumococcal cell wall; Poly(I:C), polyinosinic:polycytidylic acid; PR, protease; PRRs, pattern recognition receptors; PVM, pneumonia virus of mice; RDA, RIPK1-dependent apoptosis; RHIM, RIP homotypic interaction motif; RIG-I, cytosolic retinoic acid-inducible gene I; RIPKs, receptor interacting protein kinases; Roc, Ras-of-complex; ROS, reactive oxygen species; RR, ribonucleotide reductase; RSV, respiratory syncytial virus; SARS, severe acute respiratory syndrome; SARS-3a, SARS-CoV ORF protein 3a; SDH, succinate dehydrogenase; siRNA, small interfering RNA; SNPs (line 657) ssRNA, single-stranded RNA; T3SS, type III secretion system; T4SS, type IV secretion system; TAK1, TGFβ-activated kinase 1; TBK1, TANK-binding kinase 1; TGFβ, transforming growth factor beta; Th1, T helper 1; TLR, toll-like receptor; TNF, tumour necrosis factor; TNFR1, TNF receptor type 1; TRADD, TNFR1-associated death domain protein; TRAF, TNFR associated factors; TRAILR, TNF-related apoptosis-inducing ligand receptor; TRAM, translocating chain-associated membrane protein; TRIF, TIR-domain-containing adapter-inducing interferon-β; TRPM7, transient receptor potential cation channel subfamily M member 7; UBE2N, ubiquitin-conjugating enzyme E2N; UL45/UL48, human CMV envelope protein 45/48; UPR, unfolded protein response; vICA, viral inhibitor of caspase-8 activation; vIRA, viral inhibitor of RIP activation; vMLKL, viral MLKL-like proteins; VSV, vesicular stomatitis virus; VV, vaccinia virus; WNV, West Nile virus; WT, wild-type; XIAP, X-linked inhibitor of apoptosis protein; Yop, Yersinia outer protein; ZBP1, Z-DNA binding protein; ZIKV, Zika virus
Keywords: RIP kinase, Inflammation, Cell death, Bacterial infection, Viral infection, Pathogen
1. Introduction
Pathogen infection initiates multiple innate immune signaling pathways via the stimulation of pattern recognition receptors (PRRs) with pathogen-associated molecular patterns (PAMPs). The outcomes of PAMP-PRR signaling are diverse, and often dependent on pathogen-specific virulence factors as well as host factors including, cell type, species, existing gene polymorphisms or pre-existing co-morbidities. Nevertheless, the signal transduction that follows pathogen recognition elicits numerous defence mechanisms, including the induction of inflammatory cytokine, chemokine and interferon production, activation of programmed cell death pathways, and inevitably an adaptive immune response, all of which contribute to pathogen control and elimination [1].
There are seven members of the Receptor Interacting Protein Kinase (RIPK) family, RIPK1-7, some of which have emerged as critical effectors of immunity to infection with a diverse array of bacterial, viral and protozoal pathogens. Structurally and functionally, all members of the RIPK family share a homologous serine-threonine kinase domain (KD) with a catalytic site [2], with RIPK2 boasting additional tyrosine kinase activity [3]. RIPK1 has a C-terminal death domain (DD), and an intermediate domain (ID) which harbors a RIP homotypic interaction motif (RHIM). RIPK2 contains a C-terminal CARD (caspase activation and recruitment domain) and an ID. RIPK3 has a C-terminal RHIM domain alongside the N-terminal KD. RIPK4 and RIPK5 are characterized by C-terminal ankyrin repeats, and RIPK6 and RIPK7 each harbor a leucine-rich repeat (LRR) motif, ankyrin repeats, a Ras-of-complex (Roc) domain followed by a C-terminal of Roc (COR) domain, and a WD40 domain.
To date, the most studied RIPKs in relation to inflammation of host immune responses are RIPK1, 2 and 3, with RIPK7 following steadily behind. This rapidly moving field has brought the biochemical functions of RIPKs to the forefront of cellular immunity. Here, model pathogens including Listeria monocytogenes, Shigella flexneri, Mycobacterium tuberculosis, influenza A virus (IAV) and cytomegaloviruses (CMV) have been instrumental in dissecting the role of RIPKs in host immunity. Furthermore, research over the past decade has revealed multiple pathogenic mechanisms of bacteria and viruses that specifically target RIPKs or their downstream signaling networks for inactivation. Despite the volume of research on RIPKs and host responses to infection, there is a long way to go in understanding the physiological role of each RIPK and their functional domains in specific infection settings. This is partly due the fact that RIPK1 and RIPK3 are implicated in pro-survival transcriptional pathways as well as cell death, but also due to the ongoing discovery of novel virulence mechanisms of pathogens.
In this review, we will provide an up to date account of RIPK-mediated host responses to bacterial and viral infections as well as the mechanisms pathogens have evolved to evade RIPK signaling outcomes. Importantly, we have focused on studies that have utilised in vitro or in vivo infections with pathogenic organisms to stimulate physiological immune responses, rather than those that have used purified or synthetic stimulants, for example lipopolysaccharide (LPS) or polyinosinic:polycytidylic acid (Poly I:C). We note that although we were not able to discuss all relevant pathogens, including protozoal organisms, we have generated a comprehensive list of pathogens, the associated RIPK signaling pathways and relevant references in Table 1 .
Table 1.
List of pathogens that are controlled by RIPK signaling or those that manipulate RIPK signaling processes.
| Microorganism | Classification | RIPK | Inhibits/activates RIPK signaling | Virulence factor | Model | Reference |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Borrelia burgdorferi | Spirochete | RIPK2 | Activates | – | Mouse | [165–167] |
| Brucella abortus | Gram-negative | RIPK2 | Activates | VceC | Mouse, human | [194] |
| Chlamydia muridarum | Gram-negative | RIPK2 | Activates | – | Mouse, human | [194,195] |
| Citrobacter rodentium | Gram-negative | RIPK1, RIPK3 | Inhibits | EspL | Mouse | [113] |
| Enteropathogenic Escherichia coli | Gram-negative | RIPK1 | Inhibits | NleB1 | Human | [45,46] |
| RIPK1, RIPK3 | Inhibits | EspL | Human | [113] | ||
| Helicobacter pylori | Gram-negative | RIPK2 | Activates | – | Human | [149] |
| Klebsiella pneumoniae | Gram-negative | RIPK1, RIPK3 | Activates | – | Mouse | [17] |
| Legionella pneumophila | Gram-negative | RIPK2 | Activates | – | Mouse | [145,162] |
| RIPK1 | Inhibits | MavC | Human | [42] | ||
| Listeria monocytogenes | Gram-positive | RIPK1, RIPK3 | Activates | PFT | Mouse, human | [115] |
| RIPK2, RIPK7 | Activates | – | Mouse | [137,151,184,217] | ||
| Mycobacterium leprae | Acid-fast | RIPK2, RIPK7 | Activates | – | Human | [152,219,220] |
| Mycobacterium tuberculosis | Acid-fast | RIPK3, RIPK7 | Activates | – | Mouse, human | [119,120,222,223] |
| RIPK2 | Activates | – | Mouse | [147] | ||
| Porphyromonas gingivalis | Gram-negative | RIPK1, RIPK2 | Inhibits | Kgp | Human | [209] |
| Salmonella enterica serovar Typhimurium | Gram-negative | RIPK1, RIPK3, RIPK7 | Activates | – | Mouse | [117,118,216] |
| RIPK2 | Activates | SipA, SopE | Human | [185,188,189] | ||
| Serratia marcescens | Gram-negative | RIPK1, RIPK3 | Activates | PFT | Mouse, human | [115] |
| Shigella flexneri | Gram-negative | RIPK2 | Activates | IpgB2, OspB | Human | [177,187] |
| RIPK2 | Inhibits | – | Human | [199] | ||
| Staphylococcus aureus | Gram-positive | RIPK1, RIPK3 | Activates | PFT | Mouse, human | [114,116] |
| Streptococcus pneumoniae | Gram-positive | RIPK1, RIPK3 | Activates | PFT | Mouse, human | [115] |
| RIPK2 | Activates | – | Mouse | [157] | ||
| Uropathogenic Escherichia coli | Gram-negative | RIPK1, RIPK2 | Activates | CNF1 | Flies, human | [192] |
| RIPK1, RIPK3 | Activates | PFT | Mouse, human | [115] | ||
| Yersinia pestis | Gram-negative | RIPK1 | Activates | – | Mouse | [39] |
| Yersinia pseudotuberculosis | Gram-negative | RIPK1, RIPK3 | Activates | – | Mouse | [17,79,80] |
| Virus | ||||||
| BeAn 58058 poxvirus, Cotia poxvirus | dsDNA virus (Poxvirus) | RIPK3 | Inhibits | vMLKL | Mouse, human | [101] |
| Bovine viral diarrhoea virus | ssRNA virus (Flavivirus) | RIPK6 | Inhibits | – | Goat | [213] |
| Coxsackievirus A6 | ssRNA virus (Picornavirus) | RIPK3 | Activates | Nsp-3D | Human | [105] |
| Coxsackievirus B3 | ssRNA virus (Picornavirus) | RIPK3 | Activates non-necrotic signaling, inhibits necroptosis | Nsp-3C (3Cpro) | Human | [37] |
| Dengue virus | ssRNA virus (Flavivirus) | RIPK1 | Activates | – | Human | [130] |
| Epstein-Barr virus | dsDNA virus (γ-herpesvirus) | RIPK1, RIPK3 | Activates pro-survival signaling, inhibits necroptosis | LMP1 | Human | [35] |
| Hepatitis B virus | dsDNA virus (Hepadnavirus) | RIPK2 | Inhibits | HBeAg | Human | [208] |
| Hepatitis C virus | ssRNA virus (Flavivirus) | RIPK2 | Activates | NS5B | Mouse, human | [207] |
| Herpes simplex virus-1/2 | dsDNA virus (α-herpesvirus) | RIPK1,RIPK3 | Inhibits | ICP6/10 | Human | [73,97,98] |
| RIPK1,RIPK3 | Activates | ICP6/10 | Mouse | [98,99] | ||
| Human cytomegalovirus | dsDNA virus (β-herpesvirus) | RIPK1 | Inhibits | UL48, UL45 | Human | [30] |
| RIPK1,RIPK3 | Inhibits | IE1 | Human | [96] | ||
| Human immunodeficiency virus | ssRNA virus (Retrovirus) | RIPK1, RIPK2 | Inhibits | PR | Human | [102] |
| Influenza A virus | ssRNA virus (Orthomyxovirus) | RIPK1, RIPK3 | Activates | – | Mouse, human | [33,75,78] |
| RIPK2 | Activates | – | Mouse | [205] | ||
| Japanese encephalitis virus | ssRNA virus (Flavivirus) | RIPK3 | Activates | – | Mouse | [32] |
| Murine cytomegalovirus | dsDNA virus (β-herpesvirus) | RIPK1, RIPK3 | Inhibits | M45 (vIRA) | Mouse | [22,29,90,91,93] |
| Pneumonia virus of mice | ssRNA virus (Paramyxovirus) | RIPK1 | Activates | – | Mouse | [107] |
| Respiratory syncytial virus | ssRNA virus (Paramyxovirus) | RIPK1 | Activates | – | Human | [107] |
| SARS-CoV | ssRNA virus (Coronavirus) | RIPK3 | Inhibits | SARS 3a | Human | [110] |
| SARS-CoV-2 | ssRNA virus (Coronavirus) | RIPK1 | Unknown | Nsp-12 | Human | [109] |
| Swine influenza virus | ssRNA virus (Orthomyxovirus) | RIPK1 | Activates | – | Pig | [131] |
| Vaccinia virus | dsDNA virus (Poxvirus) | RIPK1, RIPK3 | Activates | B13R/Spi2 | Mouse | [82] |
| RIPK3 | Inhibits | E3L | Mouse, human | [87] | ||
| Vesicular stomatitis virus | ssRNA virus (Rhabdovirus) | RIPK1, RIPK3 | Activates | – | Mouse, human | [129] |
| West Nile Virus | ssRNA virus (Flavivirus) | RIPK1, RIPK3 | Activates | – | Mouse | [25] |
| Zika virus | ssRNA virus (Flavivirus) | RIPK1, RIPK3 | Activates | – | Mouse | [26] |
| Pathogens not covered in this review | ||||||
| Adenovirus type 5 | dsDNA virus (Adenovirus) | RIPK3 | Activates | – | Human | [230] |
| Murine gammaherpesvirus-68 | dsDNA virus (γ-herpesvirus) | RIPK1, RIPK3 | Activates | – | Mouse | [231] |
| Murine hepatitis virus type 3 | ssRNA virus (Coronavirus) | RIPK1 | Activates | – | Mouse | [232] |
| Murine norovirus-1 | ssRNA virus (Calicivirus) | RIPK2 | Activates | – | Mouse | [201] |
| Reovirus | dsRNA virus | RIPK1, RIPK3 | Activates | – | Mouse | [233,234] |
| Sendai virus | ssRNA virus (Paramyxovirus) | RIPK1, RIPK3 | Activates | Y1, Y2 | Mouse | [231] |
| Chlamydophila pneumoniae | Gram-negative | RIPK2 | Activates | – | Mouse | [235] |
| Pasteurella multocida | Gram-negative | RIPK1, RIPK3 | Activates | – | Chicken | [236] |
| Leishmania braziliensis | Trypanosomatid | RIPK3 | Inhibits | – | Human | [237] |
| Leishmania infantum | Trypanosomatid | RIPK1, RIPK3 | Activates | – | Mouse, human | [238,239] |
| Plasmodium falciparum | Plasmodiidae | RIPK2 | Activates | – | Mouse | [240] |
| Trypanosoma cruzi | Trypanosomatid | RIPK2 | Activates | – | Mouse | [241] |
1.1. RIPK1 and RIPK3-mediated pro-survival inflammatory signaling in pathogen infection
RIPK1 regulates inflammation in response to death receptors (DR), including tumour necrosis factor receptor 1 (TNFR1), TNF-related apoptosis-inducing ligand receptors 1 and 2 (TRAILR1/2) and Fas [[4], [5], [6]], as well as toll-like receptor (TLR)3 and TLR4 [7,8] (Fig. 1 ). RIPK3, although best known for its role in necroptosis (discussed later), can act in concert with RIPK1 to engage in pro-inflammatory, non-cell death signaling [9]. Here we will focus on research that indicate direct RIPK1 and/or RIPK3 involvement in pathways specifically described to be cell death-independent during host infection.
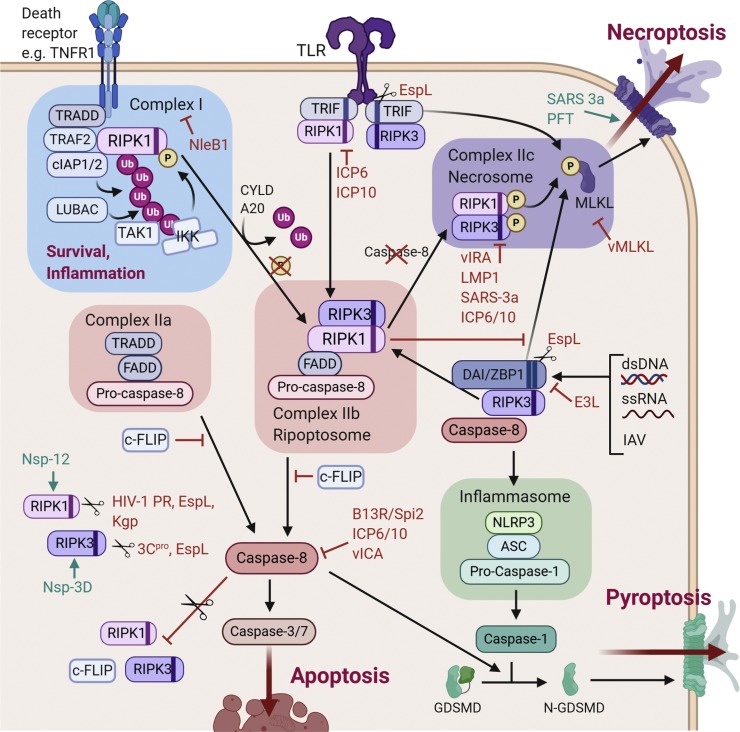
Fig. 1.
Inflammatory pathways mediated by RIPK1/3 in response to pathogen sensing. Upon stimulation with TNF, TNFR1 recruits TRADD and RIPK1 via the DD, then subsequently TRAF2/5 and cIAP1/2 to form a membrane-bound pro-inflammatory signaling complex [10,11]. Ubiquitylation of RIPK1 by cIAP1/2 and LUBAC enables recruitment of TAK1 and the IKK complex, which promotes activation of MAPK and canonical NF-κB signaling [[12], [13], [14], [15]]. Ligation of TLR3/4 reinforces these pathways through RHIM-mediated interactions between TRIF and RIPK1, with subsequent RIPK1 ubiquitylation continuing to drive NF-κB activation [7,16]. TLR4-TRIF interactions can also induce Type I IFN signaling via recruitment of RIPK1 and RIPK3, which activates TBK1 and IKKε to promote nuclear translocation of IRF3 [17]. In response to dsRNA sensing by cytosolic RIG-I or MDA5, MAVS-RIPK1 interactions additionally drive IRF3 and NF-ĸB transcription pathways [[18], [19], [20]]. Finally, cytosolic dsDNA sensing by DAI/ZBP1 promotes NF-ĸB induction via RHIM-mediated recruitment of RIPK1, with RIPK3 kinase activity also required for synergistic activation [21,22]. Virulence factors that interact with these pathways are indicated, and have been discussed in text the main body of text.
1.1.1. RIPK1/3 in inflammatory responses to viral infections
West Nile virus (WNV) and Zika virus (ZIKV) are medically important arboviruses that cause severe neurological disease in humans [23,24]. Control of WNV requires robust neuroinflammation and infiltration of peripheral leukocytes into the central nervous system (CNS), mediated by RIPK1 and RIPK3 [23,25]. Daniels et al. [25] demonstrated that Ripk3−/− and RIPK1 kinase-dead (Ripk1K45A/K45A) mice have suppressed TLR-induced chemokine expression, as well as reduced recruitment of T lymphocytes and myeloid cells into the CNS, resulting in accelerated virus-induced mortality. Similarly, direct ZIKV infection in the CNS of Ripk3−/−, Ripk1K45A/K45A and Dai−/− mice induces rapid mortality and elevated viral titres within the brain [26]. Pharmacological blockade of RIPK3 also enhances ZIKV replication in human neuroblastomas. This intrinsic neuronal cell defect results from disrupted DAI/RIPK-mediated transcription of immunoresponsive gene 1 (IRG1) required for itaconate production. Itaconate inhibits succinate dehydrogenase (SDH) activity and subsequently reprograms neuronal metabolism into an antiviral state [[26], [27], [28]]. As such, these results highlight the contribution of RIPK1/3 in defence against neurovirulent viruses, independently of cell death.
CMVs are large double-stranded DNA viruses that have the capacity to encode viral effectors that directly manipulate components of host immune pathways. Murine CMV (MCMV) expresses the M45 protein, which interacts with RIPK1 through its catalytically inactive ribonucleotide reductase (RR) domain and inhibits RIPK1 ubiquitylation [29]. This process blocks pathways dependent on polyubiquitylated RIPK1, including TNF-induced NF-ĸB and p38 MAPK activation, as well as TLR3-induced NF-ĸB activation, thus facilitating immune evasion. M45 is also shown to bind RIPK1, RIPK3 and DAI via its N-terminal RHIM domain, which interferes with DAI-induced NF-ĸB signaling [22]. Similarly, human CMV (HCMV) is able to inhibit RIPK1-mediated NF-ĸB signaling through synergistic function of its encoded UL48 and UL45 proteins [30]. During the late stages of infection, UL48 cleaves the K63-polyubiquitin chains on RIPK1, thus preventing TNF-induced NF-ĸB activation [30,31]. UL45 supports this process by enhancing UL48-RIPK1 interaction and re-localization of RIPK1 to the cytoplasmic virion assembly complex.
Conversely, RIP kinases can also be utilised to promote viral propagation. Following Japanese encephalitis virus (JEV) infection, Ripk3 gene expression is upregulated in primary neurons of wild-type (WT) mice, which corresponds to heightened viral titres [32]. RNA-sequencing of infected brain tissues revealed Ripk3−/− mice upregulate a number of IFN-stimulated genes (ISGs), independently of RIPK3 and MLKL phosphorylation. Silencing of the ISG, IFI44 L, in both WT and Ripk3−/− neurons increases viral RNA levels, suggesting that the elevated RIPK3 assists with JEV immune evasion.
RIPK3-regulated type I IFN signaling is also shown to be crucial in anti-IAV immunity. Indeed, infected Ripk3−/− mice exhibit increased pulmonary viral load and enhanced immunopathology and mortality [33]. This susceptibility is largely attributed to defective IFN production from infected Ripk3-/- macrophages, independent of IAV-associated cell death outcomes. Specifically, RIPK3 controls type I IFN signaling at the transcriptional level, where virus-induced upregulation of RIPK3 disrupts RIPK1-MAVS interaction and reduces IFN-β mRNA expression, which facilitates IAV persistence [19,33]. Interestingly, the increased RIPK3 also activates protein kinase R (PKR) that stabilises IFN-β transcripts [34], thus elevating IFN-β production and type I IFN protection [33]. This is presumed to have been counter-evolved by hosts in response to viral evasion and represents a potential avenue for therapy against pathogenic IAV.
Several other viruses are known to modulate RIPK1/3 signaling in non-cell death pathways to benefit pathogenesis. For example, the common cause for infectious mononucleosis, Epstein-Barr virus (EBV), encodes a latent membrane protein (LMP1) that binds both RIPK1 and RIPK3, and recruits E3 ligases to regulate protein ubiquitylation [35]. Consequently, LMP1 promotes RIPK1 polyubiquitylation, while RIPK3 polyubiquitylation is suppressed through yet undefined means, which switches cell fate from necroptosis to survival. Human immunodeficiency virus type 1 (HIV-1) protease (PR) cleaves RIPK1 with high specificity in human T lymphocytes, resulting in failure to activate NF-ĸB [35]. Conversely, coxsackievirus B3 (CVB) employs RIPK3 to promote autophagic flux for viral assembly [36], and as such, silencing of RIPK3 in human intestinal epithelial cells restricts early viral replication [37].
1.1.2. RIPK1/3 in inflammatory responses to bacterial infections
Yersinia pestis, the etiological agent of bubonic plague, produces an array of Yersinia outer protein (Yop) virulence effectors that are translocated into host cells via a type III secretion system (T3SS) to alter host cell signaling [38]. Infection of macrophages with Y. pestis results in TNF, IL-6 and IL-1β secretion, mediated through a RIPK1 and caspase-8 dependent, but RIPK3-independent mechanism [39]. As a result, RIPK1 deletion in murine myeloid cells causes defective cytokine release, associated with decreased IĸBα degradation and NF-ĸB nuclear translocation. In the absence of caspase activity, Y. pseudotuberculosis is also shown to induce RIPK1/RIPK3/TRIF-mediated synthesis of IFN-β in macrophages, similar to Klebsiella pneumoniae [17]. This process requires the kinase activity of both RIPKs but operates independently of MLKL-mediated necroptosis and caspase-8 apoptosis. To counter host inflammation, Y. pestis encodes YopJ, which acetylates MAPKs, IKKβ and TAK1 within their activation loops, thus dampening host MAPK and NF-ĸB signaling [40].
Legionella pneumophila is an environmental organism and accidental human pathogen that causes Legionnaires’ disease, a severe form of acute pneumonia [41]. Following infection, L. pneumophila effector MavC (also known as Lpg2147) deamidates the human ubiquitin-conjugating enzyme E2N (UBE2N), which disrupts UBE2N-mediated formation of Lys63 polyubiquitin chains [42]. This prevents RIPK1 polyubiquitylation, thus suppressing downstream NF-ĸB signaling in infected cells in vitro. Given the presence of MavC homologues in the cycle inhibiting factor (Cif) effector family of T3SS pathogens [43,44], there may be similar routes for RIPK1 inhibition in other bacterial contexts.
Bacterial effectors can also directly manipulate RIPKs. Enteropathogenic Escherichia coli (EPEC) encodes an arginine glycosyltransferase, NleB1, that modifies RIPK1 following infection of human cells [45,46]. This activity inhibits the recruitment of ubiquitinated RIPK1 to TNFR1, thus disrupting the assembly of Complex I required for TNF-induced NF-ĸB signaling [45]. Although transient expression of NleB1 inhibits NF-ĸB activation [47], NleB1 alone is not sufficient to inhibit NF-ĸB-mediated IL-8 production during EPEC infection in vitro [46], as multiple effectors are required to achieve this outcome [48].
1.2. RIPK1 and RIPK3-mediated apoptotic signaling in pathogen infection
Apoptosis is a common host response to infection that helps restrict the replicative niche of pathogens, and facilitates phagocytosis and antigen presentation through the production of apoptotic bodies [49]. Many pathogens however, have evolved mechanisms to manipulate apoptotic processes to benefit survival and virulence (Fig. 2 ).
Fig. 2.
Programmed cell death pathways regulated by RIPK1/3. Following the TNFR1-mediated assembly of pro-inflammatory complex I, deubiquitinases (CYLD or A20) remove polyubiquitin chains from RIPK1 to terminate inflammation and enable downstream death signaling [50,51]. RIPK1 (with RIPK3) interacts with FADD and pro-caspase-8 to form complex IIb (ripoptosome) [52,53], and can initiate extrinsic apoptosis [54,55]. Active caspase-8 facilitates repression of necroptosis and NF-ĸB signaling by cleaving RIPK1 and RIPK3 [[56], [57], [58]]. In the absence of caspase activity, RIPK1 and RIPK3 oligomerize to form complex IIc (necrosome) that phosphorylates MLKL and induces necroptosis [[59], [60], [61]]. TLR-driven TRIF-RIPK1 interactions can also promote ripoptosome formation [62], while TRIF-RIPK3 phosphorylates MLKL for necroptosis [63]. Following nucleic acid sensing, RHIM interactions between DAI/ZBP1 and RIPK3 induces ripoptosome formation or direct phosphorylation of MLKL [64,65]. However, the resulting necroptosis can be suppressed by RIPK1 RHIM [66,67]. DAI/ZBP1-RIPK3 complexing also promotes NLRP3 inflammasome activation and death via pyroptosis [68,69]. Virulence factors that interact with these pathways are indicated, and have been discussed in the main body of the text.
1.2.1. RIPK1/3 in apoptotic responses to viral infections
The execution of apoptosis is heavily reliant on host caspases, thus most large DNA viruses encode effectors for inhibiting caspase activity [70], however, few viral factors target RIPKs directly. Herpes simplex virus (HSV) contains numerous proteins that can interact with apoptotic signaling components, including ICP6 and ICP10 encoded by HSV-1 and HSV-2 respectively [71]. These proteins possess a C-terminal RR domain that inhibits caspase-8 function, thus suppressing TNF and Fas-induced apoptosis [72]. Curiously, this C-terminal RR, and not its N-terminal RHIM-like domain, also binds RIPK1 and prevents poly(I:C)-induced apoptosis in human cells [73]. Moreover, ICP6 and ICP10 can disrupt the RHIM-dependent interaction between TRIF and RIPK1, which may contribute to the disabling of apoptosis. Although the role of this cell death inhibition in viral pathogenesis has not been examined, it appears HSV has a strategy for managing dsRNA mediated apoptosis.
Similarly, initiation of IAV-induced PAN-optosis (pyroptosis, apoptosis, necroptosis) requires DAI sensing of viral RNA through its second Zα domain [74,75], though DAI is recently reported to also detect IAV nucleoprotein (NP) and polymerase subunit (PB1), as a collective unit of viral ribonucleoprotein [76,77]. Upon stimulation, DAI associates with RIPK3, which recruits RIPK1, FADD and MLKL to form a “ripoptosome” complex [75,78]. Here, the apoptotic arm operates under a RIPK1-FADD-caspase-8 axis, in a manner independent of RIPK1 kinase activity [78]. Neither caspase inhibition through z-VAD nor FADD deletion in mouse fibroblasts rescues cell viability, but pre-treatment of these cells with RIPK3 kinase inhibitors grants significant protection, thus highlighting the importance of RIPK3 in regulating the parallel cell death pathways. Surprisingly, DAI also triggers RIPK3-independent apoptosis, where DAI engages RIPK1 directly to recruit FADD and caspase-8, which deploys an alternative, delayed apoptotic cell death [75,78]. As inhibition of necroptosis does not affect IAV control, while Mlkl-/-Fadd-/- mice exhibit marked susceptibility to IAV-induced lethality [78], it signifies that RIPK1/3-mediated apoptosis is a major form of host protection against IAV infection.
1.2.2. RIPK1/3 in apoptotic responses to bacterial infections
In response to YopJ-mediated abrogation of inflammation during Yersinia infection [40], infected macrophages engage TLR4/TRIF and TNFR1 to induce apoptotic cell death [39]. Both cell death pathways occur in a non-redundant manner and rely on RIPK1-mediated activation of caspase-8 for downstream signaling [39,79]. The RIPK1-dependent apoptosis provides a cell-extrinsic signal required for cytokine production by inflammatory monocytes and neutrophils [80]. Consequently, Ripk1K45A/K45A mice orally infected with Y. pseudotuberculosis exhibit decreased caspase-3 staining within the mesenteric lymph nodes, as well as reduced levels of IL-12-producing monocytes and TNF-producing neutrophils. These mice are unable to restrict bacterial replication and systemic dissemination, resulting in rapid mortality.
1.3. RIPK1 and RIPK3-mediated necroptotic signaling in pathogen infection
Necroptosis has largely been studied in the context of antiviral responses, often as an alternate form of cell death for restricting viral replication. Necroptosis is a form of regulated lytic cell death that operates independent of caspases, and requires both RIPK3 and MLKL function (Fig. 2). It is important to note that although the studies discussed below have specified necroptosis as the specific cell death outcome involved in the infection process, some have only documented RIPK3 dependence but have not directly shown involvement of MLKL or pMLKL. Therefore, we have noted whether experimental evidence has been provided for dependence on MLKL within the text.
1.3.1. RIPK1/3 in necroptotic responses to viral infections
Mouse fibroblasts infected with vaccinia virus (VV), which is a poxvirus strain that encodes the caspase inhibitor B13R/Spi2, are shown to have resistance to TNF-induced apoptosis but increased sensitivity to RIPK3-dependent necroptosis [81,82]. VV-infected WT mice exhibit extensive inflammation and necrotic tissue damage within the liver, associated with formation of RIPK1-RIPK3 complexes [82]. Ripk3−/− mice fail to initiate virus-induced necroptosis, resulting in elevated viral titres and mortality [83,84]. The requirement for RIPK1 kinase activity remains unclear however, as groups have reported either increased [84] or unchanged [85] VV loads within the liver and spleen of infected Ripk1D138N/D138N mice. VV also encodes E3L, which contains an N-terminal Z-DNA binding domain that competes with DAI to inhibit DAI/RIPK3 necroptosis [86,87]. Administration of an E3L mutant lacking this domain in mouse fibroblasts and human epithelial cells results in increased IFN-induced necroptosis, featuring increased MLKL phosphorylation, and fails to elicit disease in WT mice [87].
Similarly, MCMV encodes a viral inhibitor of caspase-8 activation (vICA), which blocks DR-induced apoptosis but sensitises cells to necroptosis [88,89]. To combat this, MCMV expresses the viral inhibitor of RIP activation (vIRA), that binds RIPK1 and RIPK3 through its N-terminal RHIM domain [90]. vIRA is a potent inhibitor for MCMV-triggered necroptosis that is dependent on DAI/RIPK3 signaling [91]. MCMV mutants lacking vIRA or its RHIM domain are severely attenuated in WT mice, but unaffected in Ripk3−/− and Dai−/− mice [[91], [92], [93]]. Phosphorylation of MLKL occurs within hours during in vitro infection with vIRA RHIM mutant MCMV, and is reliant on IE3-mediated transcription of its genome [94]. Briefly, HCMV also elicits DAI-mediated production of IFNs and TNF-induced necroptosis in a RIPK-regulated manner [21,95,96]. Notably, the TNF-driven cell death is inhibited by HCMV-encoded IE1 through a yet unknown process that occurs after RIPK3 activation and MLKL phosphorylation, thus distinguishing the immunosuppressive strategies of HCMV from its murine counterpart [96].
HSV-1 and HSV-2 modulate immune responses in a manner similar to MCMV. They carry ICP6 and ICP10 respectively, which expresses an N-terminal RHIM domain that mediates RIPK1 and RIPK3 interaction, leading to anti-necroptotic signaling in human cells, but pro-necroptotic death in mice [[97], [98], [99]]. Expression of ICP6/10 or HSV challenge in human cells causes competitive RHIM binding that prevents RIPK1-RIPK3 necrosome formation and subsequent TNF-induced MLKL phosphorylation and necroptosis [97,98]. In contrast, ICP6 interaction with RIPKs in mouse cells promotes RIPK3-RIPK3 complex formation for MLKL recruitment and execution of necroptosis, independent of DR, DAI and TLR signaling [98,99]. This RIPK3-dependent necroptosis is crucial for controlling HSV-1 propagation, as Ripk3−/− mice present with markedly elevated susceptibility to infection and death, not seen in WT mice.
As mentioned previously, human γ-herpesvirus EBV binds RIPK1 and RIPK3 through the C-terminal activation region of its encoded LMP1 effector, which prevents RIPK1-RIPK3 complex formation in human nasopharyngeal cells [35]. Moreover, LMP1 promotes K63-linked polyubiquitylation of RIPK1 that forms the scaffolding required for TNF-induced NFĸB signaling [13], while inhibiting K63-linked polyubiquitylation of RIPK3 that typically supports necrosome assembly [35,100]. These post-translational modifications drive a switch from necroptotic death to a pro-survival cell fate, as indicated by the suppressed RIPK3 and MLKL phosphorylation following EBV infection in TNF-induced necroptotic cells [35].
As opposed to targeting the RHIM sequences of RIPKs, BeAn 58058 poxvirus (BAV) and Cotia poxvirus (COTV) carry viral MLKL-like proteins (vMLKL) that block necroptosis by interacting with the RIPK3 kinase domain [101]. In both human and mouse epithelial cells, vMLKL and RIPK3 bind via a pseudokinase to kinase domain interface, such that it overlaps the site typically engaged by cellular MLKL. In particular, this process in human cells prevents RIPK1 interaction, but drives RIPK3 phosphorylation despite pharmacological kinase inactivation of both RIPK1 and RIPK3 [101], which suggests that vMLKL alters RIPK3 in a manner that nulls kinase inhibitor treatment or promotes phosphorylation by another yet unknown kinase. However, in mouse cells, vMLKL inhibits RIPK3 phosphorylation, thus preventing RIPK3 activation and subsequent catalytic activity [101]. Regardless, vMLKL is capable of sequestering both mouse and human RIPK3 upon expression, which disrupts downstream TNF-induced necroptosis.
In contrast to DNA viruses, the mechanisms surrounding manipulation of necroptosis by RNA viruses are less explored. The HIV-1-encoded PR, previously described to inhibit NF-ĸB activation, also disrupts RIPK1-RIPK3 interaction in CD4 + T cells via RIPK1 cleavage [102]. This may contribute to necroptosis suppression, but no study has confirmed this observation. Additionally, necroptosis is implicated in the proliferative defect of HIV-specific CD8 + T lymphocytes in patients with progressive infection [103]. This defect is successfully reversed following pre-treatment of antigen-stimulated CD8 + T cells with necrosis inhibitor NecroX-5 or RIPK3 silencing. However, as MLKL dependence was not explored in this study, it would be appropriate to address this in further studies targeting necroptosis for HIV therapy.
Coxsackieviruses are the etiological agent of hand, foot and mouth disease [104]. Coxsackievirus A6 (CA6) infection of human cells triggers upregulation of RIPK3 via its non-structural protein (Nsp)-3D, which increases subsequent necrotic death [105]. This cell death can be inhibited by Nec-1, which dramatically reduces virus production, suggesting that necroptosis is required for CA6 infection. It should be specified that although the study attributes necroptosis to CA6 pathogenesis, no changes in MLKL nor pMLKL expression was found. In contrast, at late stages of human intestinal epithelial cell infection with coxsackievirus B3 (CVB), Nsp-3C (3Cpro) proteolytically cleaves RIPK3 and disrupts necroptosis, as determined by lack of HMGB1 release [37]. Intriguingly, the cleaved RIPK3 interacts with RIPK1 to induce a non-necrotic death [37], implying that CVB manipulates RIPK3 to redirect the host cell into a more favourable but yet undefined death pathway.
Respiratory syncytial virus (RSV) triggers necroptosis without direct modulation of RIPK1/3. RSV causes bronchiolitis in children, typically characterised by airway epithelial cell (AEC) death and massive cytokine release [106]. In response to infection, primary human AECs exhibit increased levels of pRIPK1, pMLKL and HMGB1 release, which correlates with increased necroptotic death [107]. Pharmacological inhibition of RIPK1 and MLKL reduces viral titres, suggesting that necroptosis promotes viral persistence. These results are recapitulated in a mouse model using pneumonia virus of mice (PVM). Here, inhibition of RIPK1 or MLKL lowered viral loads and prevented severe bronchiolitis, also seen in Ripk1K45A/K45A mice [107]. The necrosome is also reported to trigger neutrophil extracellular traps (NET) release in RSV-stimulated human neutrophils or neutrophils co-incubated with RSV-infected AECs [108], which facilitates RSV containment.
The topic of viral respiratory illness has gained much attention recently due to the emergence of the hypervirulent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for COVID-19. In an effort to identify potential therapeutic targets, Gordon et al. [109] have mapped the interactions between SARS-CoV-2 proteins and human proteins. Amidst the phenomenal set of results, RIPK1 was shown to associate with the viral Nsp-12, which is inferred to be an RNA polymerase. Perhaps unsurprisingly, the 2002 SARS-CoV has been shown to interact with RIPK3 via its open reading frame (ORF)-3a protein (SARS 3a) to promote SARS 3a oligomerization and subsequent necrotic death in human lung cells [110]. This process operates independently of RIPK3 kinase activity and MLKL [110], with SARS 3a likely replacing the latter as the necroptotic executioner due to its ability to function as an ion channel following membrane insertion [111]. However, SARS 3a also reduces RIPK3 and MLKL phosphorylation [110]. Infection with a SARS 3a-deletion mutant rescued mice from virus-induced mortality, suggesting an antiviral role for necroptosis in SARS-CoV infection [112].
1.3.2. RIPK1/3 in necroptotic responses to bacterial infections
Compared to viral pathogens, far fewer bacterial effectors have been identified to directly interact with RIPKs for necroptosis modulation. EPEC employs a cysteine protease, EspL, that directly cleaves the RHIM domain of RIPK1 and RIPK3, which prevents MLKL membrane complex formation during infection [113]. This inhibits TNF and TLR3/4-induced necroptosis in vitro. Furthermore, mice orally challenged with an espL deletion mutant of the EPEC-like mouse pathogen, Citrobacter rodentium, exhibit attenuated bacterial colonization in the intestine [113]. This suggests that EspL-mediated blockade of necroptosis contributes to bacterial persistence.
Staphylococcus aureus is an opportunistic pathogen that causes pneumonia and bacteraemia in immunosuppressed patients. Its pore-forming toxin (PFT) induces necroptosis in both human and murine pulmonary macrophages, and as such, inhibition of RIPK1 or MLKL in vitro, or genetic deletion of RIPK3 in vivo, significantly reduces cytotoxicity and improves S. aureus clearance in the lungs [114]. Other PFT-carrying bacterial pathogens, such as Serratia marcescens, Streptococcus pneumoniae, Listeria monocytogenes and uropathogenic E. coli (UPEC) also trigger RIPK1/RIPK3/MLKL-dependent necroptosis in macrophages, highlighting necroptosis as a promising target for PFT-associated disease intervention [115]. In contrast, in models of skin infection or sepsis, inhibition of RIPK1 or MLKL results in exacerbated disease, due to excessive IL-1β-induced inflammation [116]. This is not seen in Ripk3−/- mice, likely due to RIPK3 influence on inflammasome activation. These results indicate tissue-specific roles for RIPKs during S. aureus infection.
Similarly, Salmonella enterica serovar Typhimurium (S. Typhimurium) triggers necroptosis in macrophages to benefit bacterial survival. This process is dependent on initial type I IFN signaling following injection of S. Typhimurium in mice, as well as pathogen-mediated caspase-8 inactivation, which enables RIPK1 recruitment to IFNAR1 for phosphorylation and subsequent association with RIPK3 [117]. The resulting necroptotic cell death facilitates macrophage elimination, leading to compromised pathogen control. Alternatively, Ro et al. [118] reported that microRNA (miR)-155 upregulation following S. Typhimurium infection drives necroptotic macrophage death. Transfection of miR-155 in vitro induces RIPK1 and RIPK3 activation and subsequent necroptosis, which is partly inhibited following Nec-1 treatment. It is worth noting that although the authors describe the mode of cell death in both of the above S. Typhimurium infection studies as necroptosis, no experimental evidence showing MLKL phosphorylation or dependence on MLKL presence was provided.
Thus far, studies on mycobacterial-induced necroptosis in macrophages have been largely divisive. Mycobacterium tuberculosis triggers RIPK3-dependent necroptosis in both human and murine macrophages in a pathway reliant on reactive oxygen species (ROS) production and the mitochondrial Bcl-2 family member protein B-cell lymphoma-extra large (Bcl-xL) [119,120]. The ensuing macrophage death is suggested to assist bacterial pathogenesis, as Ripk3−/− macrophages display enhanced restriction of bacterial replication in vitro and in vivo. However, despite increased MLKL expression, pMLKL is not detected in infected macrophages, suggesting that the signaling process utilises an alternative executioner or is non-necroptotic. This is further complicated by results from Stutz et al. [121,122], which argue that deletion of MLKL or RIPK3 does not rescue macrophages from death during M. tuberculosis infection. In fact, the macrophage population and bacterial burden in infected Ripk3-/- mice are indistinguishable from WT controls. Further research is required to ascribe a role for necroptosis in mycobacterial infections.
1.4. RIPK1 and RIPK3-mediated inflammasome signaling in pathogen infection
Host cell death and inflammation can also be induced independently of death receptors through inflammasome signaling, as depicted in Fig. 2. RIPK1 and RIPK3 are primarily involved in alternative inflammasome activation pathways, which remain largely unexplored. A number of studies have used purified pathogen components such as LPS to investigate the outcomes of RIPK1/3-mediated inflammasome signaling, which appear to be largely dependent on cell type, stimulus, and the availability or functional activity of certain host signaling proteins [65,[123], [124], [125], [126], [127]]. Here we will discuss RIPK1 and RIPK3 involvement in inflammasome signaling during pathogen-specific infections.
1.4.1. RIPK1/3 in inflammasome responses to viral infections
Vesicular stomatitis virus (VSV) is a rhabdovirus that causes vesicular lesions on the mucosa of livestock [128]. VSV-infected mice exhibit NLRP3 inflammasome activation, characterised by elevated levels of cleaved caspase-1, as well as IL-1β and IL-18 secretion [129]. Here, RIPK1 complexes with RIPK3 following stimulation of a yet unidentified RNA sensor, which enables RIPK1-mediated activation of dynamin-related protein 1 (DRP1). Subsequent translocation of DRP1 to the mitochondria promotes aberrant fission and ROS production in both mouse and human cells, thus activating NLRP3 inflammasome. This DRP1-mediated inflammasome signaling is reported to also occur in response to dengue virus [130] and swine influenza virus infection [131]. Some studies contradict these observations [[132], [133], [134]], likely due to different experimental models, therefore additional research is necessary to conclusively define this form of NLRP3 signaling.
1.4.2. RIPK1/3 in inflammasome responses to bacterial infections
Yersinia is capable of inducing caspase-1 processing and cell death following infection of mouse macrophages [39,135,136]. This is driven by YopJ-mediated suppression of TAK1, which enables RIPK1, FADD and caspase-8 recruitment, and subsequent activation of caspase1. The resulting cell death exhibits significant RIPK1/caspase-8-mediated GSDMD cleavage, implicating pyroptosis in this process [136]. Notably, NLRP3 and caspase-1/11 are not involved in this pyroptotic pathway, but instead activated following potassium efflux to promote IL-1β processing and release. These observations illustrate an alternative NLRP3-independent mechanism for caspase-1 and GSDMD activation during Yersinia infection.
2. RIPK2 signaling in pathogen infection
RIPK2 (also known as RIP2, RICK and CARDIAK) is an essential scaffold for signal transduction via the nucleotide-binding oligomerization domain (NOD) proteins, NOD1 and NOD2 [[137], [138], [139], [140]], and is thus frequently implicated in innate inflammatory responses to pathogens. NOD proteins are cytosolic pattern-recognition receptors (PRRs) that activate pro-inflammatory and antimicrobial responses when exposed to pathogen associated molecular patterns (PAMPs). NOD1 recognizes Ɣ-d-glutamyl-mesodiaminopimelic acid (iE-DAP) from Gram-negative bacteria and some Gram-positive bacteria, whereas NOD2 recognizes a conserved component of bacterial peptidoglycan (PGN) consisting of muramyl dipeptide (MDP) from both Gram-positive and Gram-negative bacteria [[141], [142], [143], [144]] (Fig. 3 ). These stimulatory agents are released from bacteria upon cell wall fragmentation in bacterial killing, bacterial division, or they can be co-injected into host cells with virulence proteins by bacterial secretion systems [145]. Alternative mechanisms of NOD/RIPK2 activation will be discussed in context within the review.
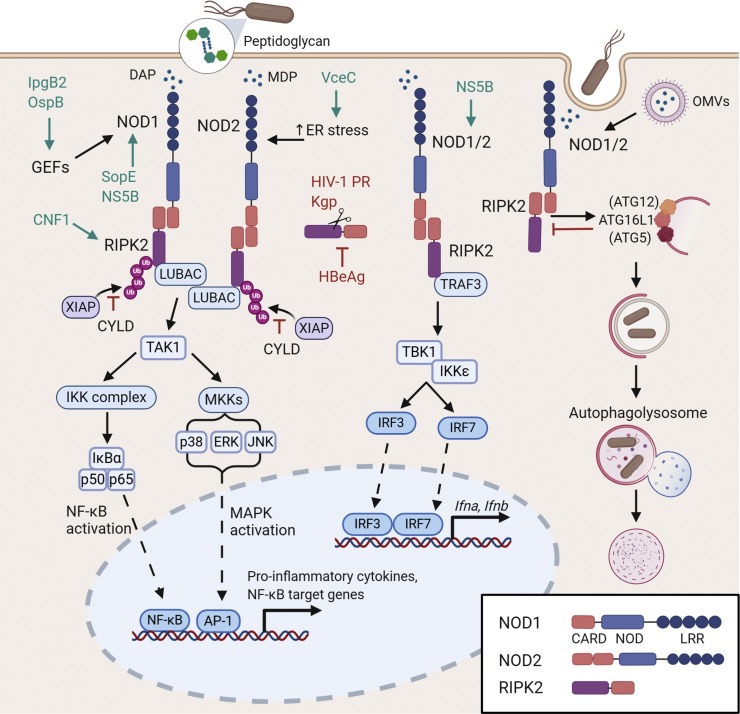
Fig. 3.
RIPK2 regulation of NOD1 and NOD2 signaling in response to PAMP sensing. Upon activation by bacterial peptidoglycan components, NOD1 and NOD2 oligomerize and interact with RIPK2 via homotypic CARD-CARD interactions [137]. Once engaged, RIPK2 is activated by autophosphorylation, then ubiquitylated by E3 ligases including XIAP and LUBAC, which further activates both NF-κB and MAPK pathways and promotes pro-inflammatory cytokine production [146]. Alternatively, TRAF3 can interact with RIPK2 following NOD1/2 ligation, redirecting signaling to TBK1 and IKKε to promote downstream IFN production [147]. Other roles of RIPK2 include mediating interactions between NOD1/2 and key autophagy protein ATG16L1, which enables autophagic bacterial clearance following NOD sensing of peptidoglycan or bacterial OMVs [[148], [149], [150]]. Virulence factors that interact with these pathways are indicated, and have been discussed in the main body of the text.
2.1. RIPK2 in host responses to bacterial pathogens
To date, much of the research on NOD1/NOD2/RIPK2 signaling mechanisms has relied on purified PGN components as a cellular stimulus. This has been useful for the identification of cellular mechanisms, however can have limitations when considering the physiological role of signaling mediators in the context of pathogen infection, especially when many pathogens encode virulence factors that can inactivate innate immune signaling. Although many studies have assessed mechanisms of NOD signaling in bacterial control, especially in the context of autophagy, this review will focus on studies with direct experimental evidence of RIPK2 involvement, and where live bacterial agents have been utilised.
Some of the earliest studies that characterised the role of RIPK2 in host responses to infections utilised L. monocytogenes, the causative agent of listeriosis. Initial studies showed macrophages from Ripk2 −/− mice were defective in NF-κB signaling and produced significantly less pro-inflammatory cytokines than WT macrophages following infection [137,151]. Furthermore, Ripk2 −/− mice were unable to control L. monocytogenes infection due to decreased NF-κB activation, and impaired IFNƔ production in T helper 1 (Th1) and natural killer (NK) cells. Overall this suggests that RIPK2 plays an important role in both innate and adaptive immunity to infection [137,151], and that RIPK2 was mediating these responses via NOD1/2 and not directly via TLR activation [140].
RIPK2 has since been shown to play a role in controlling a diverse array of bacterial pathogens, particularly those with an intracellular lifestyle (Table 1). Single nucleotide polymorphisms (SNPs) in RIPK2 increase susceptibility to mycobacterial infections, for example, multibacillary leprosy caused by M. leprae [152], and M. tuberculosis infections within the Western Chinese Han population [153]. Although the mechanisms underlying susceptibility are unclear, M. tuberculosis activates NOD2/RIPK2, which stimulates the activity of IRF5 to induce transcription of type I IFNs [147]. Furthermore, IFNγ production by Th1 cells induces macrophage maturation and anti-mycobacterial molecules important for resistance against mycobacterial infections [154,155]. Zhang et al. [152] suggested RIPK2 and NOD2 may regulate IFNγ which could explain increased susceptibility to mycobacterial infections in those with RIPK2 polymorphisms.
Streptococcus pneumoniae (pneumococcus) is a an opportunistic pathogen associated with pneumonia, ear infections, sinus infections, meningitis and bacteremia [156]. NOD2/RIPK2 are critical for anti-inflammatory signaling in response to the purified pneumococcal cell wall (PnCW) of S. pneumoniae. PnCW induces intensive inflammatory responses by macrophages and dendritic cells during systemic infection in a TLR2-dependent, NOD2/RIPK2-independent manner [157,158]. This inflammation is critically modulated by IL-10 [159], as IL-10 deficiency increases mortality in S. pneumoniae infection in vivo [160]. Curiously, this IL-10 production is TLR2, NOD2 and RIPK2-dependent, whereby RIPK2 or NOD2-deficient BMDMs have compromised IL-10 production in response to PnCW [157]. Although the mechanism is not clear, this suggests that there is cell-specific and stimulus-specific crosstalk between TLR2 and NOD2/RIPK2 pathways [161].
During L. pneumophila infection, RIPK2 mediates NF-κB activation independently of TLR/MyD88 activation, but in response to bacterial factors (likely PGN) delivered directly into the host cell cytosol by the Legionella T4SS [145]. in vivo studies have shown that RIPK2-deficiency results in poor neutrophil recruitment into the lung, a significant reduction in proinflammatory cytokine and chemokine secretion, and increased bacterial burden [162]. While one study suggested only NOD1 was involved in these responses in Ripk2−/− mice [163], another implicated both NOD1 and NOD2 [162]. Overall, it appears NOD signaling plays a role in RIPK2-mediated responses to L. pneumophila, however it doesn’t rule out involvement of other signaling pathways initiated by TLR2, IL-1R or IL-18R [137].
Borrelia burgdorferi is the causative agent of Lyme disease, a tick-borne infection that causes multi-systemic illness [164]. Early studies showed that RIPK2 expression is increased in astrocytes and microglia exposed to Borrelia spirochetes [165,166]. Subsequent work showed that peritoneal macrophages from Ripk2−/− mice exhibit a significant reduction in IL-1β, IL-6 and IL-8 production following stimulation with heat-killed Borrelia, compared to WT mice [167]. Here, through an unknown mechanism, uptake and degradation of Borrelia in lysosomes introduces PGN to the cytosol and stimulates NOD2/RIPK2-mediated NF-κB activation and inflammatory cytokine production [167].
2.1.1. RIPK2 and autophagy as a host response to bacterial infection
RIPK2 induces antibacterial autophagic responses by signaling between NODs and the autophagy factor ATG16L1 [148] (Fig. 3). Mutations in ATG16L1 disrupt an inhibitory interaction with NOD2 and consequently increase the activation of RIPK2 [150]. Excessive RIPK2 activation has been reported in pediatric Crohn’s disease (CD) [168,169] and there is a strong link between resident intestinal bacteria and CD pathogenesis; therefore, it has been proposed that ineffective bacterial clearance due to impaired anti-bacterial autophagy is an important contributor to the pathogenesis of this chronic inflammatory disease [170,171]. Autophagy has an essential role in innate immunity and the elimination of pathogens that have escaped into the cytoplasm, as it forms a double-layered membrane that envelopes cytosolic bacteria for degradation via fusion with lysosomes [[172], [173], [174]].
The invasive gastrointestinal pathogen, Shigella flexneri, is a major cause of morbidity and mortality in children under 5 years in developing countries [175], and is an emerging sexually transmitted infection of men who have sex with men [176]. S. flexneri induces NF-κB and JNK activation in a NOD1/RIPK2-dependent manner to limit bacterial replication in intestinal epithelial cells [177]. Although early studies did not investigate mechanisms of S. flexneri killing, it was pivotal in understanding the mechanisms of NOD/RIPK2-mediated autophagy in future infection studies.
The gastric pathogen Helicobacter pylori is subject to degradation by autophagy via the NOD1/RIPK2 signaling axis. H. pylori infection has long been implicated in the development of gastric cancer [[178], [179], [180]], and recently, polymorphisms in RIPK2 were found to be associated with increased susceptibility to gastric cancer in Japanese populations [181] where the prevalence of this disease is very high. Indeed, Nod1−/− mice are highly susceptible to H. pylori infection [182], and mechanistically, Irving et al. [149] demonstrated that in gastric epithelial cells, RIPK2 mediates NOD1-dependent IL-8 production and autophagosome formation in early endosomes in response to H. pylori-derived outer membrane vesicles (OMVs) containing PGN. Overall, NOD1/RIPK2 signaling protects against H. pylori infection and subsequent malignancies.
NOD2/RIPK2-mediated autophagy aids in the control of a number of pathogens including L. monocytogenes, S. Typhimurium and Shigella spp. [183]. L. monocytogenes undergoes autophagosomal degradation in phagocytic cells in mouse BMDMs and in vivo via ERK activation, in a process mediated by TLR2, NOD2 and RIPK2 [184]. In S. Typhimurium infected intestinal epithelial cells, NOD2/RIPK2 is required for autophagy induction [185]. This in vitro model of Salmonella infection demonstrated a dual role for RIPK2 tyrosine kinase activity in NOD2-dependent autophagy through activation of p38 MAPK and indirect repression of PP2A phosphatase activity [185].
2.1.2. Alternative mechanisms of NOD/RIPK2 stimulation in bacterial infections
In addition to PGN stimulation, there is increasing evidence that NOD/RIPK2 signaling can be activated by pathogen-induced modifications to the host actin cytoskeleton [[186], [187], [188], [189]]. Cytoskeletal dynamics are mediated via a balance of active GTP-bound and inactive GDP-bound forms of small Rho GTPases [190]. Salmonella and Shigella spp. utilise T3SS to translocate bacterial effector proteins into host cells, and manipulate host cytoskeletal proteins and innate immune responses [191]. Shigella infection induces the recruitment of GEF-H1, a guanidine exchange factor (GEF) for the small Rho GTPase RhoA, to the site of invasion to promote host cell entry. Following invasion, the Shigella effectors IpgB2 and OspB induce RIPK2-dependent NF-κB activation mediated by the interaction of recruited GEF-H1 with NOD1 [187]. The S. Typhimurium effector SipA drives NOD1/NOD2/RIPK2 dependent NFκB activation via an unknown mechanism [189], whereas, SopE, functions as a GEF for the small Rho GTPases Rac1 and Cdc42. In this setting, Rac1 and Cdc42 interact with the NOD1/RIPK2 signaling complex in the absence of PGN to mediate NF-κB-dependent inflammation [188]. In addition, the Escherichia coli cytotoxic necrotising factor 1 (CNF1) activates the small Rho GTPase Rac2, which then interacts with RIPK1 and RIPK2 to induce a potent inflammatory response, independent of NOD1/2 [192]. Many other bacterial pathogens have also been shown to induce changes to Rho GTPases [193], overall highlighting the role of pathogen-induced small Rho GTPase activation in NOD1/RIPK2-mediated inflammation.
Pathogen-activated endoplasmic reticulum (ER)-stress also drives NOD/RIPK2-induced inflammation [194]. The intracellular pathogen Brucella abortus induces ER stress via the IRE1⍺ pathway of the unfolded protein response (UPR). IRE1⍺ acts as a receptor that is stimulated upon binding of the Brucella T4SS effector VceC to the ER chaperone BiP, and subsequently recruits TRAF2 to activate NOD2/RIPK2-mediated NF-κB activation [194]. Although the precise mechanism is not yet established, the intracellular pathogen Chlamydia muridarum also induces NOD1/NOD2/RIPK2 signaling in response to ER stress in vitro [194], however in vivo studies have shown that RIPK2 deletion has a limited effect on chlamydial infection in terms of bacterial burden, immune responses and pathology [195]. Given that many pathogens including IAV [196] and HCMV [197] activate ER-stress followed by induction of the UPR, it could be likely that NOD/RIPK2 signaling has an underappreciated role in host immunity via this pathogen-induced mechanism.
2.1.3. Regulation of RIPK2 and implications for bacterial infection outcomes
Regulation of RIPK2-mediated inflammatory responses to infection is dependent upon the deubiquitinating enzyme CYLD [198]. During in vitro infection of mouse bone marrow-derived macrophages (BMDMs) with L. monocytogenes, CYLD binds and deubiquitylates RIPK2, resulting in decreased activation of NF-κB and ERK1/2 signaling. Thus, inhibition of RIPK2 by CYLD leads to impaired pathogen control due to a reduction in antimicrobial responses including pro-inflammatory cytokine production, ROS and nitric oxide (NO) production.
Another recent study used kinase inhibitors to demonstrate functional specificity of the kinase domain of RIPK2 in controlling bacterial pathogens. WEHI-345 is a potent inhibitor of RIPK2 that specifically targets serine/threonine kinase activity [146] and pre-treatment of CD11β+ monocytes with this inhibitor significantly reduces TNF production during in vitro infection with L. monocytogenes [146], suggesting a role for the serine/threonine kinase activity of RIPK2 in protection against bacterial infection.
RIPK2 signaling may also be regulated via the formation of RIPosomes, which are high molecular weight cytoplasmic complexes comprised of RIPK2 [199,200]. Ellwanger et al. [199] showed RIPosomes form post-NF-κB activation, and suggest that sequestration of RIPK2 in these complexes may act to dampen RIPK2 signaling. Intriguingly, RIPosomes form in the cytosol of epithelial cells upon invasion with S. flexneri, suggesting the pathogen may actively prevent RIPK2 signaling via an unknown process. Given that inhibition of XIAP was shown to promote deposition of RIPK2 in RIPosomes, it may be that Shigella encodes a virulence factor that targets XIAP for cleavage or degradation, or actively inhibits XIAP-designated sites of ubiquitylation on RIPK2 to inhibit inflammatory signaling [199].
2.2. RIPK2-mediated inflammatory responses in the control of viral pathogens
It is now established that NOD1 and NOD2 respond to viral infections, thus participating in the coordinated host defense against viruses [[201], [202], [203], [204]]. Activation of NOD1, NOD2 and RIPK2 during viral infection depends on type I IFN, synthesized as a result of activation of other PRRs. One of the first studies to assess the role of NOD2/RIPK2 activation in viral infection showed that RIPK2 was critical for dampening inflammasome activation during H1N1 IAV infection [205]. Here, Ripk2 −/− mice were highly susceptible IAV infection, whereby enhanced NLRP3 inflammasome activation and increased IL-18 secretion were potent drivers of disease progression and mortality in vivo. Negative regulation of inflammasome activity by RIPK2 is dependent its kinase-mediated activation of the mitophagy inducer, ULK1. Thus NOD2 and RIPK2 respond to IAV infection by promoting ULK1 phosphorylation and inducing mitophagy, which dampens inflammasome activation and IL-18 production. In addition, both NK cells and CD8 + T cells isolated from IAV-infected Ripk2-/- mice are highly activated and exhibit increased IFN-γ production despite the total numbers of these cells being similar in WT mice. These results indicate that increased IL-18 in Ripk2-/- mice subsequently leads to increased IFN-γ production from innate and adaptive cell populations [205].
Hepatitis B and C virus (HBV and HCV) are associated with the development of hepatocellular carcinoma [206]. NOD1/RIPK2 signaling is activated by the viral polymerase NS5B of HCV [207], thus deletion of RIPK2 in HepaRG cells expressing NS5B, results in significantly reduced MAPK activation, proinflammatory cytokine production, and IFNβ production. In HBV infection, the Hepatitis B e-antigen (HBeAg) inhibits RIPK2 expression and also interacts with RIPK2 in HepG2 cells in vitro, resulting in inhibition of NOD1/RIPK2-mediated NF-κB activation and subsequent IL-6 production [208]. These studies highlight the importance of RIPK2 in controlling chronic Hepatitis infections.
2.3. Pathogens targeting RIPK2 for inactivation
To date, the only known pathogens to directly target RIPK2 during infection are HIV-1 and the primary etiologic agent of periodontal disease, Porphyromonas gingivalis. The HIV-1 protease PR cleaves RIPK2 within the N-terminus, although its outcomes in infection have not been tested [102]. Similarly, P. gingivalis infection of human aortic endothelial cells results in rapid direct cleavage of RIPK2, and is dependent upon the lysine-specific protease, gingipain (Kgp) [209]. Given the mounting evidence for pathogens targeting RIP kinases, it would not be surprising if other pathogens were found to specifically inactivate RIPK2 in future studies.
3. The role of RIPK4, RIPK5, RIPK6 and RIPK7 in pathogen infection
There is currently relatively little published on RIPK4-6 and their role in host responses to pathogen infection, however RIPK7 is emerging as an important mediator of immunity to intracellular pathogens.
3.1. RIPK4 (DIK/PKK) and RIPK5 (SgK288)
RIPK4 has a well-described role in cellular differentiation, but also mediates proinflammatory cytokine production in keratinocytes via direct stimulation of IRF6 [210]. Furthermore, overexpression of RIPK4 leads to NF-κB and MAPK activation [211]. Although no study has reported a direct role for RIPK4 in host responses to pathogens, it would not be surprising to find RIPK4 mediates inflammation during infection. RIPK5 on the other hand, has no reported association with innate or adaptive immune responses in mammals.
3.2. RIPK6 (LRRK1) and RIPK7 (LRRK2)
Pathogenic variants of RIPK7 are one of the most prominent genetic causes of Parkinson disease (PD) [212], whereas RIPK6 variants have been shown to have no association with the development of PD. As for infection, the only reported data for RIPK6 is in relation to bovine viral diarrhoea virus (BVDV), where RIPK6 is downregulated in PBMCs infected with BVDV-2 [213]. RIPK7 however, has been shown to play a role in numerous cellular processes associated with pathogen control, including vesicular trafficking, microtubule binding, autophagy and mitophagy [214]. One of the first studies to examine the role of RIPK7 in innate immunity found that RIPK7 contributes to the restriction of S. Typhimurium by macrophages in vitro [215]. This was supported in vivo as Ripk7 −/− mice are more susceptible to S. Typhimurium intraperitoneal infection, exhibiting decreased IL-1β secretion, reduced neutrophil infiltration, high bacterial load in the peritoneal cavity and overall increased mortality [216]. Mechanistically, this study showed RIPK7 forms a complex with the NLRC4 inflammasome in a kinase dependent manner, which then promotes inflammasome activation and restriction of bacterial growth during infection [216]. Similarly, Ripk7-/- mice are more susceptible to oral infection with L. monocytogenes than WT mice [217]. This study showed that RIPK7 is highly expressed in lysozyme-positive Paneth cells and myeloid cells within the lamina propria of the ileum, suggesting RIPK7 plays a protective role at the intestinal mucosa. This is further supported by the fact that mutations in RIPK7 are associated with increased severity of inflammatory bowel disease [218].
Similar to RIPK2, polymorphisms in RIPK7 are associated with the development of multibacillary leprosy caused by M. leprae [152,219,220] and Mtb infection [221]. A recent study identified a mechanism whereby RIPK7 negatively regulates phagosome maturation in macrophages by controlling Rubicon/PI3K activity on phagosomes in a kinase dependent manner, resulting in impaired immune responses and promotion of Mtb replication [222] (Fig. 4 ). Contradictory to the requirement of RIPK7 for the control of Salmonella and Listeria infection, RIPK7-deficiency in macrophages or mice results in improved control of Mtb infection, which supports a specific role for RIPK7 in mycobacterial control via the regulation of degradative pathways [222]. Ripk7−/− mice exhibit increased transcription of type II IFN, but decreased transcription of type I IFN during Mtb infection [222], and RIPK7-deficient macrophages fail to induce type I IFN in vitro when infected with Mtb [223]. Mechanistically, RIPK7 regulates type I IFN gene expression by maintaining mitochondrial homeostasis [223]. Given that production of type I IFNs during Mtb infection have been shown to promote disease [224], this may explain why Mtb infection is limited in the absence of RIPK7. Thus, RIPK7 acts as a regulator of early clearance of Mtb and given its function is kinase dependent, there may be therapeutic potential for RIPK7-specific kinase inhibitors in tuberculosis.
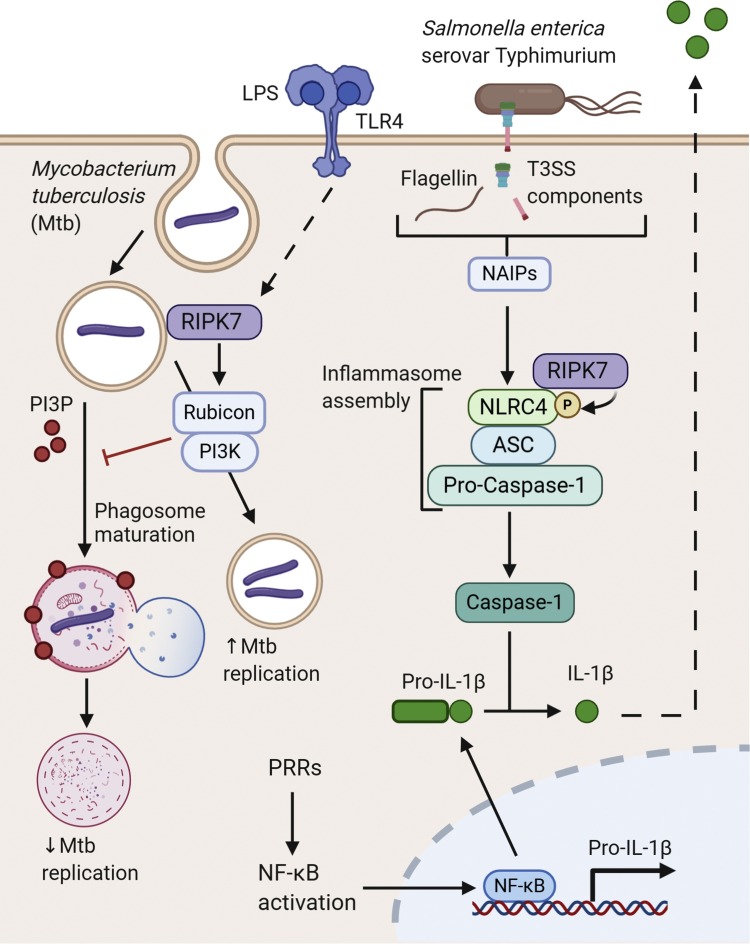
Fig. 4.
Cellular responses to bacterial infection mediated by RIPK7 (LRRK2). Sensing of LPS by TLR4 promotes localization of RIPK7 to endosomal membranes [225,226]. Here, RIPK7 can be exploited by M. tuberculosis (Mtb) to promote bacterial replication, as RIPK7 recruits Rubicon to the endosome, where this complexes with PI3K to prevent further phagosome maturation [222]. Following priming by PRRs, detection of S. Typhimurium bacterial components such as flagellin (or Type III Secretion System rod proteins) by NAIP family members induces NLRC4 activation [227]. In contrast to RIPK7’s role in Mtb infection, kinase-dependent interactions between RIPK7 and NLRC4 promote efficient inflammasome assembly and aid downstream restriction of bacterial growth [216].
Sensing of LPS by TLR4 promotes localization of RIPK7 to endosomal membranes [225,226]. Here, RIPK7 can be exploited by M. tuberculosis (Mtb) to promote bacterial replication, as RIPK7 recruits Rubicon to the endosome, where this complexes with PI3K to prevent further phagosome maturation [222]. Following priming by PRRs, detection of S. Typhimurium bacterial components such as flagellin (or Type III Secretion System rod proteins) by NAIP family members induces NLRC4 activation [227]. In contrast to RIPK7’s role in Mtb infection, kinase-dependent interactions between RIPK7 and NLRC4 promote efficient inflammasome assembly and aid downstream restriction of bacterial growth [216].
4. Concluding remarks and future perspectives
RIPK1, 2, 3 and 7 have emerged as critical mediators of inflammation and innate immunity in response to multiple diverse pathogens. Not surprisingly, many pathogens have evolved highly specific mechanisms to either directly or indirectly target RIPK signaling networks to benefit replication and survival, and have thus provided invaluable knowledge on the physiological role of RIPK signaling in the context of infection. One of the major challenges that remain in the field of host-pathogen interactions is the consistency of experimental conditions, whereby factors including genetic background of animal models, specific pathogen strains (lab adapted vs currently circulating clinical isolates), cell types (primary, site specific vs immortalised carcinoma cell lines) and the use of inhibitors (e.g. Nec-1 vs Nec-1 stable) heavily influence experimental outcomes. The more unified this becomes globally, the more reliable the data will become.
Finally, RIPKs are currently under critical review as potential therapeutic targets, as dysregulation of RIPK signaling is closely associated with hyperinflammation and pathology. Multiple studies are investigating various classes of kinase inhibitors for RIPK1-3 [228], however given the importance of kinase-dependent cell-death responses to infection, it is critical that we understand the impact of these therapeutic interventions on infection outcomes before introduction to the clinic. The same goes for the recently identified small molecule therapy, Proteolysis-Targeting Chimeras (PROTACs), for the selective degradation of RIPK2 [229]; given the importance of RIPK2 in detection of multiple intracellular pathogens, what would be the effect on infection outcome?
Overall, there has been significant progress made in the field of RIPK biology, and continued efforts on this front will help to bolster our understanding of host-pathogen interactions and potential therapeutic development for infectious diseases. To this end, we must continue to develop a comprehensive understanding the of 1) the biochemical function of each RIPK domain and the role they play in response to specific pathogens, and 2) biochemical mechanisms of virulence factors in currently circulating pathogenic organisms.
Declaration of competing interest
We, the authors declare no competing interests.
Funding
VVE is supported by a Monash Graduate Scholarship, co-funded by the Hudson Institute of Medical Research. MAW is supported by an Australian Government Research Training Program Scholarship. JSP is supported by an Australian National Health and Medical Research Council Career Development Fellowship (APP1159230).
Acknowledgements
We would like to acknowledge Medina Pell for her assistance with assembling the manuscript.
References
- 1.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.He S., Wang X. RIP kinases as modulators of inflammation and immunity. Nat. Immunol. 2018;19:912–922. doi: 10.1038/s41590-018-0188-x. [DOI] [PubMed] [Google Scholar]
- 3.Tigno-Aranjuez J.T., Asara J.M., Abbott D.W. Inhibition of RIP2’s tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev. 2010;24:2666–2677. doi: 10.1101/gad.1964410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelliher M.A., Grimm S., Ishida Y., Kuo F., Stanger B.Z., Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y., Devin A., Cook A., Keane M.M., Kelliher M., Lipkowitz S., Liu Z.G. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol. Cell. Biol. 2000;20:6638–6645. doi: 10.1128/mcb.20.18.6638-6645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanger B.Z., Leder P., Lee T.H., Kim E., Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 7.Cusson-Hermance N., Khurana S., Lee T.H., Fitzgerald K.A., Kelliher M.A. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 8.Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 9.Orozco S., Oberst A. RIPK3 in cell death and inflammation: the good, the bad, and the ugly. Immunol. Rev. 2017;277:102–112. doi: 10.1111/imr.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu H., Shu H.-B., Pan M.-G., Goeddel D.V. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/S0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 11.Micheau O., Tschopp J. Induction of TNF receptor I-Mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/S0092-8674(03)00521-X. [DOI] [PubMed] [Google Scholar]
- 12.Devin A., Lin Y., Liu Z.G. The role of the death-domain kinase RIP in tumour-necrosis-factor-induced activation of mitogen-activated protein kinases. EMBO Rep. 2003;4:623–627. doi: 10.1038/sj.embor.embor854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ea C.K., Deng L., Xia Z.P., Pineda G., Chen Z.J. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Varfolomeev E., Goncharov T., Fedorova A.V., Dynek J.N., Zobel K., Deshayes K., Fairbrother W.J., Vucic D. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J. Biol. Chem. 2008;283:24295–24299. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witt A., Vucic D. Diverse ubiquitin linkages regulate RIP kinases-mediated inflammatory and cell death signaling. Cell Death Differ. 2017;24:1160–1171. doi: 10.1038/cdd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang M., Jin W., Sun S.C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleh D., Najjar M., Zelic M., Shah S., Nogusa S., Polykratis A., Paczosa M.K., Gough P.J., Bertin J., Whalen M., Fitzgerald K.A., Slavov N., Pasparakis M., Balachandran S., Kelliher M., Mecsas J., Degterev A. Kinase activities of RIPK1 and RIPK3 can direct IFN-β synthesis induced by lipopolysaccharide. J. Immunol. 2017;198:4435–4447. doi: 10.4049/jimmunol.1601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., Yamaguchi O., Otsu K., Tsujimura T., Koh C.S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 19.Rajput A., Kovalenko A., Bogdanov K., Yang S.-H., Kang T.-B., Kim J.-C., Du J., Wallach D. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by Caspase-8-Mediated cleavage of the RIP1 protein. Immunity. 2011;34:340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser W.J., Upton J.W., Mocarski E.S. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebsamen M., Heinz L.X., Meylan E., Michallet M.-C., Schroder K., Hofmann K., Vazquez J., Benedict C.A., Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shives K.D., Tyler K.L., Beckham J.D. Molecular mechanisms of neuroinflammation and injury during acute viral encephalitis. J. Neuroimmunol. 2017;308:102–111. doi: 10.1016/j.jneuroim.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardy R.D., Richer M.J. Zika Virus Pathogenesis: from early case reports to epidemics. Viruses. 2019;11:886. doi: 10.3390/v11100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniels B.P., Snyder A.G., Olsen T.M., Orozco S., Oguin T.H., III, Tait S.W.G., Martinez J., Gale M., Jr., Loo Y.-M., Oberst A. RIPK3 restricts viral pathogenesis via cell death-independent neuroinflammation. Cell. 2017;169:301–313.e311. doi: 10.1016/j.cell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniels B.P., Kofman S.B., Smith J.R., Norris G.T., Snyder A.G., Kolb J.P., Gao X., Locasale J.W., Martinez J., Gale M., Jr., Loo Y.-M., Oberst A. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity. 2019;50:64–76.e64. doi: 10.1016/j.immuni.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy M.P., O’Neill L.A.J. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell. 2018;174:780–784. doi: 10.1016/j.cell.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Cho H., Proll S.C., Szretter K.J., Katze M.G., Gale M., Jr., Diamond M.S. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 2013;19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mack C., Sickmann A., Lembo D., Brune W. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. 2008;105:3094–3099. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon K.M., Oh S.E., Kim Y.E., Han T.-H., Ahn J.-H. Cooperative inhibition of RIP1-mediated NF-κB signaling by cytomegalovirus-encoded deubiquitinase and inactive homolog of cellular ribonucleotide reductase large subunit. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim E.T., Oh S.E., Lee Y.-O., Gibson W., Ahn J.-H. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 2009;83:12046–12056. doi: 10.1128/jvi.00411-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian P., Ye C., Zheng X., Luo C., Yang J., Li M., Wang Y., Yang J., Zhou Y., Zhang F., Lian J., Zhang Y., Jia Z., Lei Y. RIPK3 promotes JEV replication in neurons via downregulation of IFI44L. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downey J., Pernet E., Coulombe F., Allard B., Meunier I., Jaworska J., Qureshi S., Vinh D.C., Martin J.G., Joubert P., Divangahi M. RIPK3 interacts with MAVS to regulate type I IFN-mediated immunity to Influenza A virus infection. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006326. e1006326-e1006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz O., Pichlmair A., Rehwinkel J., Rogers N.C., Scheuner D., Kato H., Takeuchi O., Akira S., Kaufman R.J., Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Li Y., Peng S., Yu X., Li W., Shi F., Luo X., Tang M., Tan Z., Bode A.M., Cao Y. Epstein-Barr virus encoded latent membrane protein 1 suppresses necroptosis through targeting RIPK1/3 ubiquitination. Cell Death Dis. 2018;9 doi: 10.1038/s41419-017-0081-9. 53–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris Katharine G., Morosky Stefanie A., Drummond Coyne G., Patel M., Kim C., Stolz Donna B., Bergelson Jeffrey M., Cherry S., Coyne Carolyn B. RIP3 regulates autophagy and promotes coxsackievirus B3 infection of intestinal epithelial cells. Cell Host Microbe. 2015;18:221–232. doi: 10.1016/j.chom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philip N., Brodsky I. Cell death programs in Yersinia immunity and pathogenesis. Front. Cell. Infect. Microbiol. 2012;2 doi: 10.3389/fcimb.2012.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng D., Marty-Roix R., Ganesan S., Proulx M.K., Vladimer G.I., Kaiser W.J., Mocarski E.S., Pouliot K., Chan F.K.-M., Kelliher M.A., Harris P.A., Bertin J., Gough P.J., Shayakhmetov D.M., Goguen J.D., Fitzgerald K.A., Silverman N., Lien E. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc. Natl. Acad. Sci. 2014;111:7391–7396. doi: 10.1073/pnas.1403477111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma K.-W., Ma W. YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol. Mol. Biol. Rev. 2016;80:1011–1027. doi: 10.1128/mmbr.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton H.J., Ang D.K.Y., van Driel I.R., Hartland E.L. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 2010;23:274. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valleau D., Quaile A.T., Cui H., Xu X., Evdokimova E., Chang C., Cuff M.E., Urbanus M.L., Houliston S., Arrowsmith C.H., Ensminger A.W., Savchenko A. Discovery of ubiquitin deamidases in the pathogenic arsenal of Legionella pneumophila. Cell Rep. 2018;23:568–583. doi: 10.1016/j.celrep.2018.03.060. [DOI] [PubMed] [Google Scholar]
- 43.Cui J., Yao Q., Li S., Ding X., Lu Q., Mao H., Liu L., Zheng N., Chen S., Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taieb F., Nougayrède J.P., Oswald E. Cycle inhibiting factors (cifs): cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins (Basel) 2011;3:356–368. doi: 10.3390/toxins3040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S., Zhang L., Yao Q., Li L., Dong N., Rong J., Gao W., Ding X., Sun L., Chen X., Chen S., Shao F. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501:242–246. doi: 10.1038/nature12436. [DOI] [PubMed] [Google Scholar]
- 46.Pearson J.S., Giogha C., Ong S.Y., Kennedy C.L., Kelly M., Robinson K.S., Lung T.W.F., Mansell A., Riedmaier P., Oates C.V.L., Zaid A., Mühlen S., Crepin V.F., Marches O., Ang C.-S., Williamson N.A., O’Reilly L.A., Bankovacki A., Nachbur U., Infusini G., Webb A.I., Silke J., Strasser A., Frankel G., Hartland E.L. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature. 2013;501:247–251. doi: 10.1038/nature12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton H.J., Pearson J.S., Badea L., Kelly M., Lucas M., Holloway G., Wagstaff K.M., Dunstone M.A., Sloan J., Whisstock J.C., Kaper J.B., Robins-Browne R.M., Jans D.A., Frankel G., Phillips A.D., Coulson B.S., Hartland E.L. The type III effectors NleE and NleB from enteropathogenic E. Coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000898. e1000898-e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson J.S., Riedmaier P., Marchès O., Frankel G., Hartland E.L. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol. Microbiol. 2011;80:219–230. doi: 10.1111/j.1365-2958.2011.07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caruso S., Poon I.K.H. Apoptotic cell-derived extracellular vesicles: more than just debris. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01486. 1486–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 51.Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., Ma A., Koonin E.V., Dixit V.M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 52.Feoktistova M., Geserick P., Kellert B., Dimitrova D.P., Langlais C., Hupe M., Cain K., MacFarlane M., Häcker G., Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenev T., Bianchi K., Darding M., Broemer M., Langlais C., Wallberg F., Zachariou A., Lopez J., MacFarlane M., Cain K., Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Dondelinger Y., Aguileta M.A., Goossens V., Dubuisson C., Grootjans S., Dejardin E., Vandenabeele P., Bertrand M.J.M. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 2013;20:1381–1392. doi: 10.1038/cdd.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Du F., Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 56.Feng S., Yang Y., Mei Y., Ma L., Zhu D.-e., Hoti N., Castanares M., Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell. Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 57.Lin Y., Devin A., Rodriguez Y., Liu Z.G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinon F., Holler N., Richard C., Tschopp J. Activation of a pro-apoptotic amplification loop through inhibition of NF-κB-dependent survival signals by caspase-mediated inactivation of RIP. FEBS Lett. 2000;468:134–136. doi: 10.1016/s0014-5793(00)01212-6. [DOI] [PubMed] [Google Scholar]
- 59.Li J., McQuade T., Siemer A.B., Napetschnig J., Moriwaki K., Hsiao Y.S., Damko E., Moquin D., Walz T., McDermott A., Chan F.K., Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun L., Wang H., Wang Z., He S., Chen S., Liao D., Wang L., Yan J., Liu W., Lei X., Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 61.Wang H., Sun L., Su L., Rizo J., Liu L., Wang L.-F., Wang F.-S., Wang X. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Kaiser W.J., Offermann M.K. Apoptosis Induced by the Toll-Like Receptor Adaptor TRIF Is Dependent on Its Receptor Interacting Protein Homotypic Interaction Motif. J. Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- 63.Kaiser W.J., Sridharan H., Huang C., Mandal P., Upton J.W., Gough P.J., Sehon C.A., Marquis R.W., Bertin J., Mocarski E.S. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 2013;288:31268–31279. doi: 10.1074/jbc.M113.462341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuriakose T., Kanneganti T.D. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol. 2018;39:123–134. doi: 10.1016/j.it.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malireddi R.K.S., Kesavardhana S., Kanneganti T.-D. ZBP1 and TAK1: master regulators of NLRP3 Inflammasome/Pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front. Cell. Infect. Microbiol. 2019;9 doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J., Kumari S., Kim C., Van T.M., Wachsmuth L., Polykratis A., Pasparakis M. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540:124–128. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton K., Wickliffe K.E., Maltzman A., Dugger D.L., Strasser A., Pham V.C., Lill J.R., Roose-Girma M., Warming S., Solon M., Ngu H., Webster J.D., Dixit V.M. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature. 2016;540:129–133. doi: 10.1038/nature20559. [DOI] [PubMed] [Google Scholar]
- 68.Malireddi R.K.S., Gurung P., Kesavardhana S., Samir P., Burton A., Mummareddy H., Vogel P., Pelletier S., Burgula S., Kanneganti T.-D. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med. 2019;217 doi: 10.1084/jem.20191644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samir P., Malireddi R.K.S., Kanneganti T.-D. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naderer T., Fulcher M.C. Targeting apoptosis pathways in infections. J. Leukoc. Biol. 2018;103:275–285. doi: 10.1189/jlb.4mr0717-286r. [DOI] [PubMed] [Google Scholar]
- 71.You Y., Cheng A.-C., Wang M.-S., Jia R.-Y., Sun K.-F., Yang Q., Wu Y., Zhu D., Chen S., Liu M.-F., Zhao X.-X., Chen X.-Y. The suppression of apoptosis by α-herpesvirus. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.139. e2749-e2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dufour F., Sasseville A.M.-J., Chabaud S., Massie B., Siegel R.M., Langelier Y. The ribonucleotide reductase R1 subunits of herpes simplex virus types 1 and 2 protect cells against TNFα- and FasL-induced apoptosis by interacting with caspase-8. Apoptosis. 2011;16:256–271. doi: 10.1007/s10495-010-0560-2. [DOI] [PubMed] [Google Scholar]
- 73.Dufour F., Bertrand L., Pearson A., Grandvaux N., Langelier Y. The ribonucleotide reductase R1 subunits of herpes simplex virus 1 and 2 protect cells against poly(I · C)-induced apoptosis. J. Virol. 2011;85:8689–8701. doi: 10.1128/JVI.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kesavardhana S., Malireddi R.K.S., Burton A.R., Porter S.N., Vogel P., Pruett-Miller S.M., Kanneganti T.-D. The Zα2 domain of ZBP1 is a molecular switch regulating influenza-induced PANoptosis and perinatal lethality during development. J. Biol. Chem. 2020;295:8325–8330. doi: 10.1074/jbc.RA120.013752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thapa R.J., Ingram J.P., Ragan K.B., Nogusa S., Boyd D.F., Benitez A.A., Sridharan H., Kosoff R., Shubina M., Landsteiner V.J., Andrake M., Vogel P., Sigal L.J., tenOever B.R., Thomas P.G., Upton J.W., Balachandran S. DAI senses influenza a virus genomic RNA and activates RIPK3-Dependent cell death. Cell Host Microbe. 2016;20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuriakose T., Man S.M., Malireddi R.K.S., Karki R., Kesavardhana S., Place D.E., Neale G., Vogel P., Kanneganti T.-D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kesavardhana S., Kuriakose T., Guy C.S., Samir P., Malireddi R.K.S., Mishra A., Kanneganti T.D. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 2017;214:2217–2229. doi: 10.1084/jem.20170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nogusa S., Thapa R.J., Dillon C.P., Liedmann S., Oguin T.H., 3rd, Ingram J.P., Rodriguez D.A., Kosoff R., Sharma S., Sturm O., Verbist K., Gough P.J., Bertin J., Hartmann B.M., Sealfon S.C., Kaiser W.J., Mocarski E.S., López C.B., Thomas P.G., Oberst A., Green D.R., Balachandran S. RIPK3 activates parallel pathways of MLKL-Driven necroptosis and FADD-Mediated apoptosis to protect against influenza a virus. Cell Host Microbe. 2016;20:13–24. doi: 10.1016/j.chom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peterson L.W., Philip N.H., Dillon C.P., Bertin J., Gough P.J., Green D.R., Brodsky I.E. Cell-extrinsic TNF collaborates with TRIF signaling to promote yersinia-induced apoptosis. J. Immunol. 2016;197:4110–4117. doi: 10.4049/jimmunol.1601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peterson L.W., Philip N.H., DeLaney A., Wynosky-Dolfi M.A., Asklof K., Gray F., Choa R., Bjanes E., Buza E.L., Hu B., Dillon C.P., Green D.R., Berger S.B., Gough P.J., Bertin J., Brodsky I.E. RIPK1-dependent apoptosis bypasses pathogen blockade of innate signaling to promote immune defense. J. Exp. Med. 2017;214:3171–3182. doi: 10.1084/jem.20170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M., Beg A.A. Induction of necrotic-like cell death by tumor necrosis factor alpha and caspase inhibitors: novel mechanism for killing virus-infected cells. J. Virol. 2000;74:7470–7477. doi: 10.1128/jvi.74.16.7470-7477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho Y.S., Challa S., Moquin D., Genga R., Ray T.D., Guildford M., Chan F.K.-M. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan F.K., Shisler J., Bixby J.G., Felices M., Zheng L., Appel M., Orenstein J., Moss B., Lenardo M.J. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J. Biol. Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 84.Polykratis A., Hermance N., Zelic M., Roderick J., Kim C., Van T.-M., Lee T.H., Chan F.K.M., Pasparakis M., Kelliher M.A. Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 2014;193:1539–1543. doi: 10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Webster J.D., Kwon Y.C., Park S., Zhang H., Corr N., Ljumanovic N., Adedeji A.O., Varfolomeev E., Goncharov T., Preston J., Santagostino S.F., Patel S., Xu M., Maher J., McKenzie B.S., Vucic D. RIP1 kinase activity is critical for skin inflammation but not for viral propagation. J. Leukoc. Biol. 2020;107:941–952. doi: 10.1002/jlb.3ma1219-398r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang H.W., Jacobs B.L. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 87.Koehler H., Cotsmire S., Langland J., Kibler K.V., Kalman D., Upton J.W., Mocarski E.S., Jacobs B.L. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc. Natl. Acad. Sci. 2017;114:11506–11511. doi: 10.1073/pnas.1700999114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCormick A.L., Skaletskaya A., Barry P.A., Mocarski E.S., Goldmacher V.S. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology. 2003;316:221–233. doi: 10.1016/j.virol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Skaletskaya A., Bartle L.M., Chittenden T., McCormick A.L., Mocarski E.S., Goldmacher V.S. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Upton J.W., Kaiser W.J., Mocarski E.S. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J. Biol. Chem. 2008;283:16966–16970. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Upton J.W., Kaiser W.J., Mocarski E.S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 2012;11:290–297. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lembo D., Donalisio M., Hofer A., Cornaglia M., Brune W., Koszinowski U., Thelander L., Landolfo S. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J. Virol. 2004;78:4278–4288. doi: 10.1128/jvi.78.8.4278-4288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Upton J.W., Kaiser W.J., Mocarski E.S. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sridharan H., Ragan K.B., Guo H., Gilley R.P., Landsteiner V.J., Kaiser W.J., Upton J.W. Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 2017;18:1429–1441. doi: 10.15252/embr.201743947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeFilippis V.R., Alvarado D., Sali T., Rothenburg S., Früh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J. Virol. 2010;84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Omoto S., Guo H., Talekar G.R., Roback L., Kaiser W.J., Mocarski E.S. Suppression of RIP3-dependent necroptosis by human cytomegalovirus. J. Biol. Chem. 2015;290:11635–11648. doi: 10.1074/jbc.M115.646042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo H., Omoto S., Harris P.A., Finger J.N., Bertin J., Gough P.J., Kaiser W.J., Mocarski E.S. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe. 2015;17:243–251. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang Z., Wu S.-Q., Liang Y., Zhou X., Chen W., Li L., Wu J., Zhuang Q., Chen C.A., Li J., Zhong C.-Q., Xia W., Zhou R., Zheng C., Han J. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe. 2015;17:229–242. doi: 10.1016/j.chom.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 99.Wang X., Li Y., Liu S., Yu X., Li L., Shi C., He W., Li J., Xu L., Hu Z., Yu L., Yang Z., Chen Q., Ge L., Zhang Z., Zhou B., Jiang X., Chen S., He S. Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15438–15443. doi: 10.1073/pnas.1412767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Onizawa M., Oshima S., Schulze-Topphoff U., Oses-Prieto J.A., Lu T., Tavares R., Prodhomme T., Duong B., Whang M.I., Advincula R., Agelidis A., Barrera J., Wu H., Burlingame A., Malynn B.A., Zamvil S.S., Ma A. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 2015;16:618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petrie E.J., Sandow J.J., Lehmann W.I.L., Liang L.Y., Coursier D., Young S.N., Kersten W.J.A., Fitzgibbon C., Samson A.L., Jacobsen A.V., Lowes K.N., Au A.E., Jousset Sabroux H., Lalaoui N., Webb A.I., Lessene G., Manning G., Lucet I.S., Murphy J.M. Viral MLKL homologs subvert necroptotic cell death by sequestering cellular RIPK3. Cell Rep. 2019;28:3309–3319.e3305. doi: 10.1016/j.celrep.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 102.Wagner R.N., Reed J.C., Chanda S.K. HIV-1 protease cleaves the serine-threonine kinases RIPK1 and RIPK2. Retrovirology. 2015;12:74. doi: 10.1186/s12977-015-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaiha G.D., McKim K.J., Woods M., Pertel T., Rohrbach J., Barteneva N., Chin C.R., Liu D., Soghoian D.Z., Cesa K., Wilton S., Waring M.T., Chicoine A., Doering T., Wherry E.J., Kaufmann D.E., Lichterfeld M., Brass A.L., Walker B.D. Dysfunctional HIV-specific CD8+ T cell proliferation is associated with increased caspase-8 activity and mediated by necroptosis. Immunity. 2014;41:1001–1012. doi: 10.1016/j.immuni.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 105.Zhang S., Yu X., Meng X., Huo W., Su Y., Liu J., Liu Y., Zhang J., Wang S., Yu J. Coxsackievirus A6 induces necroptosis for viral production. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00042. 42–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lay M.K., González P.A., León M.A., Céspedes P.F., Bueno S.M., Riedel C.A., Kalergis A.M. Advances in understanding respiratory syncytial virus infection in airway epithelial cells and consequential effects on the immune response. Microbes Infect. 2013;15:230–242. doi: 10.1016/j.micinf.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 107.Simpson J., Loh Z., Ullah M.A., Lynch J.P., Werder R.B., Collinson N., Zhang V., Dondelinger Y., Bertrand M.J.M., Everard M.L., Blyth C.C., Hartel G., Van Oosterhout A.J., Gough P.J., Bertin J., Upham J.W., Spann K.M., Phipps S. RSV infection promotes necroptosis and HMGB1 release by airway epithelial cells. Am. J. Respir. Crit. Care Med. 2020;201:1358–1371. doi: 10.1164/rccm.201906-1149OC. [DOI] [PubMed] [Google Scholar]
- 108.Muraro S.P., De Souza G.F., Gallo S.W., Da Silva B.K., De Oliveira S.D., Vinolo M.A.R., Saraiva E.M., Porto B.N. Respiratory Syncytial Virus induces the classical ROS-dependent NETosis through PAD-4 and necroptosis pathways activation. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32576-y. 14166–14166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yue Y., Nabar N.R., Shi C.-S., Kamenyeva O., Xiao X., Hwang I.-Y., Wang M., Kehrl J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu W., Zheng B.-J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins e, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pearson J.S., Giogha C., Muhlen S., Nachbur U., Pham C.L., Zhang Y., Hildebrand J.M., Oates C.V., Lung T.W., Ingle D., Dagley L.F., Bankovacki A., Petrie E.J., Schroeder G.N., Crepin V.F., Frankel G., Masters S.L., Vince J., Murphy J.M., Sunde M., Webb A.I., Silke J., Hartland E.L. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat. Microbiol. 2017;2:16258. doi: 10.1038/nmicrobiol.2016.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kitur K., Parker D., Nieto P., Ahn D.S., Cohen T.S., Chung S., Wachtel S., Bueno S., Prince A. Toxin-induced necroptosis is a major mechanism of Staphylococcus aureus lung damage. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.González-Juarbe N., Gilley R.P., Hinojosa C.A., Bradley K.M., Kamei A., Gao G., Dube P.H., Bergman M.A., Orihuela C.J. Pore-forming toxins induce macrophage necroptosis during acute bacterial pneumonia. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kitur K., Wachtel S., Brown A., Wickersham M., Paulino F., Peñaloza H.F., Soong G., Bueno S., Parker D., Prince A. Necroptosis promotes Staphylococcus aureus clearance by inhibiting excessive inflammatory signaling. Cell Rep. 2016;16:2219–2230. doi: 10.1016/j.celrep.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 2012;13:954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ro Y.T., Jo G.H., Jung S.A., Lee E.H., Shin J., Lee J.H. Salmonella‑induced miR‑155 enhances necroptotic death in macrophage cells via targeting RIP1/3. Mol. Med. Rep. 2018;18:5133–5140. doi: 10.3892/mmr.2018.9525. [DOI] [PubMed] [Google Scholar]
- 119.Butler R.E., Krishnan N., Garcia-Jimenez W., Francis R., Martyn A., Mendum T., Felemban S., Locker N., Salguero F.J., Robertson B., Stewart G.R. Susceptibility of Mycobacterium tuberculosis-infected host cells to phospho-MLKL driven necroptosis is dependent on cell type and presence of TNFα. Virulence. 2017;8:1820–1832. doi: 10.1080/21505594.2017.1377881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao X., Khan N., Gan H., Tzelepis F., Nishimura T., Park S.Y., Divangahi M., Remold H.G. Bcl-x(L) mediates RIPK3-dependent necrosis in M. Tuberculosis-infected macrophages. Mucosal Immunol. 2017;10:1553–1568. doi: 10.1038/mi.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stutz M.D., Ojaimi S., Allison C., Preston S., Arandjelovic P., Hildebrand J.M., Sandow J.J., Webb A.I., Silke J., Alexander W.S., Pellegrini M. Necroptotic signaling is primed in Mycobacterium tuberculosis-infected macrophages, but its pathophysiological consequence in disease is restricted. Cell Death Differ. 2018;25:951–965. doi: 10.1038/s41418-017-0031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stutz M.D., Ojaimi S., Ebert G., Pellegrini M. Is receptor-interacting protein kinase 3 a viable therapeutic target for Mycobacterium tuberculosis infection? Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01178. 1178–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Christgen S., Place D.E., Kanneganti T.-D. Toward targeting inflammasomes: insights into their regulation and activation. Cell Res. 2020;30:315–327. doi: 10.1038/s41422-020-0295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gaidt M.M., Ebert T.S., Chauhan D., Schmidt T., Schmid-Burgk J.L., Rapino F., Robertson A.A., Cooper M.A., Graf T., Hornung V. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 125.Kang T.-B., Yang S.-H., Toth B., Kovalenko A., Wallach D. Caspase-8 blocks kinase RIPK3-Mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 126.Lawlor K.E., Khan N., Mildenhall A., Gerlic M., Croker B.A., D’Cruz A.A., Hall C., Kaur Spall S., Anderton H., Masters S.L., Rashidi M., Wicks I.P., Alexander W.S., Mitsuuchi Y., Benetatos C.A., Condon S.M., Wong W.W., Silke J., Vaux D.L., Vince J.E. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015;6:6282. doi: 10.1038/ncomms7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Netea M.G., Nold-Petry C.A., Nold M.F., Joosten L.A., Opitz B., van der Meer J.H., van de Veerdonk F.L., Ferwerda G., Heinhuis B., Devesa I., Funk C.J., Mason R.J., Kullberg B.J., Rubartelli A., van der Meer J.W., Dinarello C.A. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 2009;113:2324–2335. doi: 10.1182/blood-2008-03-146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rozo-Lopez P., Drolet B.S., Londoño-Renteria B. Vesicular stomatitis virus transmission: a comparison of incriminated vectors. Insects. 2018;9:190. doi: 10.3390/insects9040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang X., Jiang W., Yan Y., Gong T., Han J., Tian Z., Zhou R. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 2014;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 130.Hottz E.D., Lopes J.F., Freitas C., Valls-de-Souza R., Oliveira M.F., Bozza M.T., Da Poian A.T., Weyrich A.S., Zimmerman G.A., Bozza F.A., Bozza P.T. Platelets mediate increased endothelium permeability in dengue through NLRP3-inflammasome activation. Blood. 2013;122:3405–3414. doi: 10.1182/blood-2013-05-504449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park H.-S., Liu G., Liu Q., Zhou Y. Swine influenza virus induces RIPK1/DRP1-Mediated Interleukin-1 Beta production. Viruses. 2018;10:419. doi: 10.3390/v10080419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kang S., Fernandes-Alnemri T., Rogers C., Mayes L., Wang Y., Dillon C., Roback L., Kaiser W., Oberst A., Sagara J., Fitzgerald K.A., Green D.R., Zhang J., Mocarski E.S., Alnemri E.S. Caspase-8 scaffolding function and MLKL regulate NLRP3 inflammasome activation downstream of TLR3. Nat. Commun. 2015;6 doi: 10.1038/ncomms8515. 7515–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.da Costa L.S., Outlioua A., Anginot A., Akarid K., Arnoult D. RNA viruses promote activation of the NLRP3 inflammasome through cytopathogenic effect-induced potassium efflux. Cell Death Dis. 2019;10:346. doi: 10.1038/s41419-019-1579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moriwaki K., Farias Luz N., Balaji S., De Rosa M.J., O’Donnell C.L., Gough P.J., Bertin J., Welsh R.M., Chan F.K.-M. The mitochondrial phosphatase PGAM5 is dispensable for necroptosis but promotes inflammasome activation in macrophages. J. Immunol. 2016;196:407–415. doi: 10.4049/jimmunol.1501662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Philip N.H., Dillon C.P., Snyder A.G., Fitzgerald P., Wynosky-Dolfi M.A., Zwack E.E., Hu B., Fitzgerald L., Mauldin E.A., Copenhaver A.M., Shin S., Wei L., Parker M., Zhang J., Oberst A., Green D.R., Brodsky I.E. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc. Natl. Acad. Sci. 2014;111:7385–7390. doi: 10.1073/pnas.1403252111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orning P., Weng D., Starheim K., Ratner D., Best Z., Lee B., Brooks A., Xia S., Wu H., Kelliher M.A., Berger S.B., Gough P.J., Bertin J., Proulx M.M., Goguen J.D., Kayagaki N., Fitzgerald K.A., Lien E. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362:1064–1069. doi: 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kobayashi K., Inohara N., Hernandez L.D., Galan J.E., Nunez G., Janeway C.A., Medzhitov R., Flavell R.A. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 138.McCarthy J.V., Ni J., Dixit V.M. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J. Biol. Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 139.Nembrini C., Kisielow J., Shamshiev A.T., Tortola L., Coyle A.J., Kopf M., Marsland B.J. The kinase activity of Rip2 determines its stability and consequently Nod1- and Nod2-mediated immune responses. J. Biol. Chem. 2009;284:19183–19188. doi: 10.1074/jbc.M109.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Park J.H., Kim Y.G., McDonald C., Kanneganti T.D., Hasegawa M., Body-Malapel M., Inohara N., Nunez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 141.Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., Valvano M.A., Foster S.J., Mak T.W., Nuñez G., Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 142.Girardin S.E., Boneca I.G., Carneiro L.A.M., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M.-K., Labigne A., Zäthringer U., Coyle A.J., DiStefano P.S., Bertin J., Sansonetti P.J., Philpott D.J. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 143.Girardin S.E., Boneca I.G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D.J., Sansonetti P.J. Nod2 is a general sensor of peptidoglycan through Muramyl Dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 144.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S.J., Moran A.P., Fernandez-Luna J.L., Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 145.Shin S., Case C.L., Archer K.A., Nogueira C.V., Kobayashi K.S., Flavell R.A., Roy C.R., Zamboni D.S. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nachbur U., Stafford C.A., Bankovacki A., Zhan Y., Lindqvist L.M., Fiil B.K., Khakham Y., Ko H.-J., Sandow J.J., Falk H., Holien J.K., Chau D., Hildebrand J., Vince J.E., Sharp P.P., Webb A.I., Jackman K.A., Mühlen S., Kennedy C.L., Lowes K.N., Murphy J.M., Gyrd-Hansen M., Parker M.W., Hartland E.L., Lew A.M., Huang D.C.S., Lessene G., Silke J. A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat. Commun. 2015;6:6442. doi: 10.1038/ncomms7442. [DOI] [PubMed] [Google Scholar]
- 147.Pandey A.K., Yang Y., Jiang Z., Fortune S.M., Coulombe F., Behr M.A., Fitzgerald K.A., Sassetti C.M., Kelliher M.A. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Homer C.R., Richmond A.L., Rebert N.A., Achkar J.-P., McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641.e16412. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Irving Aaron T., Mimuro H., Kufer Thomas A., Lo C., Wheeler R., Turner Lorinda J., Thomas Belinda J., Malosse C., Gantier Michael P., Casillas Linda N., Votta Bartholomew J., Bertin J., Boneca Ivo G., Sasakawa C., Philpott Dana J., Ferrero Richard L., Kaparakis-Liaskos M. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014;15:623–635. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 150.Sorbara M.T., Ellison L.K., Ramjeet M., Travassos L.H., Jones N.L., Girardin S.E., Philpott D.J. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858–873. doi: 10.1016/j.immuni.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 151.Chin A.I., Dempsey P.W., Bruhn K., Miller J.F., Xu Y., Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- 152.Zhang F.-R., Huang W., Chen S.-M., Sun L.-D., Liu H., Li Y., Cui Y., Yan X.-X., Yang H.-T., Yang Rong-De, Chu T.-S., Zhang C., Zhang L., Han J.-W., Yu G.-Q., Quan C., Yu Y.-X., Zhang Z., Shi B.-Q., Zhang L.-H., Cheng H., Wang C.-Y., Lin Y., Zheng H.-F., Fu X.-A., Zuo X.-B., Wang Q., Long H., Sun Y.-P., Cheng Y.-L., Tian H.-Q., Zhou F.-S., Liu H.-X., Lu W.-S., He S.-M., Du W.-L., Shen M., Jin Q.-Y., Wang Y., Low H.-Q., Erwin T., Yang N.-H., Li J.-Y., Zhao X., Jiao Y.-L., Mao L.-G., Yin G., Jiang Z.-X., Wang X.-D., Yu J.-P., Hu Z.-H., Gong C.-H., Liu Y.-Q., Liu R.-Y., Wang D.-M., Wei D., Liu J.-X., Cao W.-K., Cao H.-Z., Li Y.-P., Yan W.-G., Wei S.-Y., Wang K.-J., Hibberd M.L., Yang S., Zhang X.-J., Liu J.-J. Genomewide association study of leprosy. N. Engl. J. Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 153.Song J., Liu T., Jiao L., Zhao Z., Hu X., Wu Q., Bai H., Lv M., Meng Z., Wu T., Chen H., Chen X., Song X., Ying B. RIPK2 polymorphisms and susceptibility to tuberculosis in a Western Chinese Han population. Infect. Genet. Evol. 2019;75:103950. doi: 10.1016/j.meegid.2019.103950. [DOI] [PubMed] [Google Scholar]
- 154.Casanova J.L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 155.Ottenhoff T.H., Verreck F.A., Hoeve M.A., van de Vosse E. Control of human host immunity to mycobacteria. Tuberculosis. 2005;85:53–64. doi: 10.1016/j.tube.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 156.Weiser J.N., Ferreira D.M., Paton J.C. Streptococcus pneumoniae: transmission, colonization and invasion. Nat. Rev. Microbiol. 2018;16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Moreira L.O., El Kasmi K.C., Smith A.M., Finkelstein D., Fillon S., Kim Y.G., Nunez G., Tuomanen E., Murray P.J. The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory and IL-10-mediated anti-inflammatory cytokine response to Gram-positive cell walls. Cell. Microbiol. 2008;10:2067–2077. doi: 10.1111/j.1462-5822.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yoshimura A., Lien E., Ingalls R.R., Tuomanen E., Dziarski R., Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 159.Orihuela C.J., Fillon S., Smith-Sielicki S.H., El Kasmi K.C., Gao G., Soulis K., Patil A., Murray P.J., Tuomanen E.I. Cell wall-mediated neuronal damage in early sepsis. Infect. Immun. 2006;74:3783–3789. doi: 10.1128/iai.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peñaloza H.F., Nieto P.A., Muñoz-Durango N., Salazar-Echegarai F.J., Torres J., Parga M.J., Alvarez-Lobos M., Riedel C.A., Kalergis A.M., Bueno S.M. Interleukin-10 plays a key role in the modulation of neutrophils recruitment and lung inflammation during infection by Streptococcus pneumoniae. Immunology. 2015;146:100–112. doi: 10.1111/imm.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Opitz B., Püschel A., Schmeck B., Hocke A.C., Rosseau S., Hammerschmidt S., Schumann R.R., Suttorp N., Hippenstiel S. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J. Biol. Chem. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 162.Frutuoso M.S., Hori J.I., Pereira M.S., Junior D.S., Sônego F., Kobayashi K.S., Flavell R.A., Cunha F.Q., Zamboni D.S. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect. 2010;12:819–827. doi: 10.1016/j.micinf.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 163.Berrington W.R., Iyer R., Wells R.D., Smith K.D., Skerrett S.J., Hawn T.R. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. Eur. J. Immunol. 2010;40:3519–3527. doi: 10.1002/eji.201040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Biesiada G., Czepiel J., Leśniak M.R., Garlicki A., Mach T. Lyme disease: review. Arch. Med. Sci. 2012;8:978–982. doi: 10.5114/aoms.2012.30948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sterka D., Jr., Marriott I. Characterization of nucleotide-binding oligomerization domain (NOD) protein expression in primary murine microglia. J. Neuroimmunol. 2006;179:65–75. doi: 10.1016/j.jneuroim.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 166.Sterka D., Jr., Rati D.M., Marriott I. Functional expression of NOD2, a novel pattern recognition receptor for bacterial motifs, in primary murine astrocytes. Glia. 2006;53:322–330. doi: 10.1002/glia.20286. [DOI] [PubMed] [Google Scholar]
- 167.Oosting M., Berende A., Sturm P., Ter Hofstede H.J., de Jong D.J., Kanneganti T.D., van der Meer J.W., Kullberg B.J., Netea M.G., Joosten L.A. Recognition of Borrelia burgdorferi by NOD2 is central for the induction of an inflammatory reaction. J. Infect. Dis. 2010;201:1849–1858. doi: 10.1086/652871. [DOI] [PubMed] [Google Scholar]
- 168.Canning P., Ruan Q., Schwerd T., Hrdinka M., Maki Jenny L., Saleh D., Suebsuwong C., Ray S., Brennan Paul E., Cuny Gregory D., Uhlig Holm H., Gyrd-Hansen M., Degterev A., Bullock Alex N. Inflammatory signaling by NOD-RIPK2 is inhibited by clinically relevant type II kinase inhibitors. Chem. Biol. 2015;22:1174–1184. doi: 10.1016/j.chembiol.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Negroni A., Stronati L., Pierdomenico M., Tirindelli D., Di Nardo G., Mancini V., Maiella G., Cucchiara S. Activation of NOD2-mediated intestinal pathway in a pediatric population with Crohn’s disease. Inflamm. Bowel Dis. 2009;15:1145–1154. doi: 10.1002/ibd.20907. [DOI] [PubMed] [Google Scholar]
- 170.Kabi A., Nickerson K.P., Homer C.R., McDonald C. Digesting the genetics of inflammatory bowel disease: insights from studies of autophagy risk genes. Inflamm. Bowel Dis. 2012;18:782–792. doi: 10.1002/ibd.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Campoy E., Colombo M.I. Autophagy in intracellular bacterial infection. Biochim. Biophys. Acta. 2009;1793:1465–1477. doi: 10.1016/j.bbamcr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 173.Deretic V., Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang J., Brumell J.H. Autophagy in immunity against intracellular bacteria. Curr. Top. Microbiol. Immunol. 2009;335:189–215. doi: 10.1007/978-3-642-00302-8_9. [DOI] [PubMed] [Google Scholar]
- 175.Khalil I.A., Troeger C., Blacker B.F., Rao P.C., Brown A., Atherly D.E., Brewer T.G., Engmann C.M., Houpt E.R., Kang G., Kotloff K.L., Levine M.M., Luby S.P., MacLennan C.A., Pan W.K., Pavlinac P.B., Platts-Mills J.A., Qadri F., Riddle M.S., Ryan E.T., Shoultz D.A., Steele A.D., Walson J.L., Sanders J.W., Mokdad A.H., Murray C.J.L., Hay S.I., Reiner R.C., Jr. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: the Global Burden of Disease Study 1990-2016. Lancet Infect. Dis. 2018;18:1229–1240. doi: 10.1016/s1473-3099(18)30475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Williamson D., Ingle D., Howden B. Extensively drug-resistant shigellosis in Australia among men who have sex with men. N. Engl. J. Med. 2019;381:2477–2479. doi: 10.1056/NEJMc1910648. [DOI] [PubMed] [Google Scholar]
- 177.Girardin S.E., Tournebize R., Mavris M., Page A.L., Li X., Stark G.R., Bertin J., DiStefano P.S., Yaniv M., Sansonetti P.J., Philpott D.J. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Forman D., Newell D.G., Fullerton F., Yarnell J.W., Stacey A.R., Wald N., Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. Br. Med. J. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Parsonnet J., Friedman G.D., Vandersteen D.P., Chang Y., Vogelman J.H., Orentreich N., Sibley R.K. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 1991;325:1127–1131. doi: 10.1056/nejm199110173251603. [DOI] [PubMed] [Google Scholar]
- 180.Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 181.Ota M., Tahara T., Otsuka T., Jing W., Nomura T., Hayashi R., Shimasaki T., Nakamura M., Shibata T., Arisawa T. Association between receptor interacting serine/threonine kinase 2 polymorphisms and gastric cancer susceptibility. Oncol. Lett. 2018;15:3772–3778. doi: 10.3892/ol.2018.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Viala J., Chaput C., Boneca I.G., Cardona A., Girardin S.E., Moran A.P., Athman R., Mémet S., Huerre M.R., Coyle A.J., DiStefano P.S., Sansonetti P.J., Labigne A., Bertin J., Philpott D.J., Ferrero R.L. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 183.Tattoli I., Sorbara M.T., Vuckovic D., Ling A., Soares F., Carneiro L.A., Yang C., Emili A., Philpott D.J., Girardin S.E. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 184.Anand P.K., Tait S.W.G., Lamkanfi M., Amer A.O., Nunez G., Pagès G., Pouysségur J., McGargill M.A., Green D.R., Kanneganti T.-D. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J. Biol. Chem. 2011;286:42981–42991. doi: 10.1074/jbc.M111.310599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Homer C.R., Kabi A., Marina-Garcia N., Sreekumar A., Nesvizhskii A.I., Nickerson K.P., Chinnaiyan A.M., Nunez G., McDonald C. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J. Biol. Chem. 2012;287:25565–25576. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bhavsar A.P., Brown N.F., Stoepel J., Wiermer M., Martin D.D.O., Hsu K.J., Imami K., Ross C.J., Hayden M.R., Foster L.J., Li X., Hieter P., Finlay B.B. The Salmonella type III effector SspH2 specifically exploits the NLR Co-chaperone activity of SGT1 to subvert immunity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fukazawa A., Alonso C., Kurachi K., Gupta S., Lesser C.F., McCormick B.A., Reinecker H.-C. GEF-H1 mediated control of NOD1 dependent NF-kappaB activation by Shigella effectors. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000228. e1000228-e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Keestra A.M., Winter M.G., Auburger J.J., Fräßle S.P., Xavier M.N., Winter S.E., Kim A., Poon V., Ravesloot M.M., Waldenmaier J.F.T., Tsolis R.M., Eigenheer R.A., Bäumler A.J. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013;496:233–237. doi: 10.1038/nature12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Keestra A.M., Winter M.G., Klein-Douwel D., Xavier M.N., Winter S.E., Kim A., Tsolis R.M., Bäumler A.J. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. mBio. 2011;2 doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Narumiya S. The small GTPase rho: cellular functions and signal transduction. J. Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 191.Deng W., Marshall N.C., Rowland J.L., McCoy J.M., Worrall L.J., Santos A.S., Strynadka N.C.J., Finlay B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 192.Boyer L., Magoc L., Dejardin S., Cappillino M., Paquette N., Hinault C., Charriere Guillaume M., Ip W.K.E., Fracchia S., Hennessy E., Erturk-Hasdemir D., Reichhart J.-M., Silverman N., Lacy-Hulbert A., Stuart Lynda M. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Keestra-Gounder A.M., Tsolis R.M. NOD1 and NOD2: beyond peptidoglycan sensing. Trends Immunol. 2017;38:758–767. doi: 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Keestra-Gounder A.M., Byndloss M.X., Seyffert N., Young B.M., Chávez-Arroyo A., Tsai A.Y., Cevallos S.A., Winter M.G., Pham O.H., Tiffany C.R., de Jong M.F., Kerrinnes T., Ravindran R., Luciw P.A., McSorley S.J., Bäumler A.J., Tsolis R.M. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532:394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Welter-Stahl L., Ojcius D.M., Viala J., Girardin S., Liu W., Delarbre C., Philpott D., Kelly K.A., Darville T. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell. Microbiol. 2006;8:1047–1057. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 196.Roberson E.C., Tully J.E., Guala A.S., Reiss J.N., Godburn K.E., Pociask D.A., Alcorn J.F., Riches D.W., Dienz O., Janssen-Heininger Y.M., Anathy V. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nakamura H., Liao H., Minami K., Toyoda M., Akutsu H., Miyagawa Y., Okita H., Kiyokawa N., Umezawa A., Imadome K., Inoue N., Fujiwara S. Human cytomegalovirus induces apoptosis in neural stem/progenitor cells derived from induced pluripotent stem cells by generating mitochondrial dysfunction and endoplasmic reticulum stress. Herpesviridae. 2013;4:2. doi: 10.1186/2042-4280-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wex K., Schmid U., Just S., Wang X., Wurm R., Naumann M., Schluter D., Nishanth G. Receptor-interacting protein Kinase-2 inhibition by CYLD impairs antibacterial immune responses in macrophages. Front. Immunol. 2016;19:650. doi: 10.3389/fimmu.2015.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ellwanger K., Briese S., Arnold C., Kienes I., Heim V., Nachbur U., Kufer T.A. XIAP controls RIPK2 signaling by preventing its deposition in speck-like structures. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gong Q., Long Z., Zhong F.L., Teo D.E.T., Jin Y., Yin Z., Boo Z.Z., Zhang Y., Zhang J., Yang R., Bhushan S., Reversade B., Li Z., Wu B. Structural basis of RIP2 activation and signaling. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-07447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kim Y.G., Park J.H., Reimer T., Baker D.P., Kawai T., Kumar H., Akira S., Wobus C., Nunez G. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9:496–507. doi: 10.1016/j.chom.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Park S.B., Hikima J.-i., Suzuki Y., Ohtani M., Nho S.W., Cha I.S., Jang H.B., Kondo H., Hirono I., Aoki T., Jung T.S. Molecular cloning and functional analysis of nucleotide-binding oligomerization domain 1 (NOD1) in olive flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 2012;36:680–687. doi: 10.1016/j.dci.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 203.Sabbah A., Chang T.H., Harnack R., Frohlich V., Tominaga K., Dube P.H., Xiang Y., Bose S. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E., Hill D.E., Regev A., Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lupfer C., Thomas P.G., Anand P.K., Vogel P., Milasta S., Martinez J., Huang G., Green M., Kundu M., Chi H., Xavier R.J., Green D.R., Lamkanfi M., Dinarello C.A., Doherty P.C., Kanneganti T.-D. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Loader M., Moravek R., Witowski S.E., Driscoll L.M. A clinical review of viral hepatitis. J. Am. Acad. Phys. 2019;32:15–20. doi: 10.1097/01.JAA.0000586300.88300.84. [DOI] [PubMed] [Google Scholar]
- 207.Vegna S., Gregoire D., Moreau M., Lassus P., Durantel D., Assenat E., Hibner U., Simonin Y. NOD1 participates in the innate immune response triggered by hepatitis C virus polymerase. J. Virol. 2016;90:6022–6035. doi: 10.1128/jvi.03230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wu S., Kanda T., Imazeki F., Nakamoto S., Tanaka T., Arai M., Roger T., Shirasawa H., Nomura F., Yokosuka O. Hepatitis B virus e antigen physically associates with receptor-interacting Serine/Threonine protein kinase 2 and regulates IL-6 gene expression. J. Infect. Dis. 2012;206:415–420. doi: 10.1093/infdis/jis363. [DOI] [PubMed] [Google Scholar]
- 209.Madrigal A.G., Barth K., Papadopoulos G., Genco C.A. Pathogen-mediated proteolysis of the cell death regulator RIPK1 and the host defense modulator RIPK2 in human aortic endothelial cells. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kwa M.Q., Scholz G.M., Reynolds E.C. RIPK4 activates an IRF6-mediated proinflammatory cytokine response in keratinocytes. Cytokine. 2016;83:19–26. doi: 10.1016/j.cyto.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 211.Meylan E., Martinon F., Thome M., Gschwendt M., Tschopp J. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 2002;3:1201–1208. doi: 10.1093/embo-reports/kvf236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Blauwendraat C., Reed X., Kia D.A., Gan-Or Z., Lesage S., Pihlstrøm L., Guerreiro R., Gibbs J.R., Sabir M., Ahmed S., Ding J., Alcalay R.N., Hassin-Baer S., Pittman A.M., Brooks J., Edsall C., Hernandez D.G., Chung S.J., Goldwurm S., Toft M., Schulte C., Bras J., Wood N.W., Brice A., Morris H.R., Scholz S.W., Nalls M.A., Singleton A.B., Cookson M.R., for the Courage-Pd Consortium, t.F.P.s.D.C, the International Parkinson’s Disease Genomics, C Frequency of loss of function variants in LRRK2 in parkinson disease. JAMA Neurol. 2018;75:1416–1422. doi: 10.1001/jamaneurol.2018.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li W., Mao L., Shu X., Liu R., Hao F., Li J., Liu M., Yang L., Zhang W., Sun M., Zhong C., Jiang J. Transcriptome analysis reveals differential immune related genes expression in bovine viral diarrhea virus-2 infected goat peripheral blood mononuclear cells (PBMCs) BMC Genomics. 2019;20:516. doi: 10.1186/s12864-019-5830-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Roosen D.A., Cookson M.R. LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol. Neurodegener. 2016;11:73. doi: 10.1186/s13024-016-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gardet A., Benita Y., Li C., Sands B.E., Ballester I., Stevens C., Korzenik J.R., Rioux J.D., Daly M.J., Xavier R.J., Podolsky D.K. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Liu W., Liu X.N., Li Y., Zhao J., Liu Z., Hu Z., Wang Y., Yao Y., Miller A.W., Su B., Cookson M.R., Li X., Kang Z. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella typhimurium infection. J. Exp. Med. 2017;214:3051–3066. doi: 10.1084/jem.20170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang Q., Pan Y., Yan R., Zeng B., Wang H., Zhang X., Li W., Wei H., Liu Z. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat. Immunol. 2015;16:918–926. doi: 10.1038/ni.3233. [DOI] [PubMed] [Google Scholar]
- 218.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fava V.M., Manry J., Cobat A., Orlova M., Van Thuc N., Ba N.N., Thai V.H., Abel L., Alcais A., Schurr E. A missense LRRK2 variant is a risk factor for excessive inflammatory responses in leprosy. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang D., Xu L., Lv L., Su L.Y., Fan Y., Zhang D.F., Bi R., Yu D., Zhang W., Li X.A., Li Y.Y., Yao Y.G. Association of the LRRK2 genetic polymorphisms with leprosy in Han Chinese from Southwest China. Genes Immun. 2015;16:112–119. doi: 10.1038/gene.2014.72. [DOI] [PubMed] [Google Scholar]
- 221.Wang Z., Arat S., Magid-Slav M., Brown J.R. Meta-analysis of human gene expression in response to Mycobacterium tuberculosis infection reveals potential therapeutic targets. BMC Syst. Biol. 2018;12:3. doi: 10.1186/s12918-017-0524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Härtlova A., Herbst S., Peltier J., Rodgers A., Bilkei-Gorzo O., Fearns A., Dill B.D., Lee H., Flynn R., Cowley S.A., Davies P., Lewis P.A., Ganley I.G., Martinez J., Alessi D.R., Reith A.D., Trost M., Gutierrez M.G. LRRK2 is a negative regulator of Mycobacterium tuberculosis phagosome maturation in macrophages. EMBO J. 2018;37 doi: 10.15252/embj.201798694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Weindel C.G., Bell S.L., Vail K.J., West K.O., Patrick K.L., Watson R.O. LRRK2 maintains mitochondrial homeostasis and regulates innate immune responses to Mycobacterium tuberculosis. eLife. 2020;9 doi: 10.7554/eLife.51071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ottenhoff T.H., Dass R.H., Yang N., Zhang M.M., Wong H.E., Sahiratmadja E., Khor C.C., Alisjahbana B., van Crevel R., Marzuki S., Seielstad M., van de Vosse E., Hibberd M.L. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ahmadi Rastegar D., Dzamko N. Leucine rich repeat kinase 2 and innate immunity. Front. Neurosci. 2020;14:193. doi: 10.3389/fnins.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schapansky J., Nardozzi J.D., Felizia F., LaVoie M.J. Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum. Mol. Genet. 2014;23:4201–4214. doi: 10.1093/hmg/ddu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Man S.M., Karki R., Kanneganti T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Martens S., Hofmans S., Declercq W., Augustyns K., Vandenabeele P. Inhibitors targeting RIPK1/RIPK3: old and new drugs. Trends Pharmacol. Sci. 2020;41:209–224. doi: 10.1016/j.tips.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 229.Mares A., Miah A.H., Smith I.E.D., Rackham M., Thawani A.R., Cryan J., Haile P.A., Votta B.J., Beal A.M., Capriotti C., Reilly M.A., Fisher D.T., Zinn N., Bantscheff M., MacDonald T.T., Vossenkamper A., Dace P., Churcher I., Benowitz A.B., Watt G., Denyer J., Scott-Stevens P., Harling J.D. Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 2020;3:140. doi: 10.1038/s42003-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Weigert M., Binks A., Dowson S., Leung E.Y.L., Athineos D., Yu X., Mullin M., Walton J.B., Orange C., Ennis D., Blyth K., Tait S.W.G., McNeish I.A. RIPK3 promotes adenovirus type 5 activity. Cell Death Dis. 2017;8:3206. doi: 10.1038/s41419-017-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schock S.N., Chandra N.V., Sun Y., Irie T., Kitagawa Y., Gotoh B., Coscoy L., Winoto A. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ. 2017;24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Farooq M., Filliol A., Simoes Eugénio M., Piquet-Pellorce C., Dion S., Raguenes-Nicol C., Jan A., Dimanche-Boitrel M.-T., Le Seyec J., Samson M. Depletion of RIPK1 in hepatocytes exacerbates liver damage in fulminant viral hepatitis. Cell Death Dis. 2019;10:12. doi: 10.1038/s41419-018-1277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Berger A.K., Danthi P. Reovirus activates a caspase-independent cell death pathway. mBio. 2013;4 doi: 10.1128/mBio.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Berger A.K., Hiller B.E., Thete D., Snyder A.J., Perez E., Jr., Upton J.W., Danthi P. Viral RNA at two stages of reovirus infection is required for the induction of necroptosis. J. Virol. 2017;91 doi: 10.1128/JVI.02404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shimada K., Chen S., Dempsey P.W., Sorrentino R., Alsabeh R., Slepenkin A.V., Peterson E., Doherty T.M., Underhill D., Crother T.R., Arditi M. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Li W., Tang Q., Dai N., Feng W., Xie C., Cheng G., Liu X., Zhang W., Hu X., Gu C. Receptor-interacting serine/threonine kinase 1- and 3-dependent inflammation induced in lungs of chicken infected with Pasteurella multocida. Sci. Rep. 2020;10:6340. doi: 10.1038/s41598-020-62042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Luz N.F., Khouri R., Van Weyenbergh J., Zanette D.L., Fiuza P.P., Noronha A., Barral A., Boaventura V.S., Prates D.B., Chan F.K.-M., Andrade B.B., Borges V.M. Leishmania braziliensis subverts necroptosis by modulating RIPK3 expression. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02283. 2283–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Barbosa L.A., Fiuza P.P., Borges L.J., Rolim F.A., Andrade M.B., Luz N.F., Quintela-Carvalho G., Lima J.B., Almeida R.P., Chan F.K., Bozza M.T., Borges V.M., Prates D.B. RIPK1-RIPK3-MLKL-Associated necroptosis drives leishmania infantum killing in neutrophils. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01818. 1818–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Farias Luz N., Balaji S., Okuda K., Barreto A.S., Bertin J., Gough P.J., Gazzinelli R., Almeida R.P., Bozza M.T., Borges V.M., Chan F.K.-M. RIPK1 and PGAM5 control leishmania replication through distinct mechanisms. J. Immunol. 2016;196:5056–5063. doi: 10.4049/jimmunol.1502492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Corbett Y., Parapini S., D’Alessandro S., Scaccabarozzi D., Rocha B.C., Egan T.J., Omar A., Galastri L., Fitzgerald K.A., Golenbock D.T., Taramelli D., Basilico N. Involvement of Nod2 in the innate immune response elicited by malarial pigment hemozoin. Microbes Infect. 2015;17:184–194. doi: 10.1016/j.micinf.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Goldenberg R.C.D.S., Iacobas D.A., Iacobas S., Rocha L.L., da Silva de Azevedo Fortes F., Vairo L., Nagajyothi F., Campos de Carvalho A.C., Tanowitz H.B., Spray D.C. Transcriptomic alterations in Trypanosoma cruzi-infected cardiac myocytes. Microbes Infect. 2009;11:1140–1149. doi: 10.1016/j.micinf.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]