Highlights
-
•
Euroimmun SARS-CoV-2 IgG, IgA tests have specificity of 99% and 86%
-
•
Specificity can be improved using competitive blocking step with recombinant spike protein.
-
•
Hospitalized COVID-19 patients median IgA seroconversion was 8 days and IgG 10 days.
-
•
Neither antibody level nor timing of antibody response correlated with days on ventilation.
Keywords: SARS CoV2, COVID, Serology, Antibody
Abstract
Objectives
Humoral immune response to SARS-CoV-2 infection has been reported in several patient cohorts with results that vary by method and population studied due to the lack of reliable commercial assays available as the pandemic initially spread. We sought to clinically assess commercial prototype SARS-CoV-2 IgG and IgA assays for use in screening for prior infection and convalescent plasma donation.
Design and Methods: Prototype SARS-CoV-2 IgG and IgA assays from Euroimmun were assessed utilizing remnant specimens. Specificity testing used specimens in their convalescent window for the common coronaviruses and other infectious diseases known to be associated with increased non-specificity in serologic assays. Sensitivity testing utilized serial specimens from molecularly confirmed SARS-CoV-2 critically ill patients to assess seroconversion. Utilizing recombinant spike protein we also developed a competitive confirmation procedure to increase assay specificity.
Results
We determined specificity to be 97% and 81%, respectively, when indeterminate samples were considered positive and 99% and 86% when indeterminate samples were considered negative. We developed a new confirmation methodology to enhance the specificity of the assays with an anticipated specificity of 98% for IgA. Valuation of hospitalized COVID-19 patients determined median IgA seroconversion to be 8 days and IgG 10 days. Neither level nor timing of antibody response correlated with days on ventilation. End titer measurements indicate that validated improved assays may be capable of semi-quantitative measurement.
Conclusions
We found these assays to be clinically acceptable for the high prevalence population tested, for instance, for convalescent plasma donation.
1. Introduction
Coronavirus infectious disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and in March of 2020 the WHO declared it a pandemic. Since the outbreak of SARS-CoV in 2002–2003 and Middle East respiratory syndrome (MERS) in 2012 our understanding of the epidemiology and pathogenesis of coronavirus infections has improved, but there are still no specific therapeutics and vaccines available for controlling and treating COVID-19 patients [1]. Persistence of measurable neutralizing antibodies for more than one year in patients recovered after SARS-CoV and MERS infections suggest that those patients may be protected from recurrent infection [2], [3]. The rapid spread of SARS-CoV-2 across the globe coupled with the paucity of effective treatments beyond the provision of supportive care make it urgent to identify patients who have developed a successful immune response to SARS-CoV-2. Serologic assays measuring disease specific immunoglobulins are a mainstay of infectious disease surveillance, diagnosis, and determination of appropriate vaccine response. However, detection of IgG, IgM, and IgA antibodies recognizing SARS-CoV-2 antigens is still poorly characterized. While many assays are now available, the dynamic and specificity of SARS-CoV-2 immune response determined by these assays is variable. There is limited information on assay validation and the role of antibodies in protective immunity has yet to be established.
WHO states that assays to detect SARS-CoV-2 specific antibodies are the next priority [4] as antibody detection provides important clinical information during the course of SARS-CoV-2 infection. Testing for antibodies in conjunction with viral RNA detection will enable evaluation of ongoing and past infection [5]. Development of both prophylactic and therapeutic vaccines is based on assessing appearance of virus-specific antibodies. Verification of anti-SARS-CoV-2 immune response is also crucial for plasma therapy using immunoglobulins from patients who have successfully recovered from COVID-19, which has emerged as a promising therapy until more specific treatments can be developed [6], [7], [8], [9]. While there is still insufficient information regarding protective immunity and infectivity after immune response it is thought that testing healthcare professionals and recovered patients for virus-specific antibodies may minimize infection spread and establish a pool of protected individuals.
The goal of this study was to characterize SARS-CoV-2 specific IgA and IgG in COVID-19 patients, determine the feasibility and specificity of these assays, and initiate description of immune response in affected individuals in a platform that would be available to other clinical laboratories as well. We have verified commercially available serologic assays, assessed the dynamics of specific immune response in critically ill hospitalized COVID-19 patients in the US, and created an antigen-specific confirmation assay for reactive specimens.
2. Materials and methods
2.1. Specimens
Patient and healthy volunteer specimens and information were utilized under the auspices of UPMC Quality Assurance for Clinical Laboratories and the University of Pittsburgh IRB #20040072. Patient samples were remnant specimens from standard of care. All specimens were kept at 4 °C following standard clinical testing and were used within two weeks of draw for these studies or were kept at −20 °C in aliquots for up to two months from draw. Serum and heparin plasma were used unless otherwise noted. Samples for specificity testing were selected for serologic positivity to listed infectious diseases (Fig. 1 ) before SARS-CoV-2 was geographically present in the city/county. Specimens used in seroconversion and other positive specimens were remnant samples from patients hospitalized for SARS-CoV-2 infection. Remnant specimens were collected and chart review performed to ensure SARS-CoV-2 molecular testing demonstrated relevant infection. All specimens for which there were serial samples with sufficient volume were included in this study for seroconversion. Specimens with SARS-CoV-2 molecular testing which demonstrated presence of antibodies but did not have serial samples available were also included to assess potential confirmation testing strategies.
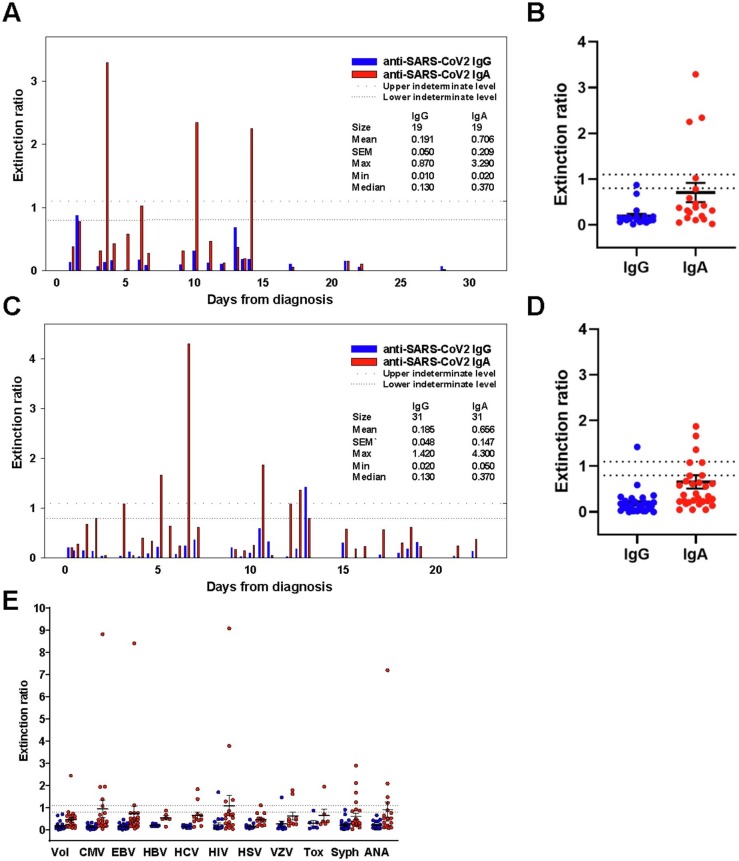
Fig. 1.
Validation of specificity testing for SARS-CoV-2 IgA and IgG antibodies in human serum samples. A. Patients tested nucleic acid positive for common coronaviruses prior to COVID-19 presence in the geographic region and had a subsequent blood sample available. X-axis: Days from nucleic acid positive test. B. Extinction ratio for all patients from (A) are plotted for comparison (n = 19). C. Patients presenting in the geographic region before COVID-19 with respiratory symptoms that prompted a nucleic acid respiratory pathogens panel test. X-axis: Days from testing indicated. D. Extinction ratio for all patients from (C) are plotted (n = 37). E. Patient cohorts positive for listed diseases prior to COVID-19 presence in the geographic region were tested for SARS-CoV-2 IgA (red dots) and IgG (blue dots) (n = 170). Vol, healthy volunteers; Syph, syphilis. Horizonal dashed/dotted lines indicate indeterminate interpretive region. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Detection of anti-SARS-CoV-2 antibodies
Prototype ELISA based tests for anti-SARS-CoV-2 IgA and IgG were from Euroimmun (Lubeck, Germany). These tests were used per manufacturer’s instructions and processed manually. These assays were later approved for use by Health Canada, CE marked, and the IgG assay was approved for use by the United States Food and Drug Administration (FDA). Results were calculated as: absorbance value of the sample divided by absorbance value of the calibrators, and expressed as extinction ratio. We utilized the manufacturers interpretation of the ratio with samples <0.8 classified as no antibody present, 0.8–<1.1 indeterminate, and ≥1.1 containing antibodies. These ELISA tests are for antibody against the S1 subunit/domain of the spike protein of SARS-CoV-2.
2.3. Other clinical laboratory testing
PCR detection of SARS-CoV-2 RNA was reported in patient charts as standard of care clinical testing. Measurement of positivity for antibodies against cytomegalovirus (CMV), Epstein-Barr virus (EBV), human immunodeficiency virus (HIV), herpes simplex virus types 1 and 2 (HSV), syphilis, and toxoplasma (Tox) was performed on the BioPlex 2200 (Bio-Rad, USA). Determination of antibodies against hepatitis B (HBV) and hepatitis C (HCV) was performed on the Centaur XP (Siemens, USA). Antinuclear antibodies (ANA) were determined by indirect immunofluorescence on the Helios (Aesku Group GmbH, Germany) and varicella zoster IgM was assessed using the Wompole ELISA (Abbott, USA). Respiratory viruses including the coronaviruses (NL63, 229E, OC43, HKU1) were tested on the ePlex respiratory pathogen panel (GenMark Dx, USA).
2.4. Blocking assay
All control and blocking proteins were obtained from Sino Biological Inc (Bejing, China) and were used at a final concentration of 25 µg/mL. Proteins used were all produced in mammalian cells for proper glycosylation and included: SARS-CoV-2 spike protein subunit 1, SARS-CoV-2 spike protein subunit 2, SARS-CoV-2 spike protein subunits 1 and 2, SARS-CoV spike protein subunit 1, and MERS spike protein. Per the manufacturers instructions patient samples are diluted10 uL in 1000 uL sample buffer prior to application to the assay plate. This diluted patient sample was mixed with the relevant protein solution (1:100 v/v) and incubated for 5 min at room temperature prior to addition to the assay plate.
2.5. Statistics
Results are expressed as mean ± SEM. Two-group analyses were performed using the unpaired or paired t test as appropriate after evaluation for normal distribution. Three or more groups were analyzed by non-parametric Kruskal-Wallis test using SigmaPlot (Systat, USA). P value < 0.05 was considered statistically significant.
3. Results
3.1. Verification of the assays
We first determined viable sample types to maximize use of sample sharing between assays to reduce patient blood draws. Using healthy volunteer serum, heparin plasma, EDTA plasma, and ACD plasma spiked with positive control we assessed plasma compared to serum. We saw a slight, but not statistically significant, increase in extinction ratio for EDTA plasma compared to serum and all sample types were equivalent for the qualitative use of SARS-CoV-2 IgA and IgG (p > 0.05). Next, we assessed intra-plate variability with kit calibrator material and found 13% variability in optical density (OD) across the plate (n = 6) for both IgA and IgG. Kit positive controls demonstrated 7% and 12% extinction ratio variability for IgG and IgA, respectively (n = 6).
Ideally to assess precision we would have preferred a 20 × 2 × 2 design per CLSI EP05 guidelines. Unfortunately, kit allocations, lot changes, and time constraints did not allow for initial evaluation using this guideline. For SARS-CoV-2 IgA inter-plate variability was assessed across 18 runs each on a separate testing day and across 4 reagent lots. The coefficient of variation (CV) for calibrator OD was 21% (n = 47), and for the kit positive control extinction ratio CV was 22% (n = 32). A positive patient sample was repeated across 4 lots of ELISA kits in 9 different runs with a CV of the extinction ratio of 18%. SARS-CoV-2 IgG inter-plate variability was assessed across 18 runs each on a separate testing day and across 2 reagent lots. The CV for calibrator OD was 28% (n = 44), and for the kit positive control extinction ratio CV was 19% (n = 29). Initial validations began before a positive patient was present in our region, limiting our ability to test positive patient samples initially. A positive patient sample was repeated across 2 lots in 9 different runs with a CV of the extinction ratio of 18%. We found this variability to be sufficient for qualitative determinations for initial testing.
A total of 226 cases without clinical COVID-19 were tested to determine assay diagnostic specificity, which was 97% (99%) for IgG and 81% (86%) for IgA if indeterminate samples were considered positive (or negative). Samples included patients that would be likely to demonstrate cross-reactivity in the assay (Fig. 1). This includes samples from patients found to be previously positive for one of the four common human coronaviruses by nucleic acid testing (Fig. 1A, B; n = 19). We found no cross-reactivity in the IgG assay (0/19 positive); the IgA assay demonstrated the slightly higher cross-reactivity than in other patient cohorts (3/19 positive). We also tested patients who presented clinically with respiratory infection symptoms before COVID-19 was present in our geographic population (Pennsylvania, Allegheny county, February 2020). We found 1 of 37 to be cross-reactive for SARS-CoV-2 IgG and 5 of 37 for IgA (Fig. 1C, D). Days from nucleic acid assessment for both non-SARS coronaviruses and other respiratory infections are shown to demonstrate the proportion of the cohort within an assumed convalescent window (Fig. 1A, C).
Many serologic assays demonstrate non-specific cross-reactivity with other infectious diseases and autoantibodies. Therefore, we assessed cohorts for several viral serology positive samples as well as ANA and a group of healthy volunteers (n = 145 total). We found higher rates of cross-reactivity in patients with antibodies reactive to CMV (4/22), syphilis (4/25), ANA (4/20) and HIV (4/20) in the IgA assay (Fig. 1E). Overall, the IgG assay demonstrated good specificity with little cross-reactivity (3/226), however, the IgA assay’s high level of cross-reactivity (31/226) made additional testing necessary to determine SARS-CoV-2 specific IgA immune response.
3.2. Immune response in COVID-19 patients
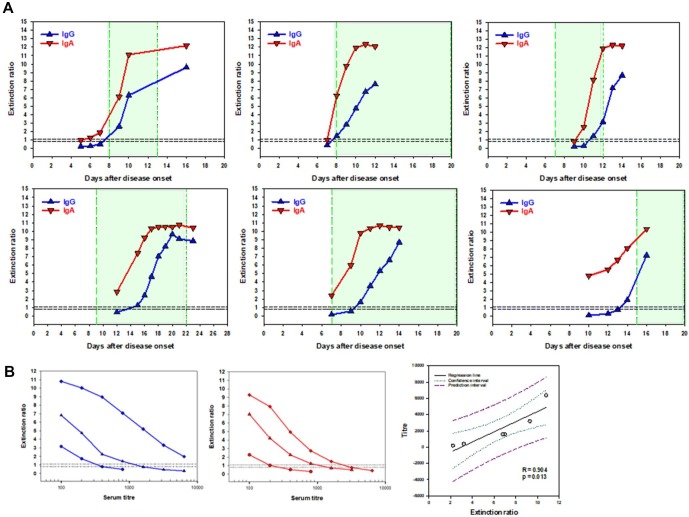
We assessed the time-course of humoral immune response in remnant clinical specimens from hospitalized patients with RT-PCR confirmed SARS-CoV-2 infection; results from the first six patients are shown in Fig. 2 A. The median time to SARS-CoV-2 spike protein specific IgA detection from symptom onset was 8 days, and 10 days for IgG. IgA preceded IgG response by 2 days on average. In our small set of patients for which we had serial measurements, we did not find that an earlier immune response was indicative of reduced ventilator duration (Fig. 2A).
Fig. 2.
Seroconversion of hospitalized nucleic acid-confirmed COVID-19 patients in the United States. A. Results of six COVID-19 patients (one per panel) with serial specimen collection during hospitalization. Green shading – days intubated: Vertical green lines indicate day of intubation (left) and day of extubation or continued intubation (right). Horizontal dashed lines indicate indeterminate interpretive region. B. Three samples for each IgG (blue) and IgA (red) were diluted to end titer to determine the correlation between extinction ratio and end titer. Left panels demonstrate the full dilution series. Right panel plots initial extinction ratio to final titer of each sample with 95% confidence intervals (green dotted lines) and predictive interval (purple dashed lines). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We chose three specimens with varying extinction ratios and performed serial titration measurements to determine the relationship between extinction ratios and serum titers. We found there to be an inverse log linear relationship with an extinction ratio between 6 and 8 corresponding to a 1:1000 titer for both IgA and IgG (Fig. 2B). The correlation analysis revealed a statistically significant relationship (p < 0.05) suggesting that the tested assay may be considered semi-quantitative.
3.3. Antigen specificity of detected antibodies
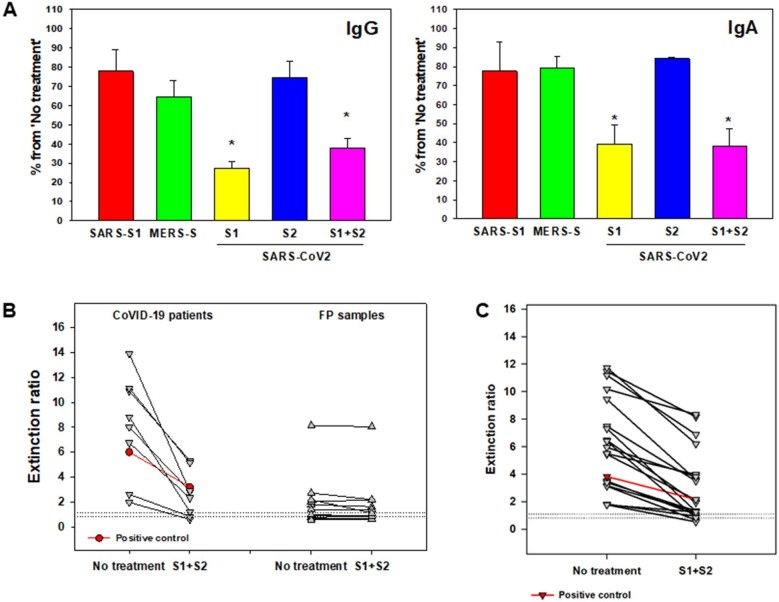
At the time of assay assessment there were no clinical laboratories performing SARS-CoV-2 antibody testing in the US and external quality controls were not available. Due to the high level of cross-reactivity seen with SARS-CoV-2 IgA and the need to confirm positivity for plasma donation, we tested for SARS-CoV-2 antibody specificity by using recombinant spike proteins S1 and S2 to competitively block the antibody binding for both IgA and IgG tests. In confirmed positive patients we found a significant competitive effect with a reduction in extinction ratios of >50% for the SARS-CoV-2 spike proteins (Fig. 3 A). As expected, there was little competition with the S2 subunit of the spike protein as the assay is specific for the S1 domain and use of the SARS-CoV-2 combined S1 and S2 subunit produced similar competition to the S1 subunit alone. Using recombinant spike proteins from SARS-CoV and MERS viruses, we did not find statistically significant competition with the antibodies produced in COVID-19 patients when considered as a group (Fig. 3A). However, we did find that some patients produced antibodies against SARS-CoV-2 that could be partially blocked by SARS-CoV and MERS spike proteins. Next, for IgA this confirmation technique demonstrated < 20% reduction in signal in 11/12 samples that had cross-reactivity (p = 0.232). And a > 50% reduction in signal in 9/9 samples that were true positives (p = 0.001). (Fig. 3B). We had too few cross-reactive IgG specimens to demonstrate the lack of competitive blocking in this cohort (repeat testing of two cross-reactive specimens gave a negative result), but we did find IgG true positive specimens to have a significant reduction in signal (n = 19, p < 0.0001 Fig. 3C).
Fig. 3.
Competitive spike proteins significantly improve assay specificity. A. Nucleic acid confirmed COVID-19 patient serum specimens (n = 3) were assayed for SARS-CoV-2 IgG and IgA antibodies. Pretreatment of the sample with SARS-CoV S1 subunit protein (SARS-S1), MERS spike protein (MERS-S), SARS-CoV-2 S1 subunit protein (S1), SARS-CoV-2 S2 subunit protein (S2), and SARS-CoV-2 S1 subunit and S2 subunit (S1 + S2) was performed to assess IgG and IgA antibody reaction specificity. Results are mean ± SEM, percent of inhibition from non-treated control sample (n = 3). B. SARS-CoV-2 IgA antibodies were assessed in nucleic acid confirmed COVID-19 patients (n = 9) and non-COVID-19 samples that displayed cross-reactivity (n = 12) with and without S1 + S2. Horizonal dashed lines indicate indeterminate interpretive region. *, p < 0.01 (paired t-test). C. SARS-CoV-2 IgG antibodies were assessed in nucleic acid confirmed COVID-19 patients (n = 19) with and without S1 + S2. Horizonal dashed lines indicate indeterminate interpretive region. *, p < 0.01 (paired t-test). Positive control(s) from the kit were used as additional controls (both in B and C) to confirm spike protein specificity.
4. Discussion
The primary humoral response to SARS/MERS-CoV infection is against the spike and nucleocapsid proteins [10]. As a consequence, most ELISA assays for antibodies against SARS-CoV-2 have likewise focused on these proteins. To aid in screening for plasma therapy candidates, vaccine response trials, and epidemiologic studies, we have validated a commercially available prototype SARS-CoV-2 IgG and IgA assays. These assays have subsequently been approved for use by Health Canada, the IgG only has been approved for use by the FDA, and both have been CE marked. We found high specificity for the IgG assay (97%, 99%) but the specificity of the IgA assay (81%, 86%) required an additional confirmatory blocking step (Fig. 1, Fig. 3). Using the specific blocking confirmation step, the putative specificity of the IgA assay is improved to 98% based on the 12 initially IgA reactive specimens tested for cross-reactivity, of which 11 displayed no change in extinction ratio with blocking proteins.
The ability to competitively block the serologic assay in true positive but not false reactive cases indicates that these are likely non-specific cross-reactivity and reduces the likelihood that the cross-reactivity could be due to other coronaviruses (Fig. 3). There is high homogeneity between the spike protein structures among MERS, SARS-CoV and SARS-CoV-2 viruses with the S1 subunits sharing about 64% amino acid homology [11]. Spike proteins from SARS-CoV-2, MERS, and SARS-CoV demonstrated that COVID-19 patient sera could be partially blocked in SARS-CoV-2 IgG and IgA assays by MERS and SARS-CoV S proteins. This highlights the variability of the antigens that elicit response in each patient in early seroconversion. This is unsurprising given the high homogeneity in the spike proteins and is in agreement with several studies demonstrating that SARS-CoV convalescent sera and spike antibody can partially neutralize SARS-CoV-2 in neutralization assays [11], [12], [13], [14], [15].
Characterization of humoral response to SARS-CoV-2 has overall demonstrated variable results in published papers. Zhao and colleagues indicated that only 83% of patients studied developed an IgM response in a spike protein ELISA, and only 65% developed an IgG response in a nucleocapsid protein ELISA [16]. Using also a nucleocapsid assay, Guo et al. found 78% of patients had detectable SARS-CoV-2 IgG levels, 85% IgM and 93% IgA [14], in keeping with subsequent work done by Liu et al. [17]. Studies to date indicate that IgM and IgA appear essentially simultaneously with more patients producing IgA than IgM [14], [16], which is in agreement with the biology of respiratory viral infections. Virions that infect mucosal surfaces encounter secretory IgA antibodies present at the apical surfaces of epithelial cells and the type of antibody that is produced can influence the outcome of viral infection. It is unclear how correctly systemic IgA reflects the specificity of mucosal IgA, which raises the question about absolute validity and utility of determining the neutralization capacity of systemic antibody in COVID-19 patients. All tested SARS-CoV and MERS patients developed neutralizing antibody detectable on day 15 [2], [3]. Preliminary work on COVID-19 demonstrates that all sera from PCR-confirmed patients demonstrate the presence of neutralizing antibodies when antibodies are detected [15].
Appearance of virus specific antibody is another important characteristic of the antiviral immune response. Although Guo et al. reported that the production of IgM, IgA, and IgG against SARS-CoV-2 were positive as early as day 1 after symptom onset [14], we found that IgA appears earlier than IgG in all tested COVID-19 patients (Fig. 2). Similar results were reported by Zhang et al. who, utilizing different ELISA kits, showed that overall the seroconversion of IgA was quicker than that of IgM and IgG [16]. We found that the median day of seroconversion for anti-S1 IgA was day 8 after symptom onset and for anti-S1 IgG day 10 after symptom onset. This is slightly later than reported for nucleocapsid protein assays for IgA [14], but in keeping with most reports [15], [16], [17]. Importantly, serum IgG against SARS-CoV-2 may rise at the same time or earlier than IgM: two reports revealed that more patients were seropositive for IgG than IgM at day 0 and day 5 of hospital admission [18] and some patients also had earlier IgG than IgM seroconversion [19]. Furthermore, more patients had earlier seroconversion for IgG than IgM for both anti-N and anti-S responses [19]. It is important to determine if these seroconversion windows are a reflection of differences in the analytical sensitivity of the IgM and IgG assays used in these studies or if they are a true reflection of the humoral response. We did observe a close seroconversion rate between IgA and IgG antibodies.
In our study, end point titration demonstrated an inverse log linear relationship between extinction ratio and titer (Fig. 2). Extinction ratios >6 were seen in all 6 patients before 14 days. We also found that extinction ratios >10 were outside the limits of detection of the assay. For plasma therapy, epidemiologic, and vaccine response purposes it will be important to determine if semi-quantitative assays can be correlated with neutralizing assay titer and ultimately a cutoff determined that is indicative of immunity. While within plate variation for this assay was acceptable, overall imprecision of this assay when processed manually would not allow for creation of a fixed cutoff value but would likely require an internal cutoff control as a comparator. It is notable that these studies were performed on the prototype kits, prior to the final kit arrangement that was subsequently approved by several country’s regulatory agencies.
Finally, we found that antibody response did not correlate with time on ventilator support, nor was day of seroconversion correlated with day of intubation (Fig. 2). As all of our cases were hospitalized patients who were eventually intubated, we cannot make a comparison with patients who had mild symptoms, though other work seems to indicate that critically ill patients will have higher antibody responses than mild cases [15], [16]. However, To et al. also reported that serum antibody levels were not correlated with clinical severity [19]. This is important to note for epidemiologic studies where less sensitive serologic methods may miss cases, and for consideration in vaccine response trials where low titers may not be indicative of lack of appropriate vaccine response.
In summary, our results demonstrate feasibility of testing SARS-CoV-2 specific IgG and IgA antibody in human blood for confirming the development of immune response and the use of these assays in a semiquantitative manner to be utilized in elucidating a potential immunity related cutoff after a natural course of the disease or as a response to vaccination. The specificity of these assays does not appear to be sufficient for large scale low prevalence screening but may be of use for high prevalence convalescent plasma screening or as an orthogonal assay in low prevalence screening. Using defined tools to measure immune responses is imperative as we work to fill the critical gaps in current knowledge. We must correlate immune responses with neutralizing titers, determine duration of immune memory, and finally assess neutralizing antibody titers for correlation with true protective immunity before we can expand appropriate use of serologic testing and understand future disease spread.
Author contributions
SEW and MRS conceived and designed the experiments, analyzed the data, prepared, and wrote the manuscript. GVS contributed to data analysis and preparing the paper. GK helped in the analysis and interpretation of the clinical data. GM, CK and JM performed the experiments, collected data and calculated the results.
Acknowledgements
We thank Dr. Alan Wells, Medical Director of UPMC Clinical Laboratories and Executive Vice-Chair for Laboratory Medicine, for his great moral and administrative support and help in the study. We also thank Mary Yost and Mary Jane Horenzy, the laboratory leading technologist and manager, respectively, for their support and assistance in organizing and protecting these studies.
References
- 1.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12(4) doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mo H., Zeng G., Ren X., Li H., Ke C., Tan Y. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11(1):49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne D.C., Iblan I., Rha B., Alqasrawi S., Haddadin A., Al Nsour M. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg. Infect. Dis. 2016;22(10):1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO, Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. Interim Guid. (2020).
- 5.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 6.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham A.C., Goh H.P., Koh D. Treatment of COVID-19: old tricks for new challenges. Crit. Care. 2020;24(1):91. doi: 10.1186/s13054-020-2818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan Y.J., Liu Q., Zhao S.Q., Huang F., Ren L., Liu L. The trial of chloroquine in the treatment of corona virus disease 2019 COVID-19 and its research progress in forensic toxicology. Fa Yi Xue Za Zhi. 2020;36(2) doi: 10.12116/j.issn.1004-5619.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Focosi D., Tang J., Anderson A., Tuccori M. Convalescent plasma therapy for Covid-19: state of the art. J. Clin. Med. 2020;Preprints 2020,;2020040097 doi: 10.1128/CMR.00072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L., Ren L., Yang S., Xiao M., Chang Y.F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okba N., Müller M., Li W., Wang C., GeurtsvanKessel C., Corman V. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]