This study summarizes drug resistance analyses in 4 recent phase 2b trials of the respiratory syncytial virus (RSV) fusion inhibitor presatovir in naturally infected adults. Adult hematopoietic cell transplant (HCT) recipients, lung transplant recipients, or hospitalized patients with naturally acquired, laboratory-confirmed RSV infection were enrolled in 4 randomized, double-blind, placebo-controlled studies with study-specific presatovir dosing. Full-length RSV F sequences amplified from nasal swabs obtained at baseline and postbaseline were analyzed by population sequencing.
KEYWORDS: fusion inhibitor, presatovir, resistance, respiratory syncytial virus
ABSTRACT
This study summarizes drug resistance analyses in 4 recent phase 2b trials of the respiratory syncytial virus (RSV) fusion inhibitor presatovir in naturally infected adults. Adult hematopoietic cell transplant (HCT) recipients, lung transplant recipients, or hospitalized patients with naturally acquired, laboratory-confirmed RSV infection were enrolled in 4 randomized, double-blind, placebo-controlled studies with study-specific presatovir dosing. Full-length RSV F sequences amplified from nasal swabs obtained at baseline and postbaseline were analyzed by population sequencing. Substitutions at RSV fusion inhibitor resistance-associated positions are reported. Genotypic analyses were performed on 233 presatovir-treated and 149 placebo-treated subjects. RSV F variant V127A was present in 8 subjects at baseline. Population sequencing detected treatment-emergent substitutions in 10/89 (11.2%) HCT recipients with upper and 6/29 (20.7%) with lower respiratory tract infection, 1/35 (2.9%) lung transplant recipients, and 1/80 (1.3%) hospitalized patients treated with presatovir; placebo-treated subjects had no emergent resistance-associated substitutions. Subjects with substitutions at resistance-associated positions had smaller decreases in viral load during treatment relative to those without, but they had similar clinical outcomes. Subject population type and dosing regimen may have influenced RSV resistance development during presatovir treatment. Subjects with genotypic resistance development had decreased virologic responses compared to those without genotypic resistance but had comparable clinical outcomes.
INTRODUCTION
Respiratory syncytial virus (RSV) can cause severe disease in elderly and immunocompromised adults. Adult hematopoietic cell transplant (HCT) recipients infected with RSV are at high risk for complications; in a representative study of 33 patients, 20 (61%) progressed to pneumonia, and 12 had a fatal outcome (1). Community-acquired respiratory viral infections, including RSV infections, are associated with chronic lung allograft dysfunction in lung transplant recipients (2). An estimated 10,000 to 14,000 deaths in people ≥65 years of age in the United States each year are attributable to RSV infection, and RSV-associated morbidity and mortality rates are comparable to those for influenza in adults with underlying cardiopulmonary conditions (3, 4).
Despite the disease burden of RSV in adults, no therapies are approved for treatment in this population. Inhaled ribavirin is approved only for treatment of severe lower respiratory tract disease in infants and young children, and its effectiveness is questionable (5, 6). Palivizumab, an anti-RSV F antibody, is approved for prevention of severe lower respiratory disease due to RSV among children <24 months of age who are considered to be at very high risk for severe RSV infection, but it is neither indicated nor effective for treatment of RSV infection (7–9). Thus, there is a significant unmet need for an effective RSV-specific antiviral therapy for both infants and adults at high risk for severe RSV disease.
Presatovir, an orally bioavailable RSV fusion inhibitor, has a favorable safety profile and significantly reduced viral load and symptoms relative to placebo in healthy adults experimentally challenged with RSV (10–12). In a phase 2a challenge study, the rate of treatment-emergent substitutions associated with presatovir resistance ranged from 7.7% to 35.3%, depending on the dosing regimen (13). Virologic response was attenuated in subjects who developed resistance-associated substitutions during treatment relative to those without such substitutions; however, clinical responses were similar between subjects with and without treatment-emergent presatovir resistance-associated substitutions (13).
Four recent phase 2b clinical trials assessed the efficacy and safety of presatovir for treatment of naturally acquired RSV infection in different adult populations at high risk of severe RSV disease, including HCT recipients with isolated RSV upper respiratory tract infection (URTI), HCT recipients with RSV lower respiratory tract infection (LRTI), lung transplant recipients, or nonimmunocompromised hospitalized patients (11, 14–16). Presatovir was generally well tolerated in all populations. Despite achieving adequate plasma exposures, presatovir treatment did not meet the prespecified primary efficacy endpoint of decrease in time-weighted average change in nasal RSV viral load from baseline compared with placebo in any study population (11, 14–16). Here, we report the genotypic resistance analyses performed for the RSV F gene across these 4 trials.
(Portions of this analysis were presented at IDWeek 2018, San Francisco, CA, 3 to 7 October 2018 [17].)
RESULTS
Subject populations.
A total of 499 subjects were randomized to the 4 phase 2b studies, of whom 441 received study drug, had detectable RSV RNA at baseline, and were included in the efficacy populations for each of the respective studies (Table 1).
TABLE 1.
Study populations, key inclusion criteria, presatovir treatment regimens, nasal swab sampling schedules, and primary endpoints in the 4 phase 2b trialsa
| Parameter | GS-US-218-0108 | GS-US-218-1502 | GS-US-218-1797 | GS-US-218-1227 |
|---|---|---|---|---|
| Population | HCT recipients | HCT recipients | Lung transplant recipients | Hospitalized patients |
| RSV infection site | Isolated URTI | LRTI | URTI or LRTI | URTI or LRTI |
| Randomization | 1:1 | 1:1 | 2:1 | 1:1 |
| Efficacy population (n) | 176 | 57 | 54 | 154 |
| Presatovir dosing | 200 mg on days 1, 5, 9, 13, and 17 | 200 mg on days 1, 5, 9, 13, and 17 | 200 mg on day 1 followed by 100 mg daily on days 2–14 | Single 200-mg dose on day 1 |
| Nasal swab days for genotypic analysisb | 1, 9, 28, or last detectable | 1, 9, 28, or last detectable | 1, 7, 28, or last detectable | 1, 5, 14, or last detectable |
| Primary endpoint | Time-weighted avg change in nasal RSV viral load from day 1–9 and proportion of subjects developing lower respiratory tract complications through day 28 (coprimary) | Time-weighted avg change in nasal RSV viral load from day 1–9 | Time-weighted avg change in nasal RSV viral load from day 1–7 in the entire efficacy population and in subjects with shorter-than-median symptom duration before start of study treatment (coprimary) | Time-weighted avg change in nasal RSV viral load from day 1–5 |
HCT, hematopoietic cell transplant; LRTI, lower respiratory tract infection; RSV, respiratory syncytial virus; URTI, upper respiratory tract infection.
Only samples with RSV RNA levels of >1,000 copies/ml were analyzed.
Genotypic analysis of RSV F from baseline nasal swabs.
Baseline RSV F gene population sequencing data were available for 373/441 subjects in the efficacy analysis populations for the 4 phase 2b studies. The most frequent reason for a lack of available baseline sequence data was an RSV RNA level below the limit of detection of the F gene sequencing assay (<1,000 copies/ml). Partial F gene sequencing data were available for some samples with viral loads close to the limit of detection of the sequencing assay. Baseline sequences were analyzed for the presence of any amino acid substitutions known to be associated with resistance to presatovir or other RSV fusion inhibitors as listed in Table 2. In the baseline analysis, the only substitution detected at any resistance-associated position in RSV F was V127A in 2.1% of presatovir-treated and 1.4% of placebo-treated subjects across all 4 studies (Table 3). Among the 8 subjects with the V127A substitution at baseline, 3 were infected with RSV subtype A and 5 with RSV subtype B.
TABLE 2.
Known amino acid substitutions in RSV F associated with reduced susceptibility to presatovir or other RSV fusion inhibitors
| Category | Substitutions |
|---|---|
| Presatovir resistance-associated substitutionsa | V127A, L138F, L138I, F140I, F140L, L141F, L141W, T323A, D338Y, S398L, K399I, K399N, T400I, T400A, T400V, I474T, D486N, E487D, F488L, F488S, F488Y, N517I |
| Fusion inhibitor resistance-associated substitutionsb | G143S, V144A, D392G, K394R, D401E, D486E, F488I, F488V, D489E, D489Y |
| Other substitutionsc | M396I, T397S |
Substitutions previously selected by presatovir treatment in a clinical study or in vitro and shown to reduce susceptibility to presatovir.
Substitutions previously selected by or shown to reduce susceptibility to RSV fusion inhibitors other than presatovir in vitro (18, 20, 29, 31, 32). The effect of these substitutions on susceptibility to presatovir is unknown.
Substitutions that developed during presatovir treatment in a clinical study for which presatovir susceptibility was unable to be characterized.
TABLE 3.
Baseline genotypic analysis of RSV F gene
| Subject group | No. (%) of subjects in study populationa |
||||
|---|---|---|---|---|---|
| HCT URTI (GS-US-218-0108) | HCT LRTI (GS-US-218-1502) | Lung transplant (GS-US-218-1797) | Hospitalized patients (GS-US-218-1227) | All | |
| Presatovir-treated subjects | 89 | 29 | 35 | 80 | 233 |
| With baseline F sequence data | 89 (100) | 28 (96.6) | 33 (94.3) | 76 (95.0) | 226 (97.0) |
| With any substitutions at resistance positionsb | 3 (3.4) | 1 (3.4) | 1 (2.9) | 0 | 5 (2.1) |
| With V127A | 3 (3.4) | 1 (3.4) | 1 (2.9) | 0 | 5 (2.1) |
| Placebo-treated subjects | 87 | 28 | 19 | 74 | 208 |
| With baseline F sequence data | 44 (50.6) | 26 (92.9) | 19 (100) | 58 (78.4) | 147 (70.7) |
| With any substitutions at resistance positions | 2 (2.3) | 1 (3.6) | 0 | 0 | 3 (1.4) |
| With V127A | 2 (2.3) | 1 (3.6) | 0 | 0 | 3 (1.4) |
HCT, hematopoietic cell transplant; LRTI, lower respiratory tract infection; URTI, upper respiratory tract infection.
Resistance substitutions are listed in Table 2.
Genotypic analysis of RSV F from postbaseline nasal swabs.
Postbaseline RSV F population sequencing results were available for 217/233 presatovir-treated and 104/208 placebo-treated subjects in the efficacy analysis populations. Failure to obtain postbaseline sequences was generally due to low RSV RNA levels; partial sequences were available for some samples with viral loads near the limit of detection (1,000 copies/ml). Across all 4 studies, substitutions at any presatovir resistance-associated position were detected by population sequencing in a total of 18 of 233 presatovir-treated subjects (Table 4); no treatment-emergent resistance-associated substitutions were detected in placebo-treated subjects. The rate of resistance development was lowest among hospitalized patients (1.3%) and lung transplant recipients (2.9%) and highest among HCT recipients with RSV LRTI (20.7%) or URTI (11.2%) (Table 4). No subject with V127A at baseline developed resistance-associated substitutions during treatment. The most frequently observed presatovir resistance-associated substitutions were T400A/I (3.0%), L141F/W (1.3%), S398L (1.3%), and F140I (0.9%) (Table 4); these substitutions were previously determined to confer high-level reduced susceptibility to presatovir. Six of 18 subjects developed multiple substitutions at presatovir resistance-associated positions during the study (Table 5).
TABLE 4.
Summary of postbaseline RSV F genotypic resistance development in presatovir-treated subjects
| Presatovir-treated subject group | No. (%) in study populationa |
||||
|---|---|---|---|---|---|
| HCT URTI (GS-US-218-0108) | HCT LRTI (GS-US-218-1502) | Lung transplant (GS-US-218-1797) | Hospitalized patients (GS-US-218-1227) | All | |
| Total | 89 | 29 | 35 | 80 | 233 |
| With available postbaseline F sequence data | 85 (95.5) | 27 (93.1) | 30 (85.7) | 75 (93.8) | 217 (93.1) |
| With any substitutions at resistance positionsb | 10 (11.2) | 6 (20.7) | 1 (2.9) | 1 (1.3) | 18 (7.7) |
| With known presatovir resistance-associated substitutions | 8 (9.0) | 4 (13.8) | 1 (2.9) | 1 (1.3) | 14 (6.0) |
| L138I | 0 | 0 | 0 | 1 (1.3) | 1 (0.4) |
| F140I | 2 (2.2) | 0 | 0 | 0 | 2 (0.9) |
| L141F/W | 2 (2.2) | 1 (3.4) | 0 | 0 | 3 (1.3) |
| L141F | 1 (1.1) | 1 (3.4) | 0 | 0 | 2 (0.9) |
| L141W | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| D338Y | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| S398L | 2 (2.2) | 1 (3.4) | 0 | 0 | 3 (1.3) |
| T400A/I | 4 (4.5) | 2 (6.9) | 1 (2.9) | 0 | 7 (3.0) |
| T400A | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| T400I | 3 (3.4) | 2 (6.9) | 1 (2.9) | 0 | 6 (2.6) |
| D486N | 0 | 1 (3.4) | 0 | 0 | 1 (0.4) |
| With known fusion inhibitor resistance-associated substitutions | 2 (2.2) | 1 (3.4) | 0 | 0 | 3 (1.3) |
| G143S | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| K394R | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| D486E | 0 | 1 (3.4) | 0 | 0 | 1 (0.4) |
| With novel substitutions at known fusion inhibitor resistance-associated positions | 2 (2.2) | 1 (3.4) | 1 (2.9) | 0 | 4 (1.7) |
| D486V | 1 (1.1) | 1 (3.4) | 1 (2.9) | 0 | 3 (1.3) |
| E487G | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| With other substitutions | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
| M396I | 1 (1.1) | 0 | 0 | 0 | 1 (0.4) |
HCT, hematopoietic cell transplant; LRTI, lower respiratory tract infection; URTI, upper respiratory tract infection.
Resistance substitutions are listed in Table 2.
TABLE 5.
Characteristics of individual subjects with development of substitutions at resistance-associated positions in RSV Fa
| Study and subject | RSV subtype | Baseline lymphocyte count (cells/μl)b | Time-weighted avg change in nasal RSV viral load (log10 copies/ml)c | Visit | RSV RNA (copies/ml) | Resistance-associated substitution(s) |
|---|---|---|---|---|---|---|
| HCT URTI (GS-US-218-0108) | ||||||
| 1 | A | <200 | −1.88 | Baseline | 7,900,000 | None |
| Day 9 | 12,000,000 | None | ||||
| Day 17 | 87,100 | None | ||||
| Day 22 | 92,400 | None | ||||
| Day 28 | 11,100 | F140F/I, T400T/I | ||||
| 2 | B | <200 | −0.06 | Baseline | 11,000,000 | None |
| Day 9 | 81,000,000 | M396M/I, S398S/L | ||||
| Day 13 | 54,700 | T400T/A | ||||
| 3 | A | 160 | −0.46 | Baseline | 83,000,000 | None |
| Day 9 | 12,000,000 | None | ||||
| Day 17 | 55,000,000 | None | ||||
| Day 22 | 532,000 | S398S/L, T400T/I | ||||
| Day 28 | 10,900 | D486D/V | ||||
| 4 | A | 480 | −0.13 | Baseline | 28,000,000 | None |
| Day 9 | 24,000,000 | None | ||||
| Day 17 | 199,000 | D338D/Y | ||||
| 5 | B | <200 | −0.66 | Baseline | 54,000,000 | None |
| Day 9 | 85,000,000 | K394K/R | ||||
| Day 13 | 18,700 | None | ||||
| Day 22 | 3,400 | Assay failure | ||||
| 6 | B | 610 | 0.28 | Baseline | 10,000,000 | None |
| Day 9 | 11,000,000 | None | ||||
| Day 17 | 10,700 | F140F/I | ||||
| Day 22 | 157,000 | E487E/G | ||||
| 7 | A | 420 | −0.25 | Baseline | 4,400,000 | None |
| Day 9 | 36,000 | None | ||||
| Day 28 | 10,600 | G143S | ||||
| 8 | A | 100 | −0.08 | Baseline | 1,300,000 | None |
| Day 9 | 4,200 | None | ||||
| Day 17 | 47,600 | L141L/F | ||||
| Day 28 | 3,400 | L141F | ||||
| 9 | B | <200 | −0.03 | Baseline | 10,000,000 | None |
| Day 9 | 93,200 | L141W | ||||
| Day 13 | 14,100 | None | ||||
| 10 | A | <200 | −0.51 | Baseline | 69,000,000 | None |
| Day 9 | 18,000,000 | None | ||||
| Day 17 | 2,900,000 | T400T/I | ||||
| Day 28 | 4,300 | None | ||||
| Day 42 | 524,000 | T400I | ||||
| Day 49 | 902,000 | None | ||||
| Day 56 | 33,900 | None | ||||
| HCT LRTI (GS-US-218-1502) | ||||||
| 1 | A | 1,140 | 1.19 | Baseline | 40,600 | None |
| Day 9 | 419,000 | None | ||||
| Day 28 | 4,200 | D486V | ||||
| 2 | A | 180 | −0.24 | Baseline | 15,000,000 | None |
| Day 9 | 8,900,000 | None | ||||
| Day 22 | 3,800,000 | None | ||||
| Day 56 | 5,400,000 | T400I | ||||
| 3 | B | 400 | −0.44 | Baseline | 28,000,000 | None |
| Day 9 | 23,000,000 | None | ||||
| Day 22 | 7,400,000 | L141L/F | ||||
| Day 28 | 54,000 | L141F | ||||
| 4 | B | 1,000 | −0.98 | Baseline | 42,000,000 | None |
| Day 9 | 7,800 | None | ||||
| Day 22 | 11,000,000 | D486E | ||||
| Day 28 | 290,000 | D486E | ||||
| 5 | A | 270 | −0.49 | Baseline | 28,000,000 | None |
| Day 9 | 1,600,000 | None | ||||
| Day 28 | 1,100,000 | D486D/N | ||||
| 6 | B | 270 | 1.44 | Baseline | 6,200 | None |
| Day 7 | 2,100,000 | S398L | ||||
| Day 17 | 134,000 | T400I | ||||
| Lung transplant (GS-US-218-1797) | ||||||
| 1 | A | 370 | 0.60 | Baseline | 13,000,000 | None |
| Day 7 | 52,000,000 | None | ||||
| Day 9 | 8,300,000 | None | ||||
| Day 21 | 307,000 | D486D/V | ||||
| Day 28 | 16,500 | T400I | ||||
| Hospital inpatients (GS-US-218-1227) | ||||||
| 1 | A | 430 | 0.11 | Baseline | 1,700,000 | None |
| Day 5 | 6,000,000 | None | ||||
| Day 14 | 5,100 | L138L/I | ||||
HCT, hematopoietic cell transplant; LRTI, lower respiratory tract infection; RSV, respiratory syncytial virus; URTI, upper respiratory tract infection.
Lymphocyte count is listed as <200 cells/μl when the patient was recorded as lymphopenic for stratification but the exact lymphocyte count was not recorded.
From baseline to day 9 for HCT recipients, day 7 for lung transplant recipients, and day 5 for hospitalized patients.
Two novel substitutions at resistance-associated positions, D486V and E487G, were detected in postbaseline sequences from presatovir-treated subjects (D486V in 3 subjects; E487G in 1 subject) (Table 4). The effects of the D486V and E487G substitutions on presatovir susceptibility have not been characterized, but other amino acid substitutions at positions 486 and 487 were previously observed to confer high-level reduced susceptibility to presatovir (D486N and E487D) or other fusion inhibitors (D486E) (18).
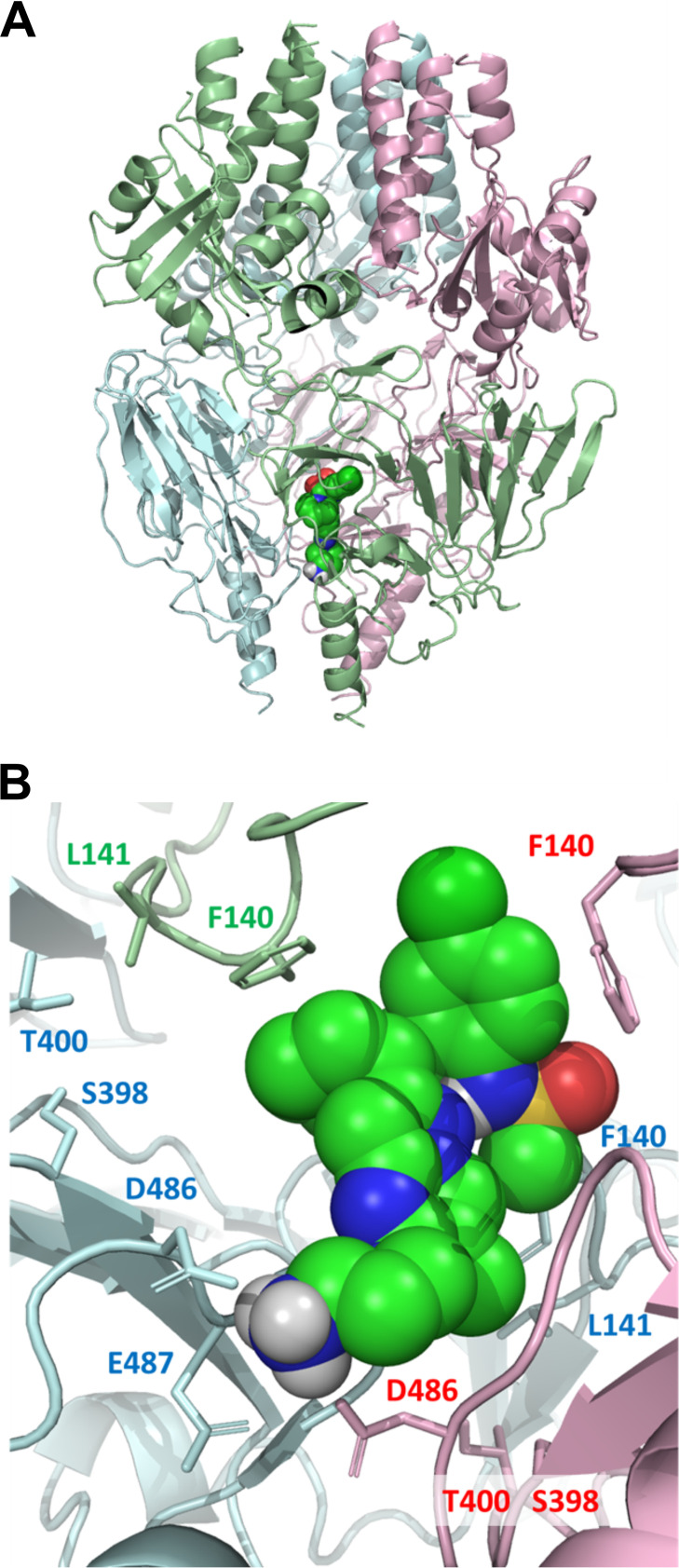
An induced-fit model (19) of presatovir bound to the prefusion RSV F trimer was developed based on the cocrystal structure of JNJ-2408068 (PDB code 5EA3) (20). The majority of treatment-emergent substitutions observed in the phase 2b studies could be mapped to the proposed interaction site, a region consistent with the location of known resistance substitutions (Fig. 1).
FIG 1.
Model of the interaction of presatovir with respiratory syncytial virus F protein. (A) Model of presatovir bound to the prefusion conformation of the RSV F protein trimer based on an X-ray crystal structure (PDB code 5EA3). A single presatovir molecule is shown in a space-filling representation. Each monomer of the trimer is shown as a ribbon diagram in a different color. (B) Detail of the modeled presatovir binding site in F protein. Key binding site residues that were mutated following presatovir treatment are highlighted.
Characteristics of subjects with treatment-emergent substitutions in RSV F.
Details of the 18 subjects who developed substitutions at resistance-associated positions during presatovir treatment are presented in Table 5. Eleven subjects were infected with RSV A and 7 with RSV B. The median time from baseline to detection of new resistance-associated substitutions was 17 days in HCT recipients with RSV URTI and 25 days in HCT recipients with LRTI.
Numerous subject characteristics were evaluated for their potential association with resistance development, including symptom duration prior to study treatment, baseline lymphocyte count, baseline viral load, RSV subtype, ribavirin use, plasma presatovir exposure, and duration of viral shedding. The only subject characteristic significantly associated with resistance development was baseline lymphocyte count in HCT recipients with URTI (study GS-US-218-0108). Relative to HCT recipients with isolated RSV URTI and without lymphopenia, patients with lymphopenia (<200 cells/μl) at baseline were significantly more likely (7/15 [47%] versus 3/74 [4%]; P < 0.001) to develop presatovir resistance-associated substitutions.
Outcomes of subjects with genotypic resistance development.
Presatovir-treated subjects with genotypic resistance development generally had smaller reductions in time-weighted average nasal RSV viral load than those without development of resistance (Table 6). Clinical outcomes were similar between HCT recipients with URTI with and without resistance development (Table 6). In HCT recipients with RSV LRTI who received presatovir, subjects with resistance development had fewer supplemental-oxygen-free days than those without resistance (median [interquartile range], 3 [0 to 6] versus 28 [25 to 28] days) (Table 6); however, this result was confounded by a higher proportion of subjects who developed resistance requiring supplemental oxygen at baseline (5 of 6 subjects with resistance versus 5 of 23 subjects without resistance). Other clinical outcomes in HCT recipients with RSV LRTI were similar between those who did and did not develop resistance (Table 6). In the lung transplant recipient and hospitalized patient populations, clinical outcomes were not compared between subjects with and without treatment-emergent resistance-associated substitutions due to low rates of genotypic resistance development.
TABLE 6.
Virologic and clinical outcomes in presatovir-treated subjects with and without genotypic resistance development in RSV F in studies of HCT recipients (GS-US-218-0108 and GS-US-218-1502)a
| Study group and outcome | Value for subjects: |
P value | Value for all subjects | |
|---|---|---|---|---|
| With resistance | Without resistance | |||
| HCT URTI (GS-US-218-0108) | ||||
| No. of subjects | 10 | 79 | 89 | |
| Change in viral load from day 1–9 (log10 copies/ml)b,c | −0.19 (−0.51, −0.06) | −1.42 (−1.96, −0.79) | 0.002d | −1.35 (−1.84, −0.51) |
| No. (%) with LRTC through day 28 | 1 (10) | 9 (11) | 1.0e | 10 (11) |
| No. (%) with respiratory failure or mortality through day 28 | 0 | 5 (6) | 1.0e | 5 (6) |
| HCT LRTI (GS-US-218-1502) | ||||
| No. of subjects | 6 | 23 | 29 | |
| Change in viral load from day 1–9 (log10 copies/ml)b,c | −0.34 (−0.49, 1.19) | −1.44 (−2.34, −0.78) | 0.019 | −1.03 (−1.84, −0.44) |
| No. of supplemental-O2-free days through day 28c | 3 (0, 6) | 28 (25, 28) | <0.001f | 26 (10, 28) |
| No. (%) with mechanical ventilation use through day 28 | 2 (33) | 1 (4) | 0.100e | 3 (10) |
| No. (%) with all-cause mortality through day 28 | 0 | 0 | NA | 0 |
HCT, hematopoietic cell transplant; LRTC, lower respiratory tract complications; LRTI, lower respiratory tract infection; NA, not applicable; URTI, upper respiratory tract infection.
Time-weighted average change in nasal RSV viral load.
Median (interquartile range).
P value for resistance versus no resistance calculated from the ANCOVA model including baseline values and stratification factors.
P value for resistance versus no resistance calculated using Fisher’s exact test.
P value for resistance versus no resistance calculated from the negative binomial model with stratification factors as covariates. Five of 6 (83%) subjects with resistance and 5 of 23 (22%) subjects without resistance required supplemental oxygen at baseline.
DISCUSSION
In the 4 phase 2b trials of presatovir efficacy and safety in naturally RSV-infected adults, substitutions at resistance-associated positions developed in 18 of 233 presatovir-treated subjects. Rates of resistance development varied among the individual phase 2b studies, with the lowest rate of resistance development observed in hospitalized patients (1.3%) and the highest in HCT recipients with LRTI (20.7%). No substitutions at resistance-associated positions developed during placebo treatment in any study; based on this finding, it is unlikely that inclusion of the remaining placebo recipients in the studies of HCT recipients with URTI and hospitalized patients would affect the results.
Most of the substitutions in RSV F that developed in presatovir-treated subjects have been previously characterized and are known to reduce susceptibility to presatovir. A small number of subjects developed novel amino acid substitutions at positions previously identified as being involved with fusion inhibitor resistance; these substitutions require further characterization to determine their impact on presatovir susceptibility. The majority of the observed treatment-emergent substitutions map to the trimer interface of the prefusion RSV F structure.
In the phase 2a study of presatovir in healthy adults experimentally challenged with RSV, the frequency of resistance development ranged from 7.7% to 35.3% among dose cohorts and was generally lowest among subjects receiving presatovir treatment at the highest doses for the longest duration (13). Dosing regimens in the phase 2b studies generally included higher doses and longer treatment durations, and resistance-associated substitutions developed during presatovir treatment at lower rates—ranging from 1.3% to 20.7%—than in the challenge study.
The phase 2b studies differed from the challenge study mainly in the subject populations enrolled and presatovir dosing regimens used; these factors likely contributed to the different rates of resistance development observed among the phase 2b studies. Hospitalized patients, who had the lowest rate of resistance development (1.3%), were generally not immunocompromised and not receiving strongly immunosuppressive therapy, as patients receiving chronic systemic immunosuppressive agents within 28 days or oral corticosteroid equivalent to prednisone at >20 mg/day for >14 days before screening were excluded. These patients received a single dose of presatovir, and this level of drug exposure likely did not impose sufficient selection pressure on the virus to favor the development of resistance. Lung transplant recipients had the next-lowest rate of resistance development (2.9%) and were immunosuppressed but had higher presatovir exposure with daily dosing for 14 days. The consistent presatovir exposure in these subjects may have efficiently suppressed RSV replication, thereby preventing the development of resistance. Relative to the other populations, HCT recipients had higher rates of resistance development (11.2% and 20.7%), and HCT recipients with URTI and lymphopenia at baseline had significantly higher rates of resistance development relative to those without lymphopenia. The HCT recipients received presatovir every 4 days through day 17, resulting in a cyclical drug exposure over several days which may have contributed to the development of drug resistance by providing the opportunity for viral replication to occur in the respiratory tracts of these immunocompromised patients during trough levels of drug exposure. While no association between plasma presatovir exposure and resistance development was identified in the phase 2b studies, it is possible that the drug level at the site of viral replication in the respiratory tract may have differed from that measured in the plasma.
In the phase 2b studies of presatovir, subjects who developed presatovir resistance-associated substitutions had smaller decreases in viral load but similar clinical outcomes relative to subjects without such substitutions, suggesting that the development of presatovir-resistant RSV variants likely did not have a significant impact on RSV clinical progression. This is consistent with results from the challenge study, in which subjects who developed presatovir resistance substitutions had increased RSV viral loads, but not significantly different symptom scores and mucus weights, relative to those without resistance substitutions (13). The RSV viral load and viral sequence data from the phase 2b and challenge studies were obtained from nasal samples and may not accurately reflect viral dynamics at the site of RSV replication within the lungs, possibly explaining the discordant effects of presatovir resistance-associated mutations on viral load and clinical outcomes. Alternately, as RSV disease severity in adults is associated with nasal cytokine levels at the time of diagnosis (21)—prior to development of resistance-associated substitutions in the phase 2b studies—early immune responses may mediate RSV symptoms and clinical outcomes. Compared with the challenge study (10), presatovir treatment in the phase 2b studies was initiated later in the course of RSV infection, likely past the peak viral load and symptomatic period in many subjects. While treatment-emergent substitutions were first detected as late as day 56 in some subjects, indicating that RSV replication and infection of new cells continue late into the disease course, the impact of this level of viral replication on clinical disease is not clear. It is possible that the development of resistance resulted from residual viral replication in subjects whose immune systems were unable to clear the virus efficiently. As a minority of subjects developed resistance-associated substitutions, and resistance in most subjects was detected at time points after the primary efficacy endpoint analysis, presatovir resistance is likely not responsible for the lack of significant virologic response in the phase 2b studies. Based on the discordant results of the clinical studies compared with the challenge study, and subgroup analyses of HCT recipients with URTI with shorter- versus longer-than-median symptom duration, initiation of treatment earlier in the course of RSV infection (as in the challenge study and GS-US-218-0108) (13, 14) is potentially needed to achieve significant virologic and clinical efficacy with presatovir.
Similar to the present study, trials of the anti-influenza drug oseltamivir had discordant virologic and clinical outcome patterns in subjects with treatment-emergent resistance. Patients who developed resistance-associated substitutions during oseltamivir treatment had extended viral shedding but comparable clinical outcomes relative to those with wild-type influenza (22). In contrast, emergence of oseltamivir resistance in immunocompromised individuals may contribute to treatment failure (23, 24); this could also be a concern for presatovir treatment. Initial infection with oseltamivir-resistant influenza virus is also associated with poor clinical outcomes (25), suggesting that transmission of RSV with resistance-associated substitutions could be a concern. In the RSV challenge study, resistance-associated substitutions that emerged during presatovir treatment were associated with reduced viral fitness in competitive outgrowth assays with wild-type RSV and are therefore not anticipated to spread in the general population. However, competitive outgrowth assays are not necessarily predictive of in vivo fitness; variants of influenza virus resistant to another drug, baloxavir marboxil, demonstrate impaired replication relative to wild-type virus in competitive cell culture assays but retain fitness and pathogenicity in animal models and have been transmitted between humans (26–28). Some RSV F variants with substitutions associated with fusion inhibitor resistance located close to those observed in the current study, such as D401E, also remain pathogenic in animal models (29). Further work in animal models could help elucidate the potential impact of presatovir resistance substitutions on RSV fitness and pathogenesis.
The RSV F V127A variant was the only known substitution at a resistance-associated position detected at baseline (8 subjects total) across the 4 phase 2b studies. This amino acid substitution is found in 2% to 45% of naturally circulating RSV isolates belonging to the dominant Buenos Aires RSV B genotype (19, 30) and may represent a polymorphism rather than a resistance-associated substitution. The combination of V127A and F140L reduces susceptibility to presatovir >410-fold (13), but it is unclear how V127A, which is not part of the RSV F trimer interface (Fig. 1), contributes to presatovir resistance, and the effect of V127A alone on RSV susceptibility to presatovir has not been evaluated. Presatovir-treated subjects with baseline V127A did not develop F140L or any other resistance-associated substitution during treatment and had a virologic response to treatment similar to that of subjects without V127A. These results indirectly suggest that V127A has little effect on presatovir susceptibility. However, it is possible that V127A could contribute indirectly to drug resistance by compensating for fitness costs of other resistance substitutions that reduce presatovir susceptibility.
Areas for future research include determining the effect on presatovir susceptibility of several substitutions observed in these studies, including V127A alone and in combination with known resistance substitutions, the novel substitutions D486V and E487G, and substitutions previously associated with resistance to other fusion inhibitors (G143S, K394R, and D486E) that emerged during presatovir treatment. Sequencing of samples at additional time points, including deep sequencing, would also be useful for longitudinal analysis of the timing of resistance emergence and patterns of resistance substitutions. Furthermore, as fusion inhibitors have a relatively low genetic barrier to resistance and can confer cross-class resistance (29), combination treatments of RSV antivirals using different mechanisms of action may be warranted to minimize resistance development.
MATERIALS AND METHODS
Trials and study design.
The 4 randomized, double-blind, placebo-controlled phase 2b studies of presatovir efficacy and safety in HCT recipients with isolated RSV URTI (GS-US-218-0108; ClinicalTrials registration no. NCT02254408), HCT recipients with RSV LRTI (GS-US-218-1502; ClinicalTrials registration no. NCT02254421), lung transplant recipients (GS-US-218-1797; ClinicalTrials registration no. NCT02534350), and hospitalized patients admitted before or after RSV infection (GS-US-218-1227; ClinicalTrials registration no. NCT02135614) were previously described (11, 14–16). Briefly, adults with naturally acquired, laboratory-confirmed RSV infection and acute onset of new or worsening respiratory symptoms prior to screening were enrolled; key study design elements are summarized in Table 1. Subjects were randomized 2:1 for lung transplant recipients and 1:1 for all other populations to receive oral presatovir or placebo. Dosing schedules were designed to maintain adequate plasma presatovir levels for the expected duration of RSV viral shedding in transplant recipients or long enough to observe significant antiviral effect in hospitalized patients; the target trough plasma presatovir concentration was >4- to 5-fold over the protein binding-adjusted effective concentration of presatovir to inhibit 95% of virus replication for wild-type RSV strains of 25 ng/ml. All HCT recipients received 200 mg presatovir on days 1 (baseline), 5, 9, 13, and 17; lung transplant recipients received 200 mg presatovir on day 1 followed by 100 mg presatovir once daily on days 2 to 14. Hospital inpatients received a single dose of 200 mg presatovir on day 1 (Table 1). Subjects were followed for 28 days after dosing with optional extended viral monitoring through day 56 for HCT and lung transplant recipients. The studies were conducted in accordance with the International Conference on Harmonisation Good Clinical Practice Guidelines and the Declaration of Helsinki, and all subjects provided written informed consent.
Sample collection.
Bilateral nasal swab specimens were collected at baseline and during and after study treatment as scheduled per each study protocol for measurement of nasal RSV viral load and genotypic analysis of RSV F.
Genotypic analyses.
Genotypic analyses were conducted for samples from subjects in the efficacy analysis population who received ≥1 dose of study drug and had baseline RSV RNA levels above the lower limit of detection of the reverse transcription quantitative real-time PCR (RT-qPCR) assay (approximately 1,000 copies/ml). All presatovir-treated subjects and a proportion of placebo-treated subjects (GS-US-218-1227, at least 25%; GS-US-218-0108, at least 50%; GS-US-218-1502 and GS-US-218-1797, 100%) were included in the resistance analyses. Respiratory syncytial virus RNA was extracted from nasal swab samples using a standard total nucleic acid extraction protocol (NucliSENS easyMAG; bioMérieux, Inc., Durham, NC, USA) and quantified by RT-qPCR at a central laboratory (Viracor Eurofins, Lee’s Summit, MO, USA) using Superscript III RT PCR master mix (Life Technologies Corporation, Carlsbad, CA, USA). The same RT-qPCR assay identified RSV subtype A or B. Genotypic analysis of RSV F was performed on samples from protocol-specific study days (Table 1), or the last sample with RSV RNA levels of >1,000 copies/ml as measured by RT-qPCR, and all extended viral monitoring samples with detectable RSV RNA. The full-length RSV F gene was amplified by reverse transcription-PCR using custom oligonucleotide primers. Population sequencing was performed on amplified F gene sequences by Sanger sequencing using custom oligonucleotide primers. Samples from lung transplant recipients were sequenced at DDL Diagnostic Laboratory (Rijswijk, the Netherlands), and all other samples were sequenced by Viracor Eurofins (Lee’s Summit, MO, USA).
Data analysis.
Baseline RSV F sequences were compared with the subtype-specific RSV reference sequences RSV A2 (GenBank accession number M74568) and RSV B1 (GenBank accession number AF013254) and analyzed for the presence of preexisting substitutions at known resistance-associated positions. Postbaseline RSV F sequences were compared with subject-specific baseline sequences to determine whether resistance-associated substitutions had developed during presatovir treatment. Known amino acid substitutions previously associated with resistance to presatovir or other RSV fusion inhibitors are listed in Table 2. Presatovir resistance-associated substitutions were previously selected by presatovir treatment in a clinical study or in vitro and have been shown to reduce susceptibility to presatovir. Fusion inhibitor resistance-associated substitutions were selected for or shown to have reduced susceptibility to RSV fusion inhibitors other than presatovir in vitro (18, 20, 29, 31, 32); the effects of these substitutions on susceptibility to presatovir are unknown. Two other substitutions, M396I and T397S, developed during presatovir treatment in clinical studies; their effect on presatovir susceptibility has not been characterized due to poor replication of the recombinant variant viruses in cell culture. Time-weighted average change in nasal RSV viral load was compared between subjects with and without treatment-emergent resistance-associated substitutions using analysis of covariance (ANCOVA) models adjusted for the baseline viral load and randomization stratification factors in each individual study. Binary clinical outcomes were compared between subjects with and without treatment-emergent resistance-associated substitutions using Fisher’s exact test. The 95% confidence intervals for numbers and percentages were calculated using the Clopper-Pearson method. Number of supplemental oxygen-free days in the study of HCT recipients with LRTI was compared between subjects with and without treatment-emergent resistance-associated substitutions using a negative binomial model with the randomization stratification factors as covariates, and an offset parameter to account for the on-study duration.
Data availability.
Sequences are available from GenBank under accession no. MT543327 to MT544300.
ACKNOWLEDGMENTS
We thank the subjects and their families for participation in the studies. Medical writing support was provided by Judy Phillips, DVM, PhD, of AlphaBioCom, LLC, and funded by Gilead Sciences, Inc.
D.P.P., Y.G., J.P., and T.R.W. are employees of Gilead Sciences, Inc., and may hold stock. D.L.G., J.W.C., and R.J. are former employees of Gilead Sciences, Inc.
This work was supported by Gilead Sciences, Inc.
REFERENCES
- 1.Whimbey E, Champlin RE, Couch RB, Englund JA, Goodrich JM, Raad I, Przepiorka D, Lewis VA, Mirza N, Yousuf H, Tarrand JJ, Bodey GP. 1996. Community respiratory virus infections among hospitalized adult bone marrow transplant recipients. Clin Infect Dis 22:778–782. doi: 10.1093/clinids/22.5.778. [DOI] [PubMed] [Google Scholar]
- 2.Khalifah AP, Hachem RR, Chakinala MM, Schechtman KB, Patterson GA, Schuster DP, Mohanakumar T, Trulock EP, Walter MJ. 2004. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med 170:181–187. doi: 10.1164/rccm.200310-1359OC. [DOI] [PubMed] [Google Scholar]
- 3.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 5.Valeant Pharmaceuticals. 2016. Virazole (ribavirin for inhalation solution, USP). Full prescribing information. Valeant Pharmaceuticals North America LLC, Bridgewater, NJ. [Google Scholar]
- 6.Randolph AG, Wang EE. 1996. Ribavirin for respiratory syncytial virus lower respiratory tract infection. A systematic overview. Arch Pediatr Adolesc Med 150:942–947. doi: 10.1001/archpedi.1996.02170340056011. [DOI] [PubMed] [Google Scholar]
- 7.MedImmune, LLC. May 2018. Synagix (palivizumab) injection, for intramuscular use. Full prescribing information. MedImmune, LLC, Gaithersburg, MD: https://www.azpicentral.com/synagis/synagis.pdf. [Google Scholar]
- 8.de Fontbrune FS, Robin M, Porcher R, Scieux C, de Latour RP, Ferry C, Rocha V, Boudjedir K, Devergie A, Bergeron A, Gluckman E, Azoulay E, Lapalu J, Socié G, Ribaud P. 2007. Palivizumab treatment of respiratory syncytial virus infection after allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 45:1019–1024. doi: 10.1086/521912. [DOI] [PubMed] [Google Scholar]
- 9.American Academy of Pediatrics Committee on Infectious Diseases, American Academy of Pediatrics Bronchiolitis Guidelines Committee. 2014. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 134:415–420. doi: 10.1542/peds.2014-1665. [DOI] [PubMed] [Google Scholar]
- 10.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O'Riordan T, Lewis SA, Li X, Toback SL, Lin S-L, Chien JW. 2014. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 11.Hanfelt-Goade D, Maimon N, Nimer A, Riviere F, Catherinot E, Ison M, Jeong SH, Walsh E, Gafter-Gvili A, Nama SR, Napora P, Chowers M, Bergeron A, Zeltser D, Moudgil H, Limaye AP, Couturaud F, Nseir W, McKevitt M, Porter D, Jordan R, Guo Y, German P, Watkins TR, Gossage DL, Chien JW, Falsey AR. 2018. A phase 2b, randomized, double-blind, placebo-controlled trial of presatovir (GS-5806), a novel oral RSV fusion inhibitor, for the treatment of respiratory syncytial virus (RSV) in hospitalized adults. Am J Respir Crit Care Med 197:A4457. [Google Scholar]
- 12.Xin Y, Weng W, Murray BP, Eisenberg EJ, Chien JW, Ling J, Silverman JA. 2018. The drug-drug interaction profile of presatovir. J Clin Pharmacol 58:771–780. doi: 10.1002/jcph.1073. [DOI] [PubMed] [Google Scholar]
- 13.Stray K, Perron M, Porter DP, Anderson F, Lewis SA, Perry J, Miller M, Cihlar T, DeVincenzo J, Chien JW, Jordan R. Drug resistance assessment following administration of RSV fusion inhibitor presatovir to participants experimentally infected with respiratory syncytial virus. J Infect Dis, in press. doi: 10.1093/infdis/jiaa028. [DOI] [PubMed] [Google Scholar]
- 14.Chemaly RF, Dadwal SS, Bergeron A, Ljungman P, Kim YJ, Cheng GS, Pipavath SN, Limaye AP, Blanchard E, Winston DJ, Stiff PJ, Zuckerman T, Lachance S, Rahav G, Small CB, Mullane KM, Patron RL, Lee DG, Hirsch HH, Waghmare A, McKevitt M, Jordan R, Guo Y, German P, Porter DP, Gossage DL, Watkins TR, Marty FM, Chien JW, Boeckh M. 3 December 2019. A phase 2, randomized, double-blind, placebo-controlled trial of presatovir for the treatment of respiratory syncytial virus upper respiratory tract infection in hematopoietic-cell transplant recipients. Clin Infect Dis doi: 10.1093/cid/ciz1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb J, Torres F, Haddad T, Dhillon G, Dilling DF, Knoop C, Rampolla R, Walia R, Ahya V, Kessler R, Mason DP, Budev M, Neurohr C, Glanville AR, Jordan R, Porter D, McKevitt MT, German P, Guo Y, Chien JW, Watkins TR, Zamora M. 2018. A phase 2b, randomized controlled trial of presatovir, an oral RSV fusion inhibitor, for the treatment of respiratory syncytial virus (RSV) in lung transplant (LT) recipients. J Heart Lung Transplant 37:S155. doi: 10.1016/j.healun.2018.01.375. [DOI] [PubMed] [Google Scholar]
- 16.Marty FM, Chemaly RF, Mullane KM, Lee DG, Hirsch HH, Small CB, Bergeron A, Shoham S, Ljungman P, Waghmare A, Blanchard E, Kim YJ, McKevitt M, Porter DP, Jordan R, Guo Y, German P, Boeckh M, Watkins TR, Chien JW, Dadwal SS. 3 December 2019. A phase 2b, randomized, double-blind, placebo-controlled multicenter study evaluating antiviral effects, pharmacokinetics, safety, and tolerability of presatovir in hematopoietic cell transplant recipients with respiratory syncytial virus (RSV) infection of the lower respiratory tract. Clin Infect Dis doi: 10.1093/cid/ciz1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter D, Guo Y, Perry J, Gossage D, Watkins T, Chien J, Jordan R. 2018. Combined resistance anayses from phase 2b studies of presatovir treatment in RSV-infected adults., abstr 1646. IDWeek 2018, San Francisco, CA, 3 to 7 October 2018.
- 18.Morton CJ, Cameron R, Lawrence LJ, Lin B, Lowe M, Luttick A, Mason A, McKimm-Breschkin J, Parker MW, Ryan J, Smout M, Sullivan J, Tucker SP, Young PR. 2003. Structural characterization of respiratory syncytial virus fusion inhibitor escape mutants: homology model of the F protein and a syncytium formation assay. Virology 311:275–288. doi: 10.1016/S0042-6822(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 19.Hause AM, Henke DM, Avadhanula V, Shaw CA, Tapia LI, Piedra PA. 2017. Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B. PLoS One 12:e0175792. doi: 10.1371/journal.pone.0175792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battles MB, Langedijk JP, Furmanova-Hollenstein P, Chaiwatpongsakorn S, Costello HM, Kwanten L, Vranckx L, Vink P, Jaensch S, Jonckers TH, Koul A, Arnoult E, Peeples ME, Roymans D, McLellan JS. 2016. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol 12:87–93. doi: 10.1038/nchembio.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh EE, Peterson DR, Kalkanoglu AE, Lee FE, Falsey AR. 2013. Viral shedding and immune responses to respiratory syncytial virus infection in older adults. J Infect Dis 207:1424–1432. doi: 10.1093/infdis/jit038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lina B, Boucher C, Osterhaus A, Monto AS, Schutten M, Whitley RJ, Nguyen-Van-Tam JS. 2018. Five years of monitoring for the emergence of oseltamivir resistance in patients with influenza A infections in the Influenza Resistance Information Study. Influenza Other Respir Viruses 12:267–278. doi: 10.1111/irv.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baz M, Abed Y, McDonald J, Boivin G. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 43:1555–1561. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 24.Memoli MJ, Hrabal RJ, Hassantoufighi A, Eichelberger MC, Taubenberger JK. 2010. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis 50:1252–1255. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito R, Sato I, Suzuki Y, Baranovich T, Matsuda R, Ishitani N, Dapat C, Dapat IC, Zaraket H, Oguma T, Suzuki H. 2010. Reduced effectiveness of oseltamivir in children infected with oseltamivir-resistant influenza A (H1N1) viruses with His275Tyr mutation. Pediatr Infect Dis J 29:898–904. doi: 10.1097/INF.0b013e3181de9d24. [DOI] [PubMed] [Google Scholar]
- 26.Checkmahomed L, M'hamdi Z, Carbonneau J, Venable M-C, Baz M, Abed Y, Boivin G. 2020. Impact of the baloxavir-resistant polymerase acid I38T substitution on the fitness of contemporary influenza A(H1N1)pdm09 and A(H3N2) strains. J Infect Dis 221:63–70. doi: 10.1093/infdis/jiz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chesnokov A, Patel MC, Mishin VP, De La Cruz JA, Lollis L, Nguyen HT, Dugan V, Wentworth DE, Gubareva LV. 2019. Replicative fitness of seasonal influenza A viruses with decreased susceptibility to baloxavir. J Infect Dis 221:367–371. doi: 10.1093/infdis/jiz472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takashita E, Ichikawa M, Morita H, Ogawa R, Fujisaki S, Shirakura M, Miura H, Nakamura K, Kishida N, Kuwahara T, Sugawara H, Sato A, Akimoto M, Mitamura K, Abe T, Yamazaki M, Watanabe S, Hasegawa H, Odagiri T. 2019. Human-to-human transmission of influenza A(H3N2) virus with reduced susceptibility to baloxavir, Japan, February 2019. Emerg Infect Dis 25:2108–2111. doi: 10.3201/eid2511.190757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan D, Lee S, Thakkar VD, Luo M, Moore ML, Plemper RK. 2014. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc Natl Acad Sci U S A 111:E3441–E3449. doi: 10.1073/pnas.1405198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trento A, Casas I, Calderón A, Garcia-Garcia ML, Calvo C, Perez-Breña P, Melero JA. 2010. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol 84:7500–7512. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cianci C, Yu KL, Combrink K, Sin N, Pearce B, Wang A, Civiello R, Voss S, Luo G, Kadow K, Genovesi EV, Venables B, Gulgeze H, Trehan A, James J, Lamb L, Medina I, Roach J, Yang Z, Zadjura L, Colonno R, Clark J, Meanwell N, Krystal M. 2004. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob Agents Chemother 48:413–422. doi: 10.1128/aac.48.2.413-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, Van Ginderen M, Verheyen N, Kim H, Willebrords R, Bonfanti JF, Bruinzeel W, Cummings MD, van Vlijmen H, Andries K. 2010. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci U S A 107:308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are available from GenBank under accession no. MT543327 to MT544300.