Infections caused by Candida glabrata have caused worldwide concern, especially when they are associated with increasing echinocandin and azole resistance. In this study, we analyzed the molecular mechanisms of azole and echinocandin resistance in C. glabrata isolates obtained from hospitalized patients in Japan from 1997 to 2019. All isolates were checked phenotypically for resistance and genotypically for mutations in PDR1, ERG11, hot spot 1 (HS1), HS2, and HS3 of FKS1, and HS1 and HS2 of FKS2, and all isolates were genotyped by multilocus sequence typing (MLST).
KEYWORDS: C. glabrata, azole resistance, echinocandin resistance, multidrug resistance, Japan
ABSTRACT
Infections caused by Candida glabrata have caused worldwide concern, especially when they are associated with increasing echinocandin and azole resistance. In this study, we analyzed the molecular mechanisms of azole and echinocandin resistance in C. glabrata isolates obtained from hospitalized patients in Japan from 1997 to 2019. All isolates were checked phenotypically for resistance and genotypically for mutations in PDR1, ERG11, hot spot 1 (HS1), HS2, and HS3 of FKS1, and HS1 and HS2 of FKS2, and all isolates were genotyped by multilocus sequence typing (MLST). Interestingly, 32.6% of the isolates were resistant to caspofungin, and 4.7% were resistant to micafungin. The isolates showed low rates of resistance to azoles, ranging from 2.3% to 9.3%, and only 4.7% of the isolates were non-wild type for flucytosine susceptibility. For the first time in Japan, 4.7% of the isolates were identified as multidrug-resistant strains. Nonsynonymous mutations in PDR1, including two novel mutations associated with azole resistance, were identified in 39.5% of the isolates, and a single nonsynonymous mutation was identified in ERG11. Nine isolates from the same patient harbored nonsynonymous mutations in HS1 of FKS2, and a single isolate harbored a single nonsynonymous mutation in HS1 of FKS1. MLST genotyping revealed 13 different sequence types (STs), with 3 new STs, and ST7 was the most prevalent among the patients (35%) and was associated with high resistance rates. Our results are of crucial clinical concern, since understanding the molecular mechanisms underlying fungal resistance is imperative for guiding specific therapy for efficient patient treatment and promoting strategies to prevent epidemic spread.
INTRODUCTION
Fungal infections represent a major worldwide threat that is associated with patients hospitalized with serious underlying diseases or immunocompromised patients (1). Furthermore, fungal infections are associated with major economic burdens on the health care system. According to a recent estimate, fungal infections in the United States alone cost $7.2 billion in 2017, with Candida and Aspergillus infections responsible for the highest total hospitalization costs of any disease (2). During the past decade, the number of Candida infections has progressively increased, and they have been associated with high morbidity and mortality rates in critically ill patients (3). Although Candida albicans is still the main cause of hospital-acquired fungal infection, an increasing incidence of infections with non-albicans Candida species, such as Candida glabrata, is reported worldwide and has gained special attention due to the emergence of echinocandin- and azole-resistant strains (3–8). For instance, in Australia, C. glabrata candidemia increased by 1.7-fold between 2004 and 2017 (6), and C. glabrata infections increased significantly, with a decrease in C. albicans infections, from 2008 to 2014 in Japan (8).
Azole resistance in C. glabrata is mediated mainly by functional mutations in the transcriptional regulator PDR1, which is responsible for the overexpression of multidrug transporters (5, 9). However, other mechanisms might be involved, such as mutations in the ERG11 gene, which is also associated with amphotericin B (AMB) resistance (10). Echinocandin resistance in C. glabrata has been basically attributed to functional mutations in the hot spot (HS) regions of both the FKS1 and FKS2 genes (9).
The worldwide emergence and spread of azole and echinocandin resistance, especially in C. glabrata, which is innately less susceptible to azoles than other species and exhibits cross-resistance to azoles, are issues of grave concern (5, 7, 11) in view of the limited antifungal options and the possibility of high mortality rates associated with infection. In Japan, little is known about the prevalence and the genetic basis of azole and echinocandin resistance in C. glabrata. Therefore, the present study was designed to elucidate the genetic mechanisms responsible for resistance in C. glabrata. In addition, the genetic relationships between the isolates were examined by multilocus sequence typing (MLST).
RESULTS
Clinical features of isolates.
Clinical information for the isolates tested in this study is listed in Table S1 in the supplemental material. In total, 58 clinical isolates of C. glabrata were recovered from 43 patients, and the median age of the 31 patients of known age was 67 years. Among the patients, 62.8% (27/43) were female, 27.9% (12/43) were male, and the sex of 9.3% (4/43) was unknown. The majority of the isolates were recovered from patients in Chiba prefecture (65.1% [28/43]), followed by Hyogo prefecture (7% [3/43]), Hokkaido prefecture (7% [3/43]), Okayama prefecture (4.7% [2/43]), and Fukuoka, Osaka, and Tokyo prefectures at 2.3% (1/43) each. Four isolates were from unconfirmed prefectures. The isolates were recovered mainly from blood (50% [29/58]), followed by catheter backflow blood (20.7% [12/58]), sputum (8.6% [5/58]), the oral cavity, urine, and vagina (3.4% [2/58] each), and the nasal cavity, skin, pleural effusion, an abdominal abscess, the cornea, and an unknown site at 1.7% (1/58) each. The majority of the isolates (39.5% [17/43]) were recovered from patients suffering from underlying diseases, including neoplasms, hematologic malignancies, and diabetes mellitus, followed by unknown conditions (25.6% [11/43]), conditions related to blood-associated diseases, congenital conditions, and genetic disorders (7% [3/43] each), oral candidiasis and respiratory disorders at 4.7% (2/43) each, and hepatic and gastric disorders at 2.3% (1/43) each. Five patients were treated with micafungin (MFG), while the antifungal treatment of the other patients is unknown.
Antifungal susceptibility profiling.
For azoles, only two isolates (4.7% [2/43]) showed resistance to fluconazole (FLC) (MIC, ≥64 μg/ml), and the rest were susceptible-dose dependent (SDD), while four isolates (9.3% [4/43]) and one isolate (2.3% [1/43]) were noted to have MIC values higher than the epidemiological cutoff values (ECVs) for voriconazole (VRC) (MIC, ≥1 μg/ml) and itraconazole (ITC) (MIC, ≥4 μg/ml), respectively (Table 1; Table S2). Resistance to MFG (MIC, ≥0.25 μg/ml) was noted only in three isolates recovered from two patients (4.7% [2/43]), while one isolate (2.3% [1/43]) was intermediate (MIC, 0.12 μg/ml) and the rest were susceptible (Table 1; Table S2). Interestingly, 26 isolates recovered from 14 patients (32.6% [14/43]) were resistant to caspofungin (CAS) (MIC, ≥0.5 μg/ml), while 31 isolates recovered from 28 patients (65.1% [28/43]) were intermediate, and only 1 isolate (2.3% [1/43]) was susceptible (Table 1; Table S2). Resistance to flucytosine (5FC) (MIC, ≥1 μg/ml) was confirmed in two isolates (4.7%). All isolates showed the wild-type (WT) phenotype for amphotericin B (AMB). The miconazole (MZ) MIC ranged from 0.03 to 1 μg/ml (Table 1; Table S2). Cross-resistance among echinocandin drugs was observed in three isolates recovered from two patients (4.7% [2/43]), and one isolate (2.3% [1/43]) was cross-resistant to all azoles. The multidrug resistance (MDR) phenotype, which confers resistance to at least two antifungal drug classes, was noted in two isolates (4.7% [2/43]). Fluconazole showed the highest geometric mean MIC value (5.46), followed by AMB (0.86), CAS (0.40), ITC (0.29), 5FC (0.15), VRC (0.12), MZ (0.09), and MFG (0.02) (Table 1).
TABLE 1.
Summary of antifungal susceptibilities of C. glabrata isolates
| Drug | No. of isolates at the following determined MIC (μg/ml): |
MIC (μg/ml): |
MIC (μg/ml) of quality control strain: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥64 | Range | GMa | C. parapsilosis ATCC 22019 | C. krusei ATCC 6258 | |
| MFG | 39 | 15 | 1 | 1 | 2 | 0.015–2 | 0.02 | 0.5 | 0.25 | ||||||||
| CAS | 1 | 31 | 18 | 5 | 1 | 2 | 0.12–8 | 0.40 | 0.25 | 0.25 | |||||||
| AMB | 13 | 45 | 0.5–1 | 0.86 | 1 | 1 | |||||||||||
| 5FC | 55 | 1 | 2 | 0.12–64 | 0.15 | 0.12 | 4 | ||||||||||
| FLC | 3 | 14 | 10 | 22 | 5 | 2 | 2 | 1–64 | 5.46 | 2 | 16 | ||||||
| ITC | 8 | 35 | 12 | 2 | 1 | 0.12–4 | 0.29 | 0.25 | 0.25 | ||||||||
| VRC | 7 | 17 | 19 | 8 | 3 | 2 | 1 | 1 | 0.03–8 | 0.12 | 0.06 | 0.12 | |||||
| MZ | 19 | 8 | 11 | 17 | 2 | 1 | 0.03–1 | 0.09 | 0.06 | 0.25 | |||||||
GM, geometric mean.
Detection of mutations in the PDR1 and ERG11 genes.
PCR testing and sequencing for the PDR1 gene revealed that 39.5% (17/43) of patients harbored isolates with nonsynonymous mutations relative to the sequence of the reference strain (GenBank accession number FJ550269.1) (Table S2 and Fig. S1), and all the isolates contained silent mutations (Table S3). With regard to the association between PDR1 mutations and azole resistance, one isolate with a nonsynonymous mutation (T370I) located in the middle homology domain showed cross-resistance to fluconazole and voriconazole (Fig. S1 and Table S2). Another nonsynonymous mutation, located in the activation domain (F817S), showed a non-WT phenotype for voriconazole, with a MIC value of 1 μg/ml (Fig. S1 and Table S2). Interestingly, one isolate (IFM 52011) without nonsynonymous mutations showed cross-resistance to three azoles (Table S2). With regard to ERG11, only one isolate (IFM 61014) harbored a nonsynonymous mutation, and all the isolates harbored silent mutations (Tables S2 and S3). The isolate with the nonsynonymous mutation in ERG11 neither harbored a PDR1 mutation nor showed a high MIC value of any azole drug.
Detection of mutations in FKS1 and FKS2 hot spot regions.
Screening of hot spot regions of the FKS1 and FKS2 genes revealed that one isolate harbored a nonsynonymous mutation (S629P) in the FKS1 HS1 region and nine isolates from the same patient harbored nonsynonymous mutations (S663P) in the FKS2 HS1 region (Table 2; Table S2). No mutations were detected in the HS2 regions of the FKS1 and FKS2 genes or the HS3 region of the FKS1 gene. Seven isolates with S663P were resistant to caspofungin only (MIC range, 0.5 to 1 μg/ml), while three isolates (two isolates with S663P and one isolate with S629P) were resistant to both caspofungin (MIC range, 2 to 8 μg/ml) and micafungin (MIC range, 1 to 2 μg/ml) (Table 2; Table S2).
TABLE 2.
FKS HS mutations in relation to caspofungin and micafungin MIC distribution
| Drug and polymorphism | % of isolates in the following MIC categorya: |
No. of isolates at the following determined MIC value (μg/ml): |
Total no. of isolates | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | ≤0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | ||
| Caspofungin | |||||||||||||||
| No HS mutations | 2.1 | 64.6 | 33.3 | 1 | 31 | 16 | 48 | ||||||||
| FKS1 S629P | 0 | 0 | 100 | 1 | 1 | ||||||||||
| FKS2 S663P | 0 | 0 | 100 | 2 | 5 | 1 | 1 | 9 | |||||||
| Micafungin | |||||||||||||||
| No HS mutations | 100 | 0 | 0 | 34 | 14 | 48 | |||||||||
| FKS1 S629P | 0 | 0 | 100 | 1 | 1 | ||||||||||
| FKS2 S663P | 66.7 | 11.1 | 22.2 | 5 | 1 | 1 | 2 | 9 | |||||||
S, susceptible; I, intermediate; R, resistant.
Genotyping of isolates using MLST.
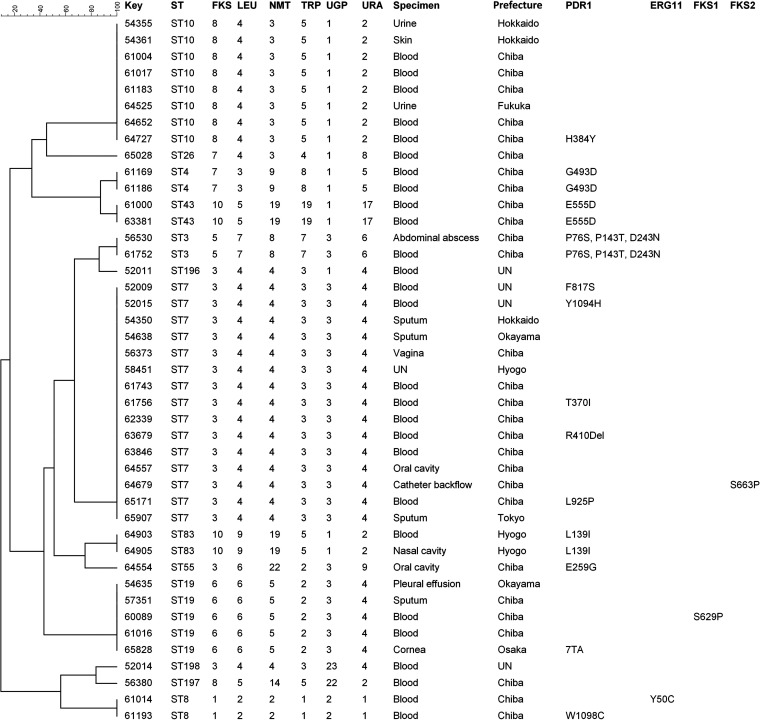
MLST analysis was performed to elucidate the putative genetic relationships among the clinical C. glabrata isolates examined. Thirteen different sequence types (STs) were identified among the isolates, including three new STs (ST196, ST197, and ST198) identified in four isolates (Tables S2, S4, and S5). The most common ST was ST7 (15/43 [35%]), followed by ST10 (8/43 [18.6%]) and ST19 (5/43 [11.6%]). Five STs (ST3, ST4, ST8, ST43, and ST83) were identified in two patients each, and five STs (ST26, ST55, ST196, ST197, and ST198) were identified in only a single patient each. All isolates from the same patient, recovered from different specimens or at different times, had the same ST. In order to determine the relationships between the STs of the isolates and gene mutations, phylogenetic analysis of the isolates was conducted by the clustering method and construction of an unrooted dendrogram using BioNumerics software, v7.6. Sequence similarity (80%) among the STs was observed only between ST43 and ST4, ST3 and ST196, and ST197 and ST198, with high diversity among the other STs identified (Fig. 1).
FIG 1.
UPGMA dendrogram showing the similarities among 43 C. glabrata isolates (a single isolate for each patient) based on MLST of six housekeeping gene loci. The groups are identified by a sequence similarity of 80%. With the exception of the similarity of three groups, consisting of ST43 and ST4, ST3 and ST196, and ST197 and ST198, all the STs identified showed high diversity. UN, unknown.
DISCUSSION
The emergence and spread of infections caused by C. glabrata, especially those associated with resistance to echinocandins and azoles, have become a major worldwide threat and have rekindled interest in revealing their prevalence, the molecular mechanisms responsible for the resistance, and their genotypes (3–8). Previous studies of C. glabrata isolates from different countries have revealed high variability in their resistance rates as well as their resistance mechanisms (5–7, 11). In Japan, rare independent studies of C. glabrata resistance have been reported (8, 12, 13), but data regarding their molecular mechanisms, as well as their genotyping, have been insufficient.
Our results demonstrated that C. glabrata infections are observed mainly in elderly people, which is in agreement with previous reports (5, 14). However, in contrast to the previous reports finding no difference between males and females in C. glabrata infections (5), our results showed a significant prevalence in females. Furthermore, in agreement with the previous reports, C. glabrata infections were observed mainly in patients suffering from underlying diseases (5, 15).
The high percentage of caspofungin-resistant isolates (32.6%) (Table 2) was astonishing. This finding poses a major clinical concern, especially considering that echinocandins are the first-line therapy for C. glabrata (5, 7). The level of caspofungin resistance in our study was much higher than the levels described previously in Germany (1.5%) (7), China (1.9%) (11), Turkey (2%) (16), and Switzerland (9.4%) (17). On the other hand, our results are in line with a previous report from Iran, where 57.7% of the isolates were non-WT for caspofungin susceptibility (5). However, caspofungin MIC values are unreliable for the determination of echinocandin resistance (18), which basically depends on the presence of nonsynonymous mutations in FKS gene hot spots (19) and noted when the isolate is resistant to at least two echinocandins (5). Our results are consistent with the previous findings (5, 18, 19), where only 4.7% of the isolates were resistant to both micafungin and caspofungin, and isolates harboring nonsynonymous FKS gene HS1 mutations were recovered from only two patients (4.7%). The low levels of FKS HS1 mutations detected in our study are in line with those detected previously in other Asian countries, including Iran and India (0%) (5, 20), in European countries, including France (1%) (21) and Germany (2.2%) (7), and in the United States (7.9%) (22). This finding is of serious clinical concern, since it has been elucidated that FKS mutation is linked with high therapeutic failure rates (7, 22). The variation in therapeutic regimens and the genetic diversity of C. glabrata isolates could explain the variation in their echinocandin resistance rates among different countries (5, 20, 23).
Our results support the previous finding that FKS mutations did not always cause the same resistance levels (24). It was also noted that both FKS1 S629P and FKS2 S663P, reported in this study, had been reported previously to be functionally equivalent and to be associated with either the low or the high end of the resistant MIC category. For instance, in our study, FKS1 S629P was associated with elevations of both micafungin and caspofungin MICs (Table 2). On the other hand, FKS2 S663P was associated with elevations of both micafungin and caspofungin MICs in two isolates and with resistance to caspofungin (MIC range, 0.5 to 1 μg/ml) in seven isolates (Table 2; Table S2). These findings clarify the complicated mechanism of echinocandin resistance and the significant roles of both phenotypic and genotypic methods in detection. The variation in MIC values among isolates with the same FKS mutations from the same patient could be attributed to variation in the relative expression of redundant FKS genes (25).
In contrast to the high caspofungin resistance level, our study confirmed the low azole resistance level, ranging from 2.3% to 9.3%. Similar results were also reported from other Asian countries, including Iran, China, India, and Korea, where azole resistance ranged from 0 to 9% (5, 11, 20, 23). Previous studies reported that some amino acid mutations in PDR1 and ERG11 are responsible for azole resistance in C. glabrata (5, 9–11). Interestingly, one novel mutation (F817S) described here, located in the activation domain and associated with a non-WT voriconazole phenotype, was recently laboratory-evolved in the presence of FLC in C. glabrata (26), supporting its significant role in azole resistance. Furthermore, we identified, for the first time, another novel PDR1 mutation associated with fluconazole resistance and a non-WT voriconazole phenotype (T370I) (Table S2). However, the association between this novel mutation and azole resistance needs to be confirmed, since the mutation appeared in a single isolate. Two fluconazole SDD isolates harboring three PDR1 mutations (P76S, P145T, D243N), previously described in fluconazole-resistant and SDD isolates in Iran and China (5, 11), were identified (Table S2). Since different strains harbored mutations in PDR1 and did not exhibit higher MIC values of azole drugs (Table S2), one could speculate that those mutations are not involved in resistance.
For ERG11, only a single fluconazole SDD isolate harbored s nonsynonymous mutation (Y50C) (Table S2). Our finding that ERG11 mutation did not induce resistance to fluconazole in C. glabrata was in agreement with a result from Iran (5) and confirms the low tendency of ERG11 mutation to induce azole resistance. A previous homology modeling study of C. glabrata showed that nonsynonymous mutations in residues 146, 243, and 246 are correlated with azole resistance (27). However, none of the azole-resistant isolates in our study harbored mutations in the neighborhood of those residues. In this study, one isolate showed cross-resistance to three azoles, and another isolate with a non-WT voriconazole phenotype did not harbor any nonsynonymous mutations in PDR1 and ERG11. Therefore, other mechanisms might be involved in their resistance (9, 10).
MDR C. glabrata is rare compared with MDR bacteria (9). In our study, 4.7% of the isolates were MDR, and to the best of our knowledge, this the first report of MDR C. glabrata in Japan. The emergence and increase of C. glabrata caspofungin resistance coupled with azole resistance could explain the increasing isolation of the MDR strains (9). The emergence of such MDR strains represents a great therapeutic challenge due to the limited antifungal options.
C. glabrata genotyping has a significant role in detecting emerging clones and in determining the correlations between certain genotypes and gene polymorphisms, mortality rates, and virulence characteristics (5, 11, 23). Based on previous findings, MLST was performed on the C. glabrata isolates. As far as we know, this is the first genotyping study of C. glabrata in Japan. Our results demonstrated that ST7 and ST10 were the most abundant STs, at 35%, and 18.6%, respectively. Furthermore, ST7 was the most prevalent ST among patients with resistant isolates; it was identified in 75% (3/4) of voriconazole-resistant isolates, 50% (1/2) each of micafungin-, fluconazole-, and 5FC-resistant isolates, and 28.6% (4/14) of caspofungin-resistant isolates (Table S6). This is the first report to confirm the important role of ST7 in resistance dissemination in C. glabrata. In agreement with our finding, ST7 was the most common ST in other Asian countries, including China (65.8%) (11) and Korea (47.8%), and was also associated with high mortality (23). These findings confirm that ST7 is the major ST circulating in the Northeast Asia region and support previous reports that STs are geographically dependent (11). For instance, ST16, ST19, and ST3 were the most prevalent STs in the United States (28).
Although this is the first study to elucidate the prevalence and molecular mechanisms of echinocandin and azole resistance in C. glabrata isolates from Japan, as well as to analyze this resistance by genotyping, it has some limitations. For instance, a small number of isolates was included in this study, and most of the isolates were recovered from a single prefecture. Therefore, other large-scale nationwide studies are needed to investigate the molecular mechanisms responsible for echinocandin and azole resistance in C. glabrata throughout Japan.
In summary, our results indicated high caspofungin resistance and low azole resistance in clinical C. glabrata isolates in Japan. Furthermore, we identified the molecular mechanisms to which their resistance could be attributed. Our results confirm that ST7 and ST10 were the most prevalent genotypes, and 11 other genotypes were also identified. The high caspofungin resistance detected in this study, together with the unique genetic characteristics of C. glabrata, is alarming and further limits therapeutic choices. Undoubtedly, higher mortality rates are very much expected for C. glabrata infections.
MATERIALS AND METHODS
C. glabrata isolate collection and identification.
A total of 58 C. glabrata isolates were provided through the National BioResource Project (NBRP), Japan (http://www.nbrp.jp/). Isolates were recovered from clinical samples of blood (n = 29), catheter backflow blood (n = 12), sputum (n = 5), urine (n = 2), the vagina (n = 2), the oral cavity (n = 2), the nasal cavity (n = 1), the cornea (n = 1), the skin (n = 1), abdominal abscess (n = 1), pleural effusion (n = 1), and unknown (n = 1) during the 23-year period from 1997 to 2019 (Table S1). Isolates were obtained from 43 patients hospitalized in different hospitals in seven prefectures across Japan (Fig. S2). The procedures of this study, including the availability of patients' clinical data, were approved by the Medical Mycology Research Center, Chiba University Ethical Committee (approval number MMRC-REC 20-19).The isolates were identified by DNA sequence analysis of the internal transcribed spacer 1 (ITS1)–5.8S rRNA–ITS2 region as described previously (29).
Antifungal susceptibility testing.
MIC values were determined by broth microdilution assays for fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), miconazole (MZ), micafungin (MFG), caspofungin (CAS), amphotericin B (AMB), and flucytosine (5FC) using dried Yeast-like Fungal DP Eiken plates (Eiken Chemicals, Tokyo, Japan) as recommended in CLSI document M27-Ed4 (30). C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were used as quality control strains, and the breakpoints were interpreted according to CLSI document M60 (31). MICs were determined visually after 24 h, with the exception of a single isolate, and were interpreted after 48 h due to the absence of growth after 24 h (31). The MIC was estimated as the lowest concentration of the antifungal inducing at least a 50% reduction (for all antifungals) or a 100% reduction (for AMB) in fungal growth from that of the control. Resistance to FLC, MFG, and CAS was observed when the MIC values were ≥64 μg/ml, ≥0.25 μg/ml, and ≥0.5 μg/ml, respectively (31). The MIC values of ITC (>2), VRC (>0.5), AMB (>2), and 5FC (>0.5) were adopted according to ECVs (32). There are neither breakpoints nor ECVs for MZ (30, 31).
DNA extraction for PCR.
DNA was extracted as described previously with minor modifications (33). Briefly, 1 to 2 loopfuls of a 24-h-to-48-h yeast culture grown on Sabouraud dextrose agar were mixed with 100 μl of lysis buffer (200 mM Tris-HCl [pH 8.0], 0.5% [wt/vol] sodium dodecyl sulfate, 30 mM EDTA). After vigorous vortexing, the solution was incubated at 100°C for 15 min, followed by mixing with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]) and centrifugation at 12,000 rpm for 5 min. The supernatant was transferred to a fresh tube and was mixed with 250 μl of 100% ethanol previously chilled at –20°C. After 10 min of incubation on ice, DNA was precipitated by centrifugation at 12,000 rpm for 20 min at 4°C. The DNA pellet was obtained after washing in cold 70% ethanol, dried, suspended in 100 μl of sterile Tris-EDTA (TE) buffer, and preserved at –20°C. One microliter of the resulting diluted solution (1:10) was used as a template for PCR experiments.
Detection of mutations associated with azole resistance.
All isolates were screened by PCR for detection of mutations in the PDR1 and ERG11 genes. GenBank sequences with accession numbers FJ550269.1 and XM_445876 were used as references for the PDR1 and ERG11 sequences, respectively (5). Seven and four primers were used for the PCR and sequencing of PDR1 and ERG11, respectively (Table S7). PCR was performed using KOD One PCR master mix (Toyobo, Osaka, Japan) as described in the manufacturer’s protocol. A PCR thermal cycler (Bio-Rad, Tokyo, Japan) was used at 98°C for 3 min, followed by 35 cycles, each consisting of 10 s at 98°C, 5 s at 53°C, and 30 s at 68°C, followed by a final step at 68°C for 2 min.
Detection of mutations associated with echinocandin resistance.
All isolates were screened by PCR for the detection of mutations in the FKS1 (HS1, HS2, HS3) and FKS2 (HS1, HS2) genes. Sequences in GenBank with accession numbers XM_446406 and XM_448401 were used for primer design and as references for the FKS1 and FKS2 sequences, respectively (34). Four primers were used for PCR and sequencing of FKS1, and four other primers were used for PCR and sequencing of FKS2 (Table S7 and Fig. S3). PCR was performed using KOD One PCR master mix at 98°C for 3 min, followed by 35 cycles, each consisting of 98°C for 10 s, 56°C for 5 s, and 68°C for 30 s, followed by a final step at 68°C for 2 min.
Sequencing and data analysis.
PCR products were purified using a FastGene Gel/PCR extraction kit (Nippon Genetics Co., Tokyo, Japan). The DNA of the PCR products was sequenced using an ABI 3130xl genetic analyzer (Thermo Fisher Scientific, Tokyo, Japan) and the BigDye Terminator cycle sequencing kit, v3.1 (Thermo Fisher Scientific), according to the manufacturer’s instructions. The sequences were assembled, aligned, and compared with the reference for mutation detection using the Benchling platform, Genetyx-win, v12 (Genetyx, Tokyo, Japan), and ATGC, v.6.0 (Genetyx).
MLST.
Genotyping of C. glabrata isolates was performed by multilocus sequence typing (MLST) using six housekeeping gene loci (FKS, LEU2, NMT1, TRP1, UGP1, URA3) (35). C. glabrata STs were identified according to the sequence-typing website (https://pubmlst.org/cglabrata/). Phylogenetic analysis of MLST results based on each of the gene variants was conducted by a clustering method using the unweighted pair group method using average linkages (UPGMA) and the minimum spanning tree algorithm of BioNumerics software, v7.6 (Applied Maths Inc., Austin, TX, USA).
Data availability.
All sequences obtained in this study for PDR1, ERG11, FKS1 HS1 and HS3, FKS1 HS2, FKS2 HS1, and FKS2 HS2 have been deposited in GenBank under accession numbers MT332856 to MT332913, MT332914 to MT332971, MT332972 to MT333029, MT333030 to MT333087, MT333088 to MT333145, and MT333146 to MT333203, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by AMED under grant numbers JP19jm0110015 and JP19fk0108094.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Procop GW. 2010. Molecular diagnostics for invasive fungal infections: a call for refinement and implementation. J Mol Diagn 12:17–19. doi: 10.2353/jmoldx.2010.090173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedict K, Jackson BR, Chiller T, Beer KD. 2019. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis 68:1791–1797. doi: 10.1093/cid/ciy776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortegiani A, Misseri G, Fasciana T, Giammanco A, Giarratano A, Chowdhary A. 2018. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J Intensive Care 6:69. doi: 10.1186/s40560-018-0342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortegiani A, Misseri G, Chowdhary A. 2019. What’s new on emerging resistant Candida species. Intensive Care Med 45:512–515. doi: 10.1007/s00134-018-5363-x. [DOI] [PubMed] [Google Scholar]
- 5.Arastehfar A, Daneshnia F, Zomorodian K, Najafzadeh MJ, Khodavaisy S, Zarrinfar H, Hagen F, Zare Shahrabadi Z, Lackner M, Mirhendi H, Salehi M, Roudbary M, Pan W, Kostrzewa M, Boekhout T. 2019. Low level of antifungal resistance in Iranian isolates of Candida glabrata recovered from blood samples in a multicenter study from 2015 to 2018 and potential prognostic values of genotyping and sequencing of PDR1. Antimicrob Agents Chemother 63:e02503-18. doi: 10.1128/AAC.02503-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman B, Slavin M, Marriott D, Halliday C, Kidd S, Arthur I, Bak N, Heath CH, Kennedy K, Morrissey CO, Sorrell TC, Van Hal S, Keighley C, Goeman E, Underwood N, Hajkowicz K, Hofmeyr A, Leung M, Macesic N, Botes J, Blyth C, Cooley L, George CR, Kalukottege P, Kesson A, McMullan B, Baird R, Robson J, Korman TM, Pendle S, Weeks K, Liu E, Cheong E, Chen S, Australian and New Zealand Mycoses Interest Group. 2017. Changing epidemiology of candidaemia in Australia. J Antimicrob Chemother 72:1103–1108. doi: 10.1093/jac/dkw422. [DOI] [PubMed] [Google Scholar]
- 7.Klotz U, Schmidt D, Willinger B, Steinmann E, Buer J, Rath PM, Steinmann J. 2016. Echinocandin resistance and population structure of invasive Candida glabrata isolates from two university hospitals in Germany and Austria. Mycoses 59:312–318. doi: 10.1111/myc.12472. [DOI] [PubMed] [Google Scholar]
- 8.Kakeya H, Yamada K, Kaneko Y, Yanagihara K, Tateda K, Maesaki S, Takesue Y, Tomono K, Kadota J-I, Kaku M, Miyazaki Y, Kamei K, Shibuya K, Niki Y, Yoshida M, Sei Y. 2018. National trends in the distribution of Candida species causing candidemia in Japan from 2003 to 2014. Med Mycol J 59:E19–E22. doi: 10.3314/mmj.17-00014. [DOI] [PubMed] [Google Scholar]
- 9.Morio F, Jensen RH, Le Pape P, Arendrup MC. 2017. Molecular basis of antifungal drug resistance in yeasts. Int J Antimicrob Agents 50:599–606. doi: 10.1016/j.ijantimicag.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Hull CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. 2012. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob Agents Chemother 56:4223–4232. doi: 10.1128/AAC.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou X, Xiao M, Wang H, Yu SY, Zhang G, Zhao Y, Xu YC. 2018. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob Agents Chemother 62:e00153-18. doi: 10.1128/AAC.00153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano R, Sakamoto Y, Kitazawa J, Yamamoto S, Kayaba H. 2018. Epidemiology, practice patterns, and prognostic factors for candidemia; and characteristics of fourteen patients with breakthrough Candida bloodstream infections: a single tertiary hospital experience in Japan. Infect Drug Resist 11:821–833. doi: 10.2147/IDR.S156633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakagami T, Kawano T, Yamashita K, Yamada E, Fujino N, Kaeriyama M, Fukuda Y, Nomura N, Mitsuyama J, Suematsu H, Watanabe H, Asai N, Koizumi Y, Yamagishi Y, Mikamo H. 2019. Antifungal susceptibility trend and analysis of resistance mechanism for Candida species isolated from bloodstream at a Japanese university hospital. J Infect Chemother 25:34–40. doi: 10.1016/j.jiac.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Moran C, Grussemeyer CA, Spalding JR, Benjamin DK Jr, Reed SD. 2010. Comparison of costs, length of stay, and mortality associated with Candida glabrata and Candida albicans bloodstream infections. Am J Infect Control 38:78–80. doi: 10.1016/j.ajic.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moretti ML, Trabasso P, Lyra L, Fagnani R, Resende MR, de Oliveira Cardoso LG, Schreiber AZ. 2013. Is the incidence of candidemia caused by Candida glabrata increasing in Brazil? Five-year surveillance of Candida bloodstream infection in a university reference hospital in southeast Brazil. Med Mycol 51:225–230. doi: 10.3109/13693786.2012.708107. [DOI] [PubMed] [Google Scholar]
- 16.Kiraz N, Dag I, Oz Y, Yamac M, Kiremitci A, Kasifoglu N. 2010. Correlation between broth microdilution and disk diffusion methods for antifungal susceptibility testing of caspofungin, voriconazole, amphotericin B, itraconazole and fluconazole against Candida glabrata. J Microbiol Methods 82:136–140. doi: 10.1016/j.mimet.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Orasch C, Marchetti O, Garbino J, Schrenzel J, Zimmerli S, Mühlethaler K, Pfyffer G, Ruef C, Fehr J, Zbinden R, Calandra T, Bille J, FUNGINOS. 2014. Candida species distribution and antifungal susceptibility testing according to European Committee on Antimicrobial Susceptibility Testing and new vs. old Clinical and Laboratory Standards Institute clinical breakpoints: a 6-year prospective candidaemia survey from the Fungal Infection Network of Switzerland. Clin Microbiol Infect 20:698–705. doi: 10.1111/1469-0691.12440. [DOI] [PubMed] [Google Scholar]
- 18.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas S, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyda ND, John J, Kilic A, Alam MJ, Lasco TM, Garey KW. 2014. FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin Infect Dis 59:819–825. doi: 10.1093/cid/ciu407. [DOI] [PubMed] [Google Scholar]
- 20.Singh A, Healey KR, Yadav P, Upadhyaya G, Sachdeva N, Sarma S, Kumar A, Tarai B, Perlin DS, Chowdhary A. 2018. Absence of azole or echinocandins resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in DNA mismatch repair pathway. Antimicrob Agents Chemother 62:e00195-18. doi: 10.1128/AAC.00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourgeois N, Laurens C, Bertout S, Balard Y, Krasteva D, Rispail P, Lachaud L. 2014. Assessment of caspofungin susceptibility of Candida glabrata by the Etest, CLSI, and EUCAST methods, and detection of FKS1 and FKS2 mutations. Eur J Clin Microbiol Infect Dis 33:1247–1252. doi: 10.1007/s10096-014-2069-z. [DOI] [PubMed] [Google Scholar]
- 22.Alexander BD, Johnson MD, Pfeiffer CD, Jiménez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byun SA, Won EJ, Kim M, Lee WG, Lee K, Lee HS, Uh Y, Healey KR, Perlin DS, Choi MJ, Kim SH, Shin JH. 2018. Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes. Front Microbiol 9:1523. doi: 10.3389/fmicb.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham CD, Iqbal N, Bolden CB, Kuykendall RJ, Harrison LH, Farley MM, Schaffner W, Beldavs ZG, Chiller TM, Park BJ, Cleveland AA, Lockhart SR. 2014. Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob Agents Chemother 58:4690–4696. doi: 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. 2012. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother 56:6304–6309. doi: 10.1128/AAC.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usher J, Haynes K. 2019. Attenuating the emergence of anti-fungal drug resistance by harnessing synthetic lethal interactions in a model organism. PLoS Genet 15:e1008259. doi: 10.1371/journal.pgen.1008259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabili M, Abdollahi Gohar A, Badali H, Mohammadi R, Moazeni M. 2016. Amino acid substitutions in Erg11p of azole-resistant Candida glabrata: possible effective substitutions and homology modelling. J Glob Antimicrob Resist 5:42–46. doi: 10.1016/j.jgar.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Lott TJ, Frade JP, Lockhart SR. 2010. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot Cell 9:619–625. doi: 10.1128/EC.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White T, Burns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th ed CLSI document M27-Ed4 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2017. Performance standards for antifungal susceptibility testing of yeasts, 1st ed CLSI document M60 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Pfaller MA, Diekema DJ. 2012. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol 50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makimura K, Mochizuki T, Hasegawa A, Uchida K, Saito H, Yamaguchi H. 1998. Phylogenetic classification of Trichophyton mentagrophytes complex strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol 36:2629–2633. doi: 10.1128/JCM.36.9.2629-2633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa-de-Oliveira S, Miranda IM, Silva RM, Silva APE, Rocha R, Amorim A, Rodrigues AG, Pina-Vaz C. 2011. FKS2 mutations associated with decreased echinocandin susceptibility of Candida glabrata following anidulafungin therapy. Antimicrob Agents Chemother 55:1312–1314. doi: 10.1128/AAC.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/jcm.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences obtained in this study for PDR1, ERG11, FKS1 HS1 and HS3, FKS1 HS2, FKS2 HS1, and FKS2 HS2 have been deposited in GenBank under accession numbers MT332856 to MT332913, MT332914 to MT332971, MT332972 to MT333029, MT333030 to MT333087, MT333088 to MT333145, and MT333146 to MT333203, respectively.