Abstract
We used metabolic modeling to computationally investigate the potential of bacterial coculture system designs for CO conversion to the platform chemical butyrate. By taking advantage of the native capabilities of wild-type strains, we developed two anaerobic coculture designs by combining Clostridium autoethanogenum for CO-to-acetate conversion with bacterial strains that offer high acetate-to-butyrate conversion capabilities: the environmental bacterium the human gut bacteriumEubacterium rectale. When grown in continuous stirred tank reactor on a 70/0/30 CO/H2/N2 gas mixture, the C. autoethanogenum-C Kluyveri co-culture was predicted to offer no mprovement in butyrate volumetric productivity compared to an engineered C. autoethanogenum monoculture despite utilizing vinyl acetate as a secondary carbon source for C. kluyveri growth enhancement. A coculture consisting of C. autoethanogenum and C. kluyveri engineered in silico to eliminate hexanoate synthesis was predicted to enhance both butyrate productivity and titer. The C. autoethanogenum-E. rectale coculture offered similar improvements in butyrate productivity without the need for metabolic engineering when glucose was provided as a secondary carbon source to enhance E. rectale growth. A bubble column model developed to assess the potential for large-scale butyrate production of the C. autoethanogenum-E. rectale design predicted that a 40/30/30 CO/H2/N2 gas mixture and a 5 m column length would be preferred to enhance C. autoethanogenum growth and counteract CO inhibitory effects on E. rectale.
Keywords: Gas fermentation, Metabolic modeling, Microbial cocultures, Clostridium autoethanogenum, Eubacterium rectale, Clostridium kluyveri
1. Introduction
The development of cost-effective technologies for sustainable production of fuels and chemicals from renewable resources remains a paramount challenge for our society. While catalytic technologies hold considerable promise for many applications, they suffer from several shortcomings including the need for costly gas pretreatment, high operating temperatures and catalyst fouling that reduces process efficiency [1]. Microbial systems represent a possible alternative for many applications due to their evolved ability to utilize diverse feedstocks and synthesize a broad array of metabolites [2]. However, the effectiveness of microbial platforms has only been demonstrated at scale for a few applications such as conversion of corn-derived glucose to ethanol [3] and the production of 1,3-propanediol from plant-derived glucose [4]. To eventually succeed in the marketplace, microbial systems must offer more flexibility with respect to the feedstock utilized as well as the product synthesized.
Butyric acid and its conjugate base butyrate are important platform chemicals for production of methyl butyrate, cellulose acetate butyrate (CAB), poly(3-hydroxybutyrate) (PHB) and other chemical intermediates [5–8] and for incorporation into animal feeds as a nutrient [9]. The global butyric acid market was USD 124.6 Million in 2014 and is estimated to more than double to USD 289.3 Million in 2020 [10]. Current butyrate production technology is based on chemical synthesis through oxidation of butyraldehyde, which is produced from crude oil derived propylene by oxosynthesis [11]. This chemical synthesis route will eventually require replacement with a renewable alternative. Due to the highly optimized nature of current butyrate production processes, competing technologies will require a relatively cheap feedstock to offer the possibility of economic competitiveness. While microbial systems have been proposed for butyrate production [5], most technologies are based solely on relatively expensive feedstocks such as glucose. Waste gas streams containing CO, CO2, H2 and N2 are produced in high volumes by several large industries including steel manufacturing, oil refining and chemical production [12–15]. LanzaTech is commercializing microbial technologies for waste gas conversion to fuels and chemicals [16]. Development efforts have focused on the use of gas fermenting acetogens such as Clostridium autoethanogenum to synthesize the native metabolic byproducts ethanol and 2,3-butanediol.
Microbial butyrate production from waste gases is challenging because wild-type strains capable of gas consumption tend to synthesize butyrate at low yields. Several bacterial genera including Butyribacterium, Butyrivibrio, Clostridium, Eubacterium, Fusobacterium, Megasphera and Sarcina are known to synthesis butyrate as a primary metabolic byproduct [8]. However, only a few butyrate-producing genera are capable of CO and CO2 uptake. Eubacterium limosum has been reported to grow on CO as a sole carbon source and to secrete acetate and trace amounts of butyrate [17]. Clostridium strains are more commonly used for butyrate and butanol synthesis due to their relatively high productivities. For example, Clostridium tyrobutyricum is capable of high butyrate synthesis from glucose and xylose but is incapable of CO consumption [18,19]. An alternative approach is to metabolically engineer the Wood-Ljungdahl pathway of gas fermenting acetogens such as Clostridium autoethanogenum and Clostridium ljungdahlii to introduce butyrate synthesis. Both in vitro [20,21] and in silico [20,21] studies have demonstrated the feasibility of engineering C. ljungdahlii to secrete butyrate while reducing the synthesis of the native products acetate and ethanol. However, these studies indicate that butyrate production was accompanied by reduced growth of the mutant strain. Furthermore, attempts to increase carbon flux to butyrate through deletion of the ack (acetate kinase) gene has the potential to reduce strain genetic stability [22]. Therefore, there is considerable motivation to develop new microbial platforms for CO-to-butyrate conversion based on wild-type strains.
In this study, we focused on in silico analysis of coculture systems in which the CO consumption and butyrate synthesis capabilities of wild-type strains were synergistically combined. Motivated by coculture designs developed for biofuel and biogas production, wastewater treatment, soil remediation and traditional food production [23–27], we sought to exploit the native metabolic capabilities of acetogens to convert CO-containing waste gases to acetate and of certain anaerobes to convert available acetate to butyrate (Fig. 1a). The two acetate-to-butyrate converting species investigated were the environmental bacterium Clostridium kluyveri and the human gut bacterium Eubacterium rectale. A coculture of C. kluyveri and C. autoethanogenum has been studied in vitro for conversion of CO-rich gas streams to butyrate, other short-chain fatty acids and higher alcohols [27,28]. A possible disadvantage of this design is that C. kluyveri can exhibit diauxic growth by consuming secreted butyrate as a substrate to synthesize other metabolic products such as hexanoate [29]. E. rectale is a common member of the human gut microbiota whose primary metabolic function in vivo is acetate-to-butyrate conversion [30]. E. rectale often secretes H2 as a secondary byproduct, thereby providing an additional energy source to support C. autoethanogenum growth. Both cocultures systems were supplied with a secondary carbon source to enhance growth of the butyrate producer. If not consumed by the butyrate-producing species, ethanol could be a valuable coproduct which is easily separated from butyrate due to their different boiling points (ethanol 78.2 °C, butyrate 163.5 °C) and inability to form an azeotrope.
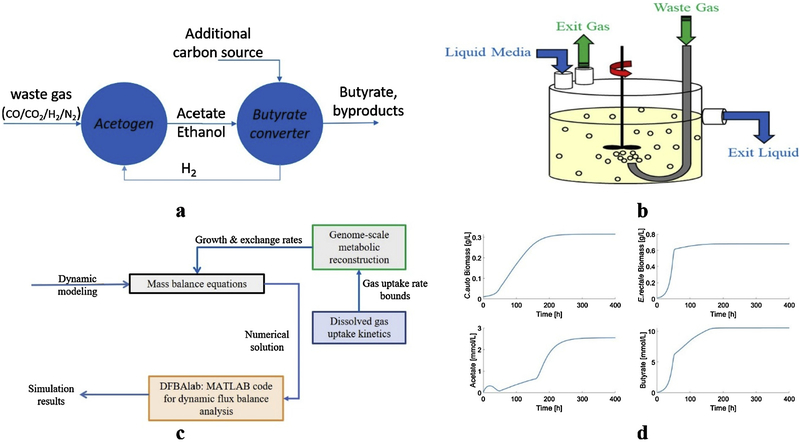
Fig. 1.
(a) Schematic representation of the coculture design concept. (b) Continuous stirred tank bioreactor (CSTBR) simulations used to evaluate system performance. (c) Dynamic flux balance model solved with DFBAlab. (d) Representative dynamic simulation results.
We developed dynamic models of coculture growth in continuous stirred tank bioreactors (CSTBRs) to assess the relative performance of the two coculture system designs along with a monoculture design based on engineered C. autoethanogenum (Fig. 1b). In addition to the usual advantage that CSTBRs offers higher volumetric productivities than batch bioreactors, continuous liquid and gas flows allowed the dissolved CO concentration to be maintained at low levels that were less likely to strongly inhibit growth of the butyrate-producing species.
The models combined genome-scale reconstructions of individual species metabolism, uptake kinetics for dissolved gas components and extracellular balances for biomass, gas-phase metabolite and liquid-phase metabolite concentrations (Fig. 1c). The MATLAB code DFBAlab was used to solve the dynamics models consisting of nonlinear ordinary differential equations with embedded linear programs (Fig. 1d). Due to its promising performance in well-mixed continuous culture, the C. autoethanogenum-E. rectale system was evaluated for coculture growth in a simulated bubble column reactor that provides more favorable gas-liquid mass transfer and the capability for large-scale production [31–33]. We believe that our study represents an important contribution towards the development of microbial platforms for waste gas-to-butyrate conversion.
2. Materials and methods
2.1. Engineered C.Autoethanogenum model
C. autoethanogenum metabolism was described by the genome-scale reconstruction iCLAU786 which accounts for 786 annotated genes, 1095 intracellular metabolites and 1108 intracellular and exchange reactions [34,35]. The reconstructed Wood-Ljungdahl pathway [36] offers the potential of synthesizing and secreting native metabolites including acetate, ethanol, lactate and 2,3-butanediol. Preliminary flux balance analysis (FBA) calculations with a maximal growth objective predicted acetate and ethanol as the primary metabolic products from CO/H2 feed mixtures. The published model contained an incomplete butyrate synthesis pathway (Table S1), consistent with the inability of the wild-type strain to produce butyrate [21]. A functional butyrate synthesis pathway was introduced by in silico insertion of the biosynthetic reactions ACACT1r, ACOAD1z, HACD1, ECOAH1, PBUTT and BUTKr and the butyrate transport reaction BUTt [20] (Table S1). OptKnock [21,37] was used to determine in silico gene deletions that would direct flux away from the native products and towards butyrate.
Dissolved CO uptake kinetics of C. autoethanogenum were specified to follow a modified Michaelis-Menten equation that accounted for CO inhibition, which experimental studies have shown is important at high dissolved CO levels [38],
| (1) |
where vCO is the CO uptake rate (mmol/gDW/h), which serves as a bound in the FBA calculation; vmax,CO is the maximum CO uptake rate (mmol/gDW/h); CL,CO is the dissolved CO concentration (mmol/L); Km,CO is the CO saturation constant (mmol/L); and KI is the CO inhibition constant (mmol/L). C. autoethanogenum uptake of dissolved H2 was assumed to follow Michaelis-Menten kinetics,
| (2) |
where vH2 is the H2 uptake rate bound; vmax,H2 is the maximum H2 uptake rate; CL,H2 is the dissolved H2 concentration; and KL,H2 is the H2 saturation constant. A literature review [39–41] yielded the CO uptake parameters and estimated H2 uptake parameters listed in Table 1.
Table 1.
Nominal parameters for the monoculture and coculture CSTBR models.
| Parameter | Symbol | Value | Source |
|---|---|---|---|
| Reactor size | V | 1 L | Specified |
| Pressure | PL | 1.013e5 Pa | Specified |
| Temperature | T | 37 °C | Specified |
| Feed gas flow rate | 1 L/h | Specified | |
| Volumetric gas-liquid mass transfer coefficient | kLa | 100 h−1 | [32,42] |
| CO mole fraction in feed gas | yCO | 70% | [43] |
| H2 mole fraction in feed gas | yH2 | 0% | [43] |
| N2 mole fraction in feed gas | yN2 | 30% | [43] |
| CO Henry’s law constant | HCO | 8e–4 mol/L/atm | [44] |
| H2 Henry’s law constant | HH2 | 6.6e–4 mol/L/atm | [44] |
| Initial biomass concentration | Xi | 0.01 mmol/L | Specified |
| Maximum CO uptake rate | vCO,m | 50 mmol/gDW/h | [39,40,41] |
| CO saturation constant | Km,CO | 0.1 mmol/L | [39,40,41] |
| CO inhibition constant | KI,CO | 5 mmol/L | [39,40,41] |
| Maximum H2 uptake rate | VH2,m | 50 mmol/gDW/h | Estimated |
| H2 uptake saturation constant | Km,H2 | 0.1 mmol/L | Estimated |
| Feed and initial glucose concentration | CG,f | 10 mmol/L | Specified |
| Feed and initial vinyl acetate concentration | CVA,f | 15 mmol/L | Specified |
| Feed and initial lactose concentration | CLac,f | 5 mmol/L | Specified |
| Maximum glucose, vinyl acetate and lactose uptake rates | vG,m (vVA,m) | 10 mmol/gDW/h | [45] |
| Glucose, vinyl acetate and lactose uptake saturation constants | Km,G (Km,VA) | 0.5 mmol/L | [45] |
| Maximum dissolved CO inhibition concentration | [CO]max | 0.8 mmol/L | [28] |
| Acetate, butyrate and ethanol maximum uptake rates | vi,m | 5 mmol/gDW/h | [45] |
| Acetate, butyrate and ethanol uptake saturation constants | Km,i | 0.5 mmol/L | [45] |
2.2. C.Autoethanogenum and C. Kluyveri coculture model
C. autoethanogenum metabolism was described by the genome-scale reconstruction iCLAU786 [34,35] without modification. The genome-scale reconstruction iCKL708 accounting for 708 genes, 804 metabolites and 994 intracellular and exchange reactions [29] was used to describe C. kluyveri metabolism. Our preliminary FBA calculations predicted a very small C. kluyveri growth rate when acetate and ethanol were the sole carbon sources. The authors of the original paper suggested supplementation with vinyl acetate or crotonate to enhance growth; we used vinyl acetate as the secondary carbon source in our design (Fig. 1a). Even with this additional carbon source, our FBA calculations predicted that hexanoate rather than butyrate would be the primary byproduct of C. kluyveri. Therefore, we applied the OptKnock algorithm [37] to the C. kluyveri genome-scale reconstruction to identify gene knockouts that would yield an engineered in silico strain with higher butyrate synthesis.
C. autoethanogenum was assumed to have the same CO and H2 uptake kinetics as in monoculture (Eqs. 1 and 2), while C. kluyveri uptake kinetics for acetate, ethanol, vinyl acetate and butyrate were assumed to follow the Michaelis-Menten equation modified to account for CO growth inhibition [46],
| (3) |
where vi is the uptake rate bound of the i -th metabolite (acetate, ethanol, vinyl acetate and butyrate); vmax,i is the maximum uptake rate, CL,i is the liquid-phase concentration; Km,i is the saturation constant; and [CO]max is the maximum dissolved CO concentration (mmol/L). Because C. kluyveri uptake parameters were not available in the literature, we assumed approximate values based on parameters reported for Escherichia coli [45].
2.3. C. Autoethanogenum and E. rectale coculture model
C. autoethanogenum metabolism was described by the genome-scale reconstruction iCLAU786 [34,35] without modification, while the iEre400 reconstruction consisting of 400 genes, 416 metabolites and 465 intracellular and exchange reactions [47] was used to model E. rectale metabolism. Our preliminary FBA calculations predicted that E. rectale would not consume ethanol or butyrate and that growth on acetate as the sole carbon source was not possible. Therefore, a secondary carbon source was necessary to support E. rectale growth. While E. rectale is known to have versatile substrate utilization capabilities that include complex carbohydrates, the genome-scale metabolic reconstruction iEre400 used in this study did not allow the uptake of complex carbohydrates. Of the simple carbohydrates (glucose, fructose, lactose and maltose) included in the model, we selected glucose due to its prevalence as a fermentation substrate. Since E. rectale is a human gut bacterium, its toxicity to dissolved CO has not been studied. However, Eubacterium limosum is known to exhibit CO tolerance and has been proposed as a model organism for CO fermentation [17,48]. Therefore, we assumed that E. rectale uptake of acetate and glucose would be inhibited by dissolved CO according to the same kinetics and parameter values as used for C. kluyveri (Eq. 3) to allow an unbiased comparison of the two coculture designs.
2.4. Continuous culture model formulation
The monoculture and coculture system designs were compared for simulated anaerobic growth in a continuous stirred tank bioreactor (CSTBR; Fig. 1b). The mass balance equation on the biomass of the i -th species was formulated as,
| (4) |
where Xi is the biomass concentration (g/L); μi is the specific growth rate (h−1) obtained from solution of the flux balance problem; D is the dilution rate (h−1); and X0 is the initial biomass concentration. Mass balances on liquid-phase metabolites had the form,
| (5) |
where Cj is the concentration (mmol/L) of j -th metabolite (glucose, vinyl acetate, butyrate and hexanoate); vi,j is the specific production (positive) or uptake (negative) rate (mmol/gDW/h) of the j -th metabolite by the i -th species; D is the dilution rate (h−1); and Cj,f is the feed concentration of j -th metabolite.
Mass balance equations on dissolved gas components had the form,
| (6) |
where CL,m is the dissolved concentration (mmol/L) of m -th gas component (CO and H2); vi,j is the specific uptake rate of the j -th component by the i -th species; kLa is the volumetric gas-liquid mass transfer coefficient, is the saturation concentration (mmol/L) of the m -th component calculated from the gas phase concentration using Henry’s law at the specified temperature and pressure; and CL,m,0 is initial liquid-phase concentration of the m -th component. Mass balance equations on gas-phase substances had the form,
| (7) |
where Cm is the gas-phase concentration (mmol/L) of the m -th gas component (CO and H2); is the feed gas volumetric flow rate (L/h); V is the liquid volume (L); and Cm,f is the feed concentration (mmol/L) of the m -th gas component. All the parameter values used in the CSTBR models are listed in the Table 1.
The monoculture and coculture reactor models consisting of ordinary differential equations (ODEs) for extracellular culture dynamics and linear programs (LPs) for intracellular steady-state metabolism were solved within MATLAB using the code DFBAlab [49,50] as shown in Fig. 1c and detailed in our previous publications [41,51–53]. DFBAlab utilized lexicographic optimization to avoid the possibility of alternative optima in the LPs [49]. The primary objective was chosen as the usual growth rate maximization [54,55], while the remaining objectives were specified according to assumed metabolic behavior of the three modeled species (Table S2). We found the directionality and ordering of these secondary objectives had a negligible effect on simulation results (Fig. 1d) with the MATLAB code ode15s used for ODE solution and Gurobi used for LP solution.
2.5. Bubble column model formulation
Based on CSTBR simulations, we identified the C. autoethanogenum and E. rectale coculture system as the most promising design based on wild-type species. This coculture design was implemented in a bubble column reactor (Figure S10) to investigate in silico performance in a more realistic reactor configuration used for large-scale production [56]. Due to space limitations and the availability of our previous publications [41,51–53] detailing the metabolic modeling framework for bubble column fermentation, the details of the coculture model are omitted here. Bubble column model parameters are provided in Table S3 for the interested reader. Moreover, all CSTBR and bubble column simulation codes used in this study can be downloaded from our website (www.ecs.umass.edu/che/henson_group/downloads.html).
3. Results and discussions
3.1. Flux balance analysis of C. Kluyveri and E. rectale metabolism
FBA with a maximal growth objective was performed as a prelude to CSTBR simulations to characterize the growth and byproduct secretion properties C. kluyveri and E. rectale. The calculations were performed with the uptake rates of carbon sources set equal to the maximum values in Table 1. When grown on glucose as the sole carbon source, E. rectale was predicted to secrete butyrate and H2 at nearly equal rates (Table 2). The addition of acetate as another carbon source only increased the growth rate slightly (+4.4%) but dramatically shifted the byproduct secretion rates by increasing butyrate (+38%) and decreasing H2 (−76%). Considering that acetate is a two-carbon molecule while butyrate is a four-carbon molecule, these predictions show that 93% of supplied acetate was converted directly to butyrate. The model predicts utilization of the acetate-to-butyrate pathway because E. rectale is able to transfer acetate back into acetyl-CoA, which can produce ATP that favors growth, and further convert acetyl-CoA into butyrate [30].
Table 2.
Flux balance analysis of E. rectale and C. kluyveri for different substrates.
| Bacterium | Uptake rate (mmol/gDW/h) | Growth rate (h−1) | Production rate (mmol/gDW/hJ | ||||
|---|---|---|---|---|---|---|---|
| Glucose (Vinyl acetate*) | Acetate | Ethanol | Butyrate | Hydrogen | Hexanoate | ||
| E. rectale | −10 | 0 | 0 | 0.6389 | 6.1192 | 7.3591 | – |
| E. rectale | −10 | −5 | 0 | 0.6671 | 8.4480 | 1.8014 | – |
| C. kluyveri* | −10 | 0 | 0 | 0.1425 | 9.0634 | 0 | 0 |
| C. kluyveri* | −10 | −5 | 0 | 0.1476 | 6.9405 | 0 | 3.0595 |
| C. kluyveri* | −10 | −5 | −5 | 0.2295 | 3.0165 | 0 | 6.9835 |
Denotes that vinyl acetate was supplied instead of glucose.
Growth of C. kluyveri on vinyl acetate as the sole carbon source was predicted to generate butyrate as the only byproduct. The predicted growth rate of C. kluyveri was about four times less than that of E. rectale on glucose. The addition of acetate shifted the byproduct secretion pattern such that hexanoate was synthesized and the butyrate secretion rate decreased substantially (−23%). A potential advantage of C. kluyveri for coculture system design was that ethanol secreted by the acetogen could be consumed. While the addition of ethanol as a third carbon source increase the growth rate (+55%), byproduct secretion was further shifted towards hexanoate and the butyrate secretion rate was substantially reduced (−57%). Collectively, these predictions suggest that E. rectale could be preferred for coculture design due to its higher growth and butyrate secretion rates.
3.2. CSTBR performance of engineered C. autoethanogenum monoculture
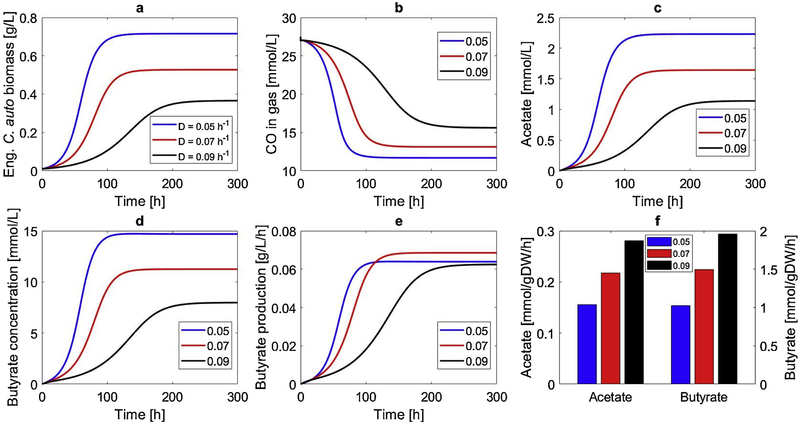
The C. autoethanogenum monoculture model was used to assess butyrate production capabilities in continuous culture. The steady-state growth rate was set by the dilution rate (D) [57] and had a strong impact on butyrate volumetric productivity. Therefore, we performed CSTBR simulations over a range of D values and presented results for three values that bracketed the optimal butyrate productivity to examine robustness of the design. Predicted responses demonstrated slow startup dynamics, with approximately 100–200 hours required to approach steady state depending on D (Fig. 2). We found that reactor washout occurred at D = 0.12h−1. The intermediate dilution rate D = 0.07h−1 yielded the highest steady-state butyrate productivity due to the favorable tradeoff between the C. autoethanogenum biomass concentration and the specific butyrate synthesis rate. However, this D value produced less CO consumption and a lower butyrate titer than rate than D = 0.05h−1. Approximately 50% of supplied CO was consumed at the optimal D value, suggesting that performance was limited by gas-liquid mass transfer of the CSTBR. The highest dilution rate D = 0.09h−1 yielded the worst performance due to inadequate biomass formation. All three D values were predicted to generate low acetate concentrations due to low secretions rates of the engineered strain. We used this monoculture system as a base case to quantify the relative performance of the coculture designs discussed below.
Fig. 2.
Effect of the dilution rate on CSTBR performance of the engineered C. autoethanogenum monoculture. (a) Dynamic response of the C. autoethanogenum biomass concentration; (b) dynamic response of the gas-phase CO concentration; (c) dynamic response of the acetate concentration; (d) dynamic response of the butyrate concentration; (e) dynamic response of the butyrate volumetric productivity; (f) steady-state acetate and butyrate secretion rates.
3.3. CSTBR performance of C. autoethanogenum and C. Kluyveri coculture
Having established that system designs based on C. autoethanogenum were expected to exhibit slow dynamics, we focused on steady-state performance of the candidate coculture designs. The highest butyrate productivity for the C. autoethanogenum-C. kluyveri system was predicted at D = 0.10h−1 as smaller D values yielded relatively low specific butyrate synthesis while higher D values generated inadequate biomass formation (Figure S1). Despite the addition of vinyl acetate as a secondary carbon source, the coculture generated a slightly lower butyrate productivity (0.06g/L/h) than the engineered C. autoethanogenum monoculture (0.07g/L/h) due to the synthesis of unutilized acetate and the undesired byproduct hexanoate. Our simulation results are difficult to compare directly to data from the original study [28] due to the experimental use of rich media containing yeast extract rather than minimal media and anaerobic bottles rather than CSTBRs. However, it is interesting to note that experiments predicted final acetate, hexanoate and butyrate concentrations of 15 mmol/L, 2 mmol/L and 15 mmol/L while our CSTBR simulation at D = 0.1 h−1 predicted the concentrations at steady state to be 15 mmol/L, 5 mmol/L and 8 mmol/L, respectively.
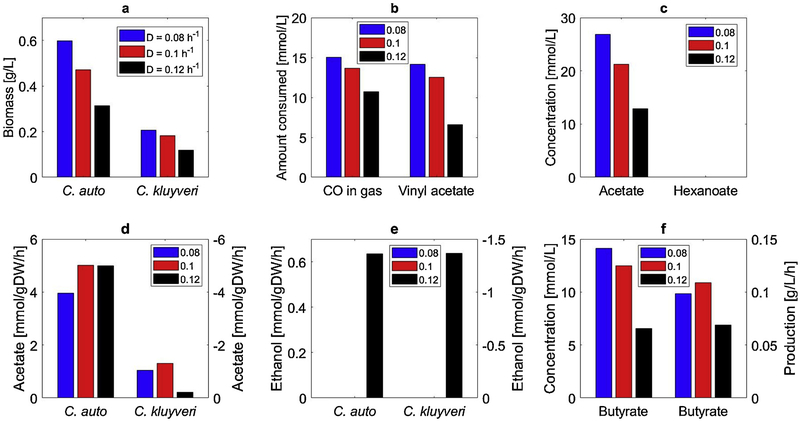
We explored the possibility of increasing butyrate production of the coculture system through in silico engineering of the C. kluyveri genome-scale reconstruction. OptKnock [37] predicted that removal of the reaction catalyzed by trans-2-enoyl-CoA reductase would completely eliminate hexanoate synthesis while simultaneously increasing butyrate secretion. C. kluyveri uses this enzyme to produce hexanoyl-CoA which is used along with butanoic acid to synthesize hexanoate. When combined with C. autoethanogenum in a simulated CSTBR, the engineered C. kluyveri strain resulted in a large increase in butyrate productivity from 0.07 g/L/h obtained with the wild-type strain to 0.11 g/L/h (Fig. 3). The optimal dilution rate remained D = 0.10h−1. Increased butyrate production was predicted to result from elimination of hexanoate synthesis rather than more effective utilization of available carbon, as the residual acetate concentration increased due to reduced acetate uptake by the engineered strain. Because genetic engineering tools for C. autoethanogenum are under development [58,59], the feasibility of realizing the mutant strain and the resulting strain performance would need to be evaluated experimentally.
Fig. 3.
Effect of the dilution rate on steady-state CSTBR performance of the coculture system consisting of C. autoethanogenum and engineered C. kluyveri. (a) C. autoethanogenum and C. kluyveri biomass concentrations; (b) amount of consumed CO and vinyl acetate; (c) acetate and hexanoate concentrations; (d) acetate secretion rate for C. autoethanogenum and uptake rate for C. kluyveri; (e) ethanol secretion rate for C. autoethanogenum and uptake rate for C. kluyveri; (f) butyrate concentration and volumetric productivity.
3.4. CSTBR performance of C. autoethanogenum and E. rectale coculture
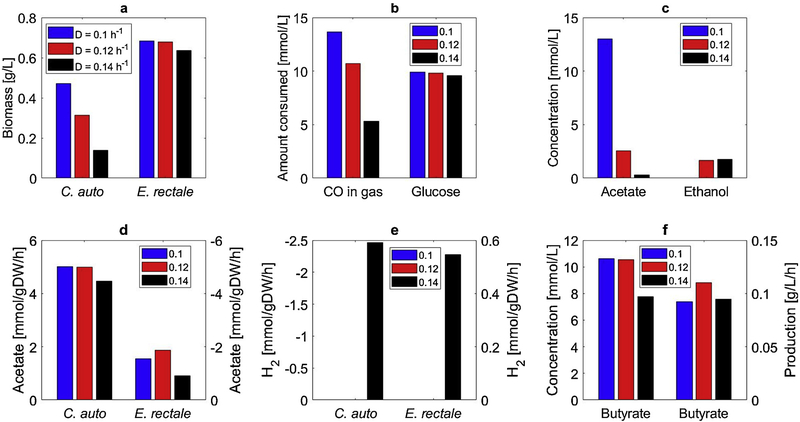
Our FBA calculations indicated that E. rectale may offer advantages to wild-type C. kluyveri for coculture system design to its higher growth rates and superior acetate-to-butyrate conversion capabilities. This hypothesis was investigated by performing CSTBR simulations for the C. autoethanogenum-E. rectale system with glucose used as the secondary carbon source. This coculture allowed the use of higher dilution rates, with washout value predicted to be D = 0.19 h−1. The optimal value D = 0.12h−1 produced a butyrate productivity of 0.11 g/L/h (Fig. 4), which was substantially higher than that achieved in engineered C. autoethanogenum monoculture (0.07 g/L/h) and cocultures containing wild-type C. kluyveri (0.07 g/L/h) and equal to that achieved in cocultures with engineered C. kluyveri. Residual acetate was reduced to 2mmol/L due to relatively high biomass concentration and specific acetate uptake of E. rectale, demonstrating more efficient carbon utilization than predicted with C. kluyveri even though slightly less CO was consumed by C. autoethanogenum due to the higher dilution rate. On the other hand, E. rectale could not consume the small amount of ethanol secreted by C. autoethanogenum. The two species engaged in H2 crossfeeding at the highest D value, which enhanced C. autoethanogenum growth and allowed coexistence at a higher dilution rate than possible with the monoculture system and the other coculture design. Due to its predicted ability to achieve relatively high butyrate productivity due to the native metabolic capabilities of the wild-type species and avoid the complications of metabolic engineering, the C. autoethanogenum-E. rectale coculture was deemed the superior design and subjected to additional investigation.
Fig. 4.
Effect of the dilution rate on steady-state CSTBR performance of the C. autoethanogenum-E. rectale coculture system. (a) C. autoethanogenum and E. rectale biomass concentrations; (b) amount of consumed CO and glucose; (c) acetate and ethanol concentrations; (d) acetate secretion rate for C. autoethanogenum and uptake rate for E. rectale; (e) H2 uptake rate for C. autoethanogenum and secretion rate for E. rectale; (f) butyrate concentration and volumetric productivity.
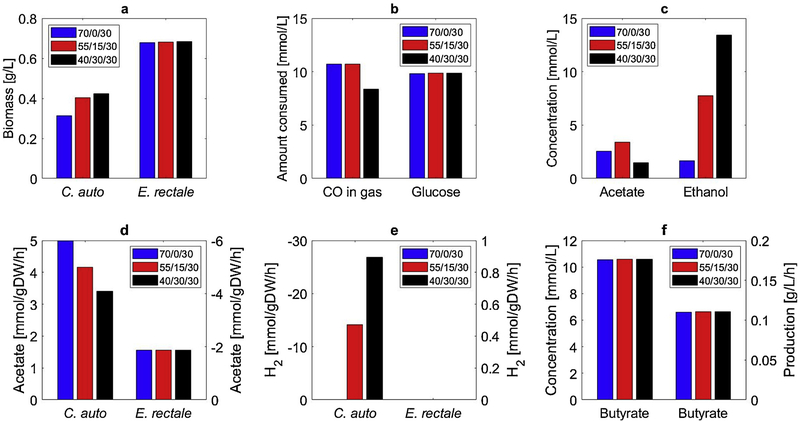
To examine system performance with respect to feed gas composition, CSTBR simulations where performed for CO/H2/N2 compositions ranging from 70/0/30 (base case) to 40/30/30. The addition of H2 to the feed gas was predicted to increase C. autoethanogenum biomass formation but shifted byproduct secretion towards ethanol rather than acetate (Fig. 5), consistent with experimental studies [28]. Despite these variations, the predicted performance of E. rectale was highly consistent with its biomass concentration and specific acetate uptake remaining effectively constant across feed compositions because the reduction in specific acetate secretion was balanced by increased C. autoethanogenum biomass. As a result, butyrate productivity and titer also were constant while acetate levels varied slightly but remained low. CO consumption by C. autoethanogenum was affected only at the highest feed H2 composition but remained less than 50% of available CO, suggesting the need for an alternative reaction configuration with higher gas-liquid mass transfer capabilities. These CSTBR simulation results suggested that the proposed coculture design was robust to feed composition but that the addition of H2 would not offer benefits for butyrate production.
Fig. 5.
Effect of gas feed composition (CO/H2/N2) on steady-state CSTBR performance of the C. autoethanogenum-E. rectale coculture system. (a) C. autoethanogenum and E. rectale biomass concentrations; (b) amount of consumed CO and glucose; (c) acetate and ethanol concentrations; (d) acetate secretion rate for C. autoethanogenum and uptake rate for E. rectale; (e) H2 uptake rate for C. autoethanogenum and secretion rate for E. rectale; (f) butyrate concentration and volumetric productivity.
Gas feeds containing H2 were predicted to result in ethanol accumulation that represented lost carbon from the perspective of butyrate production. In effort to reduce ethanol formation, we replaced C. autoethanogenum with the acetogen Clostridium ljungdahlii known to favor acetate as a byproduct [60]. Preliminary FBA calculations with the genome-scale metabolic reconstruction iHN637 [61] demonstrated the ability of C. ljungdahlii to compete with E. rectale for glucose. To avoid substrate competition, 10 mmol/L glucose was replaced with 5 mmol/L lactose as a secondary carbon source for E. rectale. This second design alternative was predicted to yield the same butyrate productivity as the C. autoethanogenum-E. rectale system when grown in a CSTBR at the nominal CO/H2/N2 feed composition of 70/0/30 (Figure S2) and required the most expensive substrate lactose. A third design alternative in which the glucose uptake ability of C. ljungdahlii was eliminated and glucose was supplied as the secondary carbon source produced a slight decrease in butyrate productivity at the nominal feed gas composition (Figure S3). However, this alternative offered some promise for H2-containing feeds as a butyrate productivity of 0.16 g/L/h was predicted for a 40/30/30 composition (Figure S4).
Analogous CSTBR simulations were performed by combining wild-type C. ljungdahlii with wild-type and engineered C. kluyveri using vinyl acetate as the secondary carbon source. The wild-type coculture design failed to generate a butyrate productivity above 0.09 g/L/h (Figures S5 and S6), while the design with engineered C. kluyveri was more promising (Figure S7) and achieved a butyrate productivity of 0.22 g/L/h for a CO/H2/N2 feed composition of 40/30/30 (Figure S8). A comparison of the butyrate productivities predicted for all four wild-type designs considered in this study shows that the C. autoethanogenum-E. rectale system offered the best performance over a range of feed gas compositions if the design with lactose as the secondary carbon source was eliminated from consideration (Figure S9a). Of the four designs involving an engineered Clostridium, the combination of C. autoethanogenum and engineered C. kluyveri was predicted to yield the best performance (Figure S9b). Given our goal of combining the native metabolic capabilities of wild-type strains and the uncertainty associated with the feasibility of the proposed genetic modifications, only the best performing wild-type system design (C. autoethanogenum-E. rectale) was considered for future investigation.
3.5. Bubble column reactor performance of C. autoethanogenum and E. rectale coculture
CSTBRs are rarely used for large-scale gas fermentation due to the prohibitive agitation costs required to achieve adequate gas-liquid mass transfer [62]. Bubble column reactors with countercurrent flow and liquid recycle (Figure S10) are used industrially to achieve high gas-liquid mass transfer rates with only liquid pumping costs [41,63]. To evaluate the performance of the C. autoethanogenum-E. rectale coculture design for large-scale production, we developed a bubble column model with simplified hydrodynamics following our previously developed framework for monoculture systems [41]. The simulated reactor had a length of 5 m, total volume of 15 m3, CO/H2/N2 feed composition of 70/0/30, feed superficial gas velocity of 150 m/h, glucose feed concentration of 200 mmol/L to sustain E. rectale growth and feed superficial liquid velocity of 50 m/h (Table S3). The gas-liquid mass transfer rate kLa was calculated from the gas bubble interfacial area a, which varied with axial position in the column. When averaged across the column, the reactor kLa= 280 h−1 which has much larger than the constant value reactor kLa = 100 h−1 used in CSTBR simulations. The interested reader should consult our previous publication for details about model formulation and solution [41].
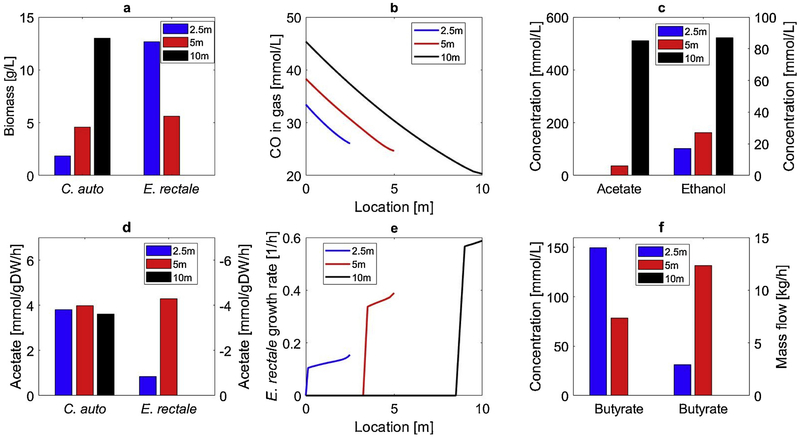
The effects of reactor length on coculture system performance were investigated as this reactor design parameter has been shown to be important in our previous monoculture modeling studies [41]. To allow meaningful comparisons when the length was changed from the nominal value L = 5m, the cross-sectional area was varied such that the reactors had the same height-to-diameter ratio and the feed gas volumetric flow rate also was scaled to maintain the same gas supply per unit volume of reactor. Simulations revealed a complex set of tradeoffs, with butyrate titer maximized for L = 2.5 m, butyrate mass production maximized for L = 5 m, and E. rectale washed out for L = 10 m (Fig. 6). The shortest column was predicted to generate low C. autoethanogenum biomass due to insufficient CO retention time but high E. rectale biomass due to reduced CO inhibition attributed to the lower hydrodynamic pressure and dissolved CO concentration at the bottom of the column. As a result, all residual acetate was consumed and butyrate titer was high but both CO utilization and butyrate production were low. The longest column generated high dissolved CO concentrations which strongly inhibited E. rectale growth across most of the column, leading to E. rectale washout. The column of intermediate length offered the most favorable tradeoff between the CO retention time and inhibition effects.
Fig. 6.
Effect of bubble column reactor length on steady-state performance of the C. autoethanogenum-E. rectale coculture system. (a) C. autoethanogenum and E. rectale biomass concentrations exiting the column; (b) gas-phase CO concentration as a function of column position; (c) acetate and ethanol concentrations exiting the column; (d) acetate secretion rates for C. autoethanogenum and uptake rates for E. rectale; (e) E. rectale growth rates as a function of column position ; (f) butyrate concentrations and mass productions based on the liquid stream exiting the column.
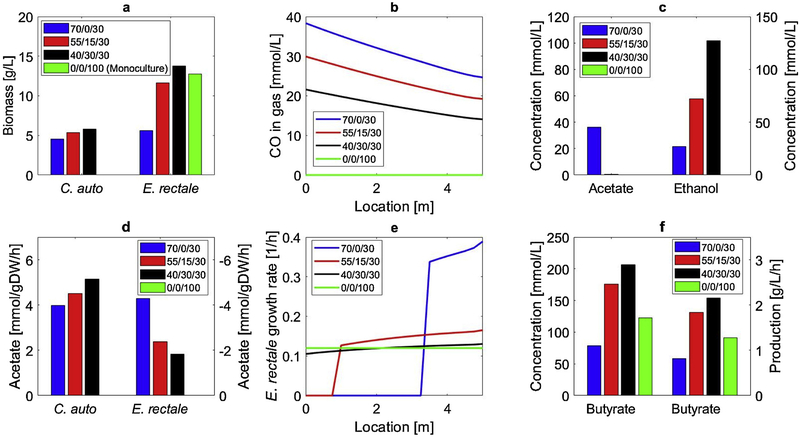
Based on these results, we simulated a reactor with L = 5 m to investigate the effects of feed gas composition on coculture system performance. Unlike predicted behavior in CSTBR, the bubble column model predicted that a CO/H2/N2 feed composition of 40/30/30 would generate the highest butyrate mass flow and titer due to reduced CO inhibition across the column (Fig. 7). In this case, E. rectale growth was almost constant across the column leading to the increased E. rectale biomass while the presence of H2 allowed enhanced C. autoethanogenum growth and biomass formation. The two species engaged in substantial acetate crossfeeding such that no residual acetate was present. Disadvantages of this feed composition were the production of ethanol as a significant byproduct due to H2 availability and the consumption of only about 35% of supplied CO because of relatively low dissolved CO concentrations. When the CO content of the feed was increased, E. rectale growth inhibition became dominant and overall system performance degraded with respect to both butyrate production and titer due to insufficient E. rectale biomass formation.
Fig. 7.
Effect of gas feed composition (CO/H2/N2) on steady-state bubble column performance of the C. autoethanogenum-E. rectale coculture system. (a) C. autoethanogenum and E. rectale biomass concentrations exiting the column; (b) gas-phase CO concentration as a function of column position; (c) acetate and ethanol concentrations exiting the column; (d) acetate secretion rates for C. autoethanogenum and uptake rates for E. rectale; (e) E. rectale growth rates as a function of column position ; (f) butyrate concentrations and mass productions based on the liquid stream exiting the column.
For sake of comparison, a monoculture of E. rectale growing solely on glucose was simulated using a CO/H2/N2 feed composition of 0/0/100. While the predicted E. rectale biomass concentration was only slightly less than that obtained with a gas mixture of 40/30/30, the two cases had strikingly different butyrate productivities and titers. The butyrate productivity obtained with pure N2 feed was predicted to be 1.2g/L/h while that obtained with the gas mixture was 2.1g/L/h, which clearly demonstrated the benefit of acetate-to-butyrate conversion in the coculture system. These results were consistent with FBA predictions that E. rectale acetate uptake only enhanced growth rate slightly but substantially increased butyrate synthesis. We calculated glucose-to-butyrate conversion to be 18% and CO-to-butyrate conversion to be 56% based on the amounts of glucose and CO actually consumed. Because essentially all supplied glucose was consumed and only about 35% of supplied CO was utilized, further design optimization would depend on increasing CO utilization while managing E. rectale inhibition by dissolved CO. Although direct comparison of bubble column and CSTBR results were difficult, the two reactor designs could be evaluated according to their CO-to-butyrate conversion efficiencies of 56% for the bubble column reactor and 22% for the CSTBR. The improved performance predicted for the bubble column was due to different volumetric mass transfer coefficients for the two reactor types along with different concentrations of C. auto and E. rectale. These predictions supported the use of bubble column reactors for CO-rich waste gas streams containing sufficiently high levels of H2.
4. Conclusions
We performed an in silico investigation of several coculture systems for conversion of CO-rich waste gases to the platform chemical butyrate in anaerobic continuous stirred tank bioreactors (CSTBRs). A monoculture system based on in silico engineering of the acetogen Clostridium autoethanogenum for butyrate synthesis was used as a basis for comparison. In addition to the difficulties in effectively engineering Clostridium strains [58,59], the monoculture system was predicted to yield the lowest butyrate productivity of all designs involving engineered strains. The combination of C. autoethanogenum for CO-to-acetate conversion with the environmental bacterium Clostridium kluyveri for acetate-to-butyrate conversion was predicted to yield relatively poor performance despite providing vinyl acetate as a secondary carbon source unless hexanoate production by C. kluyveri was eliminated through in silico metabolic engineering. An alternative coculture design based on combining C. autoethanogenum and the gut bacterium Eubacterium rectale with glucose as a secondary carbon source was predicted to provide superior performance without the need for strain engineering due to the high growth rates and acetate-to-butyrate conversion efficiency of E. rectale. An interesting design not explored in this study is a three-species system comprised of C. autoethanogenum for CO conversion to acetate and ethanol, E. rectale for acetate-to-butyrate conversion and engineered C. kluyveri for ethanol-to-butyrate conversion.
The feasibility of large-scale butyrate production with this wild-type coculture design was demonstrated through implementation in a simulated bubble column reactor, which predicted enhanced CO-to-butyrate conversion efficiency for CO-rich feeds containing sufficiently high levels of H2. Coculture performance was predicted to be limited through incomplete CO utilization by C. autoethanogenum and CO inhibition of E. rectale growth, suggesting that further optimization of bubble column design and operating parameters was possible. Overall, this study demonstrated that metabolic modeling could provide useful insights into coculture performance that can guide future experimental studies required to validate the model predictions. We intend to pursue future experimental studied aimed at testing the model predictions and providing data for model improvement, especially with respect to E. rectale growth inhibition by CO.
Supplementary Material
HIGHLIGHTS.
CO-to-butyrate conversion with coculture systems was assessed by metabolic modeling.
The two bacteria performed CO-to-acetate and acetate-to-butyrate conversion.
Clostridium autoethanogenum/Eubacterium rectale had high butyrate productivity.
E. rectale inhibition by dissolved CO has a strong effect on predicted performance.
Acknowledgements
The authors wish to acknowledge the National Science Foundation (CBET 1511346) for partial financial support of this research. This work was supported in part by a Fellowship from the University of Massachusetts to Xiangan Li as part of the Biotechnology Training Program (National Research Service Award T32 GM108556).
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.bej.2019.107338.
References
- [1].Huber GW, Iborra S, Corma A, Synthesis of transportation fuels from biomass: chemistry, catalysts, and engineering, Chem. Rev 106 (9) (2006) 4044–4098. [DOI] [PubMed] [Google Scholar]
- [2].Jiang W, Gu P, Zhang F, Steps towards ‘drop-in’biofuels: focusing on metabolic pathways, Curr. Opin. Biotechnol 53 (2018) 26–32. [DOI] [PubMed] [Google Scholar]
- [3].Martin MA, First generation biofuels compete, N. Biotechnol 27 (5) (2010) 596–608. [DOI] [PubMed] [Google Scholar]
- [4].Chandel AK, Garlapati VK, Singh AK, Antunes FAF, da Silva SS, The path forward for lignocellulose biorefineries: bottlenecks, solutions, and perspective on commercialization, Bioresour. Technol 264 (2018) 370–381. [DOI] [PubMed] [Google Scholar]
- [5].Dwidar M, Park J-Y, Mitchell RJ, Sang B-I, The future of butyric acid in industry, Scientific World J 2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang C-H, Ma Y-J, Yang F-X, Liu W, Zhang Y-D, Optimization of medium composition for butyric acid production by Clostridium thermobutyricum using response surface methodology, Bioresour. Technol 100 (18) (2009) 4284–4288. [DOI] [PubMed] [Google Scholar]
- [7].Song H, Eom M-H, Lee S, Lee J, Cho J-H, Seung D, Modeling of batch experimental kinetics and application to fed-batch fermentation of Clostridium tyrobutyricum for enhanced butyric acid production, Biochem. Eng. J 53 (1) (2010) 71–76. [Google Scholar]
- [8].Zigova J, Šturdík E, Advances in biotechnological production of butyric acid, J. Ind. Microbiol. Biotechnol 24 (3) (2000) 153–160. [Google Scholar]
- [9].Bedford A, Gong J, Implications of butyrate and its derivatives for gut health and animal production, Anim. Nutr 4 (2) (2018) 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Butyric Acid Market by Application (Animal Feed, Chemicals Intermediate, Food & Flavors, Pharmaceuticals, Perfumes, Others), by Type (Synthetic Butyric Acid, Renewable Butyric Acid) by Geography (APAC, North America, Europe, Row) - Global Analysis and Forecast to 2020, MarketsandMarketsINC, 2015. [Google Scholar]
- [11].Cascone R, Biobutanol-A replacement for bioethanol? Chem. Eng. Prog 104 (8) (2008) S4. [Google Scholar]
- [12].Liew FM, Köpke M, Simpson SD, Gas Fermentation for Commercial Biofuels Production, in Liquid, Gaseous and Solid Biofuels-conversion Techniques, IntechOpen., 2013. [Google Scholar]
- [13].Higman C, Van der Burgt M, Gasification, Gulf professional publishing, 2011. [Google Scholar]
- [14].Kirkels AF, Verbong GP, Biomass gasification: still promising? A 30-year global overview, Renew. Sustain. Energy Rev. 15 (1) (2011) 471–481. [Google Scholar]
- [15].Dürre P, Eikmanns BJ, C1-carbon sources for chemical and fuel production by microbial gas fermentation, Curr. Opin. Biotechnol 35 (2015) 63–72. [DOI] [PubMed] [Google Scholar]
- [16].Liew F, Martin ME, Tappel RC, Heijstra BD, Mihalcea C, Köpke M, Gas fermentation—a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks, Front. Microbiol 7 (2016) 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chang IS, Kim BH, Lovitt RW, Bang JS, Effect of CO partial pressure on cell-recycled continuous CO fermentation by Eubacterium limosum KIST612, Process. Biochem 37 (4) (2001) 411–421. [Google Scholar]
- [18].Huang J, Cai J, Wang J, Zhu X, Huang L, Yang S-T, Xu Z, Efficient production of butyric acid from Jerusalem artichoke by immobilized Clostridium tyrobutyricum in a fibrous-bed bioreactor, Bioresour. Technol 102 (4) (2011) 3923–3926. [DOI] [PubMed] [Google Scholar]
- [19].Wei D, Liu X, Yang S-T, Butyric acid production from sugarcane bagasse hydrolysate by Clostridium tyrobutyricum immobilized in a fibrous-bed bioreactor, Bioresour. Technol 129 (2013) 553–560. [DOI] [PubMed] [Google Scholar]
- [20].Ueki T, Nevin KP, Woodard TL, Lovley DR, Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii, MBio 5 (5) (2014) e01636–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen J, Henson MA, In silico metabolic engineering of Clostridium ljungdahlii for synthesis gas fermentation, Metab. Eng 38 (2016) 389–400. [DOI] [PubMed] [Google Scholar]
- [22].Ramos-Montañez S, Kazmierczak KM, Hentchel KL, Winkler ME, Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39, J. Bacteriol 192 (24) (2010) 6390–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hanly TJ, Henson MA, Dynamic metabolic modeling of a microaerobic yeast coculture: predicting and optimizing ethanol production from glucose/xylose mixtures, Biotechnol. Biofuels 6 (1) (2013) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bader J, Mast-Gerlach E, Popović MK, Bajpai R, Stahl U, Relevance of microbial coculture fermentations in biotechnology, J. Appl. Microbiol 109 (2) (2010) 371–387. [DOI] [PubMed] [Google Scholar]
- [25].Taniguchi M, Tanaka T, Clarification of interactions among microorganisms and development of co-culture system for production of useful substances, Recent Progress of Biochemical and Biomedical Engineering in Japan I, Springer, 2004, pp. 35–62. [DOI] [PubMed] [Google Scholar]
- [26].Chen Y, Development and application of co-culture for ethanol production by cofermentation of glucose and xylose: a systematic review, J. Ind. Microbiol. Biotechnol 38 (5) (2011) 581–597. [DOI] [PubMed] [Google Scholar]
- [27].Diender M, Uhl PS, Bitter JH, Stams AJ, Sousa DZ, High rate biomethanation of carbon monoxide-rich gases via a thermophilic synthetic coculture, ACS Sustain. Chem. Eng 6 (2) (2017) 2169–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Diender M, Stams AJ, Sousa DZ, Production of medium-chain fatty acids and higher alcohols by a synthetic co-culture grown on carbon monoxide or syngas, Biotechnol. Biofuels 9 (1) (2016) 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zou W, Ye G, Zhang J, Zhao C, Zhao X, Zhang K, Genome-scale Metabolic Reconstruction and Analysis for Clostridium kluyveri, Genome, 2018. (ja). [DOI] [PubMed] [Google Scholar]
- [30].Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla, Proc. Natl. Acad. Sci 106 (14) (2009) 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kantarci N, Borak F, Ulgen KO, Bubble column reactors, Process. Biochem 40 (7) (2005) 2263–2283. [Google Scholar]
- [32].Bredwell MD, Srivastava P, Worden RM, Reactor design issues for synthesis-gas fermentations, Biotechnol. Prog 15 (5) (1999) 834–844. [DOI] [PubMed] [Google Scholar]
- [33].Abubackar HN, Veiga MC, Kennes C, Biological conversion of carbon monoxide: rich syngas or waste gases to bioethanol, Biofuels Bioprod. Biorefining 5 (1) (2011) 93–114. [Google Scholar]
- [34].Valgepea K, Loi KQ, Behrendorff JB, de SP Lemgruber R, Plan M, Hodson MP, Köpke M, Nielsen LK, Marcellin E, Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum, Metab. Eng 41 (2017) 202–211. [DOI] [PubMed] [Google Scholar]
- [35].Valgepea K, Lemgruber RP, Meaghan K, Palfreyman RW, Abdalla T, Heijstra BD, Behrendorff JB, Tappel R, Köpke M, Simpson SD, Maintenance of ATP homeostasis triggers metabolic shifts in gas-fermenting acetogens, Cell Syst 4 (5) (2017) 505–515 e5. [DOI] [PubMed] [Google Scholar]
- [36].Ragsdale SW, Pierce E, Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation, Biochimica et Biophysica Acta (BBA)-Proteins Proteomics 1784 (12) (2008) 1873–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Burgard AP, Pharkya P, Maranas CD, Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization, Biotechnol. Bioeng 84 (6) (2003) 647–657. [DOI] [PubMed] [Google Scholar]
- [38].Daniell J, Köpke M, Simpson SD, Commercial biomass syngas fermentation, Energies 5 (12) (2012) 5372–5417. [Google Scholar]
- [39].Mohammadi M, Mohamed AR, Najafpour GD, Younesi H, Uzir MH, Kinetic studies on fermentative production of biofuel from synthesis gas using Clostridium ljungdahlii, Scientific World J 2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Valgepea K, Lemgruber RSP, Abdalla T, Binos S, Takemori N, Takemori A, Tanaka Y, Tappel R, Köpke M, Simpson SD, H 2 drives metabolic rearrangements in gas-fermenting Clostridium autoethanogenum, Biotechnol. Biofuels 11 (1) (2018) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li X, Griffin D, Li X, Henson MA, Incorporating hydrodynamics into spatiotemporal metabolic models of bubble column gas fermentation, Biotechnol. Bioeng 116 (1) (2019) 28–40. [DOI] [PubMed] [Google Scholar]
- [42].Ungerman AJ, Heindel TJ, Carbon monoxide mass transfer for syngas fermentation in a stirred tank reactor with dual impeller configurations, Biotechnol. Prog 23 (3) (2007) 613–620. [DOI] [PubMed] [Google Scholar]
- [43].Subramani V, Gangwal SK, A review of recent literature to search for an efficient catalytic process for the conversion of syngas to ethanol, Energy Fuels 22 (2) (2008) 814–839. [Google Scholar]
- [44].Linstrom P, Mallard W, NIST Chemistry WebBook, NIST Standard Reference Database Number 69 Gaithersburg MD, 20899, National Institute of Standards and Technology, 2015. [Google Scholar]
- [45].Meadows AL, Karnik R, Lam H, Forestell S, Snedecor B, Application of dynamic flux balance analysis to an industrial Escherichia coli fermentation, Metab. Eng 12 (2) (2010) 150–160. [DOI] [PubMed] [Google Scholar]
- [46].Levenspiel O, The Monod equation: a revisit and a generalization to product inhibition situations, Biotechnol. Bioeng 22 (8) (1980) 1671–1687. [Google Scholar]
- [47].Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J, Understanding the interactions between bacteria in the human gut through metabolic modeling, Sci. Rep 3 (2013) 2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chang I-S, Kim B-H, Kim D-H, Lovitt RW, Sung H-C, Formulation of defined media for carbon monoxide fermentation by Eubacterium limosum KIST612 and the growth characteristics of the bacterium, J. Biosci. Bioeng 88 (6) (1999) 682–685. [DOI] [PubMed] [Google Scholar]
- [49].Höffner K, Harwood SM, Barton PI, A reliable simulator for dynamic flux balance analysis, Biotechnol. Bioeng 110 (3) (2013) 792–802. [DOI] [PubMed] [Google Scholar]
- [50].Gomez JA, Höffner K, Barton PI, DFBAlab: a fast and reliable MATLAB code for dynamic flux balance analysis, BMC Bioinformatics 15 (1) (2014) 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen J, Gomez JA, Höffner K, Barton PI, Henson MA, Metabolic modeling of synthesis gas fermentation in bubble column reactors, Biotechnol. Biofuels 8 (1) (2015) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen J, Gomez JA, Höffner K, Phalak P, Barton PI, Henson MA, Spatiotemporal modeling of microbial metabolism, BMC Syst. Biol 10 (1) (2016) 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen J, Daniell J, Griffin D, Li X, Henson MA, Experimental testing of a spatiotemporal metabolic model for carbon monoxide fermentation with Clostridium autoethanogenum, Biochem. Eng. J 129 (2018) 64–73. [Google Scholar]
- [54].Schuetz R, Kuepfer L, Sauer U, Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli, Mol. Syst. Biol 3 (1) (2007) 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].García Sánchez CE, Torres Sáez RG, Comparison and analysis of objective functions in flux balance analysis, Biotechnol. Prog 30 (5) (2014) 985–991. [DOI] [PubMed] [Google Scholar]
- [56].Munasinghe PC, Khanal SK, Biomass-derived syngas fermentation into biofuels: opportunities and challenges, Bioresour. Technol 101 (13) (2010) 5013–5022. [DOI] [PubMed] [Google Scholar]
- [57].Villadsen J, Nielsen J, Lidén G, Bioreaction Engineering Principles, Springer Science & Business Media, 2011. [Google Scholar]
- [58].Nagaraju S, Davies NK, Walker DJF, Köpke M, Simpson SD, Genome editing of Clostridium autoethanogenum using CRISPR/Cas9, Biotechnol. Biofuels 9 (1) (2016) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liew F, Henstra AM, Köpke M, Winzer K, Simpson SD, Minton NP, Metabolic engineering of Clostridium autoethanogenum for selective alcohol production, Metab. Eng 40 (2017) 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Dürre P, Clostridium ljungdahlii represents a microbial production platform based on syngas, Proc. Natl. Acad. Sci 107 (29) (2010) 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nagarajan H, Sahin M, Nogales J, Latif H, Lovley DR, Ebrahim A, Zengler K, Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii, Microb. Cell Fact 12 (1) (2013) 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Daniell J, Köpke M, Simpson S, Commercial biomass syngas fermentation, Energies 5 (12) (2012) 5372–5417. [Google Scholar]
- [63].Hernandez-Alvarado F, Kalaga DV, Banerjee S, Kawaji M, Comparison of gas hold-up profiles in co-current, counter-current and batch bubble column reactors measured using gamma densitometry and surface of revolution method, ASME 2016 Fluids Engineering Division Summer Meeting Collocated With the ASME 2016 Heat Transfer Summer Conference and the ASME 2016 14th International Conference on Nanochannels, Microchannels, and Minichannels, American Society of Mechanical Engineers, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.