The ability to acquire azole resistance is an emblematic trait of the fungal pathogen Candida glabrata. Understanding the molecular basis of azole resistance in this pathogen is crucial for designing more suitable therapeutic strategies. This study shows that the C. glabrata transcription factor (TF) CgRpn4 is a determinant of azole drug resistance. RNA sequencing during fluconazole exposure revealed that CgRpn4 regulates the expression of 212 genes, activating 80 genes and repressing, likely in an indirect fashion, 132 genes.
KEYWORDS: CgRpn4, RNA-seq-based transcriptomics, azole resistance mechanisms, ergosterol, regulatory networks
ABSTRACT
The ability to acquire azole resistance is an emblematic trait of the fungal pathogen Candida glabrata. Understanding the molecular basis of azole resistance in this pathogen is crucial for designing more suitable therapeutic strategies. This study shows that the C. glabrata transcription factor (TF) CgRpn4 is a determinant of azole drug resistance. RNA sequencing during fluconazole exposure revealed that CgRpn4 regulates the expression of 212 genes, activating 80 genes and repressing, likely in an indirect fashion, 132 genes. Targets comprise several proteasome and ergosterol biosynthesis genes, including ERG1, ERG2, ERG3, and ERG11. The localization of CgRpn4 to the nucleus increases upon fluconazole stress. Consistent with a role in ergosterol and plasma membrane homeostasis, CgRpn4 is required for the maintenance of ergosterol levels upon fluconazole stress, which is associated with a role in the upkeep of cell permeability and decreased intracellular fluconazole accumulation. We provide evidence that CgRpn4 directly regulates ERG11 expression through the TTGCAAA binding motif, reinforcing the relevance of this regulatory network in azole resistance. In summary, CgRpn4 is a new regulator of the ergosterol biosynthesis pathway in C. glabrata, contributing to plasma membrane homeostasis and, thus, decreasing azole drug accumulation.
INTRODUCTION
Infections by fungal pathogens have become a worrying health concern worldwide. Candida spp. represent the most common cause of fungal infections and rank as the 4th leading cause of nosocomial bloodstream infections in the United States (1–3). They are responsible for localized superficial infections in healthy individuals but can cause life-threatening scenarios in immunocompromised patients. The ability to rapidly acquire azole resistance has been associated with the increased prevalence of Candida glabrata relative to the most commonly isolated species, C. albicans (1, 4–9). C. glabrata now ranks as the second or third most prevalent Candida species, depending on geographical location. While it is the second most commonly isolated species in the United States and northern Europe (7, 10), it ranks as the third most common species in southern Europe and Asia (7, 10, 11) and is less prevalent in South America (10).
Due to their safety profile and availability in both oral and intravenous formulations, azoles have been the most widely used antifungals for decades, both as a first-line treatment and as prophylaxis, especially fluconazole (12). Azoles disrupt ergosterol biosynthesis, the main sterol in fungal plasma membranes, which leads to the loss of membrane structural properties. They act by binding and inhibiting the lanosterol 14-α-demethylase, a key enzyme of the ergosterol biosynthetic pathway encoded by ERG11 (13, 14). The result is the accumulation of 14-α-methylated sterols, the depletion of ergosterol, and the production of the toxic sterol dimethylcholesta-8,24(28)-dien-3β,6α-diol (DMCDD) in the membrane (15, 16).
Investigation of the transcriptional regulation of antifungal resistance in C. glabrata is focused mostly on the role of the transcription factor (TF) CgPdr1, regarded as a master regulator of clinical acquisition of resistance to azole antifungals by activating the expression of multidrug resistance transporters (17–22). In C. albicans, CaTac1 plays a similar role by regulating the expression of multidrug resistance transporters (23–25). However, the level of sequence similarity between CaTac1 and CgPdr1 is low. Apart from these pathways, knowledge about regulatory networks involved in the response to antifungal stress is scarce, especially for C. glabrata (26).
In this study, we identified the transcription factor Rpn4 as a regulator of fluconazole resistance in C. glabrata. In Saccharomyces cerevisiae, the Rpn4 homolog regulates the expression of proteasome genes, affecting proteasome activity and ubiquitin-mediated proteolysis (27–30), and contributes to the unfolded protein response (UPR) (31). Rpn4 has also been identified as a member of the multidrug resistance network in S. cerevisiae (32, 33). Consistently, the deletion of ScRPN4 results in increased sensitivity to fluconazole (34, 35) and clotrimazole (36). Moreover, the expression of ScRPN4 and, consequently, the ubiquitin-proteasome system is regulated by the multidrug resistance regulators Pdr1 and Pdr3 (29). In C. albicans, RPN4 is also a regulator of proteasome genes, and its absence results in enhanced sensitivity to fluconazole (37). To the best of our knowledge, despite the apparent role of Rpn4 in azole resistance phenotypes, its actual role and mechanistic insights into antifungal resistance have not been addressed.
Given the identification of CgRPN4 as a new determinant of azole drug resistance in C. glabrata, its role in the transcriptome-wide response to fluconazole was assessed. The determination of Rpn4 target genes enabled the identification of a new regulatory pathway toward azole resistance, which may provide the molecular basis of Rpn4-mediated azole resistance in pathogenic and nonpathogenic yeasts.
RESULTS
The transcription factor CgRPN4 is a determinant of fluconazole resistance in Candida glabrata.
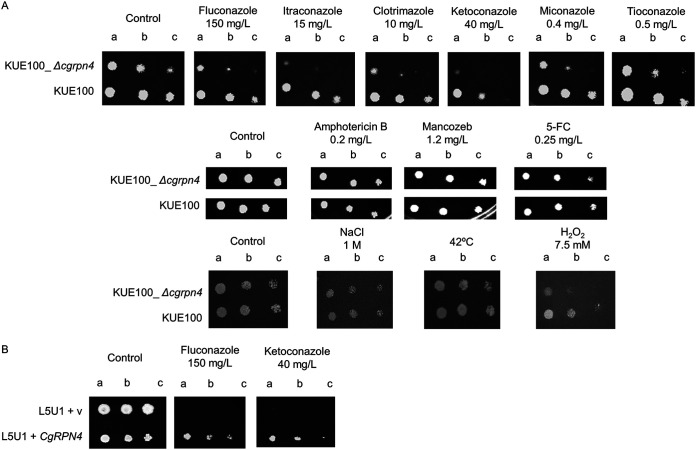
Based on the previous identification of Rpn4 as a member of the multidrug resistance network in several yeast species, the role of the uncharacterized CgRPN4 gene in azole resistance was determined. Figure 1A shows that deleting CgRPN4 increases sensitivity to 6 different azole drugs compared to the KUE100 wild-type strain. Interestingly, the overexpression of CgRPN4 in the L5U1 wild-type C. glabrata strain increased its tolerance to fluconazole and ketoconazole (Fig. 1B). Besides susceptibility to azole antifungals, phenotypes to antifungals from other families, amphotericin B and flucytosine (5-FC), were tested. Susceptibility to the fungicide mancozeb was also tested. Other than antimycotic agents, susceptibility to temperature, osmotic stress, and oxidative stress was also tested. The deletion of CgRPN4 was seen to increase susceptibility only to hydrogen peroxide (Fig. 1A). These results indicate that CgRPN4 plays a primary role in mediating azole resistance.
FIG 1.
CgRPN4 confers resistance to fluconazole in C. glabrata. (A) Comparison of the susceptibilities to antifungals and other stress agents, at the indicated concentrations, of the KUE100 and KUE100_Δcgrpn4 C. glabrata strains, in BM agar plates, by spot assays. (B) Comparison of the susceptibilities to azole antifungals, at the indicated concentrations, of the L5U1 C. glabrata strain harboring the pGREG576 cloning vector (v) or pGREG576_MTI_CgRPN4 in BM agar plates supplemented with leucine and 50 μM CuSO4 by spot assays. The inocula were prepared as described in Materials and Methods. The cell suspensions used to prepare the spots were 1:5 (lane b) and 1:25 (lane c) dilutions of the cell suspensions used in lane a. The displayed images are representative of results from at least three independent experiments.
The MICs of several antifungals were also used to quantitatively assess drug susceptibility variability between the wild-type strain and the derived Δcgrpn4 mutant (Table 1). Consistent with previous results, no MIC differences were attained for amphotericin B or 5-FC. The susceptibility profiles against the imidazole ketoconazole and the triazoles fluconazole, itraconazole, and posaconazole were also determined. The deletion of CgRPN4 led to an 8-fold increase in susceptibility to ketoconazole and posaconazole and a 2-fold increase in susceptibility to itraconazole. The clinical outcome of these data cannot be directly estimated due to the absence of breakpoints for these antifungals; however, the observed shifts in susceptibility support a relevant role of CgRpn4 in multiple-azole resistance. The deletion of the TF also resulted in a 4-fold increase in susceptibility to fluconazole. Both strains are categorized as intermediately susceptible to fluconazole, with CgRpn4 playing a measurable role in the susceptibility profile.
TABLE 1.
Susceptibility profiles determined by MIC values for the Δcgrpn4 mutant strain and the parental straina
| Drug | MIC (mg/liter) for strain |
Category (WT/mutant) | |
|---|---|---|---|
| WT | Δcgrpn4 | ||
| Fluconazole | 16 | 4 | I/I |
| Itraconazole | 0.5 | 0.25 | NA |
| Posaconazole | 1 | 0.125 | NA |
| Ketoconazole | 0.500 | 0.0625 | NA |
| Amphotericin B | 0.25 | 0.25 | S/S |
| 5-FC | 0.5 | 0.5 | NA |
NA, not applicable; I, intermediate. S, susceptible; WT, wild type.
Transcriptome-wide changes in response to fluconazole stress in C. glabrata.
We used transcriptional profiling to determine the response of the KUE100 wild-type strain to treatment with the previously determined inhibitory concentration of 150 mg/liter fluconazole. Notably, this concentration of fluconazole is higher than that attained by MIC assays, as it was found to be an inhibitory concentration under the conditions used for cell growth to perform the remaining experiments. Resistance measurements can vary according to the experimental conditions that are used (e.g., cell density, agitation, temperature, and medium), and cell density has been correlated with antifungal resistance (38). Control experiments were performed with reduced cell and fluconazole concentrations to validate the same inhibitory effect with drug concentrations more closely related to those found under physiological conditions (see Fig. S1 in the supplemental material). The fact that the remaining experiments were conducted using a much higher cell concentration than the one used in MIC assays might justify the need for a higher concentration of the antifungal. The expression levels of 44 genes were altered in C. glabrata cells following exposure to fluconazole for 1 h (log2-fold change of greater than 0.5 or less than −0.5; Benjamini-Hochberg-adjusted P value of <0.05) (Table 2; Table S1). The expression levels of 29 (66%) genes were increased, whereas the expression levels of 15 (34%) were decreased. The genes fall into 11 functional groups: drug resistance, sterol metabolism, and intracellular traffic, which are associated exclusively with the upregulated genes; lipid and fatty acid metabolism; nitrogen metabolism; carbon metabolism; heme homeostasis; mitochondrial function; the stress response; the cytoskeleton/cell cycle; and unknown function. As expected, the expression of the multidrug resistance transporter-encoding gene CDR1, a well-known biomarker of azole resistance (39–41), was upregulated 1.75-fold in fluconazole-challenged cells. Additionally, 41% of the upregulated genes are related to sterol metabolism, including components of the ergosterol biosynthetic pathway (ERG1, ERG2, ERG3, ERG4, ERG5, and ERG6) and the azole target ERG11. The upregulation of ERG11 in response to azole stress in C. glabrata and other Candida spp. has been reported previously (42–45), as has the upregulation of additional ERG genes in cases of azole resistance (46, 47). Two more genes with predicted roles in ergosterol biosynthesis, namely, HES1 and CYB5, are also present in this group. The upregulation of the ergosterol pathway is therefore likely a core response to mild fluconazole stress.
TABLE 2.
Genes differentially expressed in wild-type C. glabrata cells challenged with fluconazolea
| Functional group | C. glabrata gene | Description | Log2-fold change |
|
|---|---|---|---|---|
| WT FLC/WT control | Δcgrpn4 FLC/WT FLC | |||
| Drug resistance | CDR1 | Multidrug transporter of the ABC superfamily, involved in resistance to azoles; expression regulated by Pdr1p; increased abundance in azole-resistant strains; expression increased by loss of the mitochondrial genome | 0.81 | 0.42 |
| Sterol metabolism | ERG1b | Squalene epoxidase with role in ergosterol synthesis; involved in growth under conditions of low oxygen tension | 0.83 | −0.61 |
| ERG2b | C-8 sterol isomerase | 1.10 | −0.71 | |
| ERG3b | Delta-5,6-sterol desaturase; C-5 sterol desaturase; predicted transmembrane domain and endoplasmic reticulum binding motif; gene used for molecular typing of C. glabrata strain isolates | 1.07 | −0.63 | |
| ERG4 | Putative C-24 sterol reductase | 0.83 | −0.46 | |
| ERG5 | Putative C-22 sterol desaturase | 0.58 | −0.24 | |
| ERG6 | C-24 sterol methyltransferase; mutation confers resistance to amphotericin B and nystatin and increased sensitivity to azoles | 0.52 | −0.38 | |
| ERG11b | Putative cytochrome P-450 lanosterol 14-alpha-demethylase; target enzyme of azole antifungal drugs; increased protein abundance in azole-resistant strains | 0.96 | −0.65 | |
| ERG24 | Ortholog(s) has delta-14-sterol reductase activity and roles in cellular response to drugs, ergosterol biosynthetic processes, filamentous growth of a population of unicellular organisms in response to a biotic stimulus, and pathogenesis | 0.53 | −0.32 | |
| ERG25 | Ortholog(s) has C-4 methylsterol oxidase activity, role in ergosterol biosynthetic process, and endoplasmic reticulum membrane and plasma membrane localizations | 1.02 | −0.41 | |
| ERG29 | Ortholog(s) has roles in cellular iron ion homeostasis, ergosterol biosynthetic process, and mitochondrion organization and has endoplasmic reticulum and nuclear envelope localizations | 0.84 | −0.43 | |
| HES1b | Ortholog(s) has oxysterol binding, sterol transporter activity, and roles in endocytosis, exocytosis, maintenance of cell polarity, piecemeal microautophagy of the nucleus, and sterol transport | 1.48 | −0.96 | |
| CYB5 | Ortholog(s) has electron carrier activity, role in ergosterol biosynthetic process, and endoplasmic reticulum membrane localization | 0.73 | −0.43 | |
| Lipid and fatty acid metabolism | CSR1b | Ortholog(s) has phosphatidylinositol transporter activity | 0.59 | −0.90 |
| HBN1 | Ortholog(s) has oxidoreductase activity acting on NAD(P)H, nitrogenous group as acceptor activity, and roles in cellular response to oxidative stress and negative regulation of fatty acid metabolic process | −0.51 | −0.10 | |
| CAGL0A03740gb | Ortholog(s) has roles in fatty acid beta-oxidation and long-chain fatty acid catabolic processes and has peroxisome localization | −0.53 | 1.13 | |
| Stress response | RTA1b | Putative protein involved in 7-aminocholesterol resistance; gene is upregulated in azole-resistant strains | 0.80 | 0.51 |
| RAD14b | Ortholog(s) has damaged DNA binding, zinc ion binding activity, and roles in UV damage excision repair, nucleotide excision repair involved in interstrand cross-link repair, nucleotide excision repair, and DNA damage recognition | 0.52 | −0.53 | |
| SSA3 | Heat shock protein of the HSP70 family | −0.55 | 0.06 | |
| Nitrogen metabolism | PUT1b | Ortholog(s) has proline dehydrogenase activity, role in the proline-catabolic process to glutamate, and mitochondrion localization | 0.57 | 0.72 |
| MEP2 | Ortholog(s) has high-affinity secondary active ammonium transmembrane transporter activity and methylammonium transmembrane transporter activity | −0.50 | −0.04 | |
| Carbon metabolism | PBI1b | Has domain(s) with predicted alcohol O-acetyltransferase activity and role in alcohol metabolic process | 2.11 | −1.93 |
| ATF2 | Putative alcohol acetyltransferase involved in steroid detoxification; gene is upregulated in azole-resistant strains | 0.70 | −0.09 | |
| MLS1b | Ortholog(s) has malate synthase activity; roles in acetate catabolic process, carbon utilization, fatty acid catabolic process, and glyoxylate cycle; and cytosol, glyoxysome, and peroxisomal matrix localizations | −0.53 | 0.74 | |
| Heme biosynthesis | HEM13b | Putative coproporphyrinogen III oxidase; protein differentially expressed in azole-resistant strains | 0.81 | −0.67 |
| HEM14b | Ortholog(s) has oxygen-dependent protoporphyrinogen oxidase activity, role in heme biosynthetic process; and cytosol, mitochondrial inner membrane, and nucleus localizations | 0.51 | −0.70 | |
| HMX1 | Ortholog(s) has heme oxygenase (decyclizing) activity and roles in cellular iron ion homeostasis, heme catabolic process, response to carbon monoxide, and response to oxidative stress | −0.62 | 0.40 | |
| Cytoskeleton/cell cycle | MSC7b | Ortholog(s) has role in reciprocal meiotic recombination and cytosol, endoplasmic reticulum, and nucleus localizations | 0.88 | −0.56 |
| NCE102 | Ortholog(s) has roles in actin cytoskeleton organization, eisosome assembly, establishment of protein localization to the plasma membrane, negative regulation of protein phosphorylation, and more | −0.55 | 0.43 | |
| XBP1 | Ortholog(s) has RNA polymerase II transcription factor activity, sequence-specific DNA binding, and sequence-specific DNA binding activity | −0.69 | 0.47 | |
| FMP45 | Ortholog(s) has roles in ascospore formation and cellular response to drugs and has fungal-type cell wall organization | −0.69 | 0.47 | |
| Mitochondrial function | COX26 | Ortholog(s) has mitochondrial respiratory chain complex IV and mitochondrial respiratory chain supercomplex localization | −0.64 | 0.12 |
| Intracellular traffic | SRO7 | Ortholog(s) has Rab GTPase binding and SNARE binding activities and roles in Golgi-to-plasma membrane transport, establishment of cell polarity, exocytosis, and small GTPase-mediated signal transduction | 0.51 | −0.09 |
| Unknown function | CAGL0M11660gb | Has domain(s) with predicted hydrolase activity | 0.67 | 1.81 |
| CAGL0G05632gb | Ortholog(s) has cytoplasm localization | −0.52 | 1.32 | |
| CAGL0K07337gb | Has domain(s) with predicted ion channel activity, role in ion transport, and membrane localization | −0.63 | 0.66 | |
| CAGL0A02277g | Protein of unknown function | −0.72 | −0.11 | |
| PET10 | Ortholog(s) has lipid particle localization | −0.51 | 0.45 | |
| MUP1 | Protein of unknown function | −0.78 | 0.47 | |
| SET4 | Ortholog of S. cerevisiae SET4 and S. cerevisiae S288C YJL105W | 1.14 | −0.30 | |
| CAGL0L06776g | Has domain(s) with predicted sequence-specific DNA binding and transcription factor activities, zinc ion binding activity, and role in regulation of transcription, DNA templated | 0.85 | 0.33 | |
| CAGL0L08547g | Protein of unknown function | 0.76 | −0.05 | |
| CAGL0J00297g | Ortholog(s) has endoplasmic reticulum localization | 0.63 | −0.41 | |
| CAGL0G00594g | Ortholog(s) has Golgi apparatus and endoplasmic reticulum localization | 0.61 | −0.31 | |
The effect of the CgRpn4 deletion on the expression pattern is also shown. FLC, fluconazole; ABC, ATP-binding cassette.
Genes whose expression levels are significantly altered in the absence of CgRpn4.
In turn, genes involved in the heme biosynthetic pathway are upregulated (HEM13 and HEM14), while a gene involved in heme degradation is downregulated (HMX1). This profile suggests that the increased synthesis of heme, a vital prosthetic group of Erg11, may be required to accompany the increased expression of Erg11, aiming at increased levels of functional Erg11 molecules.
Role of CgRpn4 in transcriptome-wide changes occurring in response to fluconazole in C. glabrata.
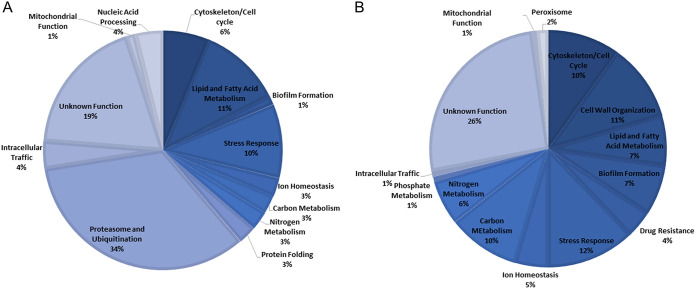
In order to study the role of CgRpn4 in the response of C. glabrata to fluconazole, gene expression changes occurring upon fluconazole exposure in the Δcgrpn4 mutant strain were compared to those observed in the wild-type strain. The expressions of 80 genes were found to be activated by CgRpn4, while 132 genes were found to be repressed, possibly in an indirect fashion, upon fluconazole exposure (Table S2). The most prevalent functional groups activated by CgRpn4 include the proteasome and ubiquitination, lipid and fatty acid metabolism, and the stress response (Fig. 2A), whereas repressed genes are enriched in cell wall organization and carbon metabolism (Fig. 2B).
FIG 2.
CgRPN4-regulated functional groups. Differentially expressed genes in exponential-phase KUE100_Δcgrpn4 C. glabrata cells compared to KUE100 cells after 1 h of fluconazole exposure. (A) CgRpn4-activated genes (downregulated in the mutant strain). (B) CgRpn4-repressed genes (upregulated in the mutant strain).
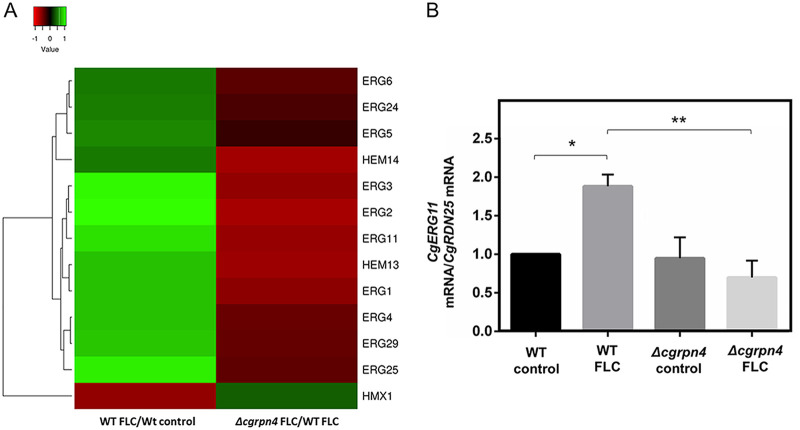
The comparison of this regulon with differentially expressed genes in fluconazole-challenged wild-type cells enabled the identification of 18 genes that respond to fluconazole only when CgRPN4 is present (Table 2). The most enriched functional group in this set comprises genes from the ergosterol biosynthetic pathway, namely, ERG1, ERG2, ERG3, and ERG11. A putative gene involved in the regulation of ergosterol synthesis in S. cerevisiae (HES1) (48, 49) is also found in this set. Interestingly, two genes involved in heme biosynthesis are also present: HEM13 and HEM14. As described previously, heme is a key prosthetic group of several ergosterol biosynthesis enzymes, and increased Hem13 protein levels were detected in an azole-resistant strain, concurrently with Erg11 (41). It is important to note that the upregulation of ergosterol biosynthesis is the most dramatic response to fluconazole stress and that CgRpn4 functions as an activator of ergosterol and heme biosynthesis (Fig. 3A). Moreover, the activation of ERG and HEM genes constitutes a specific role of this TF during fluconazole stress, as CgRpn4 does not regulate their basal expression (Table S3). Altogether, these data indicate that the activation of the ergosterol biosynthetic pathway, mediated by CgRpn4, could be a major regulatory mechanism of azole antifungal resistance. The transcriptome data were validated by real-time PCR (RT-PCR) quantification of CgERG11 expression in the wild type and the Δcgrpn4 mutant. The attained CgERG11 activation after 1 h of fluconazole exposure is comparable to the expression levels determined by the transcriptomics approach, as was the decreased fold change in the mutant strain (Fig. 3B).
FIG 3.
Comparison of ergosterol- and heme-related gene expression patterns in wild-type and Δcgrpn4 cells upon fluconazole stress. (A) Expression profiles of differentially expressed ERG and HEM genes in at least one data set. C. glabrata gene names are shown. In cases where no gene name has been assigned, the designation of its S. cerevisiae homolog is shown. (B) Comparison of the variation of CgERG11 transcript levels determined by RT-PCR in KUE100 and KUE100_Δcgrpn4 cells under control conditions or after 1 h of fluconazole exposure. Transcript levels of CgRDN25 were used for normalization. Expression values are the averages from at least three independent experiments. Error bars represent the corresponding standard deviations. *, P < 0.05; **, P < 0.01.
CgRpn4 was found to regulate the expression of several additional genes irrespective of fluconazole treatment. The largest functional group includes proteasome and ubiquitination genes. This group comprises exclusively genes activated by CgRpn4, in accordance with its role in both C. albicans and S. cerevisiae as an activator of proteasome genes (27, 28, 50). The high enrichment of proteasome subunit-encoding genes in the CgRpn4 regulon strongly indicates that its physiological function is conserved with other species.
Genes encoding five multidrug resistance transporters are repressed by CgRpn4: the major facilitator superfamily (MFS) transporters encoded by CAGL0L10912g, CAGL0B02343g, TPO1_1, and QDR2 as well as the ABC transporter YOR1. Of these, YOR1, TPO1_1, and QDR2 have been implicated in azole resistance (18, 19, 22). This indicates that CgRpn4 does not activate the expression of drug transporters, reinforcing its role as an activator of ergosterol biosynthesis as the main mechanism of fluconazole resistance.
CgRpn4 is activated upon fluconazole stress, leading to its nuclear accumulation.
In order to examine possible CgRpn4 activation mechanisms, we investigated its subcellular localization. CgRpn4 was fused to green fluorescent protein (GFP), expressed via the pGREG576_MTI_CgRPN4 plasmid in C. glabrata cells grown to mid-exponential phase in basal medium (BM) supplemented with 50 μM Cu2SO4 to induce fusion protein expression, and then transferred to fresh medium (control) or to fresh medium containing 150 mg/liter fluconazole. After 1 h of incubation, cells were inspected by fluorescence microscopy.
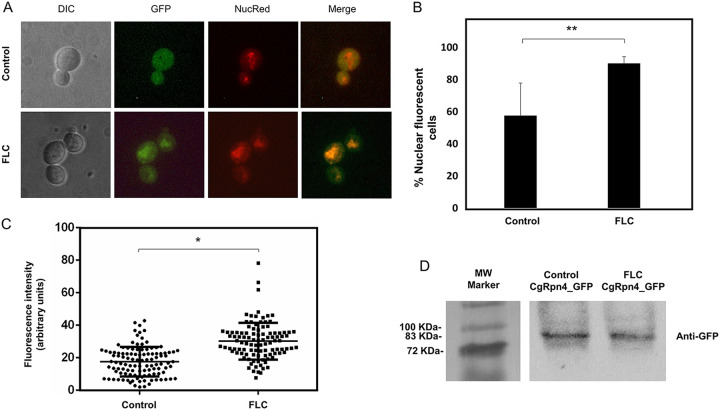
In untreated C. glabrata cells, the CgRpn4_GFP fusion protein is distributed throughout the whole cell, with some level of nuclear signal (Fig. 4A). After fluconazole stress, an enrichment of nuclear localization was observed (Fig. 4B and C). Fluconazole treatment therefore changes the relative distribution of CgRpn4 to the nucleus. The activation of its target genes could be partially dependent on the translocation of the transcription factor. In S. cerevisiae, Rpn4 protein levels are regulated by the proteasome in a negative-feedback loop. Therefore, a decrease in the CgRpn4_GFP signal could occur due to the activation of proteasome genes by the transcription factor. However, our localization data appear to show steady levels of CgRpn4 production. To evaluate if the levels of CgRpn4 are being affected by a negative-feedback loop, Western blotting was performed before and after fluconazole treatment (Fig. 4D). The CgRpn4_GFP fusion protein could be detected at similar levels under both experimental conditions, which is consistent with the localization data. This shows that CgRpn4 plasmid-driven production contributes to the protein steady state.
FIG 4.
CgRpn4 increases its relative distribution to the nucleus upon fluconazole stress. The subcellular localization of fluorescence in exponential-phase L5U1 C. glabrata cells harboring the pGREG576_MTI_CgRPN4 plasmid after 5 h of recombinant protein production under control conditions or after 1 h of fluconazole exposure was assessed. (A) Representative images of CgRpn4_GFP localization in L5U1 C. glabrata cells. DIC, differential interference contrast; GFP, green fluorescent protein; NucRed, nuclear stain; merge, GFP-NucRed overlap images. (B) Percentage of cells showing nuclear localization of the CgRpn4_GFP fusion protein under control conditions or after 1 h of fluconazole exposure. Error bars represent the corresponding standard deviations. *, P < 0.01. (C) Comparison of nuclear fluorescence intensities of CgRpn4_GFP in L5U1 C. glabrata cells under control conditions or after 1 h of fluconazole exposure. The estimation of nuclear signal intensity was calculated after deduction of the cytoplasm intensity and correction for background intensity. Error bars represent the corresponding standard deviations. *, P < 0.05. (D) Western blot detection of the CgRpn4_GFP fusion protein under control conditions or after 1 h of fluconazole exposure. Immunoblotting was carried out using a mouse anti-GFP antibody. The displayed images are representative of results from two independent experiments. MW, molecular weight.
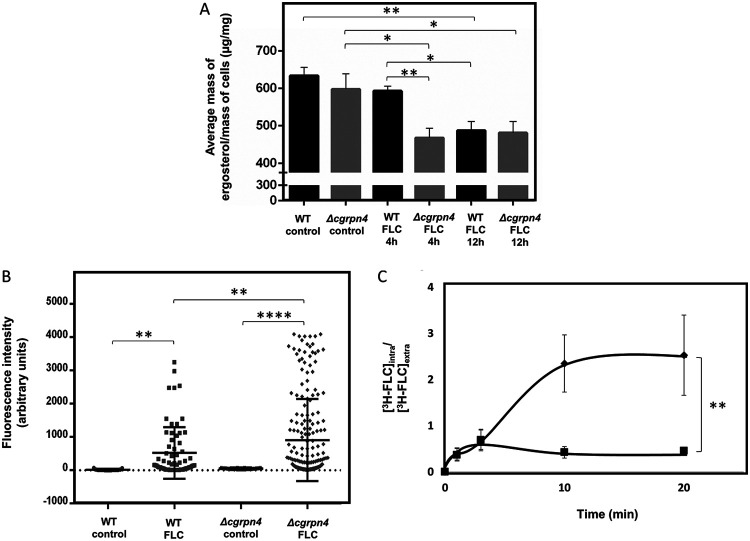
CgRPN4 plays a role in the maintenance of ergosterol levels, membrane permeability, and fluconazole accumulation.
Transcriptomics analysis of the KUE100 wild-type C. glabrata strain during fluconazole stress revealed the significant activation of the ergosterol biosynthesis pathway mediated by CgRpn4. This led us to hypothesize that CgRpn4 may contribute to preserving ergosterol levels in C. glabrata upon fluconazole exposure.
C. glabrata cells were grown to the mid-exponential phase and transferred to fresh medium (control) or fresh medium containing 150 mg/liter fluconazole. After 4 h and 12 h of incubation, cells were collected, total ergosterol was extracted, and its levels were quantified by high-performance liquid chromatography (HPLC).
Deleting CgRPN4 does not affect ergosterol levels in untreated cells (Fig. 5A). The absence of CgRPN4 reduces ergosterol levels upon fluconazole stress for 4 h or 12 h. Ergosterol levels in the wild type decreased only after 12 h and to levels comparable to those of the deletion strain, which indicates a relevant role of the transcription factor in this mechanism. Altogether, these data implicate CgRpn4 in the maintenance of the ergosterol content upon early fluconazole exposure. Moreover, these results show that CgRpn4, through transcriptional regulation of the ergosterol biosynthesis pathway, has a measurable effect on ergosterol levels during fluconazole stress, which contributes to antifungal resistance.
FIG 5.
CgRpn4 regulates ergosterol levels in C. glabrata cells during fluconazole stress, affecting permeability and drug accumulation. (A) KUE100 and KUE100_Δcgrpn4 C. glabrata cells were harvested after 15 h of growth in RPMI 1640 medium (control) or after 4 h or 12 h of fluconazole stress. Ergosterol was extracted and quantified by HPLC. Cholesterol was used as an internal standard to evaluate the yield of ergosterol extraction. The displayed ergosterol contents are representative of results from at least six independent experiments. Error bars represent the corresponding standard deviations. *, P < 0.05; **, P < 0.01. (B) Comparison of plasma membrane permeabilities of exponential-phase KUE100 (squares) and KUE100_Δcgrpn4 (diamonds) cells under control conditions or upon 1 h of fluconazole stress. The estimation of plasma membrane permeability is based on the fluorescence intensity values exhibited by yeast cells upon the passive accumulation of propidium iodide. Error bars represent the corresponding standard deviations. **, P < 0.01; ***, P < 0.001. (C) Time course accumulation of radiolabeled [3H]fluconazole in strains KUE100 (squares) and KUE100_Δcgrpn4 (diamonds) during cultivation in liquid BM in the presence of 150 mg/liter unlabeled fluconazole. Accumulation values are the averages from at least three independent experiments. Error bars represent the corresponding standard deviations. **, P < 0.01.
Cell permeability in response to fluconazole was investigated using the fluorescent probe propidium iodide (PI), and the possible participation of CgRpn4 was evaluated. Upon 1 h of exposure to fluconazole, C. glabrata cell permeability increases significantly (Fig. 5B). This observation is probably related to the inhibition of Erg11 by fluconazole, described previously to lead to the accumulation of the toxic sterol DMCDD that permeabilizes the plasma membrane (51, 52). The permeability of untreated Δcgrpn4 cells is not significantly different from that of wild-type cells. However, the permeability of Δcgrpn4 cells is significantly higher than that of the wild-type strain following fluconazole stress (Fig. 5B). These data indicate that fluconazole increases C. glabrata cell permeability and that CgRpn4 contributes to controlling its maintenance during fluconazole exposure, presumably by the upregulation of ergosterol biosynthesis.
The role of CgRPN4 in ergosterol biosynthesis and plasma membrane permeability led us to investigate if this could be related to the increased azole susceptibility observed in the Δcgrpn4 mutant. Consistent with the observed susceptibility and cell permeability phenotypes, the Δcgrpn4 deletion mutant was found to accumulate 2.5-fold more radiolabeled fluconazole than the wild-type strain after 20 min of exposure to the antifungal (Fig. 5C). These results suggest that CgRpn4 activity mediates C. glabrata resistance to fluconazole by reducing its intracellular accumulation in yeast cells, possibly as a result of its regulation of ergosterol levels and membrane permeability.
Determination of promoter recognition motifs by CgRpn4 and direct regulation of CgERG11 expression.
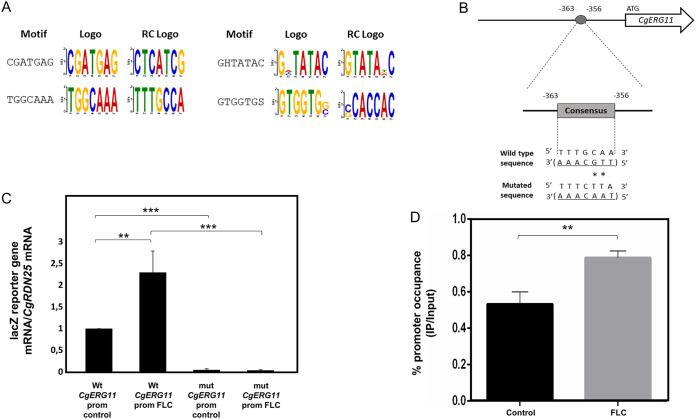
In order to identify possible CgRpn4 DNA recognition sites, the promoters of CgRpn4-activated genes during fluconazole stress were searched for enriched motifs using DREME (53). Excluding TATA box sequences, 4 overrepresented motifs were found (Fig. 6A). Interestingly, one of the identified motifs (TGGCAAA) is identical or nearly identical to a core region of the ScRpn4 and CaRpn4 consensus (GGTGGCAAA and GAAGGCAAAA, respectively) found in promoters of proteasome genes (28, 50). This indicates that there is a high level of conservation in the promoter sequences recognized by Rpn4 across these species.
FIG 6.
Possible CgRpn4 recognition sequence and direct regulation of CgERG11. (A) Motifs found to be overrepresented in the promoters of CgRpn4-activated genes, as found by the DREME tool. Sequences located upstream (bp −1000 to −1) of the genes found to be activated by CgRpn4 were retrieved from PathoYeastract (75) and submitted to DREME. The prediction was made using default parameters. The sequences shown correspond to the motifs outputted by DREME, excluding TATA box motifs. (B) The putative CgRpn4 recognition sequence in the promoter of CgERG11 is in the complementary strand. The numbers refer to the position of the consensus site relative to the first ATG of the coding region. The wild-type sequence is shown underlined below the box, with asterisks denoting the base substitutions generated by site-directed mutagenesis. The resulting mutated sequence is shown below. (C) Comparison of the variations of lacZ transcript levels determined by RT-PCR in L5U1 cells harboring the pYEP354_CgERG11prom_lacZ or pYEP354_mut_CgERG11prom_lacZ plasmid under control conditions or after 1 h of fluconazole exposure. Transcript levels of CgRDN25 were used for normalization. Expression values are the averages from at least three independent experiments. Error bars represent the corresponding standard deviations. **, P < 0.01; ***, P < 0.001. (D) ChIP–RT-PCR measurements of CgRpn4 promoter occupancy at the CgERG11 promoter element containing the possible recognition motif. ChIP experiments were performed using mouse anti-c-Myc antibody and cultures of L5U1 cells harboring the pGREG526_PDC1_CgRPN4 plasmid under control conditions or after 1 h of fluconazole exposure. The amplified amount of DNA was measured by RT-PCR and normalized to the total amount of the sample. Samples from the CgRpn4 IP were compared to the input (IP/input ratio) under each condition. To determine percent occupancy under each condition, values were calculated as the ratio of the percent precipitated under fluconazole stress to the percent precipitated under control conditions. Promoter occupancy values are the averages from three independent experiments. Error bars represent the corresponding standard deviations. **, P < 0.01.
We found a nearly identical motif (TTGCAAA) located at positions −363 to −356 (Fig. 6B) of CgERG11. To determine if this motif is required for the activation of CgERG11, the CgERG11 promoter was placed upstream of the lacZ reporter gene, and site-directed mutagenesis was used to disrupt the putative binding site.
Upon 1 h of fluconazole exposure, lacZ expression driven by the CgERG11 promoter increased 2-fold in comparison to control conditions (Fig. 6C), which is in accordance with our transcriptomics data. When the TTGCAAA motif in the CgERG11 promoter was disrupted by 2 nucleotide substitutions (from TTGCAAA to TAACAAA), lacZ expression was reduced by 20-fold compared to the wild-type promoter under control conditions and even more so after fluconazole exposure (59-fold) (Fig. 6C). These results indicate that the identified motif is required for CgERG11 basal expression and especially CgERG11 activation during fluconazole stress. Chromatin immunoprecipitation (ChIP) followed by RT-PCR was used to establish a direct link between CgRpn4 and the identified CgERG11 promoter motif using a CgRpn4_c-Myc fusion protein expressed via the pGREG526_PDC1_CgRPN4 plasmid. The results show CgRpn4 can bind the CgERG11 promoter in the region containing the TTGCAAA motif (Fig. 6D). The promoter is bound by CgRpn4 under control conditions and increases its promoter occupancy during fluconazole stress, strongly suggesting direct CgRpn4 binding to this essential motif in the CgERG11 promoter.
DISCUSSION
C. glabrata is an emerging fungal pathogen with an impressive ability to acquire resistance to azole antifungal drugs. An understanding of the molecular basis of this phenotype is crucial for designing better-suited therapeutic approaches to tackle infections caused by C. glabrata.
In this study, we present a novel pathway used by C. glabrata to respond and adapt to azole drugs under the control of the transcription factor CgRpn4. Deletion of CgRPN4 increases C. glabrata susceptibility to multiple azole drugs. According to the latest EUCAST breakpoints, the attained MIC values represent decreased susceptibility to fluconazole, although not a shift from resistant to intermediate, as the parental strain used exhibits an intermediate fluconazole susceptibility profile. In the closely related yeast S. cerevisiae and in the pathogenic species C. albicans, Rpn4 is a regulator of proteasome genes (27, 28, 50). Defects in proteasome levels can potentially impact a wide range of phenotypes and play a role in the general stress response. To test this possibility, the susceptibility of the Δcgrpn4 deletion mutant to additional antifungal drugs and other stress agents was assessed. Notably, the results indicate that CgRpn4 functions mainly as a regulator of azole resistance.
RNA sequencing (RNA-seq)-based transcriptomics revealed that Rpn4 is also a regulator of proteasome genes in C. glabrata, thus unveiling a highly conserved physiological function among yeast species. Additionally, the activation of ERG genes, including CgERG11, was identified as a key underlying mechanism of CgRpn4-mediated fluconazole resistance. These findings are supported by our data as well as data from other studies showing ERG gene upregulation during azole stress (42–47). Moreover, the upregulation of CgERG11 was found to be dependent on a promoter motif recognized and bound by CgRpn4. Together with the transcriptomics data, the essential role of the TTGCAAA motif in CgERG11 activation and the direct binding of CgRpn4 (especially under fluconazole exposure) to the CgERG11 promoter element provide evidence for a prevalent regulatory role in ergosterol biosynthesis regulation leading to fluconazole resistance.
The activation of the ergosterol biosynthesis pathways was seen to have a measurable impact on the ergosterol levels of C. glabrata during fluconazole exposure. The deletion of CgRPN4 leads to a reduction of ergosterol levels in the presence of fluconazole, indicating that the activation of ergosterol biosynthesis genes during fluconazole stress may be part of a compensatory mechanism to counteract ergosterol synthesis targeted by the antifungal. A similar mechanism has been reported for C. albicans, where fluconazole-tolerant strains present higher levels of intermediate metabolites of the sterol biosynthetic pathway and ergosterol ester, which was associated with the upregulation of ergosterol biosynthetic pathway genes (54). Furthermore, a reduction in the content of sphingolipids or ergosterol results in enhanced susceptibility to drugs (55–57). Consistent with this hypothesis, C. glabrata cells are more permeable and accumulate more intracellular fluconazole when CgRPN4 is deleted, which is in good agreement with the role of this TF in the activation of ergosterol biosynthesis. In S. cerevisiae, ScRPN4 is transcriptionally regulated under various stresses, and ScRpn4 protein levels are also controlled by proteasome activity in a negative-feedback loop (27). In our study, constant protein steady-state levels of CgRpn4 were detected, although it is possible that the efficient expression of the CgRpn4 protein by our expression system hindered the ability to detect turnover changes. As these data relate to the protein stability of CgRpn4, a possible feedback loop at the transcriptional regulation of CgRPN4 remains to be established.
Here, we show the role of the transcription factor CgRpn4 in mediating fluconazole resistance in C. glabrata. Through transcriptional control over the ergosterol biosynthesis pathway, CgRpn4 regulates ergosterol levels in the plasma membrane, thus affecting cell permeability and fluconazole accumulation. Additionally, CgRpn4 exerts direct control over CgERG11 expression, reinforcing its role as a relevant player in C. glabrata fluconazole resistance, which is likely conserved among other yeast species. Based on these results, we propose that CgRpn4 is a promising target to tackle the acquisition of azole drug resistance.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
Candida glabrata parental strains KUE100 (58) and L5U1 (cgura3Δ0 cgleu2Δ0) (59) and derived single-deletion mutants were batch cultured at 30°C at 250 rpm in basal medium (BM) (20 g/liter glucose, 2.7 g/liter ammonium sulfate, and 1.7 g/liter yeast nitrogen base [YNB] without ammonia or amino acids) or RPMI 1640–2% glucose medium at pH 7.0. For BM assays using L5U1 transformed with either the pGREG576 or pGREG526 plasmid (harboring a URA3 selection marker), the medium was supplemented with 60 mg/liter leucine. The plasmids pGREG576 and pGREG526 were obtained from the Drag&Drop collection (60). The Saccharomyces cerevisiae BY4741 strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was batch cultured at 30°C at 250 rpm in yeast extract-peptone-dextrose (YPD) medium (20 g/liter glucose, 20 g/liter peptone, 10 g/liter yeast extract).
Disruption of CgRPN4.
The deletion of C. glabrata RPN4 addressed in this study was carried out in the parental strain KUE100 using the method described previously by Ueno et al. (61). The primers used are presented in Table S4 in the supplemental material.
Cloning of the C. glabrata CgRPN4 gene (ORF CAGL0K01727g).
The pGREG576 and pGREG526 plasmids from the Drag&Drop collection were used as described previously to clone and express the C. glabrata open reading frame (ORF) CAGL0K01727g (18, 22, 62–65), giving rise to pGREG576_CgRPN4 or pGREG526_CgRPN4. The GAL1 promoter present in each plasmid was replaced by the copper-inducible MTI C. glabrata promoter (pGREG576_CgRPN4) or the constitutive PDC1 C. glabrata promoter (pGREG526_CgRPN4) (66), giving rise to the pGREG576_MTI_CgRPN4 or pGREG526_PDC1_CgRPN4 plasmid. The primers used are presented in Table S4. The recombinant plasmids were obtained through homologous recombination in S. cerevisiae and verified by DNA sequencing.
Antifungal susceptibility assays.
The susceptibility of the parental strain KUE100 to inhibitory concentrations of the selected drugs was compared to that of the deletion mutant KUE100_Δcgrpn4 by spot assays. The ability of CgRPN4 gene expression to increase wild-type resistance to the tested chemical stresses was also examined in the URA3− L5U1 C. glabrata strain using the pGREG576_MTI_CgRPN4 centromeric plasmid.
Cell suspension preparation and spot assays were carried as described previously (18, 22, 62–65). The tested drugs included the following compounds, used in the specified concentration ranges that were found to exert inhibitory growth effects: the azole antifungal drugs ketoconazole (10 to 60 mg/liter), fluconazole (100 to 250 mg/liter), miconazole (0.10 to 0.50 mg/liter), itraconazole (15 to 30 mg/liter), clotrimazole (2.5 to 15 mg/liter), and tioconazole (0.30 to 0.70 mg/liter) (all from Sigma). Antifungals from other families included the polyene amphotericin B (0.05 to 0.20 mg/liter), the pyrimidine analog 5-FC (0.10 to 0.30 mg/liter), and the broad-activity fungicide mancozeb (0.5 to 1.5 mg/liter) (all from Sigma). The following osmotic and oxidative stresses were tested at levels found to exert inhibitory growth effects: NaCl (0.5 to 2 M) (from Panreac) and H2O2 (7.5 to 15 mM) (from Sigma), respectively. The MIC values of each antifungal were determined according to the EUCAST susceptibility testing method (67). Drugs not present in the reference method (e.g., ketoconazole) were used according to the same experimental procedures. The MIC assays were performed in 96-well plates containing RPMI 1640–2% glucose medium with the appropriate drug concentrations. Cells grown for 18 h were used to create an initial inoculum containing 5 × 106 CFU/ml in distilled water, which was subsequently used to prepare a 5 × 105-CFU/ml working suspension. The plates were then inoculated with the cell suspension to a final inoculum density of 2.5 × 105 CFU/ml and incubated at 37°C for 24 h. Final cell growth was assessed by absorbance measurement at a 530-nm wavelength.
CgRpn4 subcellular localization assessment.
The subcellular localization of the CgRpn4 protein was determined based on the observation of L5U1 C. glabrata cells transformed with the pGREG576_MTI_CgRPN4 plasmid. These cells express the CgRpn4_GFP fusion protein, whose localization may be determined using fluorescence microscopy as described previously (18, 22, 62–65). C. glabrata cell suspensions were prepared in BM supplemented with leucine and 50 μM CuSO4, until a standard culture optical density at 600 nm (OD600) of 0.5 was reached, and transferred to the same medium with or without fluconazole. After 1 h of incubation, 1 drop of NucRed Live 647 was added to 1 ml of 4 × 107 cells/ml, and cell suspensions were incubated in the dark with orbital agitation (30 min at 250 rpm). Cells were centrifuged (17,500 × g for 5 min), washed twice, and resuspended in phosphate-buffered saline (PBS) for final aliquots of 107 cells/ml. The distribution of the CgRpn4_GFP fusion protein in yeast cells was determined by fluorescence microscopy with a Zeiss Axioplan microscope (Carl Zeiss MicroImaging) using excitation and emission wavelengths of 395 and 509 nm (GFP) or 541 and 686 nm (NucRed). Fluorescence images were captured using a cooled Zeiss AxioCam 503 color camera (Carl Zeiss Microscopy). For the determination of nuclear signal intensity, fluorescence intensity was defined as the average pixel-by-pixel intensity in the selected region of interest (nucleus) after the deduction of the cytoplasm pixel intensity. A minimum of 100 cells per experiment were used. The fluorescence images were background corrected by using dark-current images.
Total RNA extraction.
C. glabrata strains KUE100 and KUE100_Δcgrpn4 were grown in BM until mid-exponential phase. Subsequently, cells were transferred to fresh medium (control) or fresh medium containing 150 mg/liter fluconazole and harvested after 1 h of incubation. Total RNA was isolated using an Ambion RiboPure yeast RNA kit according to the manufacturer’s instructions.
Library preparation and gene expression analysis.
Strand-specific RNA-seq library preparation and sequencing were carried out as a paid service by the next-generation sequencing (NGS) core of the Oklahoma Medical Research Foundation, Oklahoma City, OK. Paired-end reads (Illumina HiSeq 3000 PE150, 2 by 150 bp, with 2 Gb of clean data) were obtained from KUE100 and KUE100_Δcgrpn4. Two replicates of each sample were obtained from three independent RNA isolations, which were subsequently pooled. Sample reads were trimmed using Skewer (v0.2.2) (68) and aligned to the C. glabrata CBS138 reference genome, obtained from the Candida Genome Database (CGD) (http://www.candidagenome.org/), using TopHat (v2.1.1) (69) with the parameters –p 12 (number of threads), –g 1 (maximum number of times that a read can be mapped to the genome), –b2-very-sensitive (preset option), and –library-type fr-firststrand (to account for strand specificity). HTSeq (v0.7.1) (70) was used to count mapped reads per ORF. Differentially expressed genes were identified using DESeq2 (71), with an adjusted P value threshold of 0.05 and log2-fold change thresholds of −0.5 and 0.5. Default parameters in DESeq2 were used. Candida albicans and Saccharomyces cerevisiae homologs were obtained from the Candida Genome Database and Saccharomyces Genome Database (SGD) (https://www.yeastgenome.org/), respectively.
Ergosterol quantification.
Ergosterol was extracted from cells using methods adapted from the ones described previously by Gong et al. (72) and carried out as described previously (73). Cells were cultivated in RPMI 1640–2% glucose medium with orbital agitation (250 rpm) until a standard culture OD600 of 5.0 was reached, harvested by centrifugation, and resuspended in 5 ml of methanol. One milliliter of a solution of 1 mg/ml of cholesterol (Sigma) was added as an internal standard to estimate the ergosterol extraction yield. Homogenization was carried out with glass beads for 30 s, followed by incubation at 320 rpm for 1 h. Each sample was then centrifuged, and 1.7 ml of the supernatant was collected, clarified, and stored until HPLC analysis. The extracts were separated in a 250-mm by 4-mm C18 column (LiChroCART Purospher Star RP-18 end-capped 5-μm column) at 30°C. Samples were eluted in 100% methanol at a flow rate of 1 ml/min. The detection of cholesterol and ergosterol was performed using a UV-visible (UV-Vis) detector set at 282 and 210 nm, respectively. Under the conditions used, the retention time of cholesterol was 15.4 ± 0.4 min, while ergosterol was eluted at 12.5 ± 0.2 min. Subsequent quantification of the two lipids was performed using appropriate calibration curves. The results are shown as micrograms of ergosterol per milligram of wet cell weight.
Plasma membrane permeability.
Plasma membrane permeability was assessed by the passive uptake of propidium iodide (PI) (20 mM in dimethyl sulfoxide [DMSO]; Invitrogen). C. glabrata cell suspensions from strains KUE100 and KUE100_Δcgrpn4 were prepared in BM until a standard culture OD600 of 0.5 was reached and transferred to the same medium with or without 150 mg/liter fluconazole. After 1 h of incubation, PI was added to 1 ml of 4 × 107 cells/ml to a final concentration of 20 μM, and cell suspensions were incubated in the dark with orbital agitation (15 min at 250 rpm). Cells exposed to PI were centrifuged (17,500 × g for 5 min), washed twice, and resuspended in PBS for final aliquots of 107 cells/ml. PI fluorescence was detected by fluorescence microscopy with a Zeiss Axioplan microscope (Carl Zeiss MicroImaging), using excitation and emission wavelengths of 536 and 595 nm, respectively. Fluorescence images were captured using a cooled Zeiss AxioCam 503 color camera (Carl Zeiss Microscopy), and the images were analyzed with ZEN lite software from Zeiss Microscopy. The cell-to-cell fluorescence intensity was defined as the average pixel-by-pixel intensity in the selected region of interest, and a minimum of 100 cells per experiment were used. The fluorescence images were background corrected by using dark-current images.
[3H]fluconazole accumulation assays.
[3H]fluconazole transport assays were carried out as described previously for other radiolabeled compounds (18, 22, 62–65). The internal accumulation of fluconazole was determined by calculating the ratio of the radiolabeled fluconazole measured within the yeast cells to that in the external medium (intracellular/extracellular). The parental strain KUE100 and the mutant strain KUE100_Δcgrpn4 were grown in BM until mid-exponential phase and harvested by filtration. Cells were washed and resuspended in BM to obtain dense cell suspensions (OD600 = 0.5, equivalent to approximately 1.57 mg [dry weight] ml−1). Readily, 0.1 μM [3H]fluconazole (1 mCi/ml) (Moravek Inc.) and 150 mg/liter of unlabeled fluconazole were added to the cell suspensions. Incubation proceeded for an additional 20 min. The intracellular accumulation of labeled fluconazole was monitored by filtering 200 μl of the cell suspension, at adequate time intervals, through prewetted glass microfiber filters (Whatman GF/C). The filters were washed with ice-cold TM buffer [0.1 M 2-(N-morpholino)ethanesulfonic acid (Sigma)/41 mM Tris (Sigma)], and radioactivity was measured in a Beckman LS 5000TD scintillation counter. Extracellular [3H]fluconazole was estimated by radioactivity assessment of 50 μl of the supernatant. Nonspecific [3H]fluconazole adsorption to the filters and to the cells (less than 5% of the total radioactivity) was assessed and taken into consideration. To calculate the intracellular concentration of labeled fluconazole, the internal cell volume (Vi) of the exponential cells, grown in the absence of the drug and used for accumulation assays, was considered constant and equal to 2.5 mg (dry weight) μl−1 (74).
In silico prediction of overrepresented sequences in CgRpn4-activated promoters.
The promoters (bp −1000 to −1) upstream of the coding regions of genes whose expression was found to be activated by CgRpn4 were retrieved using the Retrieve Upstream Sequence tool from PathoYeastract (75). The obtained sequences were submitted to DREME (MEME suite) (53) for the discovery of enriched sequences, using default parameters. The identified motifs were then cross-checked with ScRpn4 and CaRpn4 motifs retrieved from the TF-Consensus List tool from YEASTRACT (76, 77) and PathoYeastract (75), respectively.
Cloning of the CgERG11 promoter and site-directed mutagenesis.
The pYPE354 plasmid was used as described previously to clone and express the lacZ reporter gene (18). pYEP354 contains the yeast selectable marker URA3 and the bacterial selectable marker ampR. CgERG11 promoter DNA was generated by PCR using genomic DNA extracted from the sequenced CBS138 C. glabrata strain and primers presented in Table S4. The first primer contains a region with homology within the beginning of the CgERG11 promoter and a recognition site for the EcoRI restriction enzyme, flanked by additional bases. The second primer contains a region with homology within the end of the CgERG11 promoter and the beginning of the CgERG11 coding sequence and a recognition site for the PstI restriction enzyme, flanked by additional bases. The amplified fragment was ligated into the pYEP354 vector (T4 ligase; New England BioLabs), previously cut with the same restriction enzymes, to obtain the pYEP354_CgERG11prom_lacZ plasmid. The putative CgRpn4 consensus sequence in the CgERG11 promoter was mutated by site-directed mutagenesis using the primers listed in Table S4. The designed primers contain two mutations within the potential consensus sequence, resulting in the production of the mutated consensus sequence by PCR amplification to obtain the pYEP354_mut_CgERG11prom_lacZ plasmid. The original template was then degraded by DpnI digestion.
RT-PCR gene expression measurement.
The transcript levels of the CgERG11 or the lacZ reporter gene encoding β-galactosidase were determined by quantitative real-time PCR (RT-PCR). For CgERG11 expression measurements, the KUE100 and KUE100_Δcgrpn4 strains were grown in BM until mid-exponential phase, transferred to fresh medium (control) or fresh medium containing 150 mg/liter fluconazole (OD600 = 0.5), and harvested after 1 h of incubation. Samples were immediately frozen at −80°C until RNA extraction. For lacZ expression measurements, L5U1 cells transformed with the pYEP354_CgERG11prom_lacZ or pYEP354_mut_CgERG11prom_lacZ plasmid were grown in BM supplemented with leucine until mid-exponential phase. Fluconazole exposure, cell harvesting, and storage were performed as mentioned above. For total RNA extraction, the hot-phenol method was applied (78). RT-PCR was performed as described previously (18, 22, 62–65). The synthesis of cDNA for RT-PCR experiments, from total RNA samples, was performed using the MultiScribe reverse transcriptase kit (Applied Biosystems) and the 7500 RT-PCR thermal cycler block (Applied Biosystems) according to the manufacturer’s instructions. The quantity of cDNA for the following reactions was kept at around 10 ng. The subsequent RT-PCR step was carried out using SYBR green (NZYTech) reagents with default parameters established by the manufacturer and the primers in Table S4. The CgRDN25 gene transcript levels were used as an internal reference.
Western blot analysis.
For Western blot analysis, L5U1 C. glabrata cells transformed with the pGREG576_MTI_CgRPN4 plasmid were grown to mid-exponential phase in BM supplemented with leucine and 50 μM Cu2SO4 to induce fusion protein expression and then transferred to fresh medium (control) or fresh medium containing 150 mg/liter fluconazole. After 1 h of incubation, cells were collected and washed twice with cold PBS (pH 7.4). For protein extraction, a TCA (trichloroacetic acid) precipitation protocol was used. Briefly, cell pellets were resuspended in resuspension buffer (1.85 M NaOH, 7.5% 2-mercaptoethanol) by vortexing, followed by a 2-min incubation on ice (5 times). Proteins were precipitated with 25% TCA using 5 cycles of vortexing and 2 min of incubation on ice, followed by centrifugation. The pellets were washed with acetone and resuspended in Laemmli buffer at 65°C for 20 min (with rotation), followed by centrifugation, and the supernatant containing the protein extract was collected. Samples were separated on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Proteins were transferred to an Amersham Protran 0.2-μm nitrocellulose membrane (GE Healthcare). Immunoblotting was performed using a 1:200 dilution of mouse IgG anti-GFP antibody (catalog number sc-9996; Santa Cruz Biotechnology Inc.) and a 1:1,000 dilution of anti-mouse IgG BP-HRP (binding protein conjugated to horseradish peroxidase) (catalog number sc-516102; Santa Cruz Biotechnology Inc.). Detection was performed using Fusion Solo (Vilber) following incubation with an enhanced chemiluminescence (ECL) substrate.
CgRpn4 chromatin immunoprecipitation assays followed by RT-PCR.
L5U1 C. glabrata cells transformed with the pGREG526_PDC1_CgRPN4 plasmid were grown to mid-exponential phase in BM supplemented with leucine until 100 OD units were reached and then transferred to fresh medium (control) or fresh medium containing 150 mg/liter fluconazole. After 1 h of incubation, cells were cross-linked with 1% formaldehyde (15 min), and cross-linking was quenched with 340 mM glycine (10 min). Cells were collected, washed twice with cold PBS (pH 7.4), and frozen immediately at −80°C. Cell pellets were lysed with 0.5-mm zirconia beads (Invitrogen) in lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% NP-40, 0.5 mM dithiothreitol [DTT]) by vortexing for 20 s and incubation on ice for 2 min (4 times). Cell debris was pelleted by centrifugation, and the lysate was collected. Lysates were sonicated four times for 2 min followed by 2 min on ice using a Bioruptor water bath sonicator. Sonicated lysates were cleared by centrifugation, and 5% of the sample was collected for use as the input (sonicated chromatin, not immunoprecipitated). For immunoprecipitation (IP), chromatin was incubated overnight at 4°C (with rotation) with protein G SureBeads (Bio-Rad) coupled with IgG anti-c-Myc antibody (catalog number sc-40; Santa Cruz Biotechnology Inc.). Beads were washed according to the manufacturer’s instructions, and the samples were eluted at 65°C for 20 min in Tris-EDTA (TE) buffer with 0.5% SDS. Input and immunoprecipitated samples were incubated overnight at 65°C for cross-linking reversal. DNA was purified with the NZYGelPure kit (NZYTech). Quantification of a specific CgERG11 promoter region was performed by RT-PCR using 5 μl of the DNA preparation from each reaction mixture. The primers used are listed in Table S4. Immunoprecipitated samples were processed together with the input samples, and the amplification product was normalized to the total template used. Based on methods reported previously (79, 80), the ratio of the immunoprecipitated promoter to the input (IP/input) was calculated, and ratios in fluconazole samples were compared to the ratio in the control samples at the specific locus of the CgERG11 promoter.
Statistical analyses.
All experiments represent the averages from two or more independent experiments, with at least two technical replicates each. Error bars represent standard deviations. Statistical analyses were performed using one-way analysis of variance (ANOVA) with Tukey’s correction (comparisons of more than 2 groups) or two-tailed t tests with Welch’s correction (comparisons of 2 groups). Significance levels are indicated in the figure legends (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Data availability.
Raw data are available at the Gene Expression Omnibus (GEO) under accession number GSE129349.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge John Bennett from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD, for kindly providing the L5U1 strain.
This work was supported by the Fundação para a Ciência e a Tecnologia (FCT) (contracts PTDC/BBB-BIO/4004/2014 and PTDC/BII-BIO/28216/2017, Ph.D. grants to P.P. and M.G.) as well as by the Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-022231, the BioData.pt Research Infrastructure). We also acknowledge funding received by the iBB from the FCT (UID/BIO/04565/2013 and UIDB/04565/2020) and from the Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-007317). This study was partly supported by a Joint Usage/Research Program of the Medical Mycology Research Center, Chiba University.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no declared competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Azie N, Neofytos D, Pfaller M, Meier-Kriesche H-U, Quan S-P, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance registry and invasive fungal infections: update 2012. Diagn Microbiol Infect Dis 73:293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis 29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 4.Khatib R, Johnson LB, Fakih MG, Riederer K, Briski L. 2016. Current trends in candidemia and species distribution among adults: Candida glabrata surpasses C. albicans in diabetic patients and abdominal sources. Mycoses 59:781–786. doi: 10.1111/myc.12531. [DOI] [PubMed] [Google Scholar]
- 5.Farmakiotis D, Kyvernitakis A, Tarrand JJ, Kontoyiannis DP. 2015. Early initiation of appropriate treatment is associated with increased survival in cancer patients with Candida glabrata fungaemia: a potential benefit from infectious disease consultation. Clin Microbiol Infect 21:79–86. doi: 10.1016/j.cmi.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland AA, Harrison LH, Farley MM, Hollick R, Stein B, Chiller TM, Lockhart SR, Park BJ. 2015. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: results from population-based surveillance. PLoS One 10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl 6):5–10. doi: 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 8.Kumar K, Askari F, Sahu MS, Kaur R. 2019. Candida glabrata: a lot more than meets the eye. Microorganisms 7:39. doi: 10.3390/microorganisms7020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:409–417. doi: 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yapar N. 2014. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag 10:95–105. doi: 10.2147/TCRM.S40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan BH, Chakrabarti A, Li RY, Patel AK, Watcharananan SP, Liu Z, Chindamporn A, Tan AL, Sun PL, Wu UI, Chen YC, Xu YC, Wang H, Sun ZY, Wang LL, Lu J, Yang Q, Zhang QQ, Shao HF, Liao K, Woo PCY, Marak RSK, Kindo AJ, Wu CL, Ho MW, Lu PL, Wang LS, Riengchan P, Asia Fungal Working Group. 2015. Incidence and species distribution of candidaemia in Asia: a laboratory-based surveillance study. Clin Microbiol Infect 21:946–953. doi: 10.1016/j.cmi.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Petrikkos G, Skiada A. 2007. Recent advances in antifungal chemotherapy. Int J Antimicrob Agents 30:108–117. doi: 10.1016/j.ijantimicag.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama H, Nakayama N, Arisawa M, Aoki Y. 2001. In vitro and in vivo effects of 14alpha-demethylase (ERG11) depletion in Candida glabrata. Antimicrob Agents Chemother 45:3037–3045. doi: 10.1128/AAC.45.11.3037-3045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debnath S, Addya S. 2014. Structural basis for heterogeneous phenotype of ERG11 dependent azole resistance in C. albicans clinical isolates. Springerplus 3:660. doi: 10.1186/2193-1801-3-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Revie NM, Iyer KR, Robbins N, Cowen LE. 2018. Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol 45:70–76. doi: 10.1016/j.mib.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghannoum MA, Rice LB. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 12:501–517. doi: 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermitsky J-P, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa C, Pires C, Cabrito TR, Renaudin A, Ohno M, Chibana H, Sá-Correia I, Teixeira MC. 2013. Candida glabrata drug:H+ antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1. Antimicrob Agents Chemother 57:3159–3167. doi: 10.1128/AAC.00811-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol 61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 20.Caudle KE, Barker KS, Wiederhold NP, Xu L, Homayouni R, Rogers PD. 2011. Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot Cell 10:373–383. doi: 10.1128/EC.00073-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai H-F, Krol AA, Sarti KE, Bennett JE. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother 50:1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pais P, Costa C, Pires C, Shimizu K, Chibana H, Teixeira MC. 2016. Membrane proteome-wide response to the antifungal drug clotrimazole in Candida glabrata: role of the transcription factor CgPdr1 and the drug:H+ antiporters CgTpo1_1 and CgTpo1_2. Mol Cell Proteomics 15:57–72. doi: 10.1074/mcp.M114.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux M-E, d’Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pais P, Costa C, Cavalheiro M, Romão D, Teixeira MC. 2016. Transcriptional control of drug resistance, virulence and immune system evasion in pathogenic fungi: a cross-species comparison. Front Cell Infect Microbiol 6:131. doi: 10.3389/fcimb.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y, Varshavsky A. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A 98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett 450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 29.Owsianik G, Balzi L, Ghislain M. 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol Microbiol 43:1295–1308. doi: 10.1046/j.1365-2958.2002.02823.x. [DOI] [PubMed] [Google Scholar]
- 30.Johnson ES, Ma PC, Ota IM, Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt RM, Schessner JP, Borner GH, Schuck S. 2019. The proteasome biogenesis regulator Rpn4 cooperates with the unfolded protein response to promote ER stress resistance. Elife 8:43244. doi: 10.7554/eLife.43244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teixeira MC, Dias PJ, Monteiro PT, Sala A, Oliveira AL, Freitas AT, Sá-Correia I. 2010. Refining current knowledge on the yeast FLR1 regulatory network by combined experimental and computational approaches. Mol Biosyst 6:2471–2481. doi: 10.1039/c004881j. [DOI] [PubMed] [Google Scholar]
- 33.Teixeira MC, Dias PJ, Simões T, Sá-Correia I. 2008. Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem Biophys Res Commun 367:249–255. doi: 10.1016/j.bbrc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 34.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 35.Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wüster A, Wu J, Babu MM, Elledge SJ, Toczyski D, Lokey RS, Krogan NJ. 2010. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol 6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, Aust T, Baeriswyl L, Cerino R, Chang L, Estoppey D, Eichenberger J, Frederiksen M, Hartmann N, Hohendahl A, Knapp B, Krastel P, Melin N, Nigsch F, Oakeley EJ, Petitjean V, Petersen F, Riedl R, Schmitt EK, Staedtler F, Studer C, Tallarico JA, Wetzel S, Fishman MC, Porter JA, Movva NR. 2014. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res 169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perumal P, Mekala S, Chaffin WL. 2007. Role for cell density in antifungal drug resistance in Candida albicans biofilms. Antimicrob Agents Chemother 51:2454–2463. doi: 10.1128/AAC.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother 43:2753–2765. doi: 10.1128/AAC.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett JE, Izumikawa K, Marr KA. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob Agents Chemother 48:1773–1777. doi: 10.1128/aac.48.5.1773-1777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers PD, Vermitsky J-P, Edlind TD, Hilliard GM. 2006. Proteomic analysis of experimentally induced azole resistance in Candida glabrata. J Antimicrob Chemother 58:434–438. doi: 10.1093/jac/dkl221. [DOI] [PubMed] [Google Scholar]
- 42.Henry KW, Nickels JT, Edlind TD. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother 44:2693–2700. doi: 10.1128/aac.44.10.2693-2700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marichal P, Vanden Bossche H, Odds FC, Nobels G, Warnock DW, Timmerman V, Van Broeckhoven C, Fay S, Mose-Larsen P. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother 41:2229–2237. doi: 10.1128/AAC.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavalheiro M, Costa C, Silva-Dias A, Miranda IM, Wang C, Pais P, Pinto SN, Mil-Homens D, Sato-Okamoto M, Takahashi-Nakaguchi A, Silva RM, Mira NP, Fialho AM, Chibana H, Rodrigues AG, Butler G, Teixeira MC. 2019. A transcriptomics approach to unveiling the mechanisms of in vitro evolution towards fluconazole resistance of a Candida glabrata clinical isolate. Antimicrob Agents Chemother 63:e00995-18. doi: 10.1128/AAC.00995-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cernicka J, Subik J. 2006. Resistance mechanisms in fluconazole-resistant Candida albicans isolates from vaginal candidiasis. Int J Antimicrob Agents 27:403–408. doi: 10.1016/j.ijantimicag.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Barker KS, Crisp S, Wiederhold N, Lewis RE, Bareither B, Eckstein J, Barbuch R, Bard M, Rogers PD. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J Antimicrob Chemother 54:376–385. doi: 10.1093/jac/dkh336. [DOI] [PubMed] [Google Scholar]
- 47.Liu TT, Lee REB, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother 49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beh CT, Cool L, Phillips J, Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157:1117–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang B, Brown JL, Sheraton J, Fortin N, Bussey H. 1994. A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding protein. Yeast 10:341–353. doi: 10.1002/yea.320100307. [DOI] [PubMed] [Google Scholar]
- 50.Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol 2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanden Bossche H, Koymans L, Moereels H. 1995. P450 inhibitors of use in medical treatment: focus on mechanisms of action. Pharmacol Ther 67:79–100. doi: 10.1016/0163-7258(95)00011-5. [DOI] [PubMed] [Google Scholar]
- 52.Abe F, Usui K, Hiraki T. 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48:8494–8504. doi: 10.1021/bi900578y. [DOI] [PubMed] [Google Scholar]
- 53.Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A, Yadav V, Prasad R. 2012. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS One 7:e39812. doi: 10.1371/journal.pone.0039812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasrija R, Prasad T, Prasad R. 2005. Membrane raft lipid constituents affect drug susceptibilities of Candida albicans. Biochem Soc Trans 33:1219–1223. doi: 10.1042/BST20051219. [DOI] [PubMed] [Google Scholar]
- 56.Pasrija R, Panwar SL, Prasad R. 2008. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob Agents Chemother 52:694–704. doi: 10.1128/AAC.00861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukhopadhyay K, Prasad T, Saini P, Pucadyil TJ, Chattopadhyay A, Prasad R. 2004. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob Agents Chemother 48:1778–1787. doi: 10.1128/aac.48.5.1778-1787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueno K, Uno J, Nakayama H, Sasamoto K, Mikami Y, Chibana H. 2007. Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot Cell 6:1239–1247. doi: 10.1128/EC.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen K-H, Miyazaki T, Tsai H-F, Bennett JE. 2007. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386:63–72. doi: 10.1016/j.gene.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Jansen G, Wu C, Schade B, Thomas DY, Whiteway M. 2005. Drag&Drop cloning in yeast. Gene 344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 61.Ueno K, Matsumoto Y, Uno J, Sasamoto K, Sekimizu K, Kinjo Y, Chibana H. 2011. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS One 6:e24759. doi: 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pais P, Pires C, Costa C, Okamoto M, Chibana H, Teixeira MC. 2016. Membrane proteomics analysis of the Candida glabrata response to 5-flucytosine: unveiling the role and regulation of the drug efflux transporters CgFlr1 and CgFlr2. Front Microbiol 7:2045. doi: 10.3389/fmicb.2016.02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romão D, Cavalheiro M, Mil-Homens D, Santos R, Pais P, Costa C, Takahashi-Nakaguchi A, Fialho AM, Chibana H, Teixeira MC. 2017. A new determinant of Candida glabrata virulence: the acetate exporter CgDtr1. Front Cell Infect Microbiol 7:473. doi: 10.3389/fcimb.2017.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Costa C, Henriques A, Pires C, Nunes J, Ohno M, Chibana H, Sá-Correia I, Teixeira MC. 2013. The dual role of Candida glabrata drug:H+ antiporter CgAqr1 (ORF CAGL0J09944g) in antifungal drug and acetic acid resistance. Front Microbiol 4:170. doi: 10.3389/fmicb.2013.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costa C, Nunes J, Henriques A, Mira NP, Nakayama H, Chibana H, Teixeira MC. 2014. Candida glabrata drug:H+antiporter CgTpo3 (ORF CAGL0I10384G): role in azole drug resistance and polyamine homeostasis. J Antimicrob Chemother 69:1767–1776. doi: 10.1093/jac/dku044. [DOI] [PubMed] [Google Scholar]
- 66.Zordan RE, Ren Y, Pan S-J, Rotondo G, De Las Peñas A, Iluore J, Cormack BP. 2013. Expression plasmids for use in Candida glabrata. G3 3:1675–1686. doi: 10.1534/g3.113.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope W, EUCAST-AFST. 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18:E246–E247. doi: 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
- 68.Jiang H, Lei R, Ding S-W, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong P, Guan X, Witter E. 2001. A rapid method to extract ergosterol from soil by physical disruption. Appl Soil Ecol 17:285–289. doi: 10.1016/S0929-1393(01)00141-X. [DOI] [Google Scholar]
- 73.Santos R, Costa C, Mil-Homens D, Romão D, de Carvalho CCCR, Pais P, Mira NP, Fialho AM, Teixeira MC. 2017. The multidrug resistance transporters CgTpo1_1 and CgTpo1_2 play a role in virulence and biofilm formation in the human pathogen Candida glabrata. Cell Microbiol 19:e12686. doi: 10.1111/cmi.12686. [DOI] [PubMed] [Google Scholar]
- 74.Rosa MF, Sá-Correia I. 1996. Intracellular acidification does not account for inhibition of Saccharomyces cerevisiae growth in the presence of ethanol. FEMS Microbiol Lett 135:271–274. doi: 10.1111/j.1574-6968.1996.tb08000.x. [DOI] [PubMed] [Google Scholar]
- 75.Monteiro PT, Pais P, Costa C, Manna S, Sa-Correia I, Teixeira MC. 2017. The PathoYeastract database: an information system for the analysis of gene and genomic transcription regulation in pathogenic yeasts. Nucleic Acids Res 45:D597–D603. doi: 10.1093/nar/gkw817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monteiro PT, Oliveira J, Pais P, Antunes M, Palma M, Cavalheiro M, Galocha M, Godinho CP, Martins LC, Bourbon N, Mota MN, Ribeiro RA, Viana R, Sá-Correia I, Teixeira MC. 2020. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res 48:D642–D649. doi: 10.1093/nar/gkz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teixeira MC, Monteiro PT, Palma M, Costa C, Godinho CP, Pais P, Cavalheiro M, Antunes M, Lemos A, Pedreira T, Sá-Correia I. 2018. YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res 46:D348–D353. doi: 10.1093/nar/gkx842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Köhrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol 194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 79.Merhej J, Delaveau T, Guitard J, Palancade B, Hennequin C, Garcia M, Lelandais G, Devaux F. 2015. Yap7 is a transcriptional repressor of nitric oxide oxidase in yeasts, which arose from neofunctionalization after whole genome duplication. Mol Microbiol 96:951–972. doi: 10.1111/mmi.12983. [DOI] [PubMed] [Google Scholar]
- 80.Knutson BA, Hahn S. 2011. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol Cell Biol 31:818–831. doi: 10.1128/MCB.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are available at the Gene Expression Omnibus (GEO) under accession number GSE129349.