Analysis of the genome sequence of Yersinia mollaretii ATCC 43969 identified the blaYEM gene, encoding YEM-1, a putative subclass B2 metallo-β-lactamase. The objectives of our work were to produce and purify YEM-1 and to complete its kinetic characterization. YEM-1 displayed the narrowest substrate range among known subclass B2 metallo-β-lactamases, since it can hydrolyze imipenem, but not other carbapenems, such as biapenem, meropenem, doripenem, and ertapenem, with high catalytic efficiency.
KEYWORDS: Yersinia mollaretii, antibiotic resistance, carbapenemase, metallo-β-lactamases
ABSTRACT
Analysis of the genome sequence of Yersinia mollaretii ATCC 43969 identified the blaYEM gene, encoding YEM-1, a putative subclass B2 metallo-β-lactamase. The objectives of our work were to produce and purify YEM-1 and to complete its kinetic characterization. YEM-1 displayed the narrowest substrate range among known subclass B2 metallo-β-lactamases, since it can hydrolyze imipenem, but not other carbapenems, such as biapenem, meropenem, doripenem, and ertapenem, with high catalytic efficiency. A possible explanation of this activity profile is the presence of tyrosine at residue 67 (loop L1), threonine at residue 156 (loop L2), and serine at residue 236 (loop L3). We showed that replacement of Y67 broadened the activity profile of the enzyme for all carbapenems but still resulted in poor activity toward the other β-lactam classes.
INTRODUCTION
The genus Yersinia belongs to the Enterobacteriaceae and comprises three main groups of human-pathogenic strains: Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica. Y. pestis, the bacterial agent of bubonic plague, escapes the effects of macrophages and causes a lethal bacteremia that cannot be treated with antibiotics (1). A potential new threat is the emergence of a multidrug-resistant Y. pestis strain (2). Y. pseudotuberculosis and Y. enterocolitica are important causes of gastroenteritis in humans following the ingestion of undercooked or contaminated pork and vegetables (3, 4). Other Yersinia species, such as Y. frederiksenii, Y. kristensenii, Y. intermedia, Y. mollaretii, Y. bercovieri, and Y. rohdei, are considered nonpathogenic for humans (5, 6). A new phylogenetic and genomic classification proposes to include different representatives of the Yersinia family in the Serratia clade (7). We focused on Y. mollaretii. This species, previously known as Y. enterocolitica-like organism biogroup A, was renamed Y. mollaretii and assigned to the new biogroup 3A by Wauters et al. (8). The majority of these strains were isolated from environmental sources, such as soil, drinking water, raw vegetables, and meat, but a few were clinical isolates from the stools of patients with gastrointestinal disease. This bacterium was shown to colonize the lower ileum of humans, but the absence of human virulence markers led to the classification of this strain as a nonpathogenic saprophyte (9). The genome sequence of the reference strain, Y. mollaretii ATCC 43969 (named CCUG26331, CDC2465-87, CIP103324, and DSMZ18520 in other biobanks), was determined (NCBI reference sequence NZ_AALD02000006.1). An analysis indicated the presence of two β-lactamase genes, namely, blaB, encoding an AmpC-like enzyme, and a gene encoding a putative subclass B2 metallo-β-lactamase (MBL) called YEM-1 (for Yersinia metallo-β-lactamase) (GenBank accession no. EEQ11779.1). Notably, the blaA gene encoding an extended-spectrum class A β-lactamase was not present in other Yersinia species (10–13). This observation is consistent with phenotypic studies showing that Y. mollaretii displayed resistance to amoxicillin and intermediate susceptibility to amoxicillin-clavulanic acid and cefoxitin (14). These studies demonstrated a clear relationship between the expression of BlaA and BlaB by Yersinia strains and their antibiotic resistance patterns. To date, the impact of the production of YEM-1 on the resistance pattern is unknown.
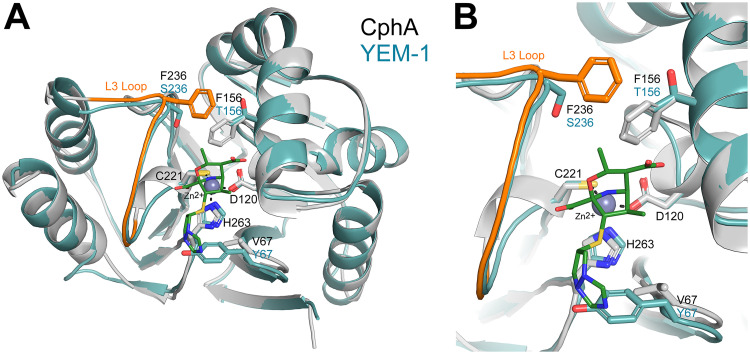
Compared to those of the characterized subclass B2 MBLs (15), such as CphA (16), Sfh-I (17), and ImiS (18), the amino acids involved in the two zinc binding sites in YEM-1 were strictly conserved (N116, H118, and H196 for the first zinc ion [Zn1] and D120, C221, and H263 for the second [Zn2]) (see Fig. 2), and the sequence identity was 54 to 60%. The value increased to 85% if YEM-1 was compared to the MBLs of Y. intermedia and Y. massiliensis. CphA is the most studied enzyme of the subclass B2 MBLs (19–21). This protein is a strict carbapenemase, and the binding of a second zinc ion inhibits its activity (22). A model of the 3-dimensional structure of YEM-1 was built using the crystallographic structure of CphA as a template (Fig. 1). As expected, the two enzymes possessed similar folds. The main difference between the two enzymes was the V67Y substitution in YEM-1 (23). V67 is conserved in all known subclass B2 members and in members of subclass B1, such as BcII, IMP-1, CcrA, and NDM-1, but not in VIM-1, where the same substitution (V67Y) was observed. This residue is part of the L1 loop and is involved in the hydrophobic wall of the active site, which plays an important role in carbapenem binding, especially for biapenem (24). Compared to CphA and the other known subclass B2 MBLs, YEM-1 has two other residues (F156 and F236) that are not conserved. In CphA, these residues are involved in an interaction between the biapenem intermediate and the enzyme (25). Moreover, F236 is part of the mobile loop L3, localized at the entrance of the active site (26). In YEM-1 and in all putative subclass B2 enzymes produced by Y. mollaretii, the residues at positions 156 and 236 are a threonine and a serine, respectively (Fig. 1). We hypothesized that the substitutions compensate for the presence of tyrosine at position 67, which reduces the size of the active site. Interestingly, for the MBLs of Y. intermedia and Y. massiliensis, the residue at position 156 is a phenylalanine. The presence of the bulky residues Y67 and F156 yields a narrow active site, which may impact the activity of the enzymes.
FIG 1.
(A) Cartoon representation of the CphA structure (gray) in complex with biapenem (green sticks) superimposed on the homology model of YEM-1 (blue). The zinc ion present in the active site is displayed as a gray sphere. (B) The residues involved in binding the zinc ion, as well as the residues at positions 67, 156, and 236, are shown as sticks.
The aims of this work were (i) to characterize the kinetic properties of wild-type (WT) YEM-1 and (ii) to evaluate the impact of the Y67, T156, and S236 substitutions on the catalytic efficiency of YEM-1 by studying various single mutants, the T156F S236F double mutant, and the Y67V T156F S236F triple mutant.
RESULTS
The blaYEM gene (747 bp) was amplified by PCR using genomic DNA from Y. mollaretii ATCC 43969 as a template. After verification of the correct nucleotide sequence of the amplimer, the gene was subcloned into pET-26b for enzyme expression. The blaYEM gene contains 15 rare codons that could affect the expression of the enzyme in Escherichia coli. Therefore, we decided to use the E. coli Rosetta(DE3) strain as a host to improve the protein yield.
Antimicrobial susceptibility tests indicated that Y. mollaretii is not resistant to all β-lactam antibiotics tested, including carbapenems. In contrast, induction of YEM-1 expression in E. coli Rosetta(DE3) resulted in resistance to imipenem (MIC, >64 μg/ml) and ertapenem (MIC, 16 μg/ml) (Table 1). We also noted that the recombinant strains remained sensitive to piperacillin, cephaloridine, and cefotaxime. Therefore, we concluded that YEM-1 is poorly active against penicillins and cephalosporins.
TABLE 1.
Comparison of the pattern of β-lactam resistance of the Yersinia mollaretii strain with those mediated by the expression of YEM-1 in E. coli DH5α and E. coli Rosetta(DE3)
| Antibiotic | MIC for: |
||||
|---|---|---|---|---|---|
| E. coli DH5α/pET-26b-blaYEM-1 | E. coli Rosetta(DE3)/pET-26b-blaYEM-1 | E. coli DH5α | E. coli Rosetta(DE3) | Yersinia mollaretii ATCC 43969 | |
| Imipenem | 0.12 | >64 | <0.12 | <0.12 | 0.5 |
| Meropenem | 0.12 | 1 | <0.12 | <0.12 | <0.12 |
| Ertapenem | 0.12 | 16 | <0.12 | <0.12 | <0.12 |
| Doripenem | 0.12 | 2 | <0.12 | <0.12 | <0.12 |
| Biapenem | <0.12 | <0.12 | <0.12 | <0.12 | <0.12 |
| Piperacillin | 1 | 32 | 0.5 | 32 | 4 |
| Cephaloridine | 2 | 4 | 0.5 | 4 | 4 |
| Cefotaxime | <0.12 | 0.5 | <0.12 | 0.5 | 0.12 |
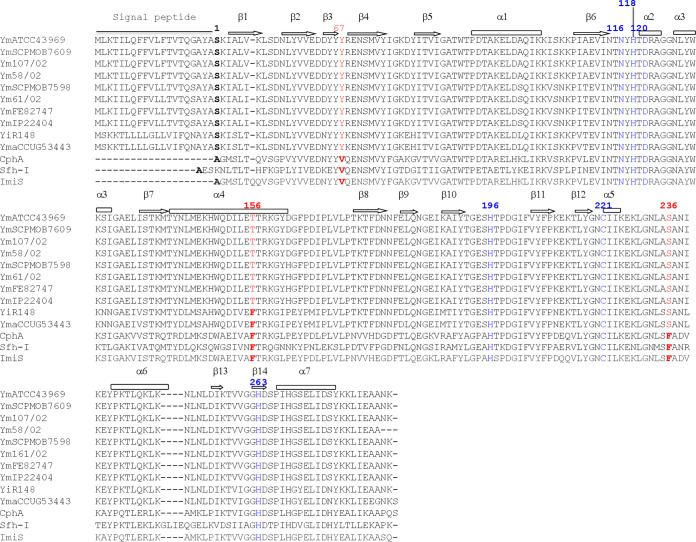
High levels of protein were produced as cytoplasmic inclusion bodies when the cultures were grown at 37°C. We succeeded in producing a soluble form of YEM-1 when the cultures were grown overnight (O/N) at 18°C in the presence of 0.1 mM (final concentration) isopropyl β-d-1-thiogalactopyranoside (IPTG). The enzyme was purified in three chromatographic steps as described in Materials and Methods, with a final yield of 6 mg/liter of culture. Analysis of the protein solution by electrospray ionization-quadrupole time of flight mass spectrometry (ESI–Q-TOF MS) confirmed the homogeneity of the protein and determined the molecular weight of the mature form of the β-lactamase. Its value (25,634 Da) indicated that the pre-β-lactamase includes a signal peptide of 20 amino acids and that the mature enzyme contains 229 amino acids, with the N-terminal sequence corresponding to NH2-SKIA. A BLAST-P analysis of the known Yersinia genomes showed the presence of a YEM-1-like MBL in other Y. mollaretii strains but also in Y. intermedia and Y. massiliensis strains. ClustalW alignment of these enzymes showed a minimal identity value of 90% (Fig. 2). We noted that Y67 and S236 are strictly conserved in all YEM-like sequences. We hypothesized that the only major difference among Y. mollaretii, Y. intermedia, and Y. massiliensis is the presence of F156 in the latter two species.
FIG 2.
ClustalW alignment of subclass B2 metallo-β-lactamases produced by Yersinia species. Shown are metallo-β-lactamase sequences for Y. mollaretii ATCC 43969 (NCBI Protein accession no. WP_004874166.1), Y. mollaretii SCPM-O-B-7609 (WP_004874166.1), Y. mollaretii 107/02 (WP_004874166.1), Y. mollaretii 58/02 (WP_050540509.1), Y. mollaretii SCPM-O-B-7598 (WP_049645673.1), Y. mollaretii 61/02 (WP_049645673.1), Y. mollaretii FE82747 (WP_049645673.1), Y. mollaretii IP22404 (WP_049645673.1), Y. intermedia R148 (WP_019211763.1), Y. massiliensis CCUG53443 (WP_019211763.1), CphA (from Aeromonas hydrophila) (WP_063844283), Sfh-I (from Serratia fonticola) (WP_071766619), and ImiS (from Aeromonas veronii) (WP_063865298). Ym, Yersinia mollaretii; Yi, Yersinia intermedia; Yma, Yersinia massiliensis. Residues at positions 67, 156, and 236 are shown in red (BBL numbering). The zinc ligand residues are shown in blue.
YEM-1 hydrolyzed carbapenems, but we could not detect any hydrolytic activities toward penicillins or cephalosporins (see Table S2 in the supplemental material). These preliminary data confirmed that YEM-1 belongs to the subclass B2 MBLs. Therefore, we could calculate only the steady-state kinetic parameters against a representative set of carbapenem antibiotics (Table 2). In contrast to CphA, YEM-1 efficiently hydrolyzed only imipenem. For the other carbapenems, we noted a large decrease in the kcat value and a major increase in Km. All these data confirmed that YEM-1 possesses a narrow activity spectrum. For example, its catalytic efficiencies against doripenem and biapenem were 100- and 430-fold lower than that against imipenem (Table 2). We concluded that YEM-1 is an imipenemase.
TABLE 2.
Kinetic parameters of WT YEM-1 and the mutants compared to those for CphA
| Antibiotic or ion | Kinetic parameter (unit)a | Valueb for the following MBL: |
||||||
|---|---|---|---|---|---|---|---|---|
| CphA | WT YEM-1 | YEM-1 Y67V | YEM-1 T156F | YEM-1 S236F | YEM-1 T156F S236F | YEM-1 Y67V T156F S236F | ||
| Imipenem | kcat (s–1) | 1,320 | 480 | 1,770 | 190 | 170 | 760 | 40 |
| Km (μM) | 310 | 160 | 270 | 32 | 90 | 20 | 60 | |
| kcat/Km (μM–1 s−1) | 3.2 (1) | 3 (1) | 7 (1) | 6 (1) | 2 (1) | 38 (1) | 0.7 (1) | |
| Meropenem | kcat (s–1) | 1,440 | >400 | 800 | >230 | >15 | >75 | >3,300 |
| Km (μM) | 630 | >2,500 | 400 | >2,500 | >2,500 | >2,500 | >2,500 | |
| kcat/Km (μM–1 s−1) | 2.23 (1.4) | 0.15 (20) | 2 (3.5) | 9 × 10–2 (66) | 10–2 (200) | 3 × 10–2 (1,260) | 1.3 (1.3) | |
| Biapenem | kcat (s–1) | 1,420 | >16 | >1,000 | >11 | >0.8 | >15 | >300 |
| Km (μM) | 1,500 | >2,500 | >2,500 | >2,500 | >2,500 | >2,500 | >2,500 | |
| kcat/Km (μM–1 s−1) | 0.96 (3) | 7 × 10–3 (430) | 0.4 (17) | 4 × 10–3 (1,500) | 3 × 10–4 (6.6 × 104) | 0.006 (6,300) | 0.13 (6) | |
| Ertapenem | kcat (s–1) | 1,410 | >400 | 1,650 | >400 | >20 | >400 | 810 |
| Km (μM) | 230 | >2,500 | 120 | >2,500 | >2,500 | >2,500 | 80 | |
| kcat/Km (μM–1 s−1) | 6.8 (0.4) | 0.15 (20) | 14 (0.5) | 0.15 (40) | 0.01 (200) | 0.15 (250) | 8 (1.1) | |
| Doripenem | kcat (s–1) | 940 | >100 | >1,300 | >20 | >5 | >10 | >50 |
| Km (μM) | 580 | >2,500 | >2,500 | >2,500 | >2,500 | >2,500 | >2,500 | |
| kcat/Km (μM–1 s−1) | 1.6 (2) | 0.03 (100) | 0.5 (14) | 0.009 (660) | 2 × 10–3 (103) | 0.004 (9,500) | 0.02 (35) | |
| Zn2 | Kd (μM) | 46 | 360 | 330 | 410 | 300 | 400 | 270 |
Kd values are the apparent dissociation constants for zinc ions.
Numbers in parentheses are ratios of the kcat/Km value against imipenem to the kcat/Km value against the indicated carbapenem for the given enzyme.
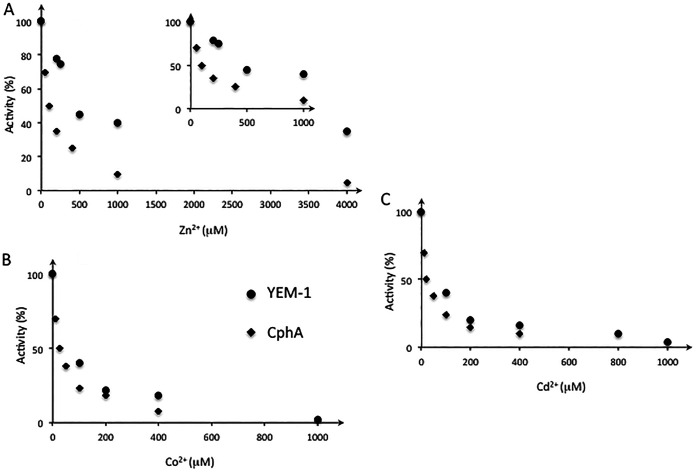
The Zn content of YEM-1 was determined at pH 6.0 by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) (Table S3). We detected only 1 Zn ion per molecule of enzyme (0.9 ± 0.2) when the buffer did not contain added metal ions. We expected that, as observed for the other members of the subclass B2 MBLs, YEM-1 had maximal activity in the monozinc form. We showed that its activity was affected by the presence of increasing concentrations of Zn2+, Co2+, and Cd2+ (Fig. 3A to C). Interestingly, YEM-1 displayed a lower affinity (apparent dissociation constant [Kd], 360 μM) for Zn2 than CphA (Kd, 46 μM), and its residual activity in the presence of 4 mM Zn2+ was equal to 40% of its activity at 1 μM Zn2+. With Co2+ and Cd2+ ions, we observed complete inactivation of the WT and mutant enzymes at a 1 mM metal ion concentration. We hypothesized that this phenomenon may be due to the binding of a second metal ion that affects MBL activity. Five YEM-1 mutants (three single mutants [the Y67V, T156F, and S236F mutants], the T156F S236F double mutant, and the Y67V T156F S236F triple mutant) were produced and purified under the same conditions defined for the WT enzyme. The kinetic results confirmed that all the mutations together do not favor broadening of the activity profile of YEM-1 against other β-lactam classes or its apparent affinity for the binding of the second zinc ion (Table 2). The Y67V substitution yielded a global increase in catalytic efficiency against carbapenems, with the exception of imipenem. Notably, the YEM-1 Y67V mutant had an activity profile similar to that of CphA. For ertapenem and meropenem, the Km values were lower than those of WT YEM-1 and comparable to those of CphA. T156 and S236 instead of phenylalanine decreased the Km value for imipenem from those of WT YEM-1 and CphA, but the Km values for the other carbapenems tested were not modified. Notably, the catalytic efficiencies of the two mutants decreased from that of the WT enzyme. Characterization of the Y67V T156F S236F triple mutant indicated that the presence of a valine residue at position 67 is essential for a broad activity profile toward carbapenems (Table 2). The results showed that the Y67V mutation resulted in improved activity toward carbapenems, except for doripenem. For the T156F and S236F mutants, and for the T156F S236F double mutant, we noted that the mutations affected the Km values and, consequently, the catalytic efficiencies of the mutants relative to those of YEM-1, except for imipenem.
FIG 3.
Plot of the residual activities (expressed as percentages) of the CphA and YEM-1 enzymes against increasing Zn(II) (A), Co(II) (B), and Cd(II) (C) concentrations. In the presence of Zn ions, the apparent dissociation constant was measured as described in Materials and Methods. Each point represents the average of three measurements made with 100 μM imipenem. The enzyme concentrations were 0.5 and 1 nM for CphA and YEM-1, respectively.
Finally, all the enzymes tested were inhibited in the presence of zinc ions. The apparent Kd values for the binding of the second zinc ion are comparable for the WT and the Y67V mutant (Kd, 400 μM). The mutations did not affect the influence of metal ions on the activity of YEM-1 or the binding of Co2+ and Cd2+.
The YEM-1 model obtained with Yasara is similar to the structure of CphA, used as a template (root mean square deviation [RMSD], 0.36 Å over 209 Cα), with a single 3-amino-acid insertion before the C-terminal helix that is not expected to affect substrate hydrolysis. The active site is also well conserved, with an RMSD of 0.32 Å for all atoms of the six residues involved in zinc binding. Compared to CphA, YEM-1 presents three important differences (25): a tyrosine at position 67 would interfere with the bulky side chain of the surrounding active site, and the replacement of V67 by a tyrosine, as well as the replacement of F156 and F236 with a threonine and a serine, respectively, were observed. According to the crystal structure of the CphA-biapenem complex (PDB code 1X8I), Y67 can participate in the stabilization of a complex between carbapenems and CphA mediated by hydrophobic contacts. On the other side of the active site, the other two mutations eliminate the hydrophobic interactions between the two phenylalanines present in CphA and widen the catalytic site cavity.
DISCUSSION
Despite the high percentage of sequence identity (58%) between YEM-1 and CphA, we noted that YEM-1 is less effective than CphA against carbapenems. The catalytic efficiency of YEM-1 decreased by at least a factor of 10, except for that against imipenem. Furthermore, for the majority of the carbapenems, we could not determine the individual kinetic constants, because the Km values were higher than the substrate concentrations tested, yielding pseudo-first-order kinetics. Analysis of the structure of the CphA-hydrolyzed biapenem complex showed the importance of H263 in the interaction with biapenem sulfur atoms and of V67 and W83 for stabilizing the carbapenem intermediate in the active site (25). H263 and W87 are conserved in YEM-1, but it contains a tyrosine at position 67 instead of a valine, as observed for all the other subclass B2 MBLs and the majority of subclass B1 MBLs. The importance of Val at position 67 was confirmed by the fact that the Y67V mutant was better able to hydrolyze carbapenems than the WT enzyme. A computational analysis of the generation of CphA variants under imipenem selection (27) showed that position 67 tolerates multiple amino acid substitutions. The different mutants retained catalytic efficiencies comparable to that of the WT enzyme. In the case of YEM-1, we showed that the presence of Y67 reduced its activity toward the other carbapenems tested. Substitution of valine at this position increased YEM-1 activity to the levels observed for the other subclass B2 MBLs. Like CphA, YEM-1 was inhibited in the presence of Zn2+ ions. Nevertheless, the apparent dissociation constant for zinc was higher for YEM-1 than for CphA. This result indicates that YEM-1 displays its maximum activity in the monozinc form and that it is less sensitive than CphA and ImiS in the presence of increasing concentrations of zinc ions. CphA and ImiS can bind up to 2 equivalents of Zn(II) or Co(II), but the maximal activity is with 1 equivalent of metal ions (28). Further verification of the higher dissociation constant of YEM-1 for Zn2+ would require the production of the YEM-1 apoenzyme form and determination of the affinity constants for Zn1 and Zn2 (29).
The lower affinity of the inhibitory Zn2+ binding site of YEM-1 could result from F156T and F236S substitutions, because they induce the loss of a hydrophobic rigidifying interaction between the two sides of the active site in the vicinity of the second Zn2+ binding site (Fig. 3A). The Y. intermedia and Y. massiliensis MBLs display the same mutation as the YEM-1 T156F mutant (Fig. 2). We concluded that they display lower catalytic efficiency toward doripenem than YEM-1.
We also noted that YEM-1 is still active at high zinc ion concentrations. Determination of the X-ray structure of the dizinc enzyme to identify the structural determinants that favor the activity of the dizinc MBL would be of interest.
In conclusion, Y. mollaretii produces two β-lactamases that could result in resistance to a large panel of β-lactam antibiotics. In 2017, Ribeiro et al. (30) identified Y. mollaretii in biofilms isolated from the dental caries of children. Therefore, the presence of β-lactamase genes in the opportunist strain Y. mollaretii should be further assessed.
MATERIALS AND METHODS
Antibiotics, bacterial strains, and plasmids.
Kanamycin was purchased from MP Biomedicals, France. Ampicillin was obtained from SA Bristol-Myers Squibb Belgium, N.V. Imipenem and ertapenem were purchased from MSD, Belgium. Meropenem was obtained from Astro Zeneca Belgium. Doripenem and biapenem were purchased from Sigma-Aldrich, Belgium. E. coli DH5α (Life Technologies, Belgium) and E. coli Rosetta(DE3) (Novagen, Inc., Madison, WI) were used as recipients for the cloned genes and for enzyme expression. The gene was initially cloned into the pJET1.2 vector (Thermo Fisher Scientific, Belgium) and then subcloned into pET-26b (Novagen, Inc., Madison, WI).
Cloning of the blaYEM-1 gene.
The blaYEM-1 gene was isolated by PCR from the purified genomic DNA of Y. mollaretii Wauters DSMZ18520 (Y. mollaretii ATCC 43969), purchased from the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures). Genomic DNA was extracted with a Wizard Genomic DNA purification kit (Promega Benelux B.V.). Two oligonucleotides, YblaUP_NdeI and YblaRP_BamHI (see Table S1 in the supplemental material), were synthesized on the basis of the sequence of the Y. mollaretii genome (NCBI reference sequence NZ_AALD02000006.1). PCR conditions were as follows: incubation at 98°C for 30 s and 35 cycles of amplification (denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s). Q5 High-Fidelity DNA polymerase (New England BioLabs, Inc.) was used in this work. The PCR product was cloned into the pJET1.2 vector. After verification of the nucleotide sequence, blaYEM-1 was subcloned into pET-26b, which had been previously digested by the NdeI and BamHI restriction enzymes.
Site-directed mutagenesis.
To change Tyr to Val at position 67, we used the oligonucleotides Y67V_fwd and Y67_rev (Table S1). The sequences of the oligonucleotides designed to generate the T156F (T156F_fwd and T156F_rev) and S236F (S236F_fwd and S236F_rev) mutants are shown in Table S1. pET-26b-blaYEM-1 was used as a template for mutagenesis, and the amplification conditions were as follows: incubation at 98°C for 30 s and 30 cycles of amplification (denaturation at 98°C for 10 s, annealing at 55°C for 5 min, and extension at 72°C for 30 s). After amplification, the PCR fragments were digested by the DpnI restriction enzyme to eliminate the template DNA. The T156F S236F double mutant and the Y67V T156F S236F triple mutant were obtained by consecutive PCR. The recombinant plasmids were selected on LB agar plates supplemented with kanamycin at 50 μg/ml. The presence of the desired mutations was confirmed by sequencing of DNA fragments based on the Sanger method with an ABI 3700 automated capillary gel electrophoresis system from Applied Biosystems. The sequences were assessed by the GIGA Genomics platform (GIGA, University of Liège).
Antimicrobial susceptibility test.
The phenotypic profile was assessed in the E. coli Rosetta(DE3)/pET-26b-blaYEM-1, E. coli DH5α/pET-26b-blaYEM-1, E. coli Rosetta(DE3), E. coli DH5α, and Y. mollaretii ATCC 43969 strains using the microdilution method and a bacterial inoculum of 5 × 105 CFU/ml according to Clinical and Laboratory Standards Institute (CLSI) performance standards (31). MIC experiments were performed in duplicate using cation-adjusted Mueller-Hinton broth (CAMHB). MICs for E. coli Rosetta(DE3)/pET-26b-blaYEM-1 were determined in CAMHB supplemented with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to maximize production of the YEM-1 enzyme.
Expression and purification of WT and mutant YEM-1.
All the enzymes were produced in E. coli Rosetta(DE3) carrying pET-26b-blaYEM-1 and pET-26b-blaYEM-1 mutants. The different recombinant strains were grown on Terrific Broth (TB) medium supplemented with kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml). The precultures were incubated O/N at 37°C under agitation, and 40 ml was used to inoculate 1 liter of fresh TB medium supplemented as described above. IPTG (final concentration, 100 μM) was added when the culture reached an A600 of 0.7, and the cultures were incubated O/N at 18°C. Cells were harvested by centrifugation (5,000 × g for 10 min at 4°C), and the pellets were resuspended in 40 ml of 50 mM morpholineethanesulfonic acid (MES) buffer (pH 6.0) (buffer A). The bacteria were disrupted at a pressure of 5,500 kPa with a cell disruptor (Emulsiflex C3; Avestin GmbH, Germany). The lysates were centrifuged at 45,000 × g for 30 min. The cleared supernatants were filtered through a 22-μm filter and then loaded onto a HiTrap SP-HP 5-ml column (GE Healthcare, Belgium) equilibrated with buffer A. The enzymes were eluted with a salt gradient using buffer A and 50 mM MES with 1 M NaCl (pH 6.0) (buffer B). The fractions containing β-lactamase activity were pooled and then dialyzed O/N in 25 mM HEPES (pH 7). The active fractions were then loaded onto a column containing a pentadentate chelator (PDC; 5 ml) equilibrated with 25 mM HEPES (pH 7.0) (buffer A). The enzymes were eluted with a salt gradient between buffer A and 25 mM HEPES with 1 M NaCl (pH 7.0) (buffer B). The active fractions were collected and concentrated on a YM-10 membrane (Amicon, Beverly, MA) to a final volume of 2 ml. Subsequently, the sample was loaded onto a Superdex 75 GL (10/300) molecular sieve column (GE Healthcare, Belgium) equilibrated in 50 mM MES buffer (pH 6.0) containing 0.2 M NaCl. At the end of each purification step, β-lactamase activity was routinely measured spectrophotometrically by following the hydrolysis of a solution of 100 μM imipenem by YEM-1. The CphA enzyme was purified previously as described in reference 15. The molecular mass of YEM-1 was determined by ESI–Q-TOF MS. Sodium ions were removed by three cycles of concentration/dilution in 25 mM ammonium acetate buffer by filtration and centrifugation using an Amicon Ultra-15 system (Millipore) with a molecular mass cutoff of 20,000 Da. Analysis was performed under acidic conditions in the presence of 0.5% (final concentration) formic acid (pH 3.0).
ICP-AES measurements.
One milliliter of each protein sample (concentrations, 12 and 24 μM) was dialyzed overnight against 1 liter of 50 mM MES (pH 6.0). Then 150 μl of each of the different samples (two aliquots for each protein concentration) was diluted in 10 ml (final volume) with MES buffer. The samples were then acid digested in DigiPREP tubes with 3 ml of HNO3 (≥65% [wt/wt]) (Sigma-Aldrich) on a DigiPREP Graphite Block Digestion system (SCP Science) as follows: 15 min at 45°C, 15 min at 65°C, and 90 min at 105°C. After cooling, the sample volumes were adjusted to 10 ml with 200 μl of HNO3 (≥65% [wt/wt]) and Milli-Q water. Metal concentrations were determined by ICP-AES (Vista AX system; Varian).
Kinetic characterization.
Kinetic experiments were performed by following the hydrolysis of each substrate at 30°C in 50 mM MES buffer (pH 6.0). The data were collected with a Specord 50 Plus spectrophotometer (Analytik Jena, Germany). Each kinetic value is the mean of three different measurements. The error was below 5%. Kinetic parameters were determined under the initial rate and by the linearization of the Michaelis-Menten equation by the Hanes-Woolf method.
The metal ion dependence of Y. mollaretii metallo-β-lactamase activity was characterized as follows. The enzyme activity was determined by monitoring the initial rate of hydrolysis of 100 μM imipenem at 30°C in 50 mM MES, pH 6, containing increasing concentrations of metal ions (ZnCl2, CoCl2, or CdCl2) from 0 to 4 mM. The diluted enzyme solutions were prepared with buffers containing 50 μg ml−1 bovine serum albumin. In our experiments, the final enzyme concentrations were equal to 5 and 2 nM. The dependence of the residual activity (expressed as a percentage) of the Y. mollaretii enzyme on increasing metal ion concentrations could be fitted using the equation 100(vi/v0) = Ki/([Zn2+] + Ki), where vi and v0 correspond to the initial rate of imipenem hydrolysis in the presence and absence of metal ion, respectively.
Molecular modeling.
A model of YEM-1 was obtained with Yasara software (32) using the CphA structure (PDB code 1X8G) as a template. YEM-1 and CphA share 58% sequence identity. The overall quality Z-score was ranked as good (–0.079), resulting from 3-dimensional packing considered good (Z-score, –0.982) and dihedrals (Z-score, 0.778) and 1-dimensional packing (Z-score, 0.679) considered optimal. Figures were prepared using PyMOL (PyMOL Molecular Graphics System; Schrödinger, LLC).
Supplementary Material
ACKNOWLEDGMENTS
R. Esposito was supported by an Erasmus student fellowship from the University of Federico II of Naples (Italy), and S. Di Costanzo was supported by an Erasmus+ fellowship for traineeship from the University of L’Aquila (Italy). This work was also supported by the University of Liège and by the Fund for Scientific Research (FRS-FNRS) Belgium.
The ICP-AES measurements were performed by M. Carnal and B. Bosman (InBioS PhytoSystems, Laboratory of Plant and Microbial Ecology, Department of Biology, Ecology, Evolution, University of Liège, Belgium).
We declare that there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pechous RD, Sivaraman V, Stasulli NM, Goldman WE. 2016. Pneumonic plague: the darker side of Yersinia pestis. Trends Microbiol 24:190–197. doi: 10.1016/j.tim.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bancerz-Kisiel A, Szweda W. 2015. Yersiniosis — a zoonotic foodborne disease of relevance to public health. Ann Agric Environ Med 22:397–402. doi: 10.5604/12321966.1167700. [DOI] [PubMed] [Google Scholar]
- 4.Mogren L, Windstam S, Boqvist S, Vågsholm I, Söderqvist K, Rosberg AK, Lindén J, Mulaosmanovic E, Karlsson M, Uhlig E, Håkansson Å, Alsanius B. 2018. The hurdle approach—a holistic concept for controlling food safety risks associated with pathogenic bacterial contamination of leafy green vegetables. A review. Front Microbiol 9:1965. doi: 10.3389/fmicb.2018.01965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulakvelidze A. 2000. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect 2:497–513. doi: 10.1016/S1286-4579(00)00311-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen PE, Cook C, Stewart AC, Nagarajan N, Sommer DD, Pop M, Thomason B, Thomason MP, Lentz S, Nolan N, Sozhamannan S, Sulakvelidze A, Mateczun A, Du L, Zwick ME, Read TD. 2010. Genomic characterization of the Yersinia genus. Genome Biol 11:R1. doi: 10.1186/gb-2010-11-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adeolu M, Alnajar S, Naushad S, Gupta R. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 8.Wauters G, Janssens M, Steigerwalt AG, Brenner DJ. 1988. Yersinia mollaretii sp. nov. and Yersinia bercovieri sp. nov., formerly called Yersinia enterocolitica biogroups 3A and 3B. Int J Syst Bacteriol 38:424–429. doi: 10.1099/00207713-38-4-424. [DOI] [Google Scholar]
- 9.McNally A, Thomson NR, Reuter S, Wren BW. 2016. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat Rev Microbiol 14:177–190. doi: 10.1038/nrmicro.2015.29. [DOI] [PubMed] [Google Scholar]
- 10.Stock I, Heisig P, Wiedemann B. 1999. Expression of β-lactamases in Yersinia enterocolitica strains of biovars 2, 4 and 5. J Med Microbiol 48:1023–1027. doi: 10.1099/00222615-48-11-1023. [DOI] [PubMed] [Google Scholar]
- 11.Pham JN, Bell SM, Martin L, Carniel E. 2000. The β-lactamases and β-lactam antibiotic susceptibility of Yersinia enterocolitica. J Antimicrob Chemother 46:951–957. doi: 10.1093/jac/46.6.951. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Mittal S, Mallik S, Virdi JS. 2006. Molecular characterization of β-lactamase genes blaA and blaB of Yersinia enterocolitica biovar 1A. FEMS Microbiol Lett 257:319–327. doi: 10.1111/j.1574-6968.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 13.Mittal S, Mallik S, Sharma S, Virdi JS. 2007. Characteristics of β-lactamases and their genes (blaA and blaB) in Yersinia intermedia and Y. frederiksenii. BMC Microbiol 7:25. doi: 10.1186/1471-2180-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stock I, Henrichfreise B, Wiedemann B. 2002. Natural antibiotic susceptibility and biochemical profiles of Yersinia enterocolitica-like strains: Y. bercovieri, Y. mollaretii, Y. aldovae and ‘Y. ruckeri’. J Med Microbiol 51:56–69. doi: 10.1099/0022-1317-51-1-56. [DOI] [PubMed] [Google Scholar]
- 15.Bottoni C, Perilli M, Marcoccia F, Piccirilli A, Pellegrini C, Colapietro M, Sabatini A, Celenza G, Kerff F, Amicosante G, Galleni M, Mercuri PS. 2016. Kinetic studies on CphA mutants reveal the role of the P158–P172 loop in activity versus carbapenems. Antimicrob Agents Chemother 60:3123–3126. doi: 10.1128/AAC.01703-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massidda O, Rossolini GM, Satta G. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol 173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saavedra MJ, Peixe L, Sousa JC, Henriques I, Alves A, Correia A. 2003. Sfh-I, a subclass B2 metallo-β-lactamase from a Serratia fonticola environmental isolate. Antimicrob Agents Chemother 47:2330–2333. doi: 10.1128/aac.47.7.2330-2333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh TR, Neville WA, Haran MH, Tolson D, Payne DJ, Bateson JH, MacGowan AP, Bennett PM. 1998. Nucleotide and amino acid sequences of the metallo-β-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob Agents Chemother 42:436–439. doi: 10.1128/AAC.42.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez Valladares M, Kiefer M, Heinz U, Soto RP, Meyer-Klaucke W, Nolting HF, Zeppezauer M, Galleni M, Frère JM, Rossolini GM, Amicosante G, Adolph HW. 2000. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett 467:221–225. doi: 10.1016/s0014-5793(00)01102-9. [DOI] [PubMed] [Google Scholar]
- 20.Vanhove M, Zakhem M, Devreese B, Franceschini N, Anne C, Bebrone C, Amicosante G, Rossolini GM, Van Beeumen J, Frère JM, Galleni M. 2003. Role of Cys221 and Asn116 in the zinc-binding sites of the Aeromonas hydrophila metallo-β-lactamase. Cell Mol Life Sci 60:2501–2509. doi: 10.1007/s00018-003-3092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bebrone C, Anne C, Kerff F, Garau G, De Vriendt K, Lantin R, Devreese B, Van Beeumen J, Dideberg O, Frère JM, Galleni M. 2008. Mutational analysis of the zinc- and substrate-binding sites in the CphA metallo-β-lactamase from Aeromonas hydrophila. Biochem J 414:151–159. doi: 10.1042/BJ20080375. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez Valladares M, Felici A, Weber G, Adolph HW, Zeppezauer M, Rossolini GM, Amicosante G, Frère JM, Galleni M. 1997. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry 36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 23.Garau G, García-Sáez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frère JM, Dideberg O. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother 48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simona F, Magistrato A, Vera DM, Garau G, Vila AJ, Carloni P. 2007. Protonation state and substrate binding to B2 metallo-β-lactamase CphA from Aeromonas hydrofila. Proteins 69:595–605. doi: 10.1002/prot.21476. [DOI] [PubMed] [Google Scholar]
- 25.Garau G, Bebrone C, Anne C, Galleni M, Frère JM, Dideberg O. 2005. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J Mol Biol 345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 26.Bebrone C, Delbrück H, Kupper MB, Schlömer P, Willmann C, Frère JM, Fischer R, Galleni M, Hoffmann KM. 2009. The structure of the dizinc subclass B2 metallo-β-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site. Antimicrob Agents Chemother 53:4464–4471. doi: 10.1128/AAC.00288-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Z, Mehta SC, Adamski CJ, Gibbs RA, Palzkill T. 2016. Deep sequencing of random mutant libraries reveals the active site of the narrow specificity CphA metallo-β-lactamase is fragile to mutations. Sci Rep 6:33195. doi: 10.1038/srep33195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crawford PA, Yang KW, Sharma N, Bennett B, Crowder MW. 2005. Spectroscopic studies on cobalt(II)-substituted metallo-β-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry 44:5168–5176. doi: 10.1021/bi047463s. [DOI] [PubMed] [Google Scholar]
- 29.De Vriendt K, Van Driessche G, Devreese B, Bebrone C, Anne C, Frère JM, Galleni M, Van Beeumen J. 2006. Monitoring the zinc affinity of the metallo-β-lactamase CphA by automated nanoESI-MS. J Am Soc Mass Spectrom 17:180–188. doi: 10.1016/j.jasms.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro AA, Azcarate-Peril MA, Cadenas MB, Butz N, Paster BJ, Chen T, Bair E, Arnold RR. 2017. The oral bacterial microbiome of occlusal surfaces in children and its association with diet and caries. PLoS One 12:e0180621. doi: 10.1371/journal.pone.0180621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Krieger E, Vriend G. 2014. YASARA View — molecular graphics for all devices — from smartphones to workstations. Bioinformatics 30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.