Abstract
Fowl adenovirus (FAdV) has posed a grave threat to the health of poultry, and the sudden outbreak highlights the importance of the new rapid diagnostic method for the control and prevention of transmission. Hence, in the present study, a novel recombinase polymerase amplification (RPA) assay, which was suitable for all 12 serotypes (FAdV-1 to 8a and 8b to 11) had been successfully launched to detect FAdV. Also, the entire amplification process could be completed in the isothermal condition when temperature ranged from 26 to 42°C within no more than 14 min, which was remarkably superior to endpoint polymerase chain reaction (98 min) with the same detecting sensitivity (as low as 0.1 fg viral DNA), avoiding sophisticated thermal cyclers with simple operation. Additionally, the same primers did not produce positive reactions with other viruses tested, demonstrating that the specificity of the RPA assay was acceptable. Moreover, this developed method could be efficiently used in the diagnosis of FAdV references and epidemic strains from different avian origins, thus making it a rapid, reliable, and point-of-care FAdV diagnostics tool, as well as an alternative to endpoint PCR.
Key words: fowl adenovirus, isothermal, recombinase polymerase amplification, rapid detection method
Introduction
Fowl adenovirus (FAdV), a part of genus Aviadenovirus, includes 5 species (FAdV-A to E) and12 serotypes, which can be grouped (FAdV-1 to 8a and 8b to 11). (Li et al., 2017). This nonenveloped virus, about 70 to 100 nm in diameter, contains double-stranded linear DNA genome of approximately 45 kb, encoding 8 structural polypeptides (e.g., hexon, penton, and fiber protein) (Chiocca et al., 1996; Mase and Nakamura, 2014). Generally, the notable clinical symptoms associated with FAdV infection include inclusion body hepatitis, gizzard erosion, and hydropericardium syndrome (HPS) (Gjevre et al., 2013; Li et al., 2016b; Schachner et al., 2018). Most serotypes can be associated with inclusion body hepatitis (Vera-Hernandez et al., 2016). But gizzard erosion, which has spread in Asia and some Europe countries recent years, is confirmed mainly induced by FAdV-1 (FAdV-A) (Lim et al., 2012; Grafl et al., 2014), and the widespread HPS which is attributed exclusively to serotype 4 (FAdV-C) is the most serious disease caused by FAdV, especially in broilers, resulting in tremendous economic losses (Mazaheri et al., 1998). In particular, the reports to date suggested that FAdV could transmit among ducks and geese, confirming that the emerging FAdV-infection imposes a potential danger to the whole poultry industry (Chen et al., 2017; Pan et al., 2017; Yu et al., 2018; Wei et al., 2019).
So far, FAdV could be diagnosed by various methods developed such as virus isolation, agar gel precipitation assay, virus seroneutralization, and enzyme-linked immunosorbent assay, and over the past decades, many endpoint PCR methods which targeting specific regions of the FAdV genome have been effectively used (Raue and Hess, 1998; Fitzgerald, 2008; Kaján et al., 2011; Shao et al., 2019). Furthermore, the application of real-time quantitative PCR assay further improves the specificity and sensitivity in FAdV detection (Günes et al., 2012). However, these methods are often limited because of its dependence on expensive specialized thermal cycling equipment or usual longer detection cycle (Daher et al., 2016). In addition, these nucleotide amplification methods concentrated mainly on the limited origins (mostly chicken) but lacked specific evaluation for other avian species (Günes et al., 2012; Junnu et al., 2014; Rajasekhar and Roy, 2014).
With the increasing advancement in molecular diagnosis, several novel techniques such as loop-mediated isothermal amplification, cross-priming amplification, fluorescent microsphere immunoassay, and droplet digital PCR assay have been successfully developed to detect FAdV and overcome most shortcomings existing in traditional methods (Niczyporuk et al., 2015; Dong et al., 2018; Feichtner et al., 2018). Apart from these approaches mentioned above, recombinase polymerase amplification (RPA), an advanced detection approach which was firstly reported in 2006, has gained increasing popularity as a simple and robust method of amplifying DNA (Hu et al., 2017). Until now, it has been reported in multiple areas, making the isothermal amplification (usually constant at 37 to 42°C) of specific DNA fragments, possible in no more than 30 min, rather than temperature cycling (Hoff, 2006; Piepenburg et al., 2006; Li et al., 2018). In RPA reaction, the phage-derived recombinase is utilized to bind with the single stranded oligonucleotide primers to scan the complementary template DNA sequence efficiently. The displaced template, interacting with the single-strand DNA binding protein, retains a stable structure and then extends as the way of PCR by the DNA polymerase when the recombinase is separated from 3′end of the oligonucleotide (Del Rio et al., 2017; Vasileva Wand et al., 2018). Since the appearance of the RPA commercial kit from TwistAmp Basic (TWISTDX Ltd., Babraham, UK) in 2014, this rapid amplification technique has been widely used in diagnosis of numerous pathogens, such as human immunodeficiency virus 1, Ebola virus, Rift Valley fever virus, dengue virus, pseudorabies virus, foot-and-mouth disease virus, middle east respiratory syndrome coronavirus, and so on (Boyle et al., 2013; Yang et al., 2017; James et al., 2018; Liu et al., 2018; Tan et al., 2018).
Here, to simplify detection conditions for FAdV, the available RPA assay has been developed. In the following study, we would describe the superiorities using this method in detail and evaluate its performance in diagnosing.
Materials and methods
FAdV References and Cells
All 12 reference serotypes of FAdV were obtained from China Veterinary Culture Collection Center (Beijing, China) and cultured on Leghorn Male-chicken Hepatocellular-carcinoma cell line (CRL-2117) which purchased from ATCC (Manassas, VA). The cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and maintained at 37°C with 5% CO2.
DNA Extraction
DNA was extracted using TIANcombi DNA Lyse&Det PCR Kit (TIANGEN, Beijing, China) according to manufacturers' instructions. A volume of 100 μL B1 was mixed with samples, then grind for moments. Then, add 100 μL B2 to the column and centrifuge it (12, 000 rpm, 2 min) for supernatants (containing viral DNA). DNA was stored at −20°C.
Endpoint PCR and Real-Time Quantitative PCR
Endpoint PCR was performed using primer pair as described in Table 1, which amplified a fragment of 564 bp (Zhao et al., 2015). The reaction mixture of 25 μL contained 12.5 μL 2 × Taq MasterMix (CWBIOTECH, Beijing, China), 10.5 μL RNase-free ddH2O, 0.5 μL (10 μmol) each primer, and 1 μL extracted DNA. The initial denaturation was set at 95°C for 10 min followed by 30 cycles of denaturation at 95°C for 1 min, 52°C for 30 s, 72°C for 30 s, and final extension at 72°C for 10 min. Amplification of target sequences was carried out in A300 Fast Thermal Cycler (LONGGENE), and the products were resolved on 2% agarose gel with SuperBuffer and stained with SuperStain (10, 000 × in Water) (CWBIOTECH) for visualization.
Table 1.
Primers used in this study.
| Num. | Sequence ID | Sequence (5' - 3′) | Length (bp) | Strain | Product length (bp) |
|---|---|---|---|---|---|
| 1 | FAdV-PCR Fw | TGCTCGTTGTGGATGGTGAA | 20 | AF083975 | 564 |
| 2 | FAdV-PCR Rev | CTCCGTGTTGGGCTGGTC | 18 | ||
| 3 | FAdV RT-PCR Fw | CACAACGTCTGCAGATCAGATTC | 23 | AAU46933 | 74 |
| 4 | FAdV RT-PCR Rev | GCGCACGCGATAGCTGTT | 18 | ||
| 5 | FAdV-Probe | FAM-ACCCGATCCAGACGGATGACACG-TAMRA | 23 | ||
| 6 | FAdV-RPA Fw | CKCCYACTCGCAATGTCACCACCGARAAGGCH | 33 | KT862807.1 | 108 |
| 7 | FAdV-RPA Rev | TKAHGCTGTASCGCACGCGRTARCTGTTGGGC | 32 | ||
| 8 | FAdV-RPA Fw | CTCCCACTCGCAATGTCACCACCGAAAAGGCA | 33 | Z67970.1 | 139 |
| 9 | FAdV-RPA Rev | CCCATGTCCAACACCCAGCTGTCCCCAACGTT | 32 | ||
| 10 | FAdV-RPA Fw | AGGCAGACGGTCGTAGCTCCCACTCGCAATGT | 32 | Z67970.1 | 155 |
| 11 | FAdV-RPA Rev | CCCATGTCCAACACCCAGCTGTCCCCAACGTT | 32 | ||
| 12 | Vector-PCR Fw | TTTTACATGAATCGCAGGGTT | 21 | Z67970.1 | 146 |
| 13 | Vector-PCR Rev | CCATAAGGTTCACCTCGGTA | 20 |
K-G/T, Y-C/T, R-A/G, S-C/G, H-A/T/C, N-A/T/C/G. PCR-(endpoint PCR). RT-PCR-(real-time quantitative PCR). Fw-Forward primer, Rev-Reverse primer.
Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
For real-time quantitative PCR, the viral DNA levels were quantified by using ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) and Premix Ex TAQ (Probe qPCR) (TAKARA, Dalian, China).The real-time quantitative PCR was conducted in 20 μL, and the initial denaturation was set at 95°C for 30 s followed by 40 cycles of denaturation at 95°C for 5 s and 60°C for 34 s. Then, the fluorescence was obtained at the end of each step at 60°C.
Primers for the RPA Assay
Using the ClustalW method, we selected a higher conserved sequence among 12 different FAdV reference sequences from GenBank and 5 epidemic strains isolated by our lab, including FAdV-A (CELO, SDJN, SDXT1603, SDGX1608), FAdV-B (340), FAdV-C (Fowl adenovirus 4, SDSG, SDSX, Fowl adenovirus 10), FAdV-D (SR48, SR49, 764, 380), and FAdV-E (CR119, SD15-21, USP-BR-453.2, SD16-116) (Table 2). To increase speed and sensitivity of the RPA assay, length of primers should be no less than 30 bp, 32 to 35 bp in general. The amplicon size was designed at a range of 100 to 200 bp to get the fastest reaction power. The primers were designed by using Primer 5 based on the criteria suggested in the TwistAmp Basic (TWISTDX Ltd., Babraham, UK) kit manual, whereas the specificity was further proved by the BLAST search in National Center for Biotechnology Information.
Table 2.
The FAdV reference strains retrieved from NCBI for RPA primers design.
| Serotype | Species | GenBank accession no. | Submitted |
|---|---|---|---|
| FAdV-1 (CELO) | A | Z67970.1 | Russia/1995 |
| FAdV-2 (SR48) | D | AF508946.1 | Belgium/2002 |
| FAdV-3 (SR49) | D | KT862807.1 | Austria/2015 |
| FAdV-4 | C | AJ431719.1 | Russia/2002 |
| FAdV-5 (340) | B | KC493646.1 | Austria/2013 |
| FAdV-6 (CR119) | E | NC_038332.1 | USA/2018 |
| FAdV-7 (SD15-21) | E | KY364398.1 | China/2017 |
| FAdV-8a (USP-BR-453.2) | E | KY229177.1 | Brazil/2018 |
| FAdV-8b (SD16-116) | E | KY426984.1 | China/2017 |
| FAdV-9 (764) | D | AF508958.2 | Belgium/2003 |
| FAdV-10 | C | U26221.1 | USA/2000 |
| FAdV-11 (380) | D | KT862812.1 | Austria/2015 |
| FAdV-1 (SDJN) | A | MF198255 | China/2016 |
| FAdV-1 (SDXT1603) | A | MN329737 | China/2016 |
| FAdV-1 (SDGX1608) | A | MN329736 | China/2016 |
| FAdV-4 (SDSG) | C | MK424834 | China/2018 |
| FAdV-4 (SDSX) | C | KT899325 | China/2015 |
Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification; NCBI, National Center for Biotechnology Information.
Test Conditions of the RPA Assay
The RPA assay was carried out by using TwistAmp Basic (TWISTDX Ltd.) and performed in a 0.2 ml reaction tube containing 50 μL dried enzyme volume, 4.8 μL (10 μmol) forward and reverse primers each, 29.5 μL rehydration buffer, and 8.4 μL template and ddH2O.
The precipitate was resuspended, and 2.5 μL (280 nm) of magnesium acetate was added to initiate the reaction. Tubes were incubated for 10 to 30 min, and the products were analyzed by agarose gel electrophoresis with SuperBuffer for 10 min (25 V/cm). Negative controls used include nontemplate control and total DNA from healthy birds.
Amplification could be performed at 37 to 42°C for about 20 to 30 min according to TwistAmp Basic (TWISTDX Ltd). Different conditions were investigated to evaluate the detection ranges of the RPA assay for adapting complicated environmental factors. Keep other factors constant, and various temperatures (22, 26, 28, 30, 37, 42°C) and time (14, 19, 24, 29, 34 min) were used in our study.
Sensitivity and Specificity of the RPA Assay
To evaluate the sensitivity of the RPA assay, a recombinant plasmid constructed with a positive specimen (GenBank Accession No. Z67970.1) was used to determine the limit. The plasmid DNA standard was quantified via the DeNovix DS-11 Spectrophotometer and diluted with serial 10-fold ranging from 1 ng to 0.1 fg in Easy dilution (TAKARA, Beijing, China). These templates were then subjected to the RPA assay in the conditions described above.
The specificity of RPA assay was assessed against other common infecting viruses in avian including Avian leukosis virus (DQ115805), Avian influenza virus (MF581307), Newcastle disease virus (MF581294), Tembusu virus (KJ740746), and Goose parvovirus (MF581304).
Evaluation of the RPA Assay
DNA from infected chickens, ducks, and geese (previously collected 28 FAdV-infected cases including 13 field variant strains) were subjected to RPA analysis to validate the diagnostic ability of the new assay. The results were further confirmed by real-time quantitative PCR, whereas endpoint PCR was also used as comparison (Table 3).
Table 3.
Detection results from the RPA assay (including field variant strains).
| Serial no. | FAdV field variant strains and infected cases | Serotype | Species | Real-time quantitative PCR results (copies/μg total DNA) | Endpoint PCR results | RPA assay results |
|---|---|---|---|---|---|---|
| 1 | Broiler/SDJN/MF198255/China/2016 | 1 | A | 7.07 E+05 | + | + |
| 2 | Broiler/SDLC/MG869820/China/2015 | 4 | C | 3.52 E+06 | + | + |
| 3 | Layer/SDJX/MF198254/China/2015 | 4 | C | 3.98 E+06 | + | + |
| 7 | Layer/SDD01/MG869819/China/2015 | 4 | C | 8.41 E+06 | + | + |
| 6 | Broiler/LC1611/MN316650/China/2016 | 4 | C | 1.00 E+06 | + | + |
| 4 | Layer/SDDZ/MH159176/China/2017 | 4 | C | 8.31 E+05 | + | + |
| 5 | Layer/SDWF/MH159177/China/2017 | 4 | C | 4.92 E+05 | + | + |
| 8 | Breeder/SDSG/MK424834/China/2018 | 4 | C | 3.61 E+07 | + | + |
| 9 | Broiler/SDRZ/MF198256/China/2011 | 8 | E | 5.97 E+07 | + | + |
| 10 | Duck/SDXT1603/MN329737/China/2016 | 1 | A | 2.45 E+06 | + | + |
| 11 | Duck/SDSX/KT899325/China/2015 | 4 | C | 1.04 E+06 | + | + |
| 12 | Goose/SDGX1608/MN329736/China/2016 | 1 | A | 4.68 E+06 | + | + |
| 13 | Goose/SDJN/MK335954/China/2016 | 4 | C | 3.46 E+06 | + | + |
| 14 | Broiler/LYFAdV013/Laiyang/China/2015 | 1 | A | 4.14 E+08 | + | + |
| 15 | Broiler/ZZFAdV125/Zaozhuang/China/2016 | 1 | A | 4.56 E+07 | + | + |
| 16 | Broiler/LYFAdV001/Linyi/China/2015 | 2 | D | 2.22 E+07 | + | + |
| 17 | Broiler/LYFAdV007/Laiyang/China/2015 | 3 | D | 7.07 E+05 | + | + |
| 18 | Ma-chicken/IMFAdV127/Inner Mongolia/China/2016 | 4 | C | 6.13 E+06 | + | + |
| 19 | Broiler/XTFAdV120/Xintai/China/2015 | 4 | C | 9.85 E+07 | + | + |
| 20 | Broiler/JXFAdV119/Juxian/China/2015 | 4 | C | 7.58 E+05 | + | + |
| 21 | Broiler/SXFAdV131/Shanxi/China/2016 | 4 | C | 1.26 E+06 | + | + |
| 22 | Pheasant/RZFAdV133/Rizhao/China/2016 | 4 | C | 4.90 E+08 | + | + |
| 23 | Layer/JXFAdV135/Juxian/China/2016 | 4 | C | 8.44 E+07 | + | + |
| 24 | Native chicken/JXFAdV138/Juxian/China/2016 | 4 | C | 1.02 E+06 | + | + |
| 25 | Cock/JXFAdV140/Juxian/China/2016 | 4 | C | 9.55 E+05 | + | + |
| 26 | Layer/HBFAdV143/Hebei/China/2017 | 4 | C | 4.27 E+06 | + | + |
| 27 | Layer/CFFAdV144/Inner Mongolia/China/2017 | 4 | C | 6.96 E+05 | + | + |
| 28 | Duck/SZFAdV118/Suzhou/China/2015 | 4 | C | 1.03 E+06 | + | + |
Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
Results
The Epidemic Status of FAdV
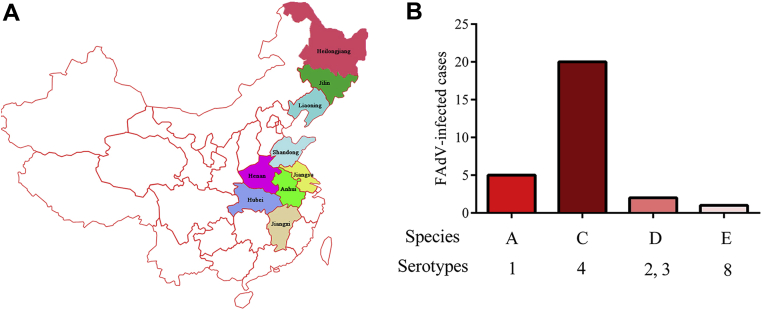
To our knowledge, since July 2015, infectious diseases caused by virulent strains of FAdV have been increased suddenly, especially the HPS in China (Figure 1A). We analyzed 28 positive FAdV-infected cases mostly focusing on 2015 to 2018 and found that they were classified into 4 types of species (4 as FAdV-A, 20 as FAdV-C, 2 as FAdV-D, and 1 as FAdV-E). Besides serotype 4 of FAdV-C, which acted as the major epidemic strain, other prevalent serotypes were also transmitted in most breeds of chickens, even waterfowls currently (Figure 1, Table 3).
Figure 1.
Geographic distribution and epidemic of FAdV. (A) The provinces affected with FAdV infection were indicated in different colors in the map of China. (B) The serotypes of FAdV were epidemic currently. Abbreviations: FadV, fowl adenovirus.
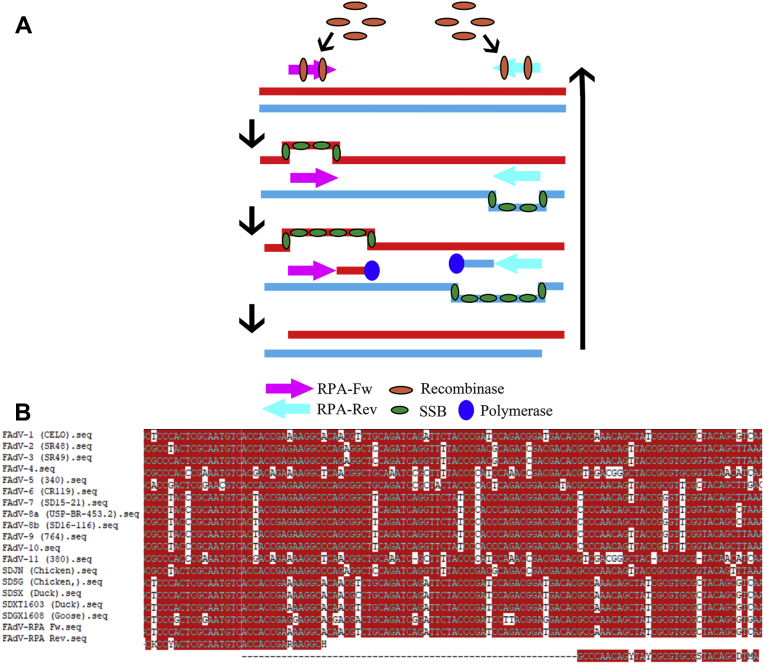
The Design of Primers
Using the ClustalW method, a short candidate region (approximately 200 bp in hexon gene) which shared a high-level identity above 95% over the 5 species were determined as the RPA target region (Figure 2B). According to this fragment, 3 pairs of primers with the target size (108, 139, 155 bp, respectively) were identified for screening out a more appropriate one for the RPA detection (Table 1).
Figure 2.
The RPA cycle sketch and the design of primers. (A) Based on the principles described above, the RPA reaction was showed by the illustration. (B) The RPA primers with the target size of 108 bp were shown according to the hexon segment alignment of different FAdV (12 reference and 5 emerging strains represent species of FAdV-A to E). Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
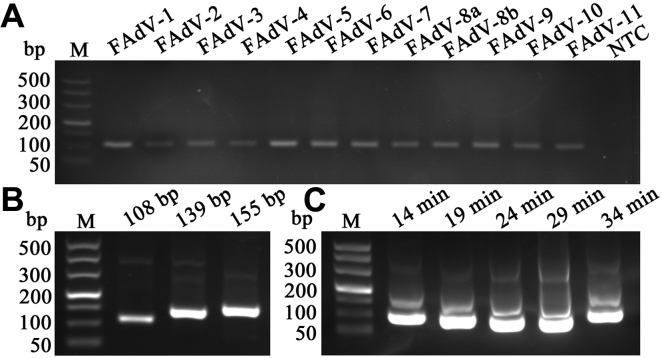
With these primers, viral DNA (FAdV-1, CELO), with the expected length, could be amplified, and sequencing results were consistent with our design. Meanwhile, owing to the more efficient amplification in short fragments by the RPA assay, the first pair of primers was selected for a universal testing. And all the serotypes of the FAdV could be detected with similar amplification efficiency, including 12 serotypes belonging to 5 species (FAdV-1 to 8a and 8b to 11) (Figures 3A and 3B). The specificity of the amplification products was confirmed and visualized by agarose gel as 108 bp bands.
Figure 3.
The universality and different test conditions of the RPA assay. (A) The RPA primers designed for the product of 108 bp could be widely used in all 12 serotypes. (B) Single right bands were successfully produced by using the 3 different primers with the targets (108, 139, 155 bp). (C) A variety of detection time (14 to 34 min) were tested and with the isothermal amplification for only14 min, the right target could be found as above. M-DL500 DNA Marker. Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
Test Conditions of the RPA Assay
Furthermore, for a more extensive application, specific reaction time (14, 19, 24, 29, and 34 min) with various temperatures (22, 26, 28, 30, 37, and 42°C) were employed to achieve the scale of detecting capability for the RPA assay, and all the test conditions showed strong positive amplification products of the expected length. It has been proved in experiments that even incubation at 37°C for only 14 min was enough to obtain the correct fragments but with a little pointless (Figure 3C). Afterward, a series of temperatures were tested and following with the incubation at such a low of 26°C, there appeared the singe right band (Figure 4A). Followingly, we further attempted to reduce the temperature to 22°C, but the activity of relevant enzymes decreased with only blurred bands, without obtaining satisfactory results (Figure 4B). Therefore, under these conditions, it has the potential to apply the RPA assay in practice, which facilitates clinical decisions more quickly.
Figure 4.
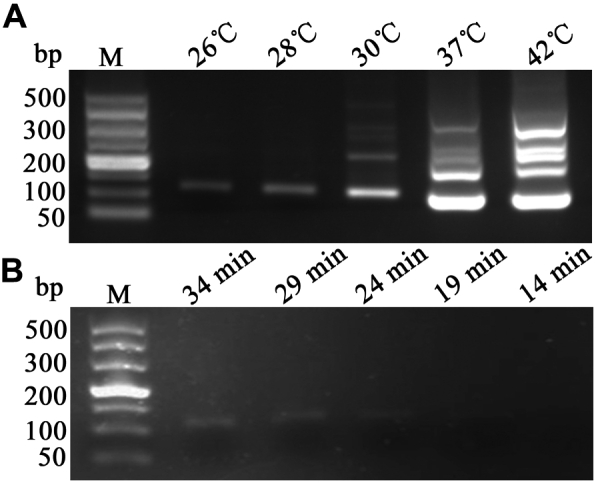
Temperature ranges for FAdV detection of the RPA assay. (A) A series of temperatures, ranging from 26 to 42°C, were used, and the right targets of 108 bp could be obtained under these tested temperatures. But some amplifications of pointiness would be found when it was more than 30°. (B) As shown above, only blurred signals could be obtained by the developed RPA assay when the tested temperature fell to 22°C. M-DL500 DNA Marker. Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
Specificity and Sensitivity of the RPA Assay
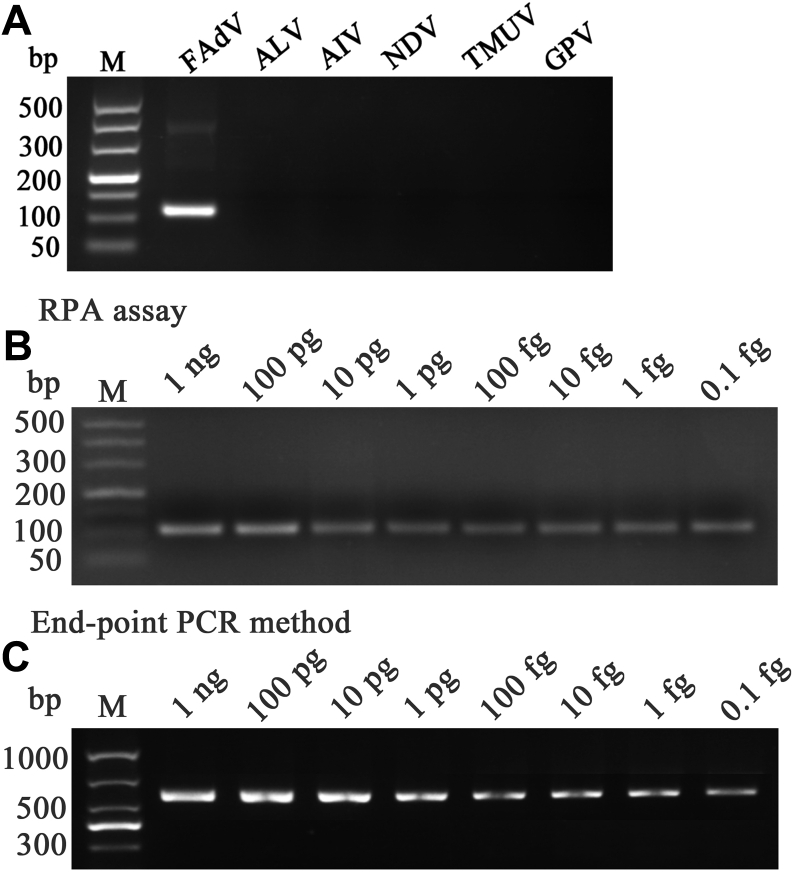
The specificity of RPA assay was performed with the other positive avian viruses which are often susceptible in clinical, including avian leukosis virus, Avian influenza virus, Newcastle disease virus, Tembusu virus, and Goose parvovirus, and no cross-reactions with these viruses were found, indicating that the primers showed a high specificity (Figure 5A).
Figure 5.
Detection specificity and sensitivity of the RPA assay. (A) Five other virus strains, including ALV, AIV, NDV, TMUV, and GPV at 10 ng per reaction, were used to check for the cross-reactions, without signals except FAdV with the RPA primers. (B, C) Ten-fold serial dilutions of the DNA standards from 1 ng to 0.1 fg were tested to determine the lowest number of target molecules detected by the RPA and endpoint PCR assay. Both methods showed the similar detection limits as low as 0.1 fg per reaction. M-DL500, 1000 DNA Marker. Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification; ALV, Avian leukosis virus; AIV, Avian influenza virus; NDV, Newcastle disease virus; TMUV, Tembusu virus; GPV, Goose parvovirus.
Additionally, the sensitivity of RPA assay was evaluated by setting a serial dilution of DNA standards (1 ng to 0.1 fg), and the results showed that as few as 0.1 fg/reaction (3.2 × 10 copies) synthetic template could be detected even at 26°C, which was comparable to the endpoint PCR (Figures 5B and 5C). Without thermal denaturation of templates, the RPA assay needs only 14 min for the whole amplification process, while as much as 98 min and 55 min were required, separately, when analyzing the same viral dose of 0.1 fg by endpoint PCR or real-time quantitative PCR.
Evaluation of the New RPA Assay
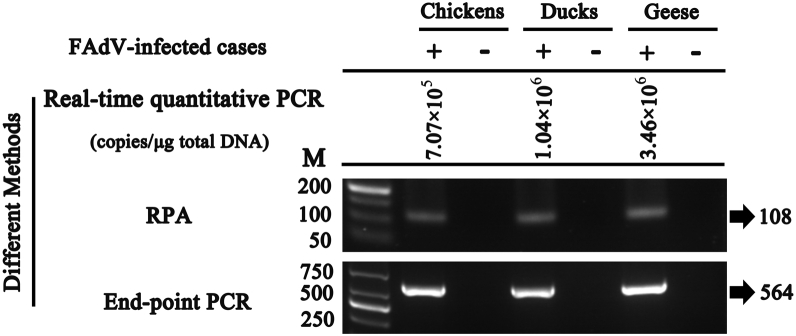
To confirm the adaptability of this method, a total of 28 FAdV-infected cases (including 13 field variant strains) mostly collected from 2015 to 2018 from different regions, which covered serotypes mainly prevalent in China, were analyzed by RPA and endpoint PCR method. All of them showed positive results, indicating that these 2 methods were completely coincident (Figure 6, Table 3). With the introduction of epidemic FAdV strains into the design of primers, the detectability, instead of being only limited in chickens, was greatly improved. Compared with endpoint PCR, the detection speed was significantly increased by the RPA assay, suggesting that it could serve as an alternative detection approach to endpoint PCR method.
Figure 6.
Evaluation of the RPA assay. Different samples from FAdV-infected or normal birds were detected by real-time quantitative PCR, endpoint PCR and RPA assay. M-DL500, 1000 DNA Marker. Abbreviations: FadV, fowl adenovirus; RPA, recombinase polymerase amplification.
Discussion
Since the year of 2015, the number of FAdV-infected cases has increased sharply, which imposed severe challenges to the poultry industry (Li et al., 2016a). What’s more, Pan et al. (2017) and Wei et al. (2019) have respectively reported that FAdV resulted in severe infections in waterfowls including ducks and geese; meanwhile new research from Li et al. (2016a) has showed multiple serotypes were prevalent currently (Li et al., 2016a; Pan et al., 2017; Wei et al., 2019). The similar epidemic status was also elaborated according to our investigate data, thus getting correct diagnoses with simple operations timely was the key component to prevent the spread of FAdV.
Up to now, many advanced assays for FAdV detection have been applied, representational methods as endpoint PCR and real-time quantitative PCR (Raue and Hess, 1998; Günes et al., 2012). Although these molecular diagnostics, comparing with traditional methods, has significantly improved in virology diagnosing accuracy, some disadvantages in respect of the operational complexity and thermocycler dependence are not well solved, limiting its use mostly in specialized and suitably equipped labs (Ma et al., 2018). Newly developed methods, like loop-mediated isothermal amplification, could detect FAdV isothermally with high sensitivity, but the temperature needed was very high (65°C) and merely suitable for the specific strain (FAdV-4) (Yuan et al., 2019; Zhai et al., 2019). While as the RPA assay putting into practice in Zika virus and other virus detection, it enables rapid detection possible (Babu et al., 2017; Vasileva Wand et al., 2018). Different from the molecular diagnostics mentioned above, RPA can be run at a constantly low temperature of 37 to 42°C and using this character, a RPA assay for detecting FAdV which was robust and capable of tolerating all the 12 serotypes was described in this study (Figure 3A) (Hoff, 2006; Piepenburg et al., 2006). After testing, this novel isothermal amplification technique was suitable for wide work conditions even at room temperature (26°C), thereby avoiding the need for sophisticated thermal cyclers (Figure 3C). In the previous study, Gunes et al developed a universal real-time quantitative PCR method and was suitable for FAdV detection in chickens, but it was time-consuming and lacked application in other avian species (Günes et al., 2012). Because of the rapidity of the RPA reaction, the time required to perform this novel molecular amplification could be considerably reduced (within 14 min), indicating that it was 85.7 and 74.5% lower compared with endpoint PCR and real-time quantitative PCR. Remarkably, with a positive identification as low as 0.1 fg viral DNA, this new RPA assay was highly sensitive, which was comparable to the sensitivity produced by endpoint PCR (Figures 5B and 5C), but a bit lower than real-time quantitative PCR, as what was established before (Günes et al., 2012). In addition, the high analytical specificity of the current RPA assay was further confirmed in subsequent experiments using a panel of extracted DNA from infected birds, thereby demonstrating that it could be applied in the identification of FAdV-detection and achieve more extensive detection ranges than before (Table 3).
The real-time quantitative PCR method is able to quantify viral loads in infected cases, but RPA is a qualitative detection method like endpoint PCR (Günes et al., 2012; Abera et al., 2014). Despite all that, however, depending on its simple operations with equivalent sensitivity and specificity, the RPA technique was believed as a perfect alternative to endpoint PCR in FAdV diagnoses. Therefore, this developed FAdV-RPA assay, coupling with the mini portable polyacrylamide gel electrophoresis equipment, is expected to be well suited for field-applicable detection which promotes the diagnosticians to obtain precise and reliable results timely as its use in human's diseases (James et al., 2018; Tan et al., 2018). In the near future, RPA assay would have the great potential in more avian viruses' diagnostics, and multiple primers would be developed to diagnose a variety of viruses in 1 reaction, not only cutting the cost but also contributing the breeders to initiating a comprehensive prevention for minimizing the risk.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFD0500106-3), China Agriculture Research System (CARS-42-19) and the funds of Shandong “Double Top” Program.
Conflict of Interest Statement: The authors did not provide any conflict of interest statement.
Contributor Information
Ziqiang Cheng, Email: czqsd@126.com.
Yi Tang, Email: Tyck288@163.com.
Youxiang Diao, Email: yxdiao@126.com.
References
- Abera T., Thangavelu A., Joy Chandran N.D., Raja A. A SYBR Green I based real time RT-PCR assay for specific detection and quantitation of Peste des petits ruminants virus. BMC Vet. Res. 2014;10:22. doi: 10.1186/1746-6148-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu B., Washburn B.K., Ertek T.S., Miller S.H., Riddle C.B., Knox G.W., Ochoa-Corona F.M., Olson J., Katircioglu Y.Z., Paret M.L. A field based detection method for Rose rosette virus using isothermal probe-based Reverse transcription-recombinase polymerase amplification assay. J. Virol. Methods. 2017;247:81–90. doi: 10.1016/j.jviromet.2017.05.019. [DOI] [PubMed] [Google Scholar]
- Boyle D.S., Lehman D.A., Lillis L., Peterson D., Singhal M., Armes N., Parker M., Piepenburg O., Overbaugh J. Rapid detection of HIV-1 Proviral DNA for Early infant diagnosis using recombinase polymerase amplification. MBio. 2013;4:e00135-13. doi: 10.1128/mBio.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Dou Y., Zheng X., Tang Y., Zhang M., Zhang Y., Wang Z., Diao Y. Hydropericardium hepatitis syndrome emerged in Cherry valley ducks in China. Transbound. Emerg. Dis. 2017;64:1262–1267. doi: 10.1111/tbed.12500. [DOI] [PubMed] [Google Scholar]
- Chiocca S., Kurzbauer R., Schaffner G., Baker A., Mautner V., Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J. Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher R.K., Stewart G., Boissinot M., Bergeron M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016;62:947–958. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio J.S., Lobato I.M., Mayboroda O., Katakis I., O'Sullivan C.K. Enhanced solid-phase recombinase polymerase amplification and electrochemical detection. Anal. Bioanal. Chem. 2017;409:3261–3269. doi: 10.1007/s00216-017-0269-y. [DOI] [PubMed] [Google Scholar]
- Dong G., Meng F., Zhang Y., Cui Z., Lidan H., Chang S., Zhao P. Development and evaluation of a droplet digital PCR assay for the detection of fowl adenovirus serotypes 4 and 10 in attenuated vaccines. J. Virol. Methods. 2018;164:691–697. doi: 10.1016/j.jviromet.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Feichtner F., Schachner A., Berger E., Hess M. Fiber-based fluorescent microsphere immunoassay (FMIA) as a novel multiplex serodiagnostic tool for simultaneous detection and differentiation of all clinically relevant fowl adenovirus (FAdV) serotypes. J. Immunol. Methods. 2018;458:33–43. doi: 10.1016/j.jim.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Fitzgerald S.D. Adenovirus infections. Dis. Poult. 2008;12:289–332. [Google Scholar]
- Gjevre A.G., Kaldhusdal M., Eriksen G.S. Gizzard erosion and ulceration syndrome in chickens and turkeys: a review of causal or predisposing factors. Avian Pathol. 2013;42:297–303. doi: 10.1080/03079457.2013.817665. [DOI] [PubMed] [Google Scholar]
- Grafl B., Prokofieva I., Wernsdorf P., Steinborn R., Hess M. Infection with an apathogenic fowl adenovirus serotype-1 strain (CELO) prevents adenoviral gizzard erosion in broilers. Vet. Microbiol. 2014;172:177–185. doi: 10.1016/j.vetmic.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Günes A., Marek A., Grafl B., Berger E., Hess M. Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E) J. Virol. Methods. 2012;183:147–153. doi: 10.1016/j.jviromet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Hoff M. DNA amplification and detection made simple (Relatively) Plos Biol. 2006;4:e222. doi: 10.1371/journal.pbio.0040222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Kalsi S., Zeimpekis I., Sun K., Ashburn P., Turner C., Sutton J.M., Morgan H. Ultra-fast electronic detection of antimicrobial resistance genes using isothermal amplification and Thin Film Transistor sensors. Biosens. Bioelectron. 2017;96:281–287. doi: 10.1016/j.bios.2017.05.016. [DOI] [PubMed] [Google Scholar]
- James A.S., Todd S., Pollak N.M., Marsh G.A., Macdonald J. Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: preparing for the future. Virol. J. 2018;15:75. doi: 10.1186/s12985-018-0985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junnu S., Lertwatcharasarakul P., Jala S., Phattanakunanan S., Moonjit P., Songserm T. Developing an Indirect ELISA based on recombinant hexon protein for serological detection of inclusion body hepatitis in chickens. J. Vet. Med. Sci. 2014;76:289–293. doi: 10.1292/jvms.13-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaján G.L., Sameti S., Benko M. Partial sequence of the DNA-dependent DNA polymerase gene of fowl adenoviruses: a reference panel for a general diagnostic PCR in poultry. Acta Vet. Hung. 2011;59:279. doi: 10.1556/AVet.2011.006. [DOI] [PubMed] [Google Scholar]
- Li C., Li H., Wang D., Wang J., Wang Y., Wang S., Li J., Ping L., Wang J., Xu S. Characterization of fowl adenoviruses isolated between 2007 and 2014 in China. Vet. Microbiol. 2016;197:62–67. doi: 10.1016/j.vetmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Li H., Wang J., Qiu L., Han Z., Liu S. Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect. Genet. Evol. 2016;45:230–241. doi: 10.1016/j.meegid.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Li J., Macdonald J., von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- Li P.H., Zheng P.P., Zhang T.F., Wen G.Y., Shao H.B., Luo Q.P. Fowl adenovirus serotype 4: Epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poult. Sci. 2017;96:2630–2640. doi: 10.3382/ps/pex087. [DOI] [PubMed] [Google Scholar]
- Lim T.H., Kim B.Y., Kim M.S., Jang J.H., Lee D.H., Kwon Y.K., Lee J.B., Park S.Y., Choi I.S., Song C.S. Outbreak of gizzard erosion associated with fowl adenovirus infection in Korea. Poult. Sci. 2012;91:1113–1117. doi: 10.3382/ps.2011-02050. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang J., Zhang R., Lin M., Shi R., Han Q., Wang J., Yuan W. Visual and equipment-free reverse transcription recombinase polymerase amplification method for rapid detection of foot-and-mouth disease virus. BMC Vet. Res. 2018;14:263. doi: 10.1186/s12917-018-1594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Cong F., Zhu Y., Wu M., Xu F., Huang R., Moore R.J., Guo P. Development of a reverse transcription recombinase polymerase amplification assay for rapid detection of Theiler's murine encephalomyelitis virus. Mol. Cell. Probes. 2018;41:27–31. doi: 10.1016/j.mcp.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Mase M., Nakamura K. Phylogenetic analysis of fowl adenoviruses isolated from chickens with gizzard erosion in Japan. J. Vet. Med. Sci. 2014;76:1535–1538. doi: 10.1292/jvms.14-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri A., Prusas C., Voss M., Hess M. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol. 1998;27:269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- Niczyporuk J.S., Wozniakowski G., Samorek-Salamonowicz E. Application of cross-priming amplification (CPA) for detection of fowl adenovirus (FAdV) strains. Arch. Virol. 2015;160:1005–1113. doi: 10.1007/s00705-015-2355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Liu L., Wang Y., Zhang Y., Qi X., Liu C., Gao Y., Wang X., Cui H. The first whole genome sequence and pathogenicity characterization of a fowl adenovirus 4 isolated from ducks associated with inclusion body hepatitis and hydropericardium syndrome. Avian Pathol. 2017;46:571–578. doi: 10.1080/03079457.2017.1311006. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. Plos Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar R., Roy P. Recombinant hexon antigen based single serum dilution ELISA for rapid serological profiling against fowl adenovirus-4 causing hydropericardium syndrome in chickens. J. Virol. Methods. 2014;207:121–127. doi: 10.1016/j.jviromet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- Raue R., Hess M. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J. Virol. Methods. 1998;73:211–217. doi: 10.1016/s0166-0934(98)00065-2. [DOI] [PubMed] [Google Scholar]
- Schachner A., Matos M., Grafl B., Hess M. Fowl adenovirus-induced diseases and strategies for their control – a review on the current global situation. Avian Pathol. 2018;47:111. doi: 10.1080/03079457.2017.1385724. [DOI] [PubMed] [Google Scholar]
- Shao H., Lu Y., Wang W., Li T., Zhang J., Wan Z., Liang G., Gao W., Qin A., Ye J. Two novel monoclonal antibodies against fiber-1 protein of FAdV-4 and their application in detection of FAdV-4/10. BMC Vet. Res. 2019;15:232. doi: 10.1186/s12917-019-1987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K.K., Azizan N.S., Yaacob C.N., Che Mat Seri N.A.A., Samsudin N.I., Teoh B.T., Sam S.S., AbuBakar S. Operational utility of the reverse-transcription recombinase polymerase amplification for detection of dengue virus. BMC Infect. Dis. 2018;18:169. doi: 10.1186/s12879-018-3065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva Wand N.I., Bonney L.C., Watson R.J., Graham V., Hewson R. Point-of-care diagnostic assay for the detection of Zika virus using the recombinase polymerase amplification method. J. Gen. Virol. 2018;99:1012–1026. doi: 10.1099/jgv.0.001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Hernandez P.F., Morales-Garzon A., Cortes-Espinosa D.V., Galiote-Flores A., Garcia-Barrera L.J., Rodriguez-Galindo E.T., Toscano-Contreras A., Lucio-Decanini E., Absalon A.E. Clinicopathological characterization and genomic sequence differences observed in a highly virulent fowl Aviadenovirus serotype 4. Avian Pathol. 2016;45:73–81. doi: 10.1080/03079457.2015.1125443. [DOI] [PubMed] [Google Scholar]
- Wei Z., Liu H., Diao Y., Li X., Zhang S., Gao B., Tang Y., Hu J., Diao Y. Pathogenicity of fowl adenovirus (FAdV) serotype 4 strain SDJN in Taizhou geese. Avian Pathol. 2019;48:477–485. doi: 10.1080/03079457.2019.1625305. [DOI] [PubMed] [Google Scholar]
- Yang Y., Qin X., Zhang W., Li Z., Zhang S., Li Y., Zhang Z. Development of an isothermal recombinase polymerase amplification assay for rapid detection of pseudorabies virus. Mol. Cell. Probes. 2017;33:32–35. doi: 10.1016/j.mcp.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Yu X., Wang Z., Chen H., Niu X., Dou Y., Yang J., Tang Y., Diao Y. Serological and pathogenic Analyses of fowl adenovirus serotype 4 (FAdV-4) strain in Muscovy ducks. Front. Microbiol. 2018;9:1163. doi: 10.3389/fmicb.2018.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wang Y., Meng K., Zhang Y., Xu H., Ai W. LAMP real-time turbidity detection for fowl adenovirus. BMC Vet. Res. 2019;15:256. doi: 10.1186/s12917-019-2015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Mei X., Wu X., Zuo L., Zhou L., Tian Y., Han X., Yang X., Wang H. A loop-mediated isothermal amplification coupling with a lateral flow dipstick for rapid and specific detection of fowl adenovirus serotype-4. J. Virol. Methods. 2019;270:79–86. doi: 10.1016/j.jviromet.2019.04.026. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhong Q., Zhao Y., Hu Y.X., Zhang G.Z. Pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS One. 2015;10:e0133073. doi: 10.1371/journal.pone.0133073. [DOI] [PMC free article] [PubMed] [Google Scholar]