Given that aminoglycosides, such as amikacin, may be used for multidrug-resistant Pseudomonas aeruginosa infections, optimization of therapy is paramount for improved treatment outcomes. This study aims to investigate the pharmacodynamics of different simulated intravenous amikacin doses on susceptible P. aeruginosa to inform ventilator-associated pneumonia (VAP) and sepsis treatment choices. A hollow-fiber infection model with two P. aeruginosa isolates (MICs of 2 and 8 mg/liter) with an initial inoculum of ∼108 CFU/ml was used to test different amikacin dosing regimens.
KEYWORDS: amikacin, Pseudomonas aeruginosa, pharmacodynamics, ventilator-associated pneumonia
ABSTRACT
Given that aminoglycosides, such as amikacin, may be used for multidrug-resistant Pseudomonas aeruginosa infections, optimization of therapy is paramount for improved treatment outcomes. This study aims to investigate the pharmacodynamics of different simulated intravenous amikacin doses on susceptible P. aeruginosa to inform ventilator-associated pneumonia (VAP) and sepsis treatment choices. A hollow-fiber infection model with two P. aeruginosa isolates (MICs of 2 and 8 mg/liter) with an initial inoculum of ∼108 CFU/ml was used to test different amikacin dosing regimens. Three regimens (15, 25, and 50 mg/kg) were tested to simulate a blood exposure, while a 30 mg/kg regimen simulated the epithelial lining fluid (ELF) for potential respiratory tract infection. Data were described using a semimechanistic pharmacokinetic/pharmacodynamic (PK/PD) model. Whole-genome sequencing was used to identify mutations associated with resistance emergence. While bacterial density was reduced by >6 logs within the first 12 h in simulated blood exposures following this initial bacterial kill, there was amplification of a resistant subpopulation with ribosomal mutations that were likely mediating amikacin resistance. No appreciable bacterial killing occurred with subsequent doses. There was less (<5 log) bacterial killing in the simulated ELF exposure for either isolate tested. Simulation studies suggested that a dose of 30 and 50 mg/kg may provide maximal bacterial killing for bloodstream and VAP infections, respectively. Our results suggest that amikacin efficacy may be improved with the use of high-dose therapy to rapidly eliminate susceptible bacteria. Subsequent doses may have reduced efficacy given the rapid amplification of less-susceptible bacterial subpopulations with amikacin monotherapy.
TEXT
Sepsis or ventilator-associated pneumonia (VAP) caused by Pseudomonas aeruginosa is associated with a mortality of between 25 and 50% (1, 2). Furthermore, patients with carbapenem-resistant P. aeruginosa infections have an increased risk of death that may be attributed to increasing illness severity and delayed administration of appropriate antibiotic therapy (3–6). Despite a potential increased mortality with aminoglycoside monotherapy, at least 80% of P. aeruginosa isolates remain susceptible to aminoglycosides such as amikacin; therefore, they may be prescribed for empirical treatment as part of combination therapy to appropriately extend the spectrum of antibiotic activity in settings with increased resistance rates (3–6).
One potential contributing factor to the apparent reduced efficacy of aminoglycosides is suboptimal dosing. Achieving an aminoglycoside maximum concentration to MIC (Cmax/MIC) ratio of ≥10 or an area under the concentration-time curve to MIC (AUC/MIC) ratio of ≥150 reduces mortality and hastens symptom resolution (7, 8). Importantly, the risk of resistance emergence and potential treatment failure may be increased when bacteria are exposed to a Cmax /MIC of <6 (9). Moreover, in patients infected with carbapenem-resistant, aminoglycoside-susceptible Klebsiella pneumoniae, aminoglycosides have been associated with favorable outcomes, particularly when a therapeutic aminoglycoside exposure may be possible at the site of infection (bloodstream, vascular catheters, soft tissues, and/or urinary tract) (10).
Aminoglycoside dose optimization must also consider the potential effect of the bacterial inoculum, the immune response, and the potential toxicity of the dosing regimen. Approximately one-third of patients with VAP have a bacterial burden exceeding 108 CFU/ml (11, 12). Reducing this bacterial burden to <1 × 106 CFU/ml may enable rapid granulocyte-mediated bacterial clearance and enhance symptom resolution (11–13). These factors may be particularly important in patients with Gram-negative bacillary pneumonia for two reasons. First, amikacin penetration into the epithelial lining fluid (ELF), the site of infection, is only approximately 10% of the plasma Cmax (14). Second, there may be limited treatment options available for multidrug-resistant bacteria should aminoglycoside therapy fail.
The aims of this study were 2-fold. First, to describe and quantify the time course of bacterial killing and emergence of resistance of two P. aeruginosa clinical isolates using the dynamic in vitro hollow-fiber infection model (HFIM) and semimechanistic mathematical modeling. Second, to determine amikacin dosing regimens that may enhance bacterial killing in both the bloodstream and ELF.
RESULTS
In vitro susceptibility and mutational frequency studies.
The modal amikacin MIC for isolates CTAP23 and CTAP40 was 2 and 8 mg/liter, respectively. The mutation frequency for isolates CTAP23 and CTAP40 in the presence of 8 and 32 mg/liter of amikacin was 6.77 × 10−7 and 1.05 × 10−7, respectively.
Hollow-fiber infection model.
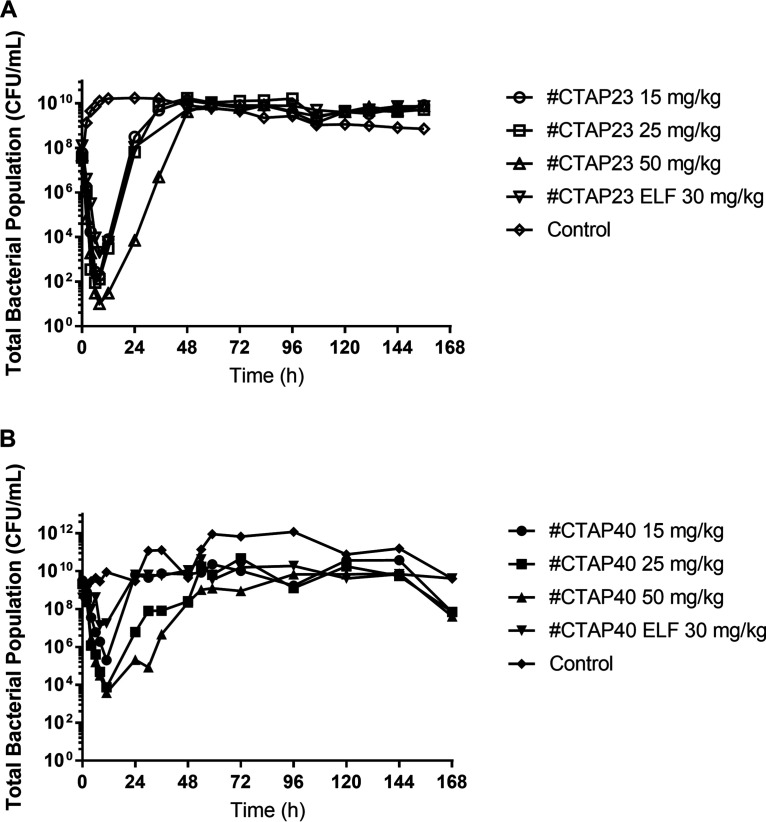
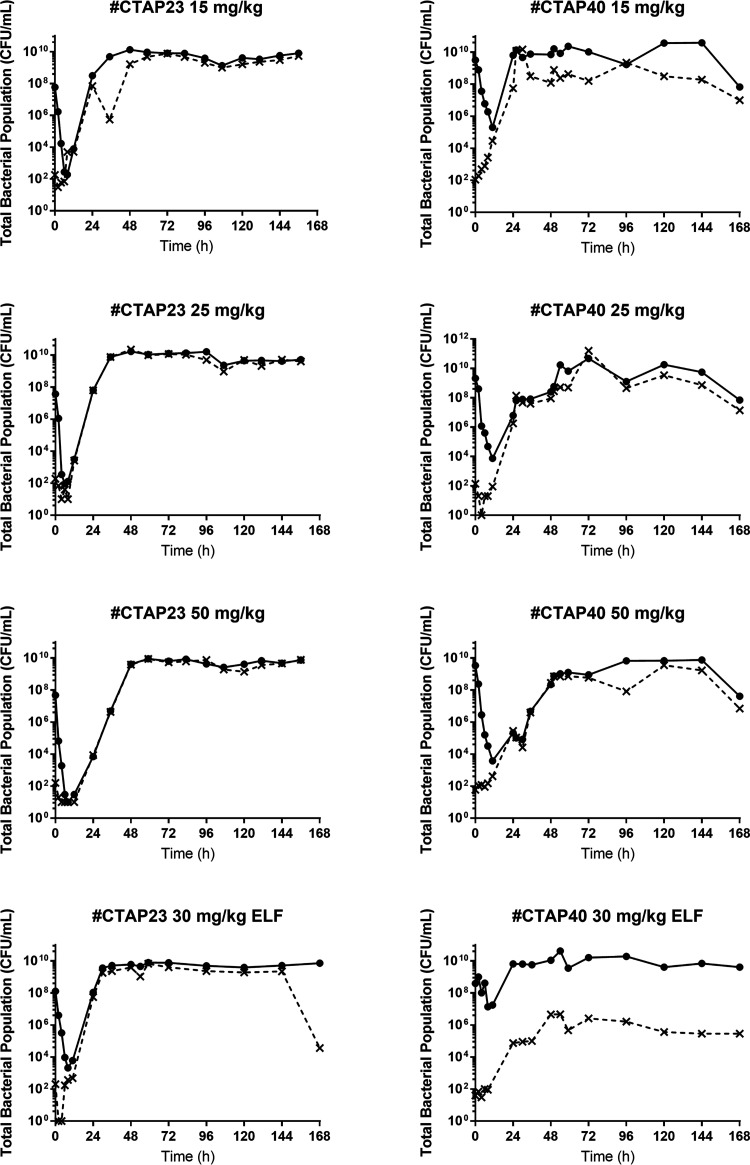
All intravenous amikacin dosing regimens against a simulated bloodstream P. aeruginosa infection resulted in a ≥4-log reduction from the starting inoculum (108 CFU/ml) during the first 8 h following the first dose of amikacin (Fig. 1). There was no appreciable difference in the rate or extent of bacterial killing between the 15, 25, and 50 mg/kg dosing regimens for isolate CTAP23 (MIC 2 mg/liter) (Fig. 1A). However, there was an approximate 1.5-log difference in the bacterial nadir between the 25 mg/kg and 50 mg/kg dosing regimens against isolate CTAP40 (MIC 8 mg/liter) (Fig. 1B). The total bacterial burden surpassed the baseline inoculum by 24 h for both isolates following administration of the 15 and 25 mg/kg dosing regimens. Only the 50 mg/kg dosing regimen for both isolates delayed the rate of bacterial regrowth, exceeding the baseline inoculum by 48 h (Fig. 1). Bacterial regrowth in the total population was mirrored by bacterial growth on amikacin-containing cation-adjusted Mueller-Hinton (CaMH) agar (Fig. 2). The MIC of the bacteria growing on amikacin-containing CaMH agar increased by a minimum of 8-fold after 7 days of amikacin administration for both isolates tested (Table 1).
FIG 1.
Total bacterial population for different amikacin dosing regimen in either blood or the epithelial lining fluid (ELF) over 168 h for isolate CTAP23 (A) and isolate CTAP40 (B).
FIG 2.
Total bacterial population (filled lines) and resistant population (dashed lines) for isolates CTAP23 and CTAP40 in blood (amikacin dosing regimens of 15, 25, and 50 mg/kg) and epithelial lining fluid (amikacin dosing regimen of 30 mg/kg).
TABLE 1.
Pseudomonas aeruginosa amikacin MICs from isolates grown on amikacin-containing (4× baseline MIC) cation-adjusted Mueller-Hinton agar after the 7-day course
| Isolate | Amikacin dose | MIC (mg/liter) |
|---|---|---|
| CTAP23 | 15 mg/kg | 32 |
| 25 mg/kg | 32 | |
| 50 mg/kg | 64 | |
| 30 mg/kg (ELF) | 16 | |
| CTAP40 | 15 mg/kg | 64 |
| 25 mg/kg | 128 | |
| 50 mg/kg | 128 | |
| 30 mg/kg (ELF) | 64 |
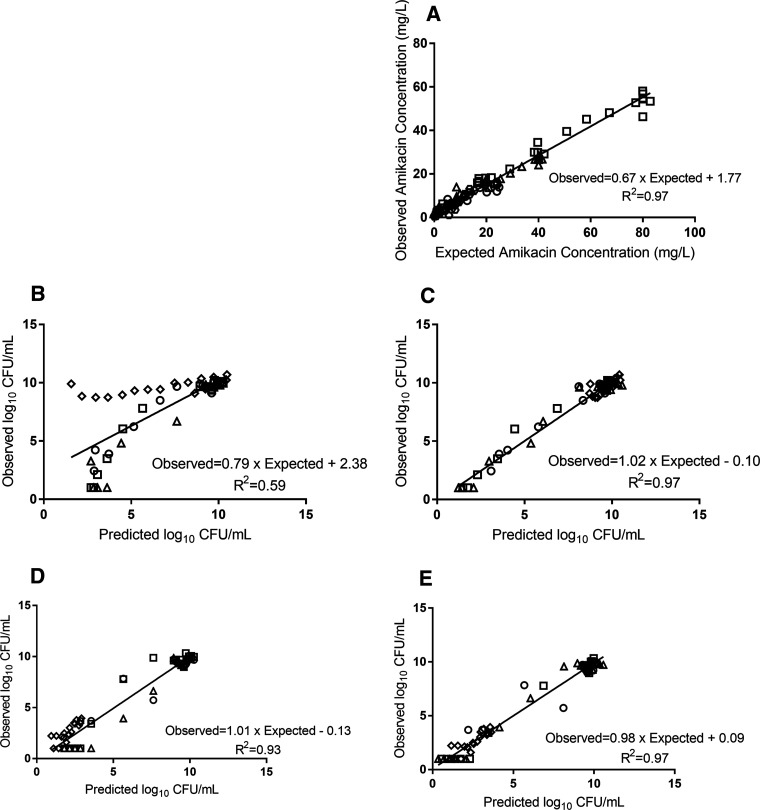
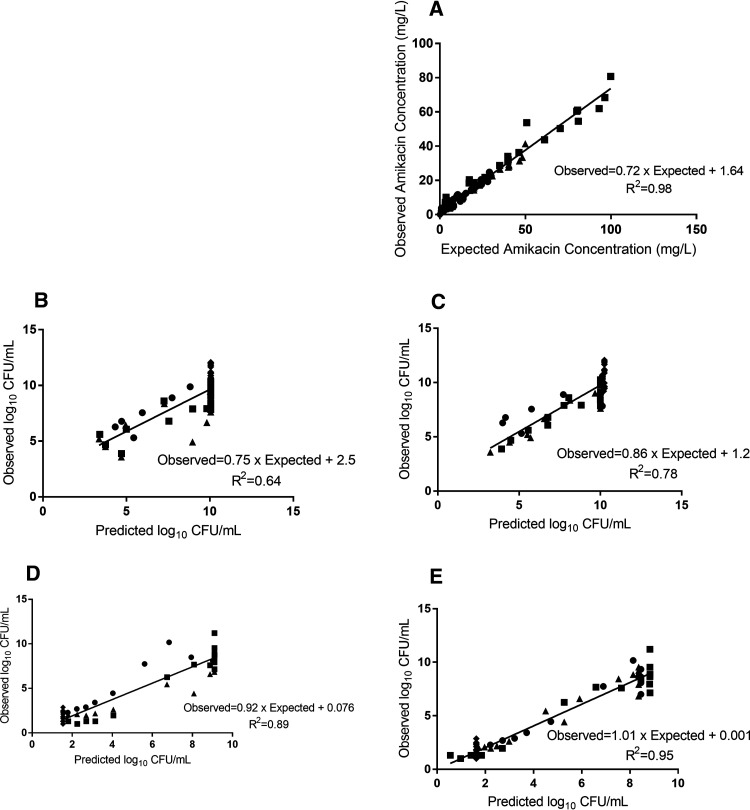
A similar pattern was observed against the simulated ELF exposure. The total bacterial population was reduced by approximately 5 logs at 8 h after the initiation of the amikacin against isolate CTAP23 (MIC 2 mg/liter), which was followed by rapid bacterial regrowth exceeding the baseline inoculum by 24 h, mirrored by growth on amikacin-containing CaMH agar (Fig. 1A; Fig. 2). Conversely, there was little appreciable bacterial killing against isolate CTAP40 (MIC 8 mg/liter), yet there was an increase in the growth on amikacin-containing CaMH agar (Fig. 2). There was no appreciable bacterial killing following subsequent dosing events after day 1 of amikacin in either the blood or ELF exposures in the HFIM. The observed amikacin concentrations for the simulated unbound plasma and ELF approximated the expected concentrations (graph A in Fig. 3 and 4).
FIG 3.
Pharmacokinetic/pharmacodynamic model observed-predicted fit for isolate CTAP23. (A) Amikacin pharmacokinetic data. (B and C) Total bacterial population observed versus predicted values for the population (B) and posterior (C) estimates. (D and E) Resistant bacterial population observed versus predicated values for the population (D) and posterior (E) estimates. Doses of 15 mg/kg (circles), 25 mg/kg (triangles), and 50 mg/kg (squares); ELF exposure (hexagons); and control (diamonds).
FIG 4.
Pharmacokinetic/pharmacodynamic model observed-predicted fit for isolate CTAP40. (A) Amikacin pharmacokinetic data. (B and C) Total bacterial population observed versus predicted values for the population (B) and posterior (C) estimates. (D and E) Resistant bacterial population observed versus predicated values for the population (D) and posterior (E) estimates. Doses of 15 mg/kg (circles), 25 mg/kg (triangles), and 50 mg/kg (squares); ELF exposure (hexagons); and control (diamonds).
Comparative genomic analysis.
There were no resistance genes or single nucleotide polymorphisms (SNPs) associated with amikacin resistance identified in the progenitor strains or isolates CTAP23 or CTAP40 prior to amikacin commencement. De novo SNPs within the fusA (FusALeu464Val) and rplB (RplBGly138Leu) genes were identified in isolates that were exposed to the 25 and 50 mg/kg daily dosing regimens, respectively, for isolate CTAP23 (Table 2). No SNPs were identified following exposure to amikacin at 15 mg/kg. SNPs were identified in the algA and tuf1 (Tuf1Val21Leu) genes for isolate CTAP40 following exposure to amikacin, with a small baseline bacterial subpopulation containing an algA (AlgAAla279Asp) SNP.
TABLE 2.
Variation identified in comparison to the initial starting strain in the isolates CTAP23 and CTAP40 lineages, where percentages reflect the prevalence of the mutation within the populationa
| Isolate and dose | fusA_2 1390 | rplB 413 | rplB 412 | algA_1 836 | tuf1_1 |
|---|---|---|---|---|---|
| Isolate CTAP23 | |||||
| Baseline | 0% | 0% | 0% | ND | ND |
| 15 mg/kg | 0% | 2% | 2% | ND | ND |
| 25 mg/kg | 57% | 4% | 4% | ND | ND |
| 50 mg/kg | 0% | 100% | 100% | ND | ND |
| Isolate CTAP40 | |||||
| Baseline | ND | ND | ND | 13% | 0% |
| 15 mg/kg | ND | ND | ND | 69% | 55% |
| 25 mg/kg | ND | ND | ND | 98% | 19% |
| 50 mg/kg | ND | ND | ND | 60% | 0% |
ND, not detected.
Pharmacokinetic/pharmacodynamic modeling.
Pharmacodynamic parameter estimates are detailed in Table 3. The average total bacterial population Bayesian posterior (model fitted estimate for each individual experimental arm) correlation coefficient (R2) was 0.97 and 0.78 for isolates CTAP23 (Fig. 3) and CTAP40 (Fig. 4) simulated blood exposures, respectively. Similar results were found for the resistant bacterial population (average Bayesian posterior R2 0.97 and 0.95 for isolates CTAP23 and CTAP40, respectively).
TABLE 3.
Pharmacodynamic model parameter estimatesa
| Parameter | Abbreviation | Mean CTAP23 (SD) | Mean CTAP40 (SD) |
|---|---|---|---|
| Susceptible growth rate constant (log10 CFU/ml/h) | Kgs | 1.31 (0.11) | 1.08 (0.20) |
| Intermediate growth rate constant (log10 CFU/ml/h) | Kgr | 0.40 (0.13) | 0.60 (0.26) |
| Resistant growth rate constant (log10 CFU/ml/h) | Kgrr | 0.69 (0.11) | 0.55 (0.13) |
| Central compartment HFIM vol (liters) | Vc | 0.32 (0.01) | 0.26 (0.05) |
| Amikacin clearance (liters/h) | Cl | 0.03 (0.00) | 0.03 (0.00) |
| Susceptible killing rate constant (log10 CFU/ml/h) | Emax,s | 5.34 (1.50) | 4.00 (3.01) |
| Intermediate killing rate constant (log10 CFU/ml/h) | Emax,r | 9.43 (3.19) | 11.20 (2.10) |
| Amikacin conc. causing 50% Emax,s (mg/liter) | EC50s | 11.61 (3.49) | 11.10 (2.53) |
| Amikacin conc. causing 50% Emax,r (mg/liter) | EC50r | 244.09 (149.73) | 349.63 (79.19) |
| Susceptible Hill coefficient | Hs | 6.00 (4.27) | 11.04 (5.80) |
| Resistant Hill coefficient | Hr | 3.42 (2.47) | 7.71 (2.61) |
| Intermediate population initial condition (CFU/ml) | ICRe | 211.05 (119.12) | 320.48 (50.57) |
| Resistant population initial condition (CFU/ml) | ICRRe | 29.46 (48.23) | 25.81 (16.70) |
| Maximum substrate consumption | Qmax | 0.81 (0.18) | 0.59 (0.29) |
| Maximum available substrate | Substrate | 3.33 × 1010 (2.22 × 1010) | 4.92 × 1010 (2.75 × 1010) |
| Substrate conc. causing 50% Qmax | Qs | 8.15 × 105 (9.61 × 104) | 5.3 × 105 (1.69 × 105) |
| Death rate constant susceptible population | Kds | 0.25 (0.15) | 0.05 (0.04) |
| Death rate constant intermediate population | Kdi | 0.24 (0.18) | 0.02 (0.03) |
| Death rate constant resistant population | Kdr | 0.03 (0.02) | 0.11 (0.32) |
The mean and standard deviation (SD) for each parameter and isolate were determined using the average and bootstrapped estimates, respectively, of the posterior model estimates for each dosing regimen.
Classification and regression tree (CART) analysis identified similar area under the concentration-time curve for the unbound fraction (fAUC) and Cmax of the unbound fraction (fCmax) thresholds for bacterial stasis for both isolates over 24 h, correlating with a difference in the fAUC/MIC and the fCmax/MIC ratio relative to the MIC of the isolate (Table 4). However, no threshold was associated with a bacterial kill in the bloodstream of 1 or 2 logs over 24 h for isolate CTAP23. Amikacin-simulated fAUC and fCmax ELF exposures were increased relative to plasma for the same bacterial kill over 24 h and was increased for isolate CTAP23 (MIC 2 mg/liter) compared with isolate CTAP40 (MIC 8 mg/liter). The probability of achieving bacterial stasis, 1- and 2-log kill after 24 h was generally high in the ELF and the bloodstream when doses of ≥30 mg/kg were used (Table 5).
TABLE 4.
Pharmacokinetic/pharmacodynamic exposures required for bacterial stasis, 1-log, and 2-log reduction in the total bacterial burden over 24 ha
| Isolate | Infection site | Exposure target | Stasis (log10 CFU/ml) | 1-log kill (log10 CFU/ml) | 2-log kill (log10 CFU/ml) |
|---|---|---|---|---|---|
| CTAP40 | Blood | fAUC | 108.81 | 124.70 | 174.95 |
| fAUC/MIC | 13.60 | 15.59 | 21.87 | ||
| fCmax | 24.73 | 25.86 | 27.15 | ||
| fCmax/MIC | 3.09 | 3.23 | 3.39 | ||
| ELF | fAUC | 328.21 | 342.69 | 366.42 | |
| fAUC/MIC | 41.03 | 42.84 | 45.80 | ||
| fCmax | 42.41 | 47.47 | 54.17 | ||
| fCmax/MIC | 5.30 | 5.93 | 6.77 | ||
| CTAP23 | Blood | fAUC | 117.54 | - | - |
| fAUC/MIC | 58.77 | ||||
| fCmax | 26.41 | - | - | ||
| fCmax/MIC | 13.21 | ||||
| ELF | fAUC | 342.92 | 688.54 | 688.82 | |
| fAUC/MIC | 171.46 | 344.27 | 344.1 | ||
| fCmax | 47.04 | 42.40 | 47.81 | ||
| fCmax/MIC | 23.52 | 21.20 | 23.91 |
-, no threshold was identified.
TABLE 5.
Probability of achieving either bacterial stasis, a 1-log reduction, or 2-log reduction in the total bacterial population within 24 h of commencing intravenous amikacin
| Isolate | Infection site | Dose in mg/kg | Renal function (ml/min) | Stasis (log10 CFU/ml) | 1-log kill (log10 CFU/ml) | 2-log kill (log10 CFU/ml) |
|---|---|---|---|---|---|---|
| CTAP40 | Blood | 15 | 60 | 1 | 1 | 1 |
| 30 | 60 | 1 | 1 | 1 | ||
| 50 | 60 | 1 | 1 | 1 | ||
| 15 | 100 | 1 | 0.99 | 0.89 | ||
| 30 | 100 | 1 | 1 | 1 | ||
| 50 | 100 | 1 | 1 | 1 | ||
| 15 | 140 | 0.90 | 0.52 | 0.16 | ||
| 30 | 140 | 1 | 1 | 1 | ||
| 50 | 140 | 1 | 1 | 1 | ||
| ELF | 15 | 60 | 0 | 0 | 0 | |
| 30 | 60 | 0.93 | 0.44 | 0.03 | ||
| 50 | 60 | 1 | 1 | 1 | ||
| 15 | 100 | 0 | 0 | 0 | ||
| 30 | 100 | 0.41 | 0.03 | 0 | ||
| 50 | 100 | 1 | 1 | 1 | ||
| 15 | 140 | 0 | 0 | 0 | ||
| 30 | 140 | 0.02 | 0 | 0 | ||
| 50 | 140 | 1 | 1 | 1 | ||
| CTAP23 | Blood | 15 | 60 | 1 | 0 | 0 |
| 30 | 60 | 1 | 0 | 0 | ||
| 50 | 60 | 1 | 0 | 0 | ||
| 15 | 100 | 0.99 | 0 | 0 | ||
| 30 | 100 | 1 | 0 | 0 | ||
| 50 | 100 | 1 | 0 | 0 | ||
| 15 | 140 | 0.69 | 0 | 0 | ||
| 30 | 140 | 1 | 0 | 0 | ||
| 50 | 140 | 1 | 0 | 0 | ||
| ELF | 15 | 60 | 0 | 0 | 0 | |
| 30 | 60 | 0.57 | 0 | 0 | ||
| 50 | 60 | 1 | 0 | 0 | ||
| 15 | 100 | 0 | 0 | 0 | ||
| 30 | 100 | 0.06 | 0 | 0 | ||
| 50 | 100 | 1 | 0 | 0 | ||
| 15 | 140 | 0 | 0 | 0 | ||
| 30 | 140 | 0 | 0 | 0 | ||
| 50 | 140 | 1 | 0 | 0 |
DISCUSSION
This study investigated the bacterial killing and emergence of resistance of two susceptible P. aeruginosa isolates exposed to the expected pharmacokinetics of amikacin in blood and ELF. Following an initial bacterial kill of ≥4 logs within the first 8 h, there was extensive bacterial regrowth for both isolates, with negligible bacterial killing following the first dose. Our results support the current European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendation that aminoglycosides may be considered for short-term use in combination with another agent until the antibiotic susceptibilities are confirmed and that aminoglycoside dose optimization may enhance bacterial killing and enhance clinical outcomes (15).
In the current study, achieving a blood and ELF amikacin fAUC exposure of approximately >175 (fAUC/MIC >21.87) and >366 mg · h/liter (fAUC/MIC >45.8), respectively, may be sufficient to reduce the bacterial burden of some P. aeruginosa isolates to <106 CFU/ml over 24 h. Such an exposure correlates to an amikacin dose of ≥30 mg/kg or ≥50 mg/kg daily for bloodstream or VAP infections, respectively, with susceptible P. aeruginosa pathogens in patients with normal creatinine clearance (∼100 ml/min). However, this threshold may also vary between bacterial isolates, as the total bacterial burden within the first 12 h appears to be, in part, mediated by reducing the burden of the susceptible and intermediate-susceptibility bacterial populations by achieving the appropriate fAUC/MIC and/or fCmax/MIC. Thereafter, a resistant bacterial population against which amikacin has no effect may emerge. The emergence of resistance is likely dependent on the relative density of the intermediate/resistant subpopulation(s) in the initial total bacterial inoculum (the mutation frequency) and the propensity for mutations to occur that mediate emergence of resistance (16). At the high inoculum used in our study, it was expected based on the mutation frequency that a resistant subpopulation existed, which was subsequently amplified following amikacin administration. This may explain the differences in the identified thresholds for a 1- or 2-log reduction between the susceptible isolates used in this study, given that the relative susceptible/intermediate-resistant bacterial populations may differ between isolates. Our results are similar to that previously described against P. aeruginosa where a simulated gentamicin plasma with a Cmax/MIC ratio of ≥36 was unable to suppress bacterial regrowth in vitro (9). However, against Acinetobacter baumannii, an amikacin Cmax/MIC ratio of 20 suppressed bacterial regrowth, highlighting the variability in response to aminoglycoside exposure that may be, in part, determined by the inoculum size and preexisting intermediate-resistant subpopulations.
Higher ELF amikacin fAUC and fCmax exposures were required to achieve stasis, 1 and 2 logs of bacterial killing over 24 h, which may be related to the delayed and lower fCmax achieved in the ELF relative to the plasma amikacin concentrations following intravenous administration, given the expected pharmacokinetic hysteresis between the bloodstream and ELF. Moreover, there was little bacterial killing against isolate CTAP40 (MIC 8 mg/liter) following a simulated intravenous 30 mg/kg dose (Fig. 2), suggesting that amikacin monotherapy will have little efficacy against higher-MIC isolates.
The identified PK/PD targets identified in our study differ to those observed in clinical studies. A previous clinical study in critically ill patients receiving intravenous amikacin demonstrated an increased chance of microbial eradication and clinical cure in patients who achieved a Cmax/MIC of >10 (9). A separate study identified a fAUC/MIC of ≥150 mg · h/liter correlated with faster symptom resolution in patients with nosocomial pneumonia (7, 8). The identified PK/PD ratios from our simulations in this study and clinical studies may be challenging to achieve with doses of <30 mg/kg (17, 18). As such, high-dose amikacin therapy (>30 mg/kg) may be considered. Limited clinical data exist for such dosing regimens, but doses of ≥60 mg/kg have been used as part of salvage therapy in conjunction with renal replacement therapy to minimize the probability of toxicity in a small case series (19). Furthermore, the use of a single dose of amikacin in patients with severe sepsis or septic shock may mitigate the risk of nephrotoxicity, which is unlikely to occur for an aminoglycoside duration of <3 days (20). Nevertheless, the use of such high doses would place the patient within an amikacin fAUC exposure that has previously been associated with a significant probability of developing nephrotoxicity; however, this is confounded by the different aminoglycosides used and a prolonged treatment duration (21). This approach should be evaluated in a clinical trial to ensure that both the target PK/PD exposures are met and to assess the potential clinical utility of high dose, short duration therapy in terms of patient morbidity and mortality.
Despite the achievement of these targets, resistance may still emerge with amikacin monotherapy. Amikacin resistance was identified for both isolates receiving doses up to 50 mg/kg within 48 h of amikacin initiation. Mutations affecting the ribosomal binding unit (RplBGly138Leu), elongation factors (FusALeu464Val and Tuf1Val21Leu), and mucoidal phenotype (AlgAAla279Asp) appear to mediate this resistance, which is consistent with a previous study with tobramycin with similar SNPs within the rplB and fusA genes that likely inhibit aminoglycoside binding to the 30S ribosomal subunit (22). The relevance of the AlgA mutant is not currently known; however, alteration of alginate production may modify biofilm formation, a known potentiator of antibiotic resistance emergence (23). These mutations were associated with an increased MIC; however, the relative MIC increase was similar following each dosing regimen. Furthermore, a specific mutation was not often consistently identified for all resistant bacterial populations following a specific amikacin dosing regimen. This would suggest that there are either multiple smaller subpopulations or that alternative resistance mechanisms, such as amikacin efflux, exist (24). Nonetheless, given the likely de novo emergence of resistance, it is unlikely that subsequent amikacin doses will achieve appreciable further bacterial killing (24). These results would support the notion that amikacin may enhance initial bacterial killing but should be combined with a second agent either empirically or as directed therapy to ensure bacterial eradication and minimize the probability of treatment failure.
Our study is not without limitations. First, the lack of a simulated immune response in vitro limits the external validity when applying our results to clinical practice. Nonetheless, as previously discussed, optimizing bacterial killing in vitro may generalize to optimal clinical outcomes (25). Moreover, our in vitro model and subsequent dosing simulations may best represent an immunocompromised patient. Second, only two clinical P. aeruginosa isolates were tested, therefore our results may not generalize to other infecting isolates. Third, the amikacin ELF concentration-time curve is estimated from other aminoglycosides, which may not reflect the exposures achieved for amikacin. This approach may be reasonable given the lack of amikacin-specific data and similar chemical structures between aminoglycosides. Nonetheless, further research detailing the ELF pharmacokinetics of amikacin over a dosing interval are required; thus our results should be considered useful for generating hypotheses. Fifth, we did not perform whole-genome sequencing (WGS) on the various phenotypically distinct colonies. This may mean that specific resistance mechanisms may not be appropriately identified if they are present in a sparsely represented bacterial subpopulation. Last, we did not simulate the ELF milieu, which is known to contain mucin and an acidic pH, factors that are known to impact aminoglycoside-mediated bacterial killing (26–29). The impact of mucin was considered by simulating the estimated unbound amikacin fraction.
Future amikacin intravenous administration may be with the use of a single high dose (≥30 mg/kg) of the antibiotic for patients with either bloodstream infections or VAP from multiresistant pathogens, such as P. aeruginosa, to improve the probability of bacterial eradication. However, this must be balanced with ongoing review of the amikacin doses required for clinical effectiveness against P. aeruginosa, where doses may result in unacceptable toxicity and combinations with other active antipseudomonal agents are preferred. Given the likely low efficacy of bacterial killing in the ELF following intravenous administration, alternate amikacin administration routes, such as nebulized therapy, may be considered. Clinical trials are required to define the optimal dosing regimen of amikacin for difficult-to-treat infections, such as VAP.
MATERIALS AND METHODS
Antimicrobial agents.
Amikacin analytical reference standards (Sigma-Aldrich, batch LRAA5755) were used for in vitro MIC susceptibility testing and preparing amikacin-containing CaMH agar plates. Commercially available amikacin vials (DBL amikacin sulfate 500 mg/2 ml, batch CO61221AA) stored at 4°C were used for hollow-fiber infection model (HFIM) dosing. Amikacin stock solutions were aseptically prepared in a class II biosafety cabinet by diluting amikacin with sterile distilled water and storing at –80°C.
Bacterial isolates.
Two clinical P. aeruginosa isolates (CTAP40 and CTAP23) were sourced from critically ill patients. Isolates were stored in CaMH broth with 20% glycerol vol/vol at –80°C and were grown on CaMH agar and incubated at 37°C for 24 h prior to in vitro susceptibility testing and HFIM studies. A 0.5 McFarland bacterial suspension was prepared in sterile water using morphologically similar colonies and diluted in CaMH broth to the desired inoculum. For HFIM studies, bacteria were suspended in 40 ml of CaMH broth and incubated at 37°C with constant agitation for a duration of time based on previous growth curves to achieve a final inoculum of approximately 108 CFU/ml.
In vitro susceptibility testing.
Broth microdilution was performed in accordance with Clinical & Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (30, 31). Briefly, a volume of bacteria suspended in CaMH broth (final inoculum 5.5 × 105 CFU/ml) was added to a 96-well flat- or round-bottom plate containing serial 2-fold dilutions of amikacin in CaMH broth. Inoculated 96-well plates were incubated for 16 to 24 h at 37°C. Round-bottom plates were visually inspected for growth; the lowest amikacin concentration with no apparent growth was defined as the MIC. The MIC for the flat-bottom plates was determined using a Multiskan FC microplate photometer (Thermo Fisher Scientific, Finland), and defined as the concentration with an optical density (OD) ratio of <0.1 of the growth control. The modal MIC of four replicates within an individual experiment for each method (CLSI and EUCAST) was selected as the isolate MIC.
Mutation frequency.
A 10-ml culture of a 102 CFU/ml inoculum was incubated in CaMH broth for 24 h at 37°C. Quantitative culturing methods with diluted and undiluted samples were performed on the resultant bacterial growth using both standard CaMH agar and amikacin-containing CaMH agar (4-fold baseline MIC). The mutation frequency was taken as the ratio of the bacterial concentration growing on amikacin-containing plates to the initial inoculum after incubating for 48 h at 37°C.
Hollow-fiber infection model.
The HFIM was assembled as described previously using FiberCell Systems polysulfone cartridges (C2011) in all experiments and conducted over 7 days (32, 33). One HFIM experiment was conducted for each dosing regimen and isolate combination with an initial bacterial concentration of 1 × 108 CFU/ml.
Unbound amikacin blood exposures were simulated using the pharmacokinetic model derived by Romano et al., assuming an 80-kg patient with sepsis, a creatinine clearance of 100 ml/min, and 17% protein binding (34, 35). Amikacin dosing regimens of 15, 25, and 50 mg/kg once daily infused over 30 min were tested. High 50 mg/kg doses were also tested given that these doses have been previously used clinically (36). The ELF amikacin concentrations and resultant half-life in the HFIM apparatus were approximated using previous aminoglycoside ELF:serum ratios in conjunction with the established concentration-time curves for the blood amikacin exposure (14, 37, 38). In brief, the estimated unbound plasma concentration of amikacin was multiplied by the average ELF:serum penetration ratio (0.12, 0.3, 0.85, and 1.14) identified for other aminoglycosides (gentamicin and tobramycin) at the corresponding time points (0.5, 1, 2, and 4 h) (14, 37, 38). The ELF half-life (1.92 h) was derived from a noncompartmental analysis of the resultant concentration-time curve over the course of 24 h, which approximates that identified previously (39, 40). A mucin-bound fraction of 50% was assumed, representing a likely worst-case scenario (26). An ELF amikacin exposure following an intravenous dose of 30 mg/kg once daily administered over 30 min was simulated.
Samples were periodically removed from the central compartment outlet at 0.25, 0.5, 0.45, 1, 2, 3, 4, 6, 8, 10, 12, 24, 25, 30, 36, 48, 49, 54, 60, 72, 73, 78, 84, 96, 120, 144, 145, and 156 h to determine the amikacin concentration for pharmacokinetic analysis. As the central compartment contents rapidly equilibrate with the hollow-fiber cartridge, the concentrations obtained in the central compartment reflect that in the hollow-fiber cartridge. Bacterial quantification was performed with periodic sampling at 0, 2, 4, 6, 8, 11, 24, 35, 48, 59, 72, 96, 120, 144, and up to 168 h from the cartridge extracapillary space. Samples were washed twice in phosphate-buffered saline to minimize antibiotic carry-over. A 100-μl aliquot of an appropriately diluted bacterial suspension was manually plated onto CaMH agar and amikacin-containing CaMH agar (4-fold baseline isolate MIC). The limit of quantification was 2-log10 CFU/ml.
Drug assay.
Amikacin was measured in CaMH broth by a validated liquid chromatography-mass spectrometry (LC-MS) method. Briefly, 50 μl of a CaMH broth sample (neat or diluted) was combined with 50 μl of water and 20 μl of vancomycin (50 mg/liter) added as the internal standard. Amikacin was extracted using protein precipitation with 50 μl of trichloroacetic acid (15%, vol/vol). Samples were centrifuged at 12,000 × g for 5 min and an aliquot of the supernatant (0.5 μl) was injected onto a Nexera2 UHPLC system coupled to an 8030+ triple quadrupole MS detector (Shimadzu, Kyoto, Japan). Chromatographic separation was achieved using a Poroshell 120 HILIC column (Agilent, Santa Clara, USA) and a gradient of formic acid (0.2% [vol/vol]) and acetonitrile with 0.2% formic acid (vol/vol). Detection of amikacin and the internal standard was performed using an electrospray source in positive mode with optimized multiple reaction monitoring conditions for each analyte. Amikacin was monitored at three fragmentation ions (586.25 → 163.10, 586.25 → 264.15, and 586.25 → 425.15) and vancomycin was monitored at two fragmentation ions (725.60 →144.10 and 746.10 → 144.20).
Calibration lines of amikacin were quadratic with 1/concentration2 weighting from 0.2 to 10 mg/liter with a maximum deviation from the nominal concentration of 2.1%. Mean intrabatch accuracy and precision values were −6.2% and 8.3% at 0.8 and 8 mg/liter, respectively.
Whole-genome sequencing.
Bacterial isolates for whole-genome sequencing were subcultured onto amikacin-containing (4× baseline MIC) CaMH agar, as the resistant bacterial population profile may be transient without the presence of amikacin. Bacterial DNA was extracted without single colony purification to capture population diversity using the DNeasy UltraClean DNA extraction kit in accordance with the manufacturer’s directions and quantified using spectrophotometry (NanoDrop; Thermo Fisher) and fluorometry (Qubit; Thermo Fisher). Paired-end DNA libraries were prepared using the Nextera kit (Illumina; Australia) in accordance with the manufacturer’s directions. Sequencing was performed using the Illumina Mini-Seq (150-bp paired ends). Improved draft genome assemblies were constructed for the two progenitor strains, isolates CTAP23 and CTAP40, using the microbial genome assembler pipeline (MGAP v1.1) (41), and annotated using Prokka v1.12 (42). The comparative genomics pipeline, SPANDx v3.2.1 (43), was used to determine genomic variation using either the isolate CTAP23 or isolate CTAP40 as the reference genome, depending on the lineage analyzed. Within-species mixtures were analyzed using the GATK v4.1.0.0 (44) to identify mutations with less than 100% allele frequency using the method outlined in Aziz et al. (45).
Mathematical pharmacokinetic/pharmacodynamic modeling.
All HFIM data from simulated bloodstream exposures were comodeled using Pmetrics for R version 1.5.2 considering the results of the whole-genome sequencing study (46, 47). The final structural model is described by equations 1 to 5 that describe amikacin pharmacokinetics and bacterial growth of three subpopulations. Model diagnostics, including the Akaike-information-criteria, log-likelihood, coefficient of determination (R2) from the observed versus expected plots, and visual-predictive-checks were used to evaluate and compare models.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Equation 1 describes amikacin elimination. Equations 2, 3, and 4 describe the bacterial growth, including the theoretical maximal bacterial density and amikacin-mediated killing of the susceptible, intermediate and resistant bacterial populations, respectively. Equation 5 describes the consumption of an artificial substrate (Sub) required for sustained bacterial growth.
Amk, amount of amikacin (mg); R(1), amikacin infusion rate (mg/h); CL, amikacin clearance; Vc HFIM, circuit volume; CFUs, CFUi, and CFUr represent the bacterial burden for the susceptible, intermediate, and resistant P. aeruginosa subpopulations, respectively (CFU/ml); Kgmax,s, Kgmax,i, Kgmax,r, maximal growth rate constants for the susceptible, intermediate, and resistant P. aeruginosa subpopulations, respectively (log10 CFU/ml/h); Kkillmax,s and Kkillmax,i, the maximum rate of amikacin-mediated bacterial killing (log10 CFU/ml/h); Kds, Kdi, and Kdr, intrinsic bacterial death rate constants for the susceptible, intermediate, and resistant subpopulations, respectively (log10 CFU/ml/h); EC50s and EC50i amikacin concentration producing half-maximal bacterial killing for the susceptible and intermediate subpopulations, respectively; Sub, amount of a fictitious substance required for bacterial growth; Qmax, maximum rate of substance use; Qs, 50% of maximal substance use; Hs and Hi, slope functions for the susceptible and intermediate subpopulations, respectively.
Monte Carlo dosing simulation studies (n = 1,000) were performed using Pmetrics. Mean pharmacokinetic parameter estimates, as well as standard deviations of the clearance and volume of distribution, were obtained from the study conducted by Romano et al. (34) and applied to the simulations for the pharmacodynamic model. Mean value pharmacodynamic model parameters were estimated for specific isolates and were used for simulations. Moreover, different creatinine clearance values were used to describe patients with low, normal, and high renal amikacin clearance. The fAUC within the first 24 h was calculated employing Pmetrics, which included both the period of infusion and the monoexponential decay. Classification and regression tree analyses (CART) were used to determine the amikacin fAUC (mg · h/liter) achieving stasis, 1-log, and 2-log reduction in the bacterial concentration within the first 24 h.
ACKNOWLEDGMENTS
A.J.H. would like to acknowledge funding from a Griffith School of Medicine Research Higher degree scholarship. F.B.S. would like to acknowledge funding from a University of Queensland postdoctoral fellowship. J.A.R. would like to recognize funding from the Australian National Health and Medical Research Council for a Centre of Research Excellence (APP1099452) and a Practitioner Fellowship (APP1117065) grant. D.S.S. is funded by an Advanced Queensland Fellowship (AQRF13016‐17RD2).
We thank Hanna Sidjabat, Centre for Clinical Research, University of Queensland, for kindly providing the isolates used for this study.
REFERENCES
- 1.Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG Jr, van Duin D. 2017. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by Pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother 61:e02671-16. doi: 10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumbarello M, De Pascale G, Trecarichi EM, Spanu T, Antonicelli F, Maviglia R, Pennisi MA, Bello G, Antonelli M. 2013. Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive Care Med 39:682–692. 2013 doi: 10.1007/s00134-013-2828-9. [DOI] [PubMed] [Google Scholar]
- 3.Britt NS, Ritchie DJ, Kollef MH, Burnham CA, Durkin MJ, Hampton NB, Micek ST. 2018. Importance of site of infection and antibiotic selection in the treatment of carbapenem-resistant Pseudomonas aeruginosa sepsis. Antimicrob Agents Chemother 62:e02400-17. doi: 10.1128/AAC.02400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buehrle DJ, Shields RK, Clarke LG, Potoski BA, Clancy CJ, Hong Nguyen M. 2017. Carbapenem-resistant Pseudomonas aeruginosa bacteremia: risk factors for mortality and microbiologic treatment failure. Antimicrob Agents Chemother 61:e01243-16. doi: 10.1128/AAC.01243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis 78:443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Zilberberg MD, Shorr AF, Micek ST, Vazquez-Guillamet C, Kollef MH. 2014. Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit Care 18:596. doi: 10.1186/s13054-014-0596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore RD, Lietman PS, Smith CR. 1987. Clinical response to aminoglycoside therapy: importance of the ratio of peak concentration to minimal inhibitory concentration. J Infect Dis 155:93–99. doi: 10.1093/infdis/155.1.93. [DOI] [PubMed] [Google Scholar]
- 8.Kashuba AD, Nafziger AN, Drusano GL, Bertino JS. 1999. Optimizing aminoglycoside therapy for nosocomial pneumonia caused by Gram-negative bacteria. Antimicrob Agents Chemother 43:623–629. doi: 10.1128/AAC.43.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tam VH, Ledesma KR, Vo G, Kabbara S, Lim T, Nikolaou M. 2008. Pharmacodynamic modeling of aminoglycosides against Pseudomonas aeruginosa and Acinetobacter baumannii: identifying dosing regimens to suppress resistance development. Antimicrob Agents Chemother 52:3987–3993. doi: 10.1128/AAC.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields RK, Clancy CJ, Press EG, Hong Nguyen M. 2016. Aminoglycosides for treatment of bacteremia due to carbapenem-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3187–3192. doi: 10.1128/AAC.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drusano GL, Liu W, Fikes S, Cirz R, Robbins N, Kurhanewicz S, Rodriquez J, Brown D, Baluya D, Louie A. 2014. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210:1319–1324. doi: 10.1093/infdis/jiu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drusano GL, Fregeau C, Liu W, Brown DL, Louie A. 2010. Impact of burden on granulocyte clearance of bacteria in a mouse thigh infection model. Antimicrob Agents Chemother 54:4368–4372. doi: 10.1128/AAC.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opota O, Croxatto A, Prod'hom G, Greub G. 2015. Blood culture-based diagnosis of bacteraemia: state of the art. Clin Microbiol Infect 21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Najmeddin F, Shahrami B, Azadbakht S, Dianatkhah M, Rouini MR, Najafi A, Ahmadi A, Sharifnia H, Mojtahedzadeh M. 2020. Evaluation of epithelial lining fluid concentration of amikacin in critically ill patients with ventilator-associated pneumonia. J Intensive Care Med 35:400–404. doi: 10.1177/0885066618754784. [DOI] [PubMed] [Google Scholar]
- 15.European Committee for Antimicrobial Susceptibility Testing. 2020. Guidance document on implementation and use of the revised aminoglycoside breakpoints. https://eucast.org/ast_of_bacteria/guidance_documents/.
- 16.Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roger C, Nucci B, Louart B, Friggeri A, Knani H, Evrard A, Lavigne J, Allaouchiche B, Lefrant J, Roberts JA, Muller L. 2016. Impact of 30 mg/kg amikacin and 8 mg/kg gentamicin on serum concentrations in critically ill patients with severe sepsis. J Antimicrob Chemother 71:208–212. doi: 10.1093/jac/dkv291. [DOI] [PubMed] [Google Scholar]
- 18.Roger C, Nucci B, Molinari N, Bastide S, Saissi G, Pradel G, Barbar S, Aubert C, Lloret S, Elotmani L, Polge A, Lefrant J, Roberts JA, Muller L. 2015. Standard dosing of amikacin and gentamicin in critically ill patients results in variable and subtherapeutic concentrations. Int J Antimicrob Agents 46:21–27. doi: 10.1016/j.ijantimicag.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Brasseur A, Hites M, Roisin S, Cotton F, Vincent J-L, De Backer D, Jacobs F, Taccone FS. 2016. A high-dose aminoglycoside regimen combined with renal replacement therapy for the treatment of MDR pathogens: a proof-of-concept study. J Antimicrob Chemother 71:1386–1394. doi: 10.1093/jac/dkv491. [DOI] [PubMed] [Google Scholar]
- 20.Picard W, Bazin F, Clouzeau B, Bui H, Soulat M, Guilhon E, Vargas F, Hilbert G, Bouchet S, Gruson D, Moore N, Boyer A. 2014. Propensity-based study of aminoglycoside nephrotoxicity in patients with severe sepsis or septic shock. Antimicrob Agents Chemother 58:7468–7474. doi: 10.1128/AAC.03750-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drusano GL, Ambrose PG, Bhavnani SM, Bertino JS, Nafziger AN, Louie A. 2007. Back to the future: using aminoglycosides again and how to dose them optimally. Clin Infect Dis 45:753–760. doi: 10.1086/520991. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Jonker MJ, Moustakas I, Brul S, Ter Kuile BH. 2016. Dynamics of mutations during development of resistance by Pseudomonas aeruginosa against five antibiotics. Antimicrob Agents Chemother 60:4229–4236. doi: 10.1128/AAC.00434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Høiby N. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob Agents Chemother 48:1175–1187. doi: 10.1128/aac.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barclay ML, Begg EJ, Chambers ST, Thornley PE, Pattemore PK, Grimwood K. 1996. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J Antimicrob Chemother 37:1155–1164. doi: 10.1093/jac/37.6.1155. [DOI] [PubMed] [Google Scholar]
- 25.Gumbo T, Pasipanodya JG, Romero K, Hanna D, Nuermberger E. 2015. Forecasting accuracy of the hollow fiber model of tuberculosis for clinical therapeutic outcomes. Clin Infect Dis 61:S25–S31. doi: 10.1093/cid/civ427. [DOI] [PubMed] [Google Scholar]
- 26.Huang JX, Blaskovich MA, Pelingon R, Ramu S, Kavanagh A, Elliott AG, Butler MS, Montgomery AB, Cooper MA. 2015. Mucin binding reduces colistin antimicrobial activity. Antimicrob Agents Chemother 59:5925–5931. doi: 10.1128/AAC.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bataillon V, Lhermitte M, Lafitte JJ, Pommery J, Roussel P. 1992. The binding of amikacin to macromolecules from the sputum of patients suffering from respiratory diseases. J Antimicrob Chemother 29:499–508. doi: 10.1093/jac/29.5.499. [DOI] [PubMed] [Google Scholar]
- 28.van 't Veen A, Mouton JW, Gommers D, Kluytmans JA, Dekkers P, Lachmann B. 1995. Influence of pulmonary surfactant on in vitro bactericidal activities of amoxicillin, ceftazidime, and tobramycin. Antimicrob Agents Chemother 39:329–333. doi: 10.1128/aac.39.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodem CR, Lampton LM, Miller DP, Tarka EF, Everett ED ED. 1983. Endobronchial pH. Relevance of aminoglycoside activity in Gram-negative bacillary pneumonia. Am Rev Respir Dis 127:39–41. doi: 10.1164/arrd.1983.127.1.39. [DOI] [PubMed] [Google Scholar]
- 30.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 31.European Committee for Antimicrobial Susceptibility Testing. 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect doi: 10.1046/j.1469-0691.2003.00790.x. [DOI] [Google Scholar]
- 32.Cadwell J. 2012. The hollow fiber infection model for antimicrobial pharmacodynamics and pharmacokinetics. Adv in Pharmacoepidem & Drug Safety doi: 10.4172/2167-1052.S1-007. [DOI] [Google Scholar]
- 33.Cadwell J. 2015. The hollow fiber infection model: principles and practice. Advances in Antibiotics and Antibodies 1:101–106. [Google Scholar]
- 34.Romano S, de Gatta MDF, Calvo V, Mendez E, Domínguez-Gil A, Lanao JM. 1998. Influence of clinical diagnosis in the population pharmacokinetics of amikacin in intensive care unit patients. Clin Drug Invest 15:435–444. doi: 10.2165/00044011-199815050-00008. [DOI] [PubMed] [Google Scholar]
- 35.Brunnemann SR, Segal JL. 1991. Amikacin serum protein binding in spinal-cord injury. Life Sci 49:PL1–PL5. doi: 10.1016/0024-3205(91)90030-F. [DOI] [PubMed] [Google Scholar]
- 36.Layeux B, Taccone FS, Fagnoul D, Vincent JL, Jacobs F. 2010. Amikacin monotherapy for sepsis caused by panresistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:4939–4941. doi: 10.1128/AAC.00441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panidis D, Markantonis SL, Boutzouka M, Karatzas S, Baltopoulos G. 2005. Penetration of gentamicin into the alveolar lining fluid of critically ill patients with ventilator-associated pneumonia. Chest 128:545–552. doi: 10.1378/chest.128.2.545. [DOI] [PubMed] [Google Scholar]
- 38.Boselli E, Breilh D, Djabarouti S, Guillaume C, Rimmelé T, Gordien J, Xuereb F, Saux M, Allaouchiche B. 2007. Reliability of mini-bronchoalveolar lavage for the measurement of epithelial lining fluid concentrations of tobramycin in critically ill patients. Intensive Care Med 33:1519–1523. doi: 10.1007/s00134-007-0688-x. [DOI] [PubMed] [Google Scholar]
- 39.Bowker KE, Noel AR, Tomaselli S, Attwood M, MacGowan AP. 2018. Pharmacodynamics of inhaled amikacin (BAY 41–6551) studied in an in vitro pharmacokinetic model of infection. J Antimicrob Chemother 73:1305–1313. doi: 10.1093/jac/dky002. [DOI] [PubMed] [Google Scholar]
- 40.Stass H, Willmann S, Windl T. 2014. Risk assessment for amikacin inhale in ICU patients using whole-body physiologically based PK-models, abstr P-926, p 232 Abstr 43rd Critical Care Congress. Society of Critical Care Medicine, San Francicso, CA. [Google Scholar]
- 41.Sarovich D. 2017. MGAP—microbial-genome-assembler-pipeline Zenodo doi: 10.5281/zenodo.825368. [DOI] [Google Scholar]
- 42.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 43.Sarovich DS, Price EP. 2014. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz A, Currie BJ, Mayo M, Sarovich DS, Price P. 2020. Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality. Microb Genom 6:e000326. doi: 10.1099/mgen.0.000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neely MN, van Guilder MG, Yamada WM, Schumitzky A, Jelliffe RW. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. 2004. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis 190:1642–1651. doi: 10.1086/424849. [DOI] [PubMed] [Google Scholar]