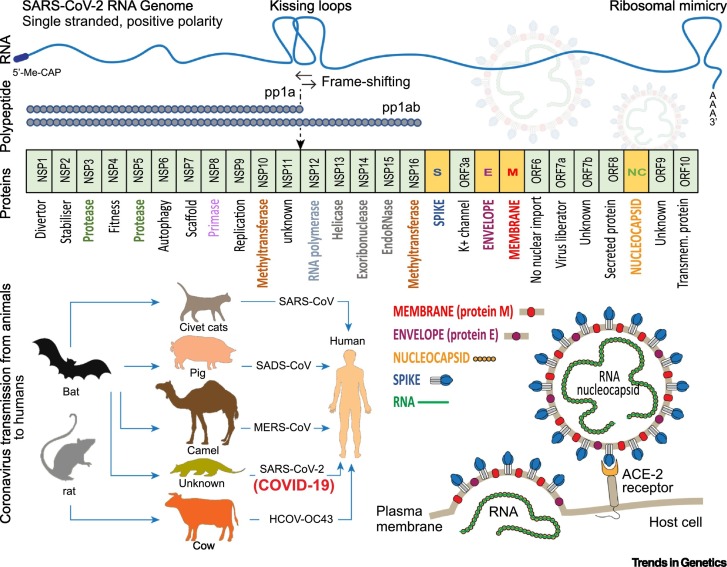
Figure 1.
General Schematic Overview of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)-2 Genome (top) Indicating Loop Formations That Aid Codon Frame-Shifting or Translational Machinary Diversion via Ribosomal Mimicry.
Polyproteins (pp1a and pp1ab) holding nonstructural proteins (NSPs) are translated as well as accessory proteins from open reading frames (ORFs) and structural proteins (represented in a modular format). Potential function of the proteins are indicated. Coronavirus transmission from animal to humans is shown including SARS-CoV-2 with unknown intermediate host that causes coronavirus disease 2019 (COVID-19) (in brackets). Adapted with modifications from Loey et al Symmetry 12(4) 651, 2020. Simplified illustration of SARS-CoV-2 structural components and assembled virus which binds to the constitutively expressed angiotensin-converting enzyme (ACE) 2 receptor enabling entry with release of the RNA genome into the host.
Lessons Learned from the Genome
Equivalent to a thousandth of the diameter of a human hair, severe acute respiratory syndrome coronavirus 2 (SARS-CoV)-2 is an average size for an enveloped virus. Its structural proteins Spike, Envelope, and Membrane, embedded in a host-derived lipid edifice, enclose a nucleocapsid, holding one of the longest RNA viral genomes. Hijacking host translational machinery, this single-stranded genome encodes two polyproteins, with self-cleavage capacity for release of 16 nonstructural proteins representing an enzymatic medley that forms an impressive armoury. While distinct from SARS-CoV and Middle East respiratory syndrome coronavirus, the SARS-CoV-2 genome shares major identity with bat-SL-CoVZC45, although not enough for direct ancestry. A formidable invader, SARS-CoV-2 binds to a constitutively expressed angiotensin-converting enzyme 2 receptor for multiorgan infection and host defence short-circuiting ability. Harbouring one of the hardest coronavirus membrane proteins, this novel virion maintains exclusive resilience in bodily fluids and external environments, necessitating complex strategies for its eradication.
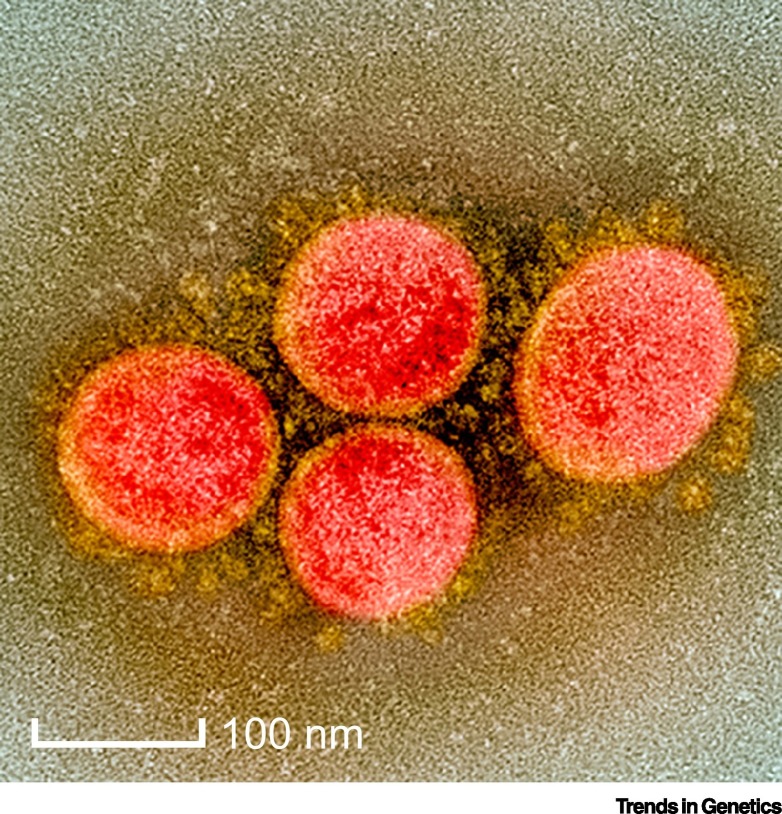
Figure 2.
Transmission Electron Micrograph of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)-2 Virus Particles, Isolated from a Patient.
Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Copyright credit: NIAID.
GENOME FACTS:
SARS-CoV-2 isolate Wuhan-Hu-1 [(NCBI accession number NC_045512 (version 2)] represents a 29 903 base pair single-stranded RNA with 5′ to 3′ positive polarity orientation.
The nucleotide proof-reading ability of SARS-CoV-2 enables its capacity for maintaining the largest known genome of any RNA virus.
Like SARS-CoV, the RNA genome of SARS-CoV-2 most likely has a 5′-methylated cap due to the action of 2′-O-ribose methyltransferase that it encodes and a 3′ poly A+ tail (length 33 bp).
Over 70% of the genome encodes one gene, ORF1ab, capable of encoding both a small (pp1a) and large (pp1ab) polyprotein. Other genes S, E, M, and N encode the structural proteins Spike, Envelope, Membrane, and Nucleocapsid, respectively. Ten open reading frames (ORF1–ORF10) encode accessory proteins with speculated functionality.
A frameshifting stimulation element in ORF1ab tricks the host’s ribosomal machinery into translating two polyproteins from a single mRNA. Not unique to SARS-CoV-2, this process occurs in other coronaviruses and retroviruses. The ribosome effectively skids on a ‘slippery’ sequence after formation of pseudoknots and ‘kissing’ stem loops.
Just upstream from the 3′ untranslated region in the SARS-CoV-2 genome, a conserved stem-loop II motif (s2m) forms a tertiary structure comparable to the 16S rRNA in an act of macromolecular mimicry to hijack the host translational machinery for its own usage.
The reduced levels of ‘slow-codons’ in the SARS-CoV-2 genome may ensure its higher protein translational rate compared to other coronavirus groups which have the ability to infect humans.
SPECIES FACTS:
Named upon isolation as 2019-nCoV due to the initial appearance of SARS-CoV-2 in patients in 2019, this localised issue in China escalated into the first worldwide coronavirus pandemic. By mid-July 2020, 10.9 million people worldwide were infected.
The World Health Organization characterised the infection of SARS-CoV-2 as COVID-19 (i.e., coronavirus disease 2019). While primarily considered a pulmonary disease due to SARS-CoV-2 being mainly distributed in the lower respiratory tract, infection also causes damage to the heart, blood vessels, liver, and kidneys, and degeneration of the gut–blood barrier.
Children are particularly vulnerable to developing dermatological lesions on their feet due to SARS-CoV-2, resembling those caused by blood clots from measles, chickenpox, or frostbite.
Over 80% of COVID-19 patients experience ageusia or anosmia at the beginning of the infection, indicative that the central nervous system may be accessible to SARS-CoV-2.
Worldwide SARS-CoV-2 sequence analysis identified the L37F mutation in nonstructural protein NSP6 that could significantly modify COVID-19 pathogenicity in populations in Asia, America, Oceania, and Europe by altering the host autophagic lysosomal antiviral machinery.
Fun fact about the Genome
The SARS-CoV-2 nonstructural protein 3 (Nsp3) has a low 46% similarity to a C-Jun kinase-interacting protein also found in a ray-finned fish Labrus bergylta, a protogynous hermaphrodite, that begins life as a female yet with territorial dominance becomes male.
TAXONOMIC AND CLASSIFICATION INFORMATION
REALM: Riboviria
EMPIRE: Orthornavirae
STRAIN: Pisuviricota
CLASS: Pisoniviricetes
ORDER: Nidovirales
FAMILY: Coronaviridae
SUBFAMILY: Orthocoronavirinae
GENUS: Betacoronavirus
SUBGENUS: Sarbecovirus
SPECIES: Severe acute respiratory syndrome related coronavirus
Literature
- 1.de Groot R.J. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; 2012. Virus taxonomy; pp. 806–828. [Google Scholar]
- 2.Anand K. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu T. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baranov P.V. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson M.P. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:e5. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C.W., Chen M.F. Composition of human-specific slow codons and slow di-codons in SARS-CoV and 2019-nCoV are lower than other coronaviruses suggesting a faster protein synthesis rate of SARS-CoV and 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;53:419–424. doi: 10.1016/j.jmii.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan S. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12:360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]