Highlights
-
•
We proposed a novel framework, CHP-Net, to differentiate and localize COVID-19 from community acquired pneumonia.
-
•
We used excessive data augmentation to extend the available dataset and optimize the CHP-Net generalization capability.
-
•
Comparing to other ConvNet, CHP-Net works much more efficiently to extract feature information on chest X-Ray.
-
•
All metrics, including categorical loss, accuracy, precision, recall and F1-score, proved CHP-Net fits good for the task.
-
•
CHP-Net are better than the previous methods tested in detecting COVID-19 and exceeding to radiologist.
Keywords: COVID-19, Computer-aided detection (CAD), Community-acquired pneumonia (CAP), Deep learning (DL), Chest X-ray (CXR)
Abstract
The COVID-19 outbreak continues to threaten the health and life of people worldwide. It is an immediate priority to develop and test a computer-aided detection (CAD) scheme based on deep learning (DL) to automatically localize and differentiate COVID-19 from community-acquired pneumonia (CAP) on chest X-rays. Therefore, this study aims to develop and test an efficient and accurate deep learning scheme that assists radiologists in automatically recognizing and localizing COVID-19. A retrospective chest X-ray image dataset was collected from open image data and the Xiangya Hospital, which was divided into a training group and a testing group. The proposed CAD framework is composed of two steps with DLs: the Discrimination-DL and the Localization-DL. The first DL was developed to extract lung features from chest X-ray radiographs for COVID-19 discrimination and trained using 3548 chest X-ray radiographs. The second DL was trained with 406-pixel patches and applied to the recognized X-ray radiographs to localize and assign them into the left lung, right lung or bipulmonary. X-ray radiographs of CAP and healthy controls were enrolled to evaluate the robustness of the model. Compared to the radiologists’ discrimination and localization results, the accuracy of COVID-19 discrimination using the Discrimination-DL yielded 98.71%, while the accuracy of localization using the Localization-DL was 93.03%. This work represents the feasibility of using a novel deep learning-based CAD scheme to efficiently and accurately distinguish COVID-19 from CAP and detect localization with high accuracy and agreement with radiologists.
1. Introduction
At the end of 2019, a novel coronavirus pneumonia, COVID-19, began to rapidly propagate due to widespread person-to-person transmission [1]. By April 5th, 2020, there were over 1 million confirmed COVID-19 cases, and tens of thousands of people lost their lives. The epidemic of 2019-nCoV pneumonia poses an enormous threat and challenge to the global population.
Key to fighting against COVID-19 is diagnosing infected patients as soon as possible and providing them with proper treatment and care. Reverse-transcription polymerase chain reaction (RT-PCR) is considered to be the gold standard for diagnosing the disease [2]. Some respiratory specialists have suggested that a nucleic acid test was not required to confirm the disease if typical imaging findings of coronavirus were present [3]. Volumetric CT chest images of the lungs and soft tissues have been investigated in recent studies for detecting COVID-19 [4]. However, the high radiation doses and costs limit the use of CT, especially for pregnant women and children [5]. Chest X-ray, a non-invasive chest exam with a low radiation dose, can completely image the lungs and is more time and cost efficient than CT. Therefore, X-rays can serve as an effective method for the early detection of COVID-19. However, COVID-19 may share some common radiographic features with other pneumonias, making its discriminability difficult for radiologists.
Currently, artificial intelligence using deep learning plays an important role in the medical image area due to its excellent feature extraction ability. Deep learning models (DLs) accomplish tasks by automatically analysing multi-modal medical images. Computer vision refers to using DLs in processing images or videos and how a computer might gain information and understanding from this method. Advanced computer-aided diagnosis schemes are mostly based on state-of-the-art methods, such as fully convolutional neural networks (FCNNs) [6], VGGNet [7], ResNet [8], Inception [9], and Xception [10]. Some examples of the application of AI include cancer detection and classification [11], the diagnosis of diabetic retinopathy [12], multi-classification of multi-modality skin lesions [13], polyp detection during colonoscopy [14], etc.
Since COVID-19 has become widespread, many researchers have dedicated their efforts to the application of machine vision and deep learning in the diagnosis of the disease based on medical images and have achieved good results. Ozturk et.al. [15] proposed a model that uses DarkNet as a classifier and obtained a classification accuracy of 98.08% for binary classification (COVID-19 vs. no findings) and 87.02% for multiple classification (COVID-19 vs. no findings vs. pneumonia). Khan et.al. [16] proposed a deep model based on the Xception architecture to detect COVID-19 cases from chest X-ray images, which achieved an overall accuracy of 89.6% and a recall rate for COVID-19 cases of 93% in classifying COVID-19, healthy conditions, bacterial pneumonia and viral pneumonia. Narin et al. [17] employed three state-of-the-art deep learning models (ResNet50, InceptionV3 and Inception-ResNetV2) and obtained the best accuracy of 98% with a pre-trained ResNet50 model for 2-class classification. However, they did not include pneumonia cases in their experiment. Moreover, there are several deep learning models that use CT images to detect COVID-19. For example, ResNet-18 was used as the CNN to diagnose the diease from chest CT, which achieved an area under the curve of 0.92 and had equal sensitivity to a senior radiologist [18].
Computer-aided detection (CAD) is a common occurrence in hospitals. Bai et al. [19] found that artificial intelligence (AI) can help clinicians distinguish COVID-19 from other pneumonia on chest CT. Moreover, it is not easy to distinguish soft tissue with poor contrast on X-ray. To overcome these limitations, CAD systems have been implemented to assist clinicians in automatically detecting and quantifying suspected diseases of vital organs on X-rays [20]. Notably, deep learning can automatically detect clinical abnormalities from chest X-rays at a level exceeding practising radiologists [21]. Due to the lack of available data, however, almost all previous study data originated from open datasets on the net, and thus it is unknown how well their corresponding models would perform with real-world data. To the best of our knowledge, there are few studies about localizing the disease. In this paper, we developed an efficient and accurate deep learning model-based computer-aided detection scheme for automatically localizing COVID-19 from CAP on chest X-rays. The novel CAD scheme comprised two different DLs: the first DL is utilized to automatically recognize and collect X-rays belonging to COVID-19 patients (i.e., the X-rays differentiated from CAP patients), and the second DL is utilized for detecting localization of left lung, right lung or bipulmonary in each X-ray radiograph. The details of this work, including the structure of the CAD scheme and performance evaluation, are manifested in the following sections.
2. Methodology
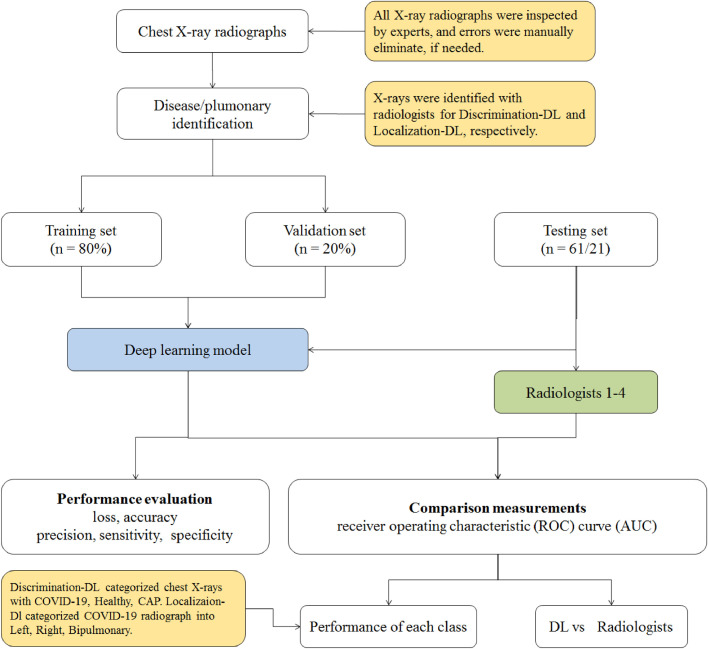
Our proposed CAD framework comprises two deep learning models: the Discrimination-DL and the Localization-DL. The whole process of our proposed scheme is demonstrated in Fig. 1 . The first Discrimination-DL is used to automatically recognize and collect X-rays of COVID-19 patients. The acquired X-rays are then applied as input to the second Localization-DL for automated detection of left lung, right lung or bipulmonary. The lung localized from all X-rays of interest is combined to detect and localize the lung parts of the particular patient at the end step. The DL architectures, training process and measurement methods for the two DLs are discussed in the next two sections.
Fig. 1.
Flow diagram of the proposed CAD system illustrating the discrimination and localization of COVID-19 from CAP on chest X-ray radiographs. We utilized the Discrimination-DL to distinguish COVID-19 from CAP on chest X-rays, and the Localization-DL was trained to detect lung localization (i.e., left lung or right lung or bipulmonary). Abbreviations: Healthy: healthy controls; CAP: community-acquired pneumonia; Left: left lung; Right: right lung.
2.1. Patient cohorts
There are openly available annotated chest X-ray databases with recorded patients. The dataset used in this study comprises a total of 3545 chest X-ray images and is referred to as COVID19-DB. To generate the COVID19-DB dataset, for example, chest X-ray radiographs, we combined and modified two different openly available datasets: 1) chest X-ray radiographs of the pneumonia cohort were collected from the RSNA Pneumonia Detection Challenge [22], and 2) the COVID-19 cohort with chest X-ray radiographs comprised [23]. Four radiologists from Xiangya Hospital manually verified the COVID-19 from CAP on chest X-rays. This study was approved by the Ethics Committee of the Xiangya Hospital of Central South University. Informed consent was obtained from all participants.
The rescale strategy is important in applying DL to large images since the resolution is limited by the GPU memory. In terms of our task, there are few data available, and we use extensive data augmentation [24] (such as random clipping, flipping, shifting, tilting and scaling) to extend the available dataset as well as optimize the CAD scheme generalization capability. To optimize and improve the proposed CAD scheme, we will consistently expand the data of COVID19-DB.
More specifically, we collected 2004 radiographs of CAP, 1314 radiographs of healthy controls, and 204 radiographs of COVID-19 that were randomly split into two independent subsets for the Discrimination-DL, where the training subset used 80%, and the validation subset used 20%. The training and validation subsets were together employed for model fitting and preventing model over-fitting in the training phase. There was no patient overlap between phases. Moreover, the objects of the Xiangya Hospital of Central South University were patients diagnosed with Covid-19 and CAP from January 25, to May 1, 2020. The enrolled Covid-19 patients were confirmed as positive by RT-PCR on nasopharyngeal swabs and throat swabs. Clinical manifestations, laboratory and X-rays of patients were collected. All chest X-rays (CXRs) were acquired as computed or digital radiographs following usual local protocols. CXRs were acquired in the posteroanterior (PA) or anteroposterior (AP) projection. X-ray images were collected from 21 Covid-19 patients, 20 CAP patients and 20 controls, which were mainly used as test data. The testing subset was performed in the testing phase to prove model generalization.
For each of the COVID-19 patients in the COVID19-DB, the non-pulmonary region was divided into the left lung and right lung. By doing so, we can acquire a sample of 200 X-ray radiographs as the training set and 21 X-rays from the Xiangya Hospital as the testing for the Localization-DL. Among them, 157 and 183 were “positive” infected in the left pulmonary region and right pulmonary region, respectively. We randomly assembled 200 X-rays of healthy controls as “negative” infections located in both pulmonary regions. Next, to localize the pulmonary region of COVID-19, we conducted experiments to identify the infected region located in the left lung, right lung or bipulmonary region.
2.2. Deep learning architectures
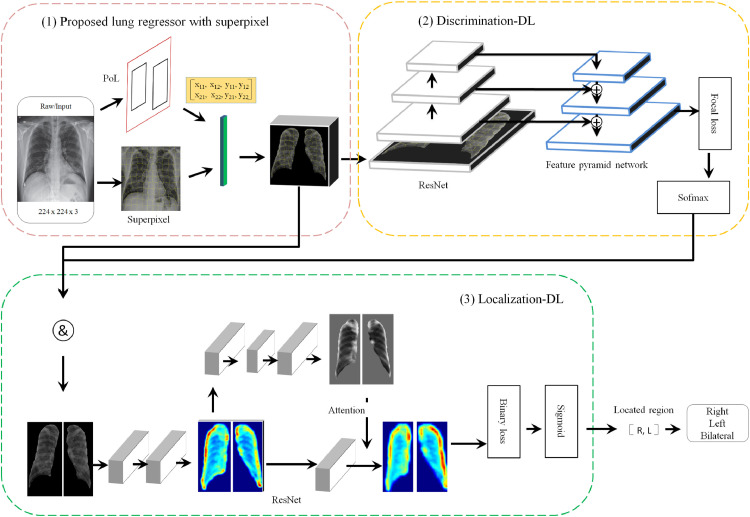
The structure of the proposed CAD scheme is a single, unified network composed of three parts, as illustrated in Fig. 2 : (1) a bounding-box regression that encodes lung features into each superpixel, (2) a discrimination deep learning model that predicts a differentiating probability distribution, and (3) a localizing deep learning model that distinguishes the corresponding pulmonary and generates the location probability distribution.
Fig. 2.
Deep learning architectures. (1). We utilized the proposal of a lung regressor (PoL) with superpixel to generate the Discrimination-DL input. The PoL matrix is a 2 × 4 vector, which illustrates the bipulmonary region coordinates. (2). The Discrimination-DL adopts a feature pyramid network as a backbone network on top of a ResNet architecture and generates a differentiated probability across cohort categories. (3). The Localization-DL constructed attention modules use a state-of-the-art residual attention network basic unit. The located region is defined as a 1 × 2 vector and represents all potential pulmonary locations. Abbreviations: PoL: proposal of lung regressor; CAP: community-acquired pneumonia; Left/L: left lung; Right/R: right lung; Bilateral: bipulmonary.
2.2.1. Proposed pulmonary regressor with superpixel
The task of the Discrimination-DL is to distinguish COVID-19 from CAP on chest X-rays, which are acquired for detection of the infected pulmonary region. We formulated this task as a categorical classification problem. Specifically, we utilized each X-ray radiograph as the input of a classifier (Discrimination-DL) and discriminated whether the X-ray radiograph belonged to the COVID-19, CAP or healthy.
In this way, each X-ray radiograph excluded non-pulmonary regions, and the lungs were first segment with a proposal of lung regressor (namely, PoL, a minimum bounding-box approach [25]). The minimum bounding-box approach (Eq. (1)) takes the non-zero region, which corresponds to lung regions, and computes the vector representing the 4 parameterized coordinates for each part as follows:
| (1) |
where x and y are the centre coordinates of the lung region, and w and h are its width and height, respectively. Each variable * and *a (where * is one of x, y, w, h) denote the predicted lung region and ground-truth label, respectively. The output of the PoL consists of two vectors and is illustrated in Fig. 2 (1). The X-ray radiograph is divided into 100 superpixels (a set of pixels with similar representations), and the calculated features are stored in each superpixel. Manual lung segmentation is often dependent on the expertise of the clinician and is time-consuming. Therefore, we present the VGG [7]-based PoL network for lung segmentation.
2.2.2. Discrimination structure
In the Discrimination-DL, we employed the feature pyramid network as the backbone, as shown in Fig. 2 (2). The backbone computes a convolutional feature map over an entire input image. First, the original X-ray radiographs are resized to 224 × 224 using cubic interpolation of OpenCV. This step can be interpreted as image resampling processing, and the Discrimination-DL greatly reduces the input size and parameters, which promotes training efficiency and reduces the risk of over-fitting. Based on the resampled X-ray radiographs, we build a feature pyramid network (FPN) [26] basic unit on top of the ResNet [8] architecture as a COVID-19 discriminator to determine a preliminary probability distribution. In general, FPN utilizes a top-down structure and skip connection to improve multi-scale prediction from a single X-ray radiograph. We constructed an FPN with pyramid level l ∈ [5, 8], which indicates a resolution of 2l.
In COVID19-DB, we discovered that the COVID-19 cohort, CAP cohort and pneumonia cohort encounter class imbalance. Lin et al. [27] introduced the focal loss starting for class imbalance of binary classification. With this method, we extend the binary focal loss function to the multi-focal loss function for the Discrimination-DL as
| (2) |
Here, i is the index of a class in a chest X-ray, and pi is the predicted probability of class i. By default, the weight factor α is 0.25, N is the number of classes, and the tuneable focusing parameter γ is 2. Finally, we note that the multi-focal loss function (Eq. (2)) combines the softmax operation [28]. This energy function is the approximate maximum-function and is defined as
| (3) |
where x takes as input a vector, and the individual zi(x) values are the elements of the input vector and can take any real value. The term in the denominator is the normalization term, which ensures that the sum of the output values of the function will equal 1, thus constituting a valid probability distribution. With the help of the softmax classifier, the Discrimination-DL architecture maps each X-ray radiograph to a vector of three continuous numbers between 0, 1 and 2, indicating the probabilities of the input radiograph belonging to COVID-19, healthy or CAP classes. In Discrimination-DL, the predictions of a class do not compete with those of another class. The batch size and number of epochs were experimentally set to 16 and 20, respectively, for all experiments. The pseudocode for Discrimination-DL is shown in Algorithm 1 .
Algorithm 1.
Training procedure for Discrimination-DL.
2.2.3. Localization structure
After X-ray radiographs belonging to COVID-19 are identified, a deep learning-based scheme is used to locate the pulmonary region. This task is formulated as a binary classification issue. In this study, we developed the Localization-DL architecture, as shown in Fig. 2 (4). The objective of Localization-DL is to identify the infected pulmonary region as belonging to the left lung, right lung or bipulmonary region. We employ a residual attention network [29] as the basic unit to achieve this goal. In the attention residual learning, the output H of the attention module is modified as
| (4) |
where attention mask M ∈ [0, 1] with M approximates 0, H approximates original features F, and κ ranges over the corresponding spatial location (i.e., 0 for right pulmonary, 1 for left pulmonary). The final layer computes the binary cross-entropy function (5) combined with the sigmoid classifier function, which is defined as follows:
| (5) |
where yi and specify the predicted class and ground-truth class, respectively. The input of the Localization-DL comprises the left pulmonary and right pulmonary, which is a 64 × 128 rectangular patch obtained by downsampling a 224 × 224 patch at the extracted lung pixel depicted on the X-ray radiograph. The located region part is a one-dimensional vector that contained 1 for the infected region and 0 for the non-infected region on both pulmonary regions (e.g., [1, 1] illustrates that bipulmonary are infected).
The proposed CAD scheme uses 3 × 3 convolutions, which are activated by a rectified linear unit (ReLU, [30]) and is trained end-to-end using the Adam [31] optimizer, derived from Eq. (7):
| (6) |
where α is the initial learning rate, dw is the gradient at time t along w, vt is the exponential average of the gradients along w and st is the exponential average of the squares of the gradients along w. Here, β 1 is 0.9, β 2 is 0.999 and ϵ is . We used an initial learning rate of for Adam that was decayed by a factor of 5 each time when the validation loss plateaus after an epoch. The optimization was performed for 50 epochs through COVID-DB, and the batch size was experimentally set to 16. To prevent over-fitting, we used the EarlyStopping function from the Keras framework, which stopped training when a monitored quantity stopped improving. The pseudocode for Localization-DL is shown in Algorithm 2 .
Algorithm 2.
Training procedure for Localization-DL.
2.3. Loss function
For training the DL functions, we assign a multi-focal loss function to each category in the Discrimination-DL and a binary class label to each pulmonary part. We assign a positive label to the COVID-19 cohort and assign a negative label to non-COVID-19 cohorts. Cohorts that are negative do not contribute to the Localization-DL objective. With these definitions (2) and (5), we define an objective function to minimize the multitask loss for a chest X-ray radiograph as
| (7) |
Here, i is the index of a category in a mini-batch and pi is the discriminated probability of category i. yi is the predicted category, and is the ground truth. In the term the multi-focal loss is activated for a positive label () and is activated otherwise (). In the term the binary loss is activated only for a positive label () and is deactivated otherwise (). The outputs of the Discrimination-DL and the Localization-DL layers consist of p and y, respectively.
3. Performance
The proposed work was implemented using the publicly available Keras [32] framework with the TensorFlow [33] backend. Training and testing of each model used a single NVIDIA GTX1080Ti GPU with hexa-core 3.20 GHz processor and 16 GB memory. To measure the experiments in a fair setting (inspired by [30]), all DLs were designed and implemented using the same principles. The pseudocode for the testing procedure is shown in Algorithm 3 .
Algorithm 3.
Testing procedure for the computer-aided diagnosis scheme.
3.1. Patient characteristics
Our testing cohort consisted of 61 chest X-ray radiographs, of which 21 were with COVID-19 and 20 were with CAP and 20 were with non-infection (as depicted in Table 1 ). The average age with COVID-19 was lower than that with non-COVID-19 (45 vs. 64 years). The COVID-19 cohort was less likely to have an evaluated C-reactive protein (CRP) than the non-COVID-19 cohort (31 vs. 41) or a reduced erythrocyte sedimentation rate (ESR) (52 vs 66).
Table 1.
Composition of the number of allocated CXRs.
| Dataset | Covid19 | Healthy | CAP |
|---|---|---|---|
| RSNA pneumonia detection challenge | 0 | 1314 | 2004 |
| COVID-19 CXR dataset | 204 | 0 | 0 |
| Total | 225 | 1334 | 2024 |
| Training subset | 160 | 1050 | 1600 |
| Validation subset | 44 | 264 | 404 |
| Testing subset (Xiangya Hospital) | 21 | 20 | 20 |
CXR: chest X-rays; RSNA: Radiological Society of North America.
We evaluated the proposed CAD scheme with respect to the discrimination of COVID-19 from CAP and the localization of CAP parts separately. For the CAD scheme, we developed two DLs, one for COVID-19 discrimination and one for infected pulmonary localization, and proposed a lung box as well as a superpixel approach as preprocessing methods.
3.2. Discrimination influences
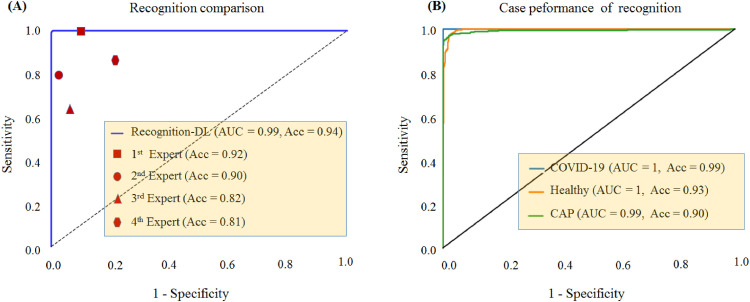
First, we evaluated the model w. r. t. its ability to discriminate a given chest X-ray radiograph into COVID-19 or non-COVID-19 regardless of localization. The Discrimination-DL achieved a training loss of 2.29% and training accuracy of 98.71% and yielded a validation loss of 4.58% and validation accuracy of 95.12%. Fig. 3 shows the receiver operating characteristic (ROC) curve (AUC) comparison with the radiologists’ performance and the performance of each class case. The result shows that the most accurate DL demonstrated a performance that outperformed four radiologists. However, the CAP accuracy was poor in the remaining cases.
Fig. 3.
Performance of discriminating COVID-19 from CAP on testing subset. (A). ROC curve for the Discrimination-DL with radiologists for performance comparison. The area under the ROC of the Discrimination-DL was 99%. (B). ROC curves for COVID-19 from CAP on the testing subset trained with Discrimination-DL were 1 for COVID-19, 1 for healthy controls, and 0.99 for CAP. Abbreviations: receiver operating characteristic (ROC) curve (AUC); accuracy (Acc); community-acquired pneumonia (CAP); healthy controls (Healthy).
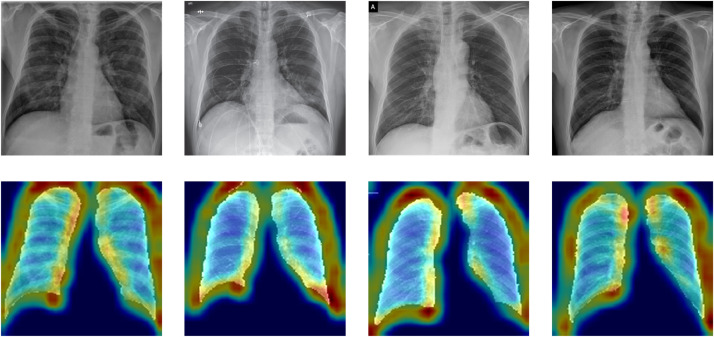
Furthermore, we compared the average predictions of the Discrimination-DL and present a summary of studies conducted on the automatic diagnosis of COVID-19 from chest X-rays. Narin et al. [17] achieved an accuracy of 98% with a pre-trained ResNet50 model but did not include pneumonia cases in their experiment. Hemdan et al. [34] achieved an accuracy of 90% on a 2-class problem based on the VGG19 architecture pre-trained on ImageNet. Ozturk et al. [15] proposed a CNN model based on the DarkNet architecture to classify COVID-19 cases from X-ray images and achieved a binary and 3-class classification accuracy of 87%. Khan et al. [16] introduced CoroNet, which is based on the Xception structure pre-trained on the ImageNet dataset, and achieved an accuracy of 89%. Table 2 shows that the performances of Discrimination-DL are better than those of the compared methods. A Grad-CAM [35] for important features of the lung that lead the Discrimination-DL to classify a case as COVID-19 from CAP was generated using gradient-weighted class activation mapping [36], which on representative chest X-ray radiographs from the test set demonstrates that the model focused on the area of abnormality (Fig. 4 ).
Table 2.
Automated predictions on a testing subset of chest X-Rays.
| Method | Architecture | Pre-trained | Predicted performance |
|||
|---|---|---|---|---|---|---|
| Covid19 | Healthy | CAP | Average | |||
| Narin et al. [17] | ResNet50 | 100% | 100% | × | 98% | |
| Hemdan et al. [34] | VGG19/DenseNet201 | 100% | 80% | × | 90% | |
| Ozturk et al. [15] | DarkNet | × | 98% | 86% | 85% | 87% |
| Khan et al. [16] | Xception | 89% | 85% | 95% | 89% | |
| Discrimination-DL | ResNet50 + FPN | × | 99% | 93% | 90% | 94% |
Pre-trained: the model was loaded with pre-trained weights from ImageNet database; CAP: community-acquired pneumonia; Healthy: healthy controls.
Fig. 4.
Representative chest X-ray radiographs corresponding to Grad-CAM images.
3.3. Localization results
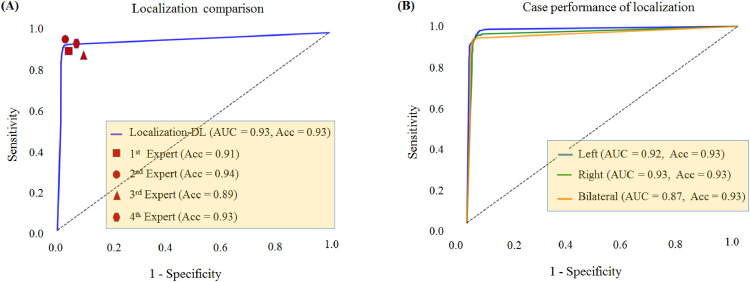
The Localization-DL yielded a training loss of 3.73% and training accuracy of 96.95%, while the Localization-DL achieved a validation loss of 5.26% and validation accuracy of 94.62%. We assessed the ability to localize the presence of CAD on the infected pulmonary tissue, as shown in the upper panel of Fig. 5 , which illustrated the ROC curve comparing the Localization-DL with radiologists’ performance and the performance of the infected pulmonary system. To assess localization performance, the model agreed best with the human observer. The result depicts the Localization-DL results rival that of four radiologists. Note here that the performance of the single pulmonary system is clearly superior to that of the bipulmonary system.
Fig. 5.
Performance of localizing infected pulmonary on the testing subset. (A). The ROC curve for Localization-DL compared with radiologist performance. The area under the curve was 93%. (B). The ROC curve of each case with the trained Localization-DL was 0.92 for left pulmonary, 0.93 for right pulmonary and 0.87 for bipulmonary. Abbreviations: receiver operating characteristic (ROC) curve (AUC); accuracy (Acc); left pulmonary (left); right pulmonary (right); bipulmonary (Bilaterel).
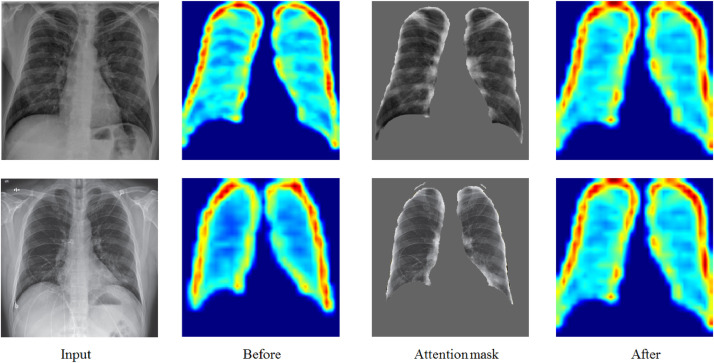
The attention mechanism brought a more discriminative feature representation. Fig. 6 shows several examples of attention residual learning. The pulmonary instance mask highlights the potentially infected part of the pulmonary system.
Fig. 6.
Several examples illustrating that the high-level part features with attention masks.
3.4. CAD performance
Our final CAD scheme achieved a test accuracy of 93.65%, sensitivity of 90.92% and specificity of 92.62%. Compared to the average radiologist, our CAD scheme had higher test accuracy (93.65% vs. 88.14%), sensitivity (90.92 vs. 77.54%) and specificity (92.65% vs. 86.24%). The testing time was 272 ms per image on the testing subset. The above results suggest that our CAD scheme enjoys high efficiency and good performance.
4. Discussion
Since COVID-19 was first detected at the end of 2019, it has spread all over the world. Since the vaccine is estimated to be years away [37], it is critical to diagnose the disease at an early stage so that patients infected with COVID-19 receive treatment and isolation to prevent virus spread. Mass testing has been successful in controlling the outbreak of COVID-19 in numerous places, such as China [38]. PCR is considered the gold standard to confirm the disease. However, it is time consuming and limited by the number of test kits.
In contrast, chest X-ray (CXR) machines are available in all hospitals to produce projection images of the patient’s thorax. Usually, CXR is the first choice for radiologists to detect chest pathology and plays a pivotal role in the diagnosis of CAP and epidemiological studies. It has been recently reported that ground-glass opacification, consolidation, peripheral and diffuse distribution, and bilateral involvement are found in the chest X-ray findings in COVID-19 A recent study found that radiologists detect COVID-19 from CAP with high specificity but moderate sensitivity, which means there are missed diagnoses of COVID-19. This reminds us that there is an urgent demand for developing applications to aid radiologists in detecting COVID-19.
In this study, we developed an efficient and accurate CAD scheme and demonstrated its feasibility to recognize and localize COVID-19 from CAP on chest X-ray radiographs. This work and the novel CAD scheme have a number of unique characteristics. First, a Discrimination-DL with proposed lung boxes was developed for the automatic detection of chest X-rays belonging to COVID-19 patients. This DL-based process can generate high detection accuracy compared to results from radiologists. Therefore, the Discrimination-DL for automatic accuracy detection of COVID-19 chest X-rays is reliable and can be employed to replace manual diagnoses, which provides the capability of managing large-scale datasets with high efficiency.
Second, we developed a Localization-DL-based scheme to identify infected pulmonary (i.e., left pulmonary, right pulmonary or bipulmonary), which considers the location coordinates and neighbourhood information. After sufficient training and optimization, the Localization-DL provides optimal identification results without human intervention. Therefore, the Localization-DL was a reliable CAD scheme for fully automatic identification of pulmonary infections from single COVID-19 chest X-rays.
Third, the two challenges in this study (i.e., discrimination and localization) were both formulated as classification problems, and a deep learning model was used as the classifier to address the problems. Recently, deep learning has been successfully used to diagnose lung abnormalities at the radiologist level, consuming much less time [21]. Following the success of DLs in many medical image analysis communities, this work demonstrated that DL models are effective for recognizing X-rays that belong to COVID-19 and localizing infected pulmonary tissue from a single chest X-ray.
In addition, based on the experimental results, we also observed several obstacles. For example, (1) due to the lack of resources, we were not able to compare COVID-19 chest X-ray images to pneumonia caused by other types of viruses. Instead, we collected 2004 radiographs of CAP, which in our view is sufficient to represent the typical distribution of CAP. (2) Almost all deep learning methods lack interpretability, which makes it difficult to determine the exact image features for generating the output. (3) This study focuses on whether a radiograph is COVID-19 but has not addressed classifying the disease according to severity. The next goal of our team will be to make efforts to predict not only whether COVID-19 exists but also the degree of severity to further the administration of patients. In all, future improvements would be to collect more data, perhaps hundreds or thousands of images of both COVID-19 and other viral pneumonia. The new dataset should also consider geographic diversity, which will increase its applicability worldwide.
5. Conclusions
The COVID-19 pandemic continues to threaten the health and lives of billions of people. Early discrimination and localization of the disease is the key to winning the battle against the virus. With this in mind, we developed and tested a novel CAD scheme for COVID-19 detection and localization based on a sequential two-step process including (1) detecting chest X-ray radiographs belonging to COVID-19 and (2) identifying infected pulmonary tissue from each of the detected chest X-rays. We used two open-source datasets that contained 225 and 2024 images from patients infected with COVID-19 and pneumonia, respectively as well as 1334 images from healthy people for training and validation. We demonstrated that this novel deep learning-based CAD scheme enables the differential diagnosis of COVID-19 from pneumonia on chest X-rays from a different data source, Xiangya Hospital, and can localize infected pulmonary tissue from each recognized X-ray with high accuracy or agreement with manual results from radiologists.
Although much work is still required to create a production-ready solution, the hope is that the promising results achieved by our two-step DL scheme on the test dataset and an increasing number of open datasets will lead to it being implemented to accelerate the development of highly accurate and practical deep learning solutions for detecting and localizing COVID-19 from chest radiographs. Due to the lack of corresponding images from the data source, radiographs of other types of viral pneumonia were not included. As new data are collected, our goal is to extend the proposed DL to the classification of four groups of radiographs (COVID-19, healthy, bacterial pneumonia and viral pneumonia), risk stratification for survival analysis, and more detailed localization of infected pulmonary tissue. Overall, our study provides a novel and reliable CAD tool for assisting in the processing of large-scale chest X-ray data and in accurately identifying COVID-19-infected pulmonary tissue in future clinical practice.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgement
This work was supported by the Projects of the National Social Science Foundation of China under Grant 19BTJ011, also funded bythe National Natural Science Foundation of China (grant no. 81770584 and no. 81570504) and by the Graduate Student Innovation Foundation of Central South University (2019zzts213).
Biographies
Zheng Wang received the B.S. degree in Computational mathematics and Applied Mathematics of Xiangtan University, China in 1998, and received the M.S. Computer Science of Karlsruhe Technology University, Germany in 2005. He is now a Ph.D. student of college of Mathematics and Statistics, Central South University, China. His research interests are Her machine learning and deep learning, computer-aided diagnose in medicine.
Ying Xiao received the B.S. and M.S. degrees in Internal Medicine of Central South University in 2015 and 2018 separately. She is now a Ph.D. student of XiangYa Medicine school of Central South University. Since 2016, she worked with Professor Hous team on the project of computer aided diagnosis in medicine, Her research interests are machine learning and image processing.
Yong Li received the B.S. in Internal Medicine of Xiamen University in 2015. He is now a postgraduate student of Xiangya Medicine school of Central South University. His research interests are machine learning on digestive disease and pattern recognition.
Jie Zhang received the B.S., M.S., and Ph.D. degrees in Internal Medicine of Central South University. She is now the associate professor of gastroenterology department of Second XiangYa Hospital. Her research interests are machine learning on CT images and inflammatory bowel disease.
Fanggeng Lu received the B.S., M.S., and Ph.D. degrees in Internal Medicine of Central South University. He is now the professor of gastroenterology department of Second XiangYa Hospital. His research interests are digestive disease, machine learning and image processing.
Muzhou Hou received the B.S. degree from Hunan Normal University, Changsha, China, the M.S. degree and the Ph.D. degree from Central South University, Changsha, China, in 1985, 2002 and 2009, respectively. He is currently a Professor and doctorial advisor in college of Mathematics and Statistics, Central South University, China. His current research interests include machine learning, neural networks, data mining and numerical approximation.
Xiaowei Liu received the B.S., M.S., and Ph.D. degrees in Internal Medicine of Central South University. He is now the director and professor of gastroenterology department. His research area is artificial intelligence applications in medicine images, immune related digestive disease and intestine homeostasis. He has obtained several honors and awards, including Second Prize of Hunan Science and Technology Progress Award and Leading Talent in the ”225” Engineering Medicine Discipline of High-level Health Talents in Hunan Province.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in wuhan, china, of novel coronavirus infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. A novel coronavirus from patients with pneumonia in china, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. Ct imaging features of 2019 novel coronavirus (2019-ncov) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y., Shin H.J., Kim M.-J., Lee M.-J. Comparison of effective radiation doses from x-ray, ct, and pet/ct in pediatric patients with neuroblastoma using a dose monitoring program. Diagn. Int. Radiol. 2016;22(4):390–394. doi: 10.5152/dir.2015.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shelhamer E., Long J., Darrell T. ully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2016;39 doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]; 1–1
- 7.Simonyan K., Zisserman A. International Conference on Learning Representations. 2015. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- 8.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 9.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 10.Chollet F. 30th IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1800–1807. [Google Scholar]
- 11.Gecer B., Aksoy S., Mercan E., Shapiro L., Weaver D., Elmore J. Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks. Pattern Recognit. 2018;84 doi: 10.1016/j.patcog.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chudzik P., Majumdar S., Caliva F., Al-Diri B., Hunter A. Medical Imaging 2018: Image Processing. Vol. 10574. SPIE; 2018. Exudate segmentation using fully convolutional neural networks and inception modules; pp. 785–792. [Google Scholar]
- 13.Bi L., Feng D.D.F., Fulham M., Kim J. Multi-label classification of multi-modality skin lesion via hyper-connected convolutional neural network. Pattern Recognit. 2020;107:107502. [Google Scholar]
- 14.Zhang R., Zheng Y., Poon C.C., Shen D., Lau J.Y. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recognit. 2018;83:209–219. doi: 10.1016/j.patcog.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozturk T., Talo M., Yildirim A., Baloglu U.B., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;28:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Narin, C. Kaya, Z. Pamuk, Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks, 2020. [DOI] [PMC free article] [PubMed]
- 18.Mei X., Lee H.-C., yue Diao K., Huang M., Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with covid-19. Nat. Med. 2020:1–5. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai H., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., Tran T., Choi J., Wang D.-C., Shi L.-B., Mei J., Jiang X.-L., Pan I., Zeng Q.-H., Hu P.-F., Li Y.-H., Fu F.-X., Huang R., Sebro R., Liao W.-H. Ai augmentation of radiologist performance in distinguishing covid-19 from pneumonia of other etiology on chest ct. Radiology. 2020:201491. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasr-Esfahani E., Samavi S., Karimi N., Soroushmehr S.M.R., Najarian K. 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) Vol. 2016. 2016. Vessel extraction in x-ray angiograms using deep learning; pp. 643–646. [DOI] [PubMed] [Google Scholar]
- 21.Rajpurkar P., Irvin J., Ball R.L., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Langlotz C.P. Deep learning for chest radiograph diagnosis: aretrospective comparison of the chexnext algorithm to practicing radiologists. PLoS Med. 2018;15(11):1002686. doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R.S. of North America, Rsna pneumonia detection challenge, 2019, https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. [DOI] [PubMed]
- 23.J.P. Cohen, P. Morrison, L. Dao, Covid-19 image data collection, 2020.
- 24.Dosovitskiy A., Springenberg J., Riedmiller M., Brox T. Discriminative unsupervised feature learning with exemplar convolutional neural networks. IEEE Trans. Pattern Anal. Mach. Intell. 2014;1 doi: 10.1109/TPAMI.2015.2496141. [DOI] [PubMed] [Google Scholar]
- 25.Girshick R., Donahue J., Darrell T., Malik J. 2014 IEEE Conference on Computer Vision and Pattern Recognition. 2014. Rich feature hierarchies for accurate object detection and semantic segmentation; pp. 580–587. [Google Scholar]
- 26.Lin T., Dollár P., Girshick R., He K., Hariharan B., Belongie S. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Feature pyramid networks for object detection; pp. 936–944. [Google Scholar]
- 27.Lin T., Goyal P., Girshick R., He K., Dollár P. 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Focal loss for dense object detection; pp. 2999–3007. [Google Scholar]
- 28.Zeiler M.D., Fergus R. Computer Vision – ECCV 2014. 2014. Visualizing and understanding convolutional networks; pp. 818–833. [Google Scholar]
- 29.F. Wang, M. Jiang, C. Qian, S. Yang, C. Li, H. Zhang, X. Wang, X. Tang, Residual attention network for image classification, in: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6450–6458.
- 30.C. Szegedy, A. Toshev, D. Erhan, Deep neural networks for object detection, in: Proceedings of the 26th International Conference on Neural Information Processing Systems, volume 2, pp. 2553–2561.
- 31.Kingma D., Ba J. Adam: a method for stochastic optimization. Int. Confer. Learn.Represent. 2014;1 [Google Scholar]
- 32.N. Ketkar, Deep Learning with Python: A Hands-on Introduction, pp. 97–111.
- 33.N. Ketkar, Deep Learning with Python: A Hands-on Introduction, pp. 159–194.
- 34.E.E.-D. Hemdan, M. Shouman, M. Karar, Covidx-net: A framework of deep learning classifiers to diagnose COVID-19 in x-ray images, 2020.
- 35.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 36.Das A., Agrawal H., Zitnick L., Parikh D., Batra D. Human attention in visual question answering: do humans and deep networks look at the same regions? Comput. Vision Image Understanding. 2017;163:90–100. [Google Scholar]
- 37.Chen W.-H., Strych U., Hotez P., Bottazzi M. The sars-cov-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]