Abstract
SARS-CoV-2 neurotropism has been increasingly recognized by its imaging and syndromic manifestations in the literature. The purpose of this report is to explore the limited yet salient current evidence that SARS-CoV-2′s host genomic targets PTBP1 and the 14-3-3 protein isoform encoding genes YWHAE and YWHAZ may be hold the key to understanding how neurotropism triggers neurodegeneration and how it may contribute to the onset of neurodegenerative disease. Considering that PTBP1 silencing in particular has recently been shown to reverse clinical parkinsonism and induce neurogenesis, as well as the known interactions of PTBP1 and YWHAE/Z with coronaviruses – most notably 14-3-3 and SARS-CoV, recent studies reinvigorate the infectious etiology hypotheses on major neurodegenerative disease such as AD and iPD. Considering that human coronaviruses with definite neurotropism have been shown to achieve long-term latency within the mammalian CNS as a result of specific accommodating mutations, the corroboration of genomic-level evidence with neuroimaging has vast potential implications for neurodegenerative disease.
Keywords: SARS-CoV-2, PTBP1, 14-3-3 proteins, Epigenetics, Neurotropism, Neurodegeneration
Background
Currently, multiple studies have identified a wide range of neurological deficits associated with COVID-19 [1], reflecting both immune-mediated process involving the development [2] and parenchymal injury [3]. Furthermore, a recent study by Lu and colleagues demonstrated that microstructural changes in the cerebral parenchyma can occur during the recovery phase from COVID-19 [4], indicating that the onset of CNS damage can be asynchronous with systemic manifestations and the typically salient severe respiratory disease. Asynchronous latency within the CNS may also be surmised by hyposmia, an early symptom of SARS-CoV-2 infection, that may denote presymptomatic (i.e. prior to febrile disease or fulminant respiratory symptoms) transcribial neuroinvasion [5]. The latter concept is supported both by imaging findings involving the frontal lobes [4], and hypometabolism along the olfactory pathways as detected by 18FDG-PET/CT [6].
Hypothesis
A recent translatomics experiment [7] revealed that SARS-CoV-2 infection perturbed the expression of three genes crucial for neuronal survival in its host cells: (i) the gene encoding the RNA binding polypyrimidine tract binding protein, PTBP1 and (ii) the genes encoding the isoforms ζ and ε of the 14-3-3 protein, YWHAZ and YWHAE correspondingly.
Considering that aberrant expression of either gene may trigger neurodegeneration, the hypothesis presented herein aims to synthesize the contribution of their physiological roles and conversely, their perturbations, in host – virus interactions and neurodegenerative disease.
The RNA binding polypyrimidine tract binding protein PTBP1, viruses and neurodegeneration
PTBP1 knockdown or depletion has recently been shown to induce neurogenesis and reversal of disease phenotypes in idiopathic Parkinson’s disease (iPD) [8]. PTB proteins have been shown to interact with several coronaviruses (CoV) by via their recruitment in regulatory complexes [9]. From α physiological view, the PTB/nPTB spliceosomic switch is a master regulator of neuronal fate and maturation [10]; thus, interactions that would lead to aberrant sequestration, as shown in the extranuclear shuttling of PTBP1 during TMEV infection [11], would be reasonably adequate to induce dysregulate a critical neuronal survival pathway.
In addition to PTBP1, YWHAZ and YHWAE (encoding 14-3-3ε and 14-3-3ζ correspondingly) proteins are also potential contributors to neurodegeneration. In a similar fashion to PTBP1, 14-3-3ε/ζ have been shown to regulate neuronal migration and differentiation in murine models [12]. YWHAZ and YWHAE expression has been linked directly to neurodegenerative disease such as Alzheimer’s Disease (AD) [13] and iPD [14], indicating that perturbations introduced by SARS-CoV-2 may be critical for both long term and short-term neuronal survival. Interestingly, 14-3-3ζ regulates autophagy-lysosomal trafficking and ER stress response/proteostasis on the epigenetic level (i.e. via the regulation of histone post-translational modifications), furthermore providing a common mechanism between viral infection, neuronal survival and neurodegeneration. Notably, 14-3-3 proteins were shown to mediate phosphorylation dependent shuttling of SARS-CoV’s N protein between cellular compartments, indicating that its differential expression during SARS-CoV-2 infection may be indispensable for its lifecycle [15].
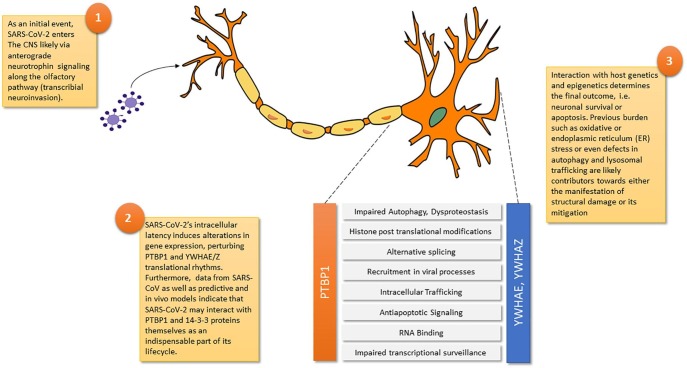
Considering that accommodating mutations may enable long term CNS latency within the CNS [16], neurodegeneration and cerebral structural damage in general may represent the combinatory effects of the viral lifecycle and its interaction with host genetics as the substrate. Considering that both PTBP1 and YWHAE/Z are increasingly recognized as SARS-CoV-2′s targets even by conceptually different studies [17], [18], the question arising is what other host factors mitigate or enhance these perturbations (Fig. 1 ).
Fig. 1.
An integrated model of SARS-CoV-2 neuroinvasion and the perturbations introduced to neurons by targeting PTBP1 and YWHAE and YWHAZ.
These observations offer both retrospective and prospective insight into what might be the basis for a renewed understanding of the neuroimmune bases of neurodegenerative disease, with a central role for PTBP1, YHWAE/Z and their role in epigenetics and mRNA surveillance.
Declarations
-
•
Ethics approval and consent to participate: Not applicable.
-
•
Consent for publication: Not applicable/Single Author
-
•
Availability of data and materials: Not applicable.
-
•
Competing interests: None declared.
-
•
Funding: No funding source.
-
•
Authors' contributions: Single Author.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
To Mrs Maria Pasmali, for proofreading this manuscript. Fig. 1 was created via a modification of a publicly available neuron schema (available from https://en.wikipedia.org/wiki/Neuron#/media/File:Neuron_Hand-tuned.svg, available under CC BY-SA 3.0)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.110212.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ellul MA, Benjamin L, Singh B, et al. Neurological associations of COVID-19. The Lancet Neurology. 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed]
- 2.Panariello A., Bassetti R., Radice A. Anti-NMDA receptor encephalitis in a psychiatric Covid-19 patient: a case report. Brain Behav Immun. 2020;87:179–181. doi: 10.1016/j.bbi.2020.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kremer S., Lersy F., de Seze J. Brain MRI findings in severe COVID-19: a retrospective observational study. Radiology. 2020 doi: 10.1148/radiol.2020202222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Y, Li X, Geng D, et al. Cerebral Micro-Structural Changes in COVID-19 Patients; An MRI-based 3-month Follow-up Study. EClinicalMedicine. [DOI] [PMC free article] [PubMed]
- 5.Vavougios G.D. Potentially irreversible olfactory and gustatory impairments in COVID-19: Indolent vs. fulminant SARS-CoV-2 neuroinfection. Brain Behav Immun. 2020;87:107–108. doi: 10.1016/j.bbi.2020.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karimi-Galougahi M., Yousefi-Koma A., Bakhshayeshkaram M., Raad N., Haseli S. (18)FDG PET/CT scan reveals hypoactive orbitofrontal cortex in anosmia of COVID-19. Acad Radiol. 2020;27(7):1042–1043. doi: 10.1016/j.acra.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bojkova D., Klann K., Koch B. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PubMed] [Google Scholar]
- 8.Qian H., Kang X., Hu J. Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature. 2020;582(7813):550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sola I., Galan C., Mateos-Gomez P.A. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol. 2011;85(10):5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J., Qian H., Xue Y., Fu X.D. PTB/nPTB: master regulators of neuronal fate in mammals. Biophys Rep. 2018;4(4):204–214. doi: 10.1007/s41048-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maranon D.G., Anderson J.R., Maranon A.G., Wilusz J. The interface between coronaviruses and host cell RNA biology: Novel potential insights for future therapeutic intervention. Wiley Interdiscip Rev RNA. 2020:e1614. doi: 10.1002/wrna.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyo-oka K., Wachi T., Hunt R.F. 14-3-3epsilon and zeta regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J Neurosci. 2014;34(36):12168–12181. doi: 10.1523/JNEUROSCI.2513-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F., Diao X., Wang F. Identification of key regulatory genes and pathways in prefrontal cortex of Alzheimer's disease. Interdiscip Sci. 2020;12(1):90–98. doi: 10.1007/s12539-019-00353-8. [DOI] [PubMed] [Google Scholar]
- 14.Kelly J., Moyeed R., Carroll C., Albani D., Li X. Gene expression meta-analysis of Parkinson's disease and its relationship with Alzheimer's disease. Mol Brain. 2019;12(1):16. doi: 10.1186/s13041-019-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol. 2005;79(17):11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vavougios G.D. Host proteases as determinants of coronaviral neurotropism and virulence. Brain Behav Immun. 2020;87:27. doi: 10.1016/j.bbi.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Li P, Ju X, et al. In vivo structural characterization of the whole SARS-CoV-2 RNA genome identifies host cell target proteins vulnerable to re-purposed drugs. bioRxiv 2020: 2020.07.07.192732. [DOI] [PMC free article] [PubMed]
- 18.Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master Regulator Analysis of the SARS-CoV-2/Human Interactome. J Clin Med. 2020;9(4) doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.