Members of the Enterobacter cloacae complex are important opportunistic human pathogens capable of causing a wide variety of infections. During recent decades, aminoglycoside-resistant E. cloacae complex isolates have increasingly been reported and have become a major concern. Here, we employed high-throughput sequencing in combination with specific PCR assays to investigate the prevalence of aminoglycoside resistance genes among 170 isolates of the E. cloacae complex collected from a teaching hospital in Wenzhou, China.
KEYWORDS: AAC(3)-IIg, aminoglycoside resistance, aminoglycoside-modifying enzyme, 3-N-acetyltransferase, Enterobacter cloacae complex
ABSTRACT
Members of the Enterobacter cloacae complex are important opportunistic human pathogens capable of causing a wide variety of infections. During recent decades, aminoglycoside-resistant E. cloacae complex isolates have increasingly been reported and have become a major concern. Here, we employed high-throughput sequencing in combination with specific PCR assays to investigate the prevalence of aminoglycoside resistance genes among 170 isolates of the E. cloacae complex collected from a teaching hospital in Wenzhou, China. A total of 12 known genes [aphA-1, strA, strB, aac(6′)-IIc, aadA2, aac(3)-IId, aadB, aadA1, rmtB, armA, aadA5, and aac(6′)-Ie–aph(2′')-Ia] and 1 novel gene [aac(3)-IIg] were identified, with aphA-1 (71.18%), strA (55.29%), and strB (52.35%) being the most prevalent, and aac(3)-IIg was detected with a positive rate of 21.76% (37/170). The aac(3)-IIg gene was 810 bp in length and encoded a protein that shared 72 to 78% identities with previously known AAC(3)-II aminoglycoside 3-N-acetyltransferases. The MICs of gentamicin and tobramycin were 512 μg/ml and 64 μg/ml, respectively, when aac(3)-IIg was cloned into Escherichia coli DH5α. All aac(3)-IIg-positive isolates exerted broad aminoglycoside resistance profiles, mediated by the coexistence of multiple resistance genes. Moreover, aminoglycoside resistance and resistance genes were found to be transferable in most strains (24/37). Nevertheless, pulsed-field gel electrophoresis (PFGE) and dendrogram analysis showed clonal diversity among these isolates. S1 nuclease PFGE, Southern hybridization, and whole-genome sequencing indicated that aac(3)-IIg was located on transferable as well as nontransferable plasmids of various sizes. The analysis of the genetic environment suggested that aac(3)-IIg is embedded within a class 1 integron, with IS26 playing an important role in its mobility.
INTRODUCTION
Aminoglycosides are highly potent, broad-spectrum antibiotics that act through inhibition of bacterial protein synthesis and have been utilized for the treatment of life-threatening infections for almost 80 years (1). In the clinical setting, resistance to aminoglycosides is most commonly mediated by the presence of various aminoglycoside-modifying enzymes (AMEs), including acetyltransferases (AACs), nucleotidyltransferases (ANTs), and phosphotransferases (APHs) (2, 3). To date, over 100 AMEs have been described, and AACs represent the largest group of AMEs (4, 5). AACs catalyze the acetylation of —NH2 groups in the aminoglycoside antibiotics using acetyl coenzyme A as the acetyl donor (3, 5). Based on their position specificities for aminoglycoside modifications, these enzymes are further divided into subtypes, whose nomenclature consists of the three-letter abbreviation AAC, as an identifier for the type of enzymatic modification, followed by the site of modification enclosed between parentheses, a roman number particular to the resistance profile that it confers, and, in some cases, a lowercase letter when multiple enzymes that modify the same position exist (3, 4).
For the AAC(3) enzymes, a number of different proteins with different substrate specificities have been identified (3, 4). The subclass AAC(3)-II confers resistance to gentamicin (GEN), netilmicin (NET), tobramycin (TOB), sisomicin (SIS), 2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin, and dibekacin and is widespread among Enterobacteriaceae and other Gram-negative clinical isolates (3, 4). Furthermore, the aac(3)-II alleles are usually found on mobile genetic elements (i.e., plasmids and transposons) and, thereby, can be horizontally transferred among different pathogens (6–8).
Species of the Enterobacter cloacae complex (including E. cloacae, Enterobacter asburiae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter ludwigii, and Enterobacter nimipressuralis) are widely distributed in nature (9). They can occur in terrestrial and aquatic environments and also in the intestinal tracts of humans and animals (10). Over recent decades, these microorganisms have taken on clinical significance and have emerged as troublesome pathogens that are frequently involved in nosocomial infections (9–12), especially for E. cloacae and E. hormaechei, which are most frequently isolated from human clinical specimens (10, 13). From the antibiotic resistance point of view, most isolates of the E. cloacae complex constitutively produce the AmpC β-lactamase and are intrinsically resistant to ampicillin, amoxicillin, amoxicillin-clavulanic acid, first- and second-generation cephalosporins, and cefoxitin, while they are generally susceptible to fluoroquinolones, trimethoprim-sulfamethoxazole, chloramphenicol, aminoglycosides, tetracyclines, piperacillin-tazobactam, and carbapenems (9, 10, 12). Nevertheless, these organisms are capable of acquiring genes encoding resistance to multiple classes of antibiotics (12), and clinical isolates resistant to aminoglycosides by producing AMEs have frequently been reported in recent years (10, 14–16). The rapid acquisition of resistance phenotypes is most often plasmid mediated and is also associated with the dissemination of transposable elements (9).
Here, we investigated the distribution of aminoglycoside resistance genes among 170 clinical E. cloacae complex isolates from a Chinese teaching hospital. Novel to this study, we observed and identified a new AAC(3)-II determinant, named aac(3)-IIg, that represents a seventh evolutionary lineage in this group of aminoglycoside resistance genes. In addition, we characterized aac(3)-IIg and analyzed the aminoglycoside susceptibility profiles of aac(3)-IIg-harboring isolates and the association of aac(3)-IIg with other aminoglycoside resistance determinants. Moreover, the molecular epidemiology of these aac(3)-IIg-producing isolates as well as the genetic environment of the aac(3)-IIg gene were analyzed.
RESULTS
Prevalence of aminoglycoside resistance genes.
To investigate the prevalence of the aminoglycoside resistance genes among clinical isolates of E. cloacae, we screened pooled DNA of 170 strains collected from a teaching hospital in Wenzhou, China, by next-generation sequencing (NGS). A total of 34 million reads were obtained and mapped onto the previously known aminoglycoside resistance gene sequences collected from the database. As a result, a total of 13 aminoglycoside resistance genes were identified (Table 1). The most abundant were 3 APH genes, strB [aph(6)-Id], strA [aph(3′')-Ib], and aphA-1 [aph(3′)-Ia], which had an average sequencing depth of over 4,000 times (Table 1). Of the remaining 10 genes, 3 [aac(3)-IIg, aac(3)-IId, and aac(6′)-IIc] were AAC genes (3); 4 {aadA2, aadA5, aadB [ant(2′')-Ia], and aadA1 (ant(3′')-Ia)} were ANT genes (3); both armA (17) and rmtB (18) were 16S rRNA methylase genes; and aac(6′)-Ie–aph(2′')-Ia, which encodes a bifunctional enzyme, had the lowest abundance (55.5 times).
TABLE 1.
Characteristics of the aminoglycoside resistance genes identified in the pooled samples and their distribution among 170 clinical E. cloacae complex isolates
| Gene name | GenBank accession no. | Aminoglycoside resistance profilea | Depth | Length (ntb) | Identity (%) | Coverage (%) | No. (%) of isolates |
|---|---|---|---|---|---|---|---|
| aac(3)-IIg | M97172 [aac(3)-IIb] | GEN, KAN, TOB, SIS, MCR, NET | 332.81 | 810 | 78.81 | 100 | 37 (21.76) |
| aac(3)-IId | EU022314.1 | GEN, KAN, TOB, SIS, MCR, NET | 1,177.12 | 861 | 100.00 | 100 | 35 (20.59) |
| aac(6')-Ie–aph(2'')-Ia | GU565967.1 | GEN, KAN, TOB, SIS, MCR, NET, RIB | 55.50 | 1,440 | 100.00 | 100 | 1 (0.59) |
| aac(6')-IIc | AF162771 | TOB, RIB | 3,236.76 | 582 | 100.00 | 100 | 49 (28.82) |
| aadA2 | AF156486 | STR, SPE | 1,743.47 | 780 | 100.00 | 100 | 39 (22.94) |
| aadA5 | AF137361 | STR, SPE | 281.97 | 789 | 100.00 | 100 | 4 (2.35) |
| aadB [ant(2'')-Ia] | AF078527 | GEN, KAN, TOB, SIS, MCR | 1,006.94 | 534 | 100.00 | 100 | 18 (10.59) |
| aadA1 [ant(3'')-Ia] | X02340.1 | STR, SPE | 176.58 | 972 | 99.62 | 97.94 | 12 (7.06) |
| aphA-1 [aph(3')-Ia] | BX664015.1 | KAN, TOB, NEO, RIB | 4,496.58 | 816 | 98.89 | 100 | 121 (71.18) |
| strA [aph(3'')-Ib] | AF313472 | STR | 7,357.34 | 804 | 99.25 | 100 | 94 (55.29) |
| strB [aph(6)-Id] | AF024602 | STR | 7,679.38 | 837 | 99.64 | 100 | 89 (52.35) |
| armA | GU437214.1 | GEN, KAN, AMK, TOB, SIS, MCR, NET | 209.67 | 774 | 100.00 | 100 | 5 (2.94) |
| rmtB | AM886293.1 | GEN, KAN, AMK, TOB, SIS, MCR, NET | 671.63 | 756 | 100.00 | 100 | 9 (5.29) |
Aminoglycoside resistance profiles of recombinant E. coli DH5α producing each resistance gene detected in this study. Abbreviations: GEN, gentamicin; KAN, kanamycin; TOB, tobramycin; SIS, sisomicin; MCR, micronomicin; NET, netilmicin; RIB, ribostamycin; STR, streptomycin; SPE, spectinomycin; NEO, neomycin; AMK, amikacin.
nt, number of nucleotides.
Next, we examined the presence of 13 aminoglycoside resistance genes among the 170 isolates by PCR with specific primers. A total of 162 (95.3%) isolates were positive for these AME-encoding and/or 16S rRNA methylase genes. Despite repetitive PCR, 8 isolates (4.7%) did not yield a positive result for any of these resistance genes. This is in agreement with the MIC results indicating that the 8 isolates were susceptible to all of the aminoglycosides tested in this study (data not shown). Moreover, consistent with the relative abundance of these genes in the pooled samples, PCR detection of the 13 genes in the 170 individual isolates revealed a similar trend of detection rates (Table 1), suggesting that the pooled sequencing data were of a sufficient depth to reflect the genetic structure of the samples. However, as the pooled whole-genome sequencing (WGS) of 170 isolates generated only 34 million 150-bp reads, we cannot rule out the possibility that the sequencing data may have missed some antibiotic resistance genes in these strains. Taken together, these results demonstrate the high prevalence of aminoglycoside resistance genes, especially AME-encoding genes, among clinical E. cloacae complex isolates from this teaching hospital. Remarkably, we also observed an aac(3)-II-like gene [aac(3)-IIg] that showed relatively low homology (78.8% identity) with the reference aac(3)-IIb gene, indicating the potential presence of a novel AAC(3)-II determinant among these isolates.
Cloning and functional analysis of aac(3)-IIg.
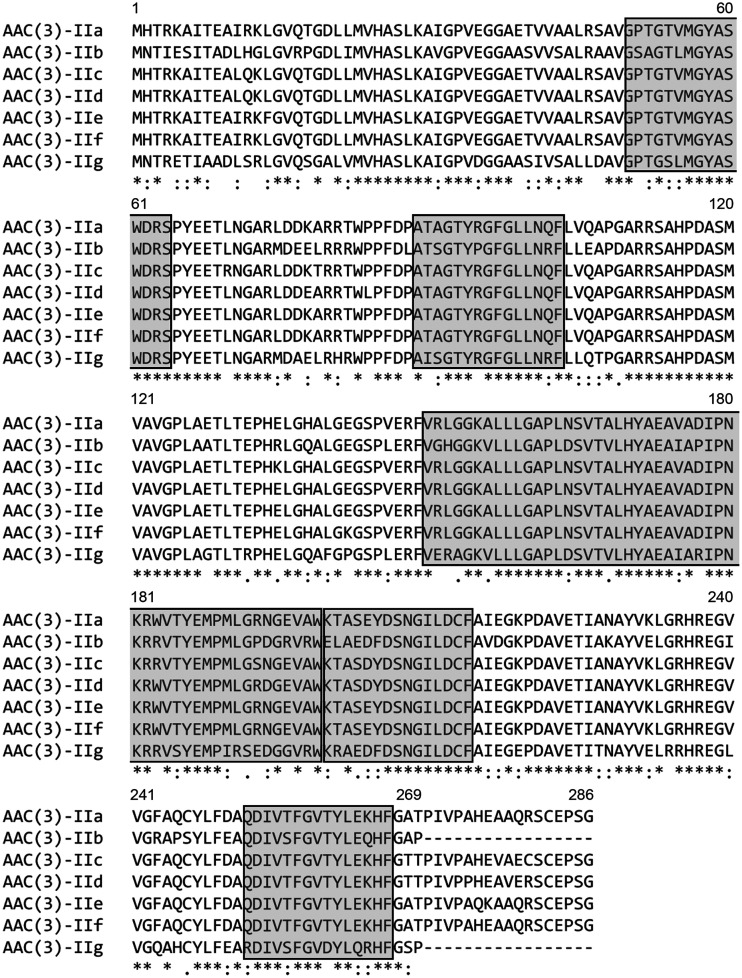
To gain further information on the potential aac(3)-II gene, we first performed bioinformatics analysis of its molecular sequence identified in this study. Nucleotide sequence comparisons showed that the full-length open reading frame (ORF) was identical to the mapping result for the pooled sequencing reads, and all 37 strains harbored the same aac(3)-II-like gene, which was 810 bp in length and which encoded 269 amino acids. Phylogenetic analysis of the predicted protein and 176 aminoglycoside resistance determinants obtained from the Comprehensive Antibiotic Resistance Database (CARD) showed that this protein was clustered together with 6 AAC(3)-II enzymes (see Fig. S1 in the supplemental material), indicating that this protein was closely related to the subclass AAC(3)-II. The sequence identities between the protein and the 6 previously known AAC(3)-II enzymes, AAC(3)-IIa, AAC(3)-IIb, AAC(3)-IIc, AAC(3)-IId, AAC(3)-IIe, and AAC(3)-IIf, were 72.49%, 78.81%, 72.86%, 73.61%, 72.12%, and 72.49%, respectively. Multiple-sequence alignment of the protein sequences (Fig. 1) showed that this protein contains a number of conserved residues and motifs characteristic of the AAC(3)-II enzymes (4), suggesting that the protein is a member of the subclass AAC(3)-II and was designated AAC(3)-IIg (GenBank accession no. MT090547) in this study. A BLASTp search using the AAC(3)-IIg amino acid sequence as a query against the NCBI database showed that a putative aminoglycoside 3-N-acetyltransferase present in plasmid pH11 from Klebsiella pneumoniae (GenBank accession no. ALP55389.1) and on the chromosomes of Gammaproteobacteria (GenBank accession no. WP_012695485.1) has the same sequence; their antibiotic resistance functions, however, were not investigated.
FIG 1.
Sequence alignment of AAC(3)-IIg with other AAC(3)-II proteins. The GenBank accession numbers were as follows: AAC(3)-IIa, X51534.1; AAC(3)-IIb, M97172.1; AAC(3)-IIc, X54723.1; AAC(3)-IId, EU022314.1; AAC(3)-IIe, EU022315.1; and AAC(3)-IIg, MT090547 (this study). Protein sequence alignment was performed using the Clustal Omega program. The Clustal Omega program determined the conservation of residues. Dashes, amino acids that are absent; asterisks, fully conserved residues; colons, residues with strongly similar properties; periods, residues with low similarity. The conserved motif sites predicted by the MEME program are boxed. Numbers correspond to the amino acid residues in each full-length protein.
To detect the resistance activities of AAC(3)-IIg, a 944-bp fragment containing the complete ORF and its putative promoter region was amplified from the genomic DNA of three randomly selected strains (strains Y108, Y315, and Y2152) using PCR. The DNA fragment was then sequenced, cloned into pMD19T, and transformed into Escherichia coli DH5α. The MICs of a variety of aminoglycosides for the donors, the transformants, and the recipient controls are shown in Table 2. Clearly, aac(3)-IIg expression conferred greatly reduced susceptibility to gentamicin, micronomicin, sisomicin, tobramycin, kanamycin (KAN), and netilmicin. On the other hand, AAC(3)-IIg did not confer resistance to streptomycin (STR), spectinomycin, amikacin, neomycin (NEO), ribostamycin, or apramycin. These results confirmed that the aac(3)-IIg gene may contribute to an aminoglycoside resistance profile typical of AAC(3)-II enzymes (3, 4). The original isolates Y108, Y315, and Y2152 had different levels of resistance to and a wider spectrum of resistance to the antibiotics examined (Table 2; Table S3), indicating that aac(3)-IIg may be expressed at different levels and other resistance genes may have been present in these strains.
TABLE 2.
MICs of various aminoglycosides for individual recombinant E. coli DH5α isolates producing AAC(3)-IIg and the corresponding 3 aac(3)-IIg-positive isolates
| Strain | MIC (μg/ml)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | KAN | TOB | SIS | MCR | NET | STR | SPE | AMK | NEO | RIB | APR | |
| DH5α | ≤0.25 | 1 | 0.5 | ≤0.5 | ≤0.5 | 0.5 | 2 | 4 | ≤1 | 1 | 2 | 4 |
| DH5α/pMD19T-aac(3)-IIg (Y108) | 512 | 8 | 32 | 512 | 1,024 | 2 | 2 | 8 | ≤1 | 1 | 2 | 4 |
| DH5α/pMD19T-aac(3)-IIg (Y315) | 512 | 8 | 64 | 512 | 1,024 | 4 | 2 | 8 | ≤1 | 1 | 2 | 4 |
| DH5α/pMD19T-aac(3)-IIg (Y2152) | 512 | 8 | 64 | 512 | 1,024 | 4 | 2 | 4 | ≤1 | 1 | 2 | 4 |
| Y108 | 128 | 64 | 64 | 256 | 512 | 128 | 128 | 16 | 8 | 2 | 512 | 4 |
| Y315 | 256 | 256 | 128 | 1,024 | 1,024 | 64 | 128 | 16 | 8 | 16 | 1,024 | 4 |
| Y2152 | 64 | 128 | 4 | 128 | 256 | 1 | 128 | 4 | ≤1 | 16 | 512 | 4 |
Abbreviations: GEN, gentamicin; KAN, kanamycin; TOB, tobramycin; SIS, sisomicin; MCR, micronomicin; NET, netilmicin; STR, streptomycin; SPE, spectinomycin; AMK, amikacin; NEO, neomycin; RIB, ribostamycin; APR, apramycin.
Antibiotic susceptibility to aminoglycosides and aminoglycoside resistance gene profiles.
To gain a better understanding of the associations between phenotypic and genotypic aminoglycoside resistance patterns in the 37 aac(3)-IIg-positive isolates, we evaluated their aminoglycoside susceptibilities and associated resistance genes within individual isolates (Table 3; Table S3). Of these, all strains (100%) were resistant to gentamicin, kanamycin, and streptomycin; 34 (91.9%) were tobramycin and netilmicin resistant; 30 (81.1%) were neomycin resistant; 3 (8.1%) were resistant to spectinomycin; and 2 (5.4%) were resistant to both spectinomycin and amikacin. The most frequently observed aminoglycoside resistance pattern was GEN-KAN-STR-TOB-NEO-NET (64.9%, 24/37). Apart from this, the GEN-KAN-STR-TOB-NET and GEN-KAN-STR-NEO resistance patterns were detected in 18.9% (7/37) and 8.1% (3/37) of the isolates, respectively (Table 3). As AAC(3)-IIg did not confer resistance to neomycin, streptomycin, spectinomycin, or amikacin, these results indicate that other genetic determinants are involved in the nonsusceptibility to these aminoglycosides.
TABLE 3.
Antimicrobial susceptibility and genotypic and epidemiologic characteristics of the 37 aac(3)-IIg-positive E. cloacae complex isolates
| Straina | Resistance profileb | Aminoglycoside resistance genesc | Pulsotype |
|---|---|---|---|
| Y3 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aphA-1 | 3 |
| Y4 | GEN, KAN, STR, NEO | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 18 |
| Y7 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 3 |
| Y8 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 9 |
| Y10 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 13 |
| Y24 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 18 |
| Y40 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 23 |
| Y43 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 23 |
| Y59 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 23 |
| Y67 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 21 |
| Y75 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 7 |
| Y81 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aphA-1 | 17 |
| Y88 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 16 |
| Y108 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 4 |
| Y118 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 21 |
| Y129 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 5 |
| Y130 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB | 5 |
| Y131 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aphA-1 | 5 |
| Y137 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 15 |
| Y150 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 22 |
| Y165 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 2 |
| Y176 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 11 |
| Y178 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 22 |
| Y184 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 19 |
| Y233 | GEN, KAN, STR, TOB, NEO, NET, SPE, AMK | aac(3)-IIg , strA , strB , aac(6')-IIc , aadA1 , aphA-1 , armA | 1 |
| Y243 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 22 |
| Y249 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 6 |
| Y261 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 9 |
| Y274 | GEN, KAN, STR, TOB, NEO, NET, SPE, AMK | aac(3)-IIg, strA, strB, aac(6')-IIc, aadA1, aphA-1, armA | 1 |
| Y295 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 10 |
| Y308 | GEN, KAN, STR, TOB, NET | aac(3)-IIg, strA, strB, aac(6')-IIc | 20 |
| Y315 | GEN, KAN, STR, TOB, NEO, NET | aac(3)-IIg , strA , strB , aac(6')-IIc , aphA-1 | 8 |
| Y320 | GEN, KAN, STR, NEO | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1, aac(3)-IId | 10 |
| Y323 | GEN, KAN, STR, TOB, NEO, NET, SPE | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1, aac(3)-IId, aadA2 | 14 |
| Y324 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 12 |
| Y327 | GEN, KAN, STR, TOB, NET | aac(3)-IIg , strA , strB , aac(6')-IIc | 19 |
| Y2152 | GEN, KAN, STR, NEO | aac(3)-IIg, strA, strB, aac(6')-IIc, aphA-1 | 10 |
Isolates with transconjugants are underlined.
The resistance phenotypes transferred to the recipient by conjugation are underlined. Abbreviations: GEN, gentamicin; KAN, kanamycin; STR, streptomycin; TOB, tobramycin; SPE, spectinomycin; AMK, amikacin; NEO, neomycin; NET, netilmicin.
Genes that were cotransferred by conjugation are underlined.
The aac(3)-IIg-positive isolates were then analyzed for the presence of other aminoglycoside resistance genes which encode various individual resistance profiles when expressed in E. coli (Table 1). As shown in Table 3, among 24 GEN-, KAN-, STR-, TOB-, NEO-, and NET-resistant isolates, 79.2% (19/24) were positive for aac(3)-IIg, strA, strB, aac(6′)-IIc, and aphA-1, which was the most prevalent resistance gene profile. Other resistance gene profiles included aac(3)-IIg, strA, strB, and aphA-1; aac(3)-IIg, strA, strB, and aac(6′)-IIc; and aac(3)-IIg, strA, and strB, with positive rates of 12.5% (3/24), 4.2% (1/24), and 4.2% (1/24), respectively. For these strains, positive associations between phenotypic resistance and the presence of the corresponding resistance genes were detected, although the phenotype or the genotype alone did not accurately predict the other (Tables 1 and 3). Also, the discrepancy between genotypes and phenotypes may have implications for the complexity of the mechanisms underlying this resistance phenotype, which can emerge from many different genetic determinants. In addition, all (100%, 7/7) of the GEN-, KAN-, STR-, TOB-, and NET-resistant strains carried the aac(3)-IIg strA strB aac(6′)-IIc gene profile, implying that the presence of aphA-1 in GEN-, KAN-, STR-, TOB-, NEO-, and NET-resistant strains correlated with NEO resistance (Table 3). With regard to the 3 GEN-, KAN-, STR-, and NEO-resistant isolates, 2 were found to harbor aac(3)-IIg, strA, strB, aac(6′)-IIc, and aphA-1, and 1 isolate harbored aac(3)-IIg, strA, strB, aac(6′)-IIc, aphA-1, and aac(3)-IId. It is a bit odd that the 3 strains were not resistant to tobramycin, which suggests that the presence or absence of a specific gene associated with a particular resistance phenotype does not necessarily mean that the strain is resistant or susceptible. One possible explanation for this discrepancy is the variable expression levels of resistance genes, which may affect the particular resistance phenotypes within individual isolates. It is also possible that this susceptibility phenotype may be caused by point mutations which alter the general uptake and/or efflux of tobramycin (permeability). Additionally, there may be other mechanisms beyond those listed here which may also explain aspects of the phenotype (e.g., changes in metabolism or the response to tobramycin). The aadB, aadA5, aac(6′)-Ie–aph(2″)-Ia, and rmtB genes were not detected in any of the isolates tested (Table 3).
Molecular epidemiology of the 37 aac(3)-IIg-positive E. cloacae complex isolates.
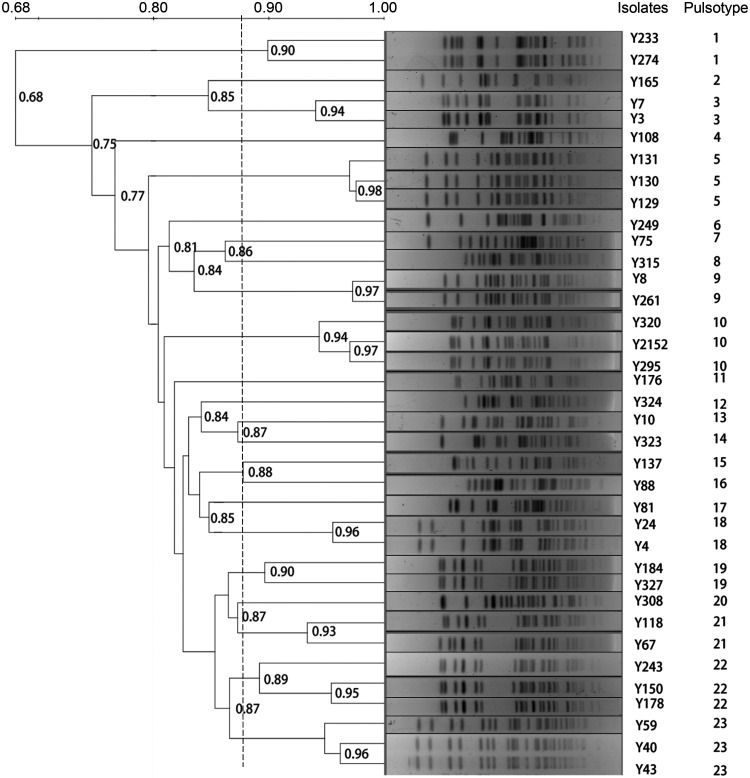
To investigate the clonal relatedness between the 37 aac(3)-IIg-positive isolates, all strains were genotyped by pulsed-field gel electrophoresis (PFGE) analysis, and 23 major PFGE types, named PFGE types 1 to 23, were identified (Fig. 2). Of these, PFGE types 1, 3, 9, 18, 19, and 21 were comprised of 2 isolates each and PFGE types 5, 10, 22, and 23 were comprised of 3 isolates each (Fig. 2). The similar profiles in the PFGE patterns seen indicated that these strains were highly homologous and that a small clonal outbreak might have occurred. The remaining 13 isolates showed distinct individual patterns (Fig. 2), suggesting considerable molecular heterogeneity among these isolates.
FIG 2.
PFGE patterns of the 37 aac(3)-IIg-positive E. cloacae complex isolates. Genomic DNA from each isolate was digested with XbaI and subsequently subjected to PFGE to generate diagnostic genomic DNA fingerprints. The dendrogram of the PFGE profiles was clustered by UPGMA, and a genetic similarity index scale is shown on the right of the dendrogram. The strain number and PFGE types are included along each PFGE lane.
Transfer of the aac(3)-IIg gene and plasmid analysis.
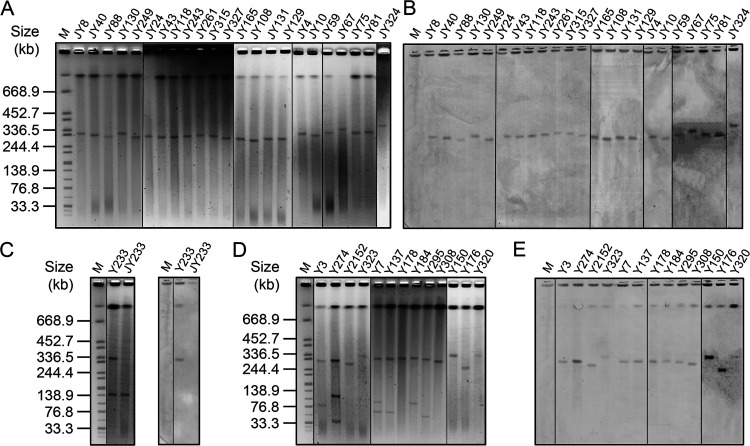
To investigate the molecular basis of the prevalence of aac(3)-IIg, the 37 E. cloacae complex strains were subjected to assessment of their ability to undergo conjugative transfer of the aac(3)-IIg gene to E. coli strain C600. Plasmids from 24 isolates were successfully transferred to recipients by conjugation (Fig. 3). S1 nuclease PFGE (S1-PFGE) and Southern hybridization were then performed on transconjugants or on E. cloacae complex strains to determine the range of transmissible and nontransmissible aac(3)-IIg-positive elements harbored by the test strains. S1-PFGE showed that the 24 transconjugants carried only one plasmid ranging from 140 kb to ∼340 kb in size (Fig. 3A and C). Isolate Y233 harbored two plasmids (Fig. 3C); however, only the plasmid with a medium size of about 140 kb was transferred to the recipient (Fig. 3C). Sequencing of the PCR products against plasmids extracted from the 24 transconjugants revealed that the aac(3)-IIg gene was located on a conjugative plasmid, except for that extracted from the transconjugant of Y233 (Table 3), which was further confirmed by Southern hybridization with the probe specific for aac(3)-IIg (Fig. 3B and C). In contrast, the hybridization signal for the aac(3)-IIg-specific probe in isolate Y233 was obtained on the larger nonconjugative plasmid (∼320 kb) (Fig. 3C). Nevertheless, it should be noted that the chromosome of isolate Y233 also had hybridization signals with the aac(3)-IIg-specific probe (Fig. 3C).
FIG 3.
S1-PFGE and Southern hybridization analysis of aac(3)-IIg-bearing conjugative and nonconjugative plasmids. (A, B) S1-PFGE analysis of plasmids from 23 transconjugants (A) and Southern hybridization with an aac(3)-IIg-specific probe (B). (C) S1-PFGE (left) and Southern blotting (right) of plasmid DNA of isolate Y233 and its transconjugant, JY233. (D, E) S1-PFGE patterns of 13 E. cloacae complex isolates that could not transfer aminoglycoside resistance to recipients by conjugation (D) and Southern hybridization with an aac(3)-IIg-specific probe (E). Lanes M, molecular size markers.
On the other hand, 24 of the 37 aac(3)-IIg-positive E. cloacae isolates were found to successfully transfer aminoglycoside resistance to the recipient strains by conjugation (Table 3; Table S3). The patterns of resistance of the transconjugants carrying aac(3)-IIg to selected aminoglycoside antimicrobials were universally wider than those of isolates carrying the cloned aac(3)-IIg (Table 2; Table S3), indicating that other resistance determinants were located on the respective conjugative plasmids. Consistent with this interpretation, the presence of additional resistance genes was demonstrated by sequencing of the PCR products and comparison of the sequences with those of plasmids extracted from the transconjugants (Table 3).
We next examined the plasmid profiles of the remaining 13 isolates with aac(3)-IIg that could not transfer aminoglycoside resistance to recipients by conjugation under the experimental conditions used in this study. The results of S1-PFGE revealed the presence of 1 to 3 visible plasmids in the 13 isolates, and in all cases there was a large plasmid with a size of approximately 310 kb which hybridized with the aac(3)-IIg-specific probe (Fig. 3D and E), indicating that aac(3)-IIg was located on the ∼310-kb nonconjugative plasmids. Interestingly, hybridization analyses also revealed that these strains may also harbor a chromosomal aac(3)-IIg gene (Fig. 3E).
Genetic environment of the aac(3)-IIg gene.
To further determine the location of aac(3)-IIg in strains that could not transfer resistance phenotypes to the recipient strains by conjugation, as well as to investigate the genetic features of aac(3)-IIg, three isolates (isolates Y233, Y323, and Y2152) were selected for whole-genome sequencing. The general genomic features of the three genomes are summarized in Table S4. Their genome sizes were all about 4.7 Mbp, similar in length to the lengths of other completed Enterobacter genomes (4.5 to 5.4 Mbp) (19, 20). The three isolates were identified as E. hormaechei subsp. steigerwaltii (Y233), E. hormaechei subsp. oharae (Y323), and E. hormaechei subsp. oharae (Y2152), based on both average nucleotide identity (ANI) analysis and the hsp60 and rpoB gene sequences (20–22). In silico multilocus sequence typing (MLST) revealed that the sequence types (ST) were ST461, ST303, and ST303 for Y233, Y323, and Y2152, respectively. Furthermore, each of the three strains contained three circular plasmids, varying in size from ∼2.5 to 394 kb (Table S4). Although some resistance genes were found to be on the chromosome, the majority of the antibiotic resistance determinants were located on plasmids encoding resistance to multiple classes of antibiotics (Table S4). It should be noted that the plasmids ranging in size from 2.495 to 5.976 kb did not carry any resistance genes (Table S4), and the aac(3)-IIg genes in these strains were present only in a plasmid context, indicating that the hybridization signals observed at the location of the chromosome may be due to nonspecific binding (Fig. 3C and E). However, the possibility that the aac(3)-IIg gene was probably located on the chromosome of other unsequenced isolates in this study cannot be excluded.
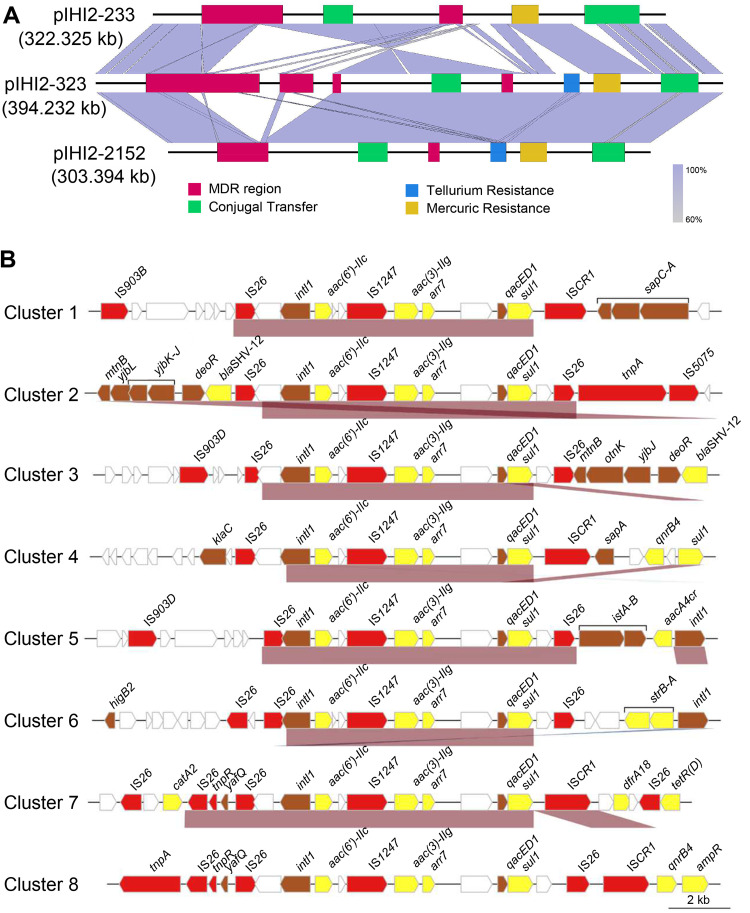
Next, the sequences of the corresponding plasmids containing aac(3)-IIg in Y233, Y323, and Y2152 were analyzed. The results showed that the plasmids in Y233, Y323, and Y2152 had circular DNA sequences of 322,325, 394,232, and 303,394 bp in length containing 386, 495, and 382 predicted ORFs, respectively. These plasmids shared core IncHI2 backbone markers and were designated pIHI2-233 (GenBank accession no. CP049047.1), pIHI2-323 (GenBank accession no. CP049189.1), and pIHI2-2152 (GenBank accession no. CP049193.1), respectively (Fig. 4A). Comparative genome analyses showed that there was an inversion of a 20-kb fragment containing the conjugative transfer region 1 (Tra1) in pIHI2-233 compared with the corresponding sequences in plasmids pIHI2-323 and pIHI2-2152. Moreover, pIHI2-323 had the largest multidrug resistance (MDR) area, which was divided into 4 parts, followed by pIHI2-233, which was divided into 2 parts, and pIHI2-2152 had the smallest MDR region, which was also divided into two parts (Fig. 4A).
FIG 4.
Comparative genomics analysis of plasmids and the genetic environment of the aac(3)-IIg gene. (A) Comparison of the genome structures of plasmids pIHI2-233 (GenBank accession no. CP049047.1), pIHI2-323 (GenBank accession no. CP049189.1), and pIHI2-2152 (GenBank accession no. CP049193.1). Boxes are colored based on the gene function classification. Orthologous regions are connected and color coded. (B) Structure of the aac(3)-IIg gene-related regions. Eight representative sequences from the eight clusters (one sequence from each cluster) are shown. The arrows represent sequence units or genes and are color coded, with the arrowheads indicating the direction of transcription. The names of the sequence units are indicated above the arrows, with the sequence units of unknown function left blank.
To identify the potential mobile genetic elements associated with aac(3)-IIg, 34 DNA sequences of about 20 kb in length with the aac(3)-IIg gene in the center were retrieved from all aac(3)-IIg-containing sequences in the NCBI nucleotide sequence database. Of these sequences, 28 were from plasmid sequences and 6 were from complete or partial bacterial chromosomes. The species distribution of these sequences is shown in Table S5, and 12 of these originated from the E. cloacae complex. By multiple-sequence alignment, 8 clusters with more than 85% identity were obtained, and among these, pIHI2-233, pIHI2-323, and pIHI2-2152 were divided into the first cluster (Table S6). The results of homologous analysis revealed that an approximately 9-kb class 1 integron carrying the gene cassettes aac(6′)-IIc–IS1247–aac(3)–IIg–arr7 was conserved among all clusters (Fig. 4B). Most of these integrons were bounded with two copies of IS26 (cluster 2, cluster 3, cluster 5, cluster 6, and cluster 8), and cluster 5 and cluster 6 shared the same direct repeats (DRs) and IS26, suggesting an IS26-mediated segment insertion. In cluster 1, cluster 4, and cluster 7, the integron was found downstream of IS26 and upstream of ISCR1 to form a complex class 1 integron (Fig. 4B).
DISCUSSION
In the present study, by combining high-throughput sequencing and PCR screening, we investigated the prevalence of aminoglycoside resistance genes in 170 E. cloacae complex isolates collected from a teaching hospital in Wenzhou, China. We found an extremely high prevalence of AME-encoding genes (162/170, 95.3%) and a relatively low prevalence of two 16S rRNA methylase genes, armA (5/170, 2.94%) and rmtB (9/170, 5.29%). Previous studies have found AAC(6′)-Ib and its variant, AAC(6′)-Ib-cr, as well as AAC(3)-IIa, to be the most prevalent AMEs conferring resistance to aminoglycosides in clinical isolates of Enterobacteriaceae spp. (1, 14, 23). Intriguingly, none of these enzymes were able to be identified in our isolate group (Table 1). The difference may be due to different bacterial samples or geographic locations in these epidemiologic surveys. Regardless, our data highlight the widespread occurrence of AMEs in clinical E. cloacae complex isolates in the hospital from which the isolates were recovered. In addition, unlike AMEs, 16S rRNA methylases have emerged as a novel mechanism for high-level resistance to almost all clinically important aminoglycosides (17). These genes are mostly located on transferable plasmids carrying bacterial recombination systems, like transposons and integrons, and the global spread of such resistance determinants has become a great concern (24). Fortunately, only armA and rmtB have been detected in clinical isolates in China, until the sampling period of the study (25–28). In agreement with these reports, we found armA and rmtB to be the only 16S rRNA methylase genes detected among the isolates analyzed in this study (Table 1). However, due to their ability to confer high levels of resistance to aminoglycosides (e.g., see the results for strains Y233 and Y274 in Table S3 in the supplemental material), further experiments are needed to determine the function of AMEs in armA- and rmtB-positive strains, as well as the mechanisms driving the dissemination of these 16S rRNA methylase genes.
A major finding of our study is the characterization of a novel gene, aac(3)-IIg, encoding an AAC(3)-II aminoglycoside 3-N-acetyltransferase that significantly increases the MICs of gentamicin, micronomicin, sisomicin, tobramycin, kanamycin, and netilmicin when expressed in E. coli. Previously, a total of 6 aac(3)-II variants (3, 29), some with a proven function [aac(3)-IIa and aac(3)-IIb] (7, 30) and others with a putative function based on amino acid sequence similarity or resistance phenotype [aac(3)-IIc, aac(3)-IId, aac(3)-IIe, and aac(3)-IIf] (6, 29, 31), have been identified. The AAC(3)-IIg protein shares <80% amino acid identity with all of the previously known AAC(3)-II enzymes (3, 29) and is notably divergent from AAC(3)-IIa–AAC(3)-IIf (Fig. 1). This determinant therefore represents a seventh evolutionary lineage of aac(3)-II genes. The evaluation of resistance to aminoglycoside antibiotics revealed that this gene conferred an AAC(3)-II resistance profile (3), thus confirming the result of the phylogenetic analysis. However, the enzymatic activity, key amino acid residues, as well as mechanistic and structural aspects of AAC(3)-IIg remain to be elucidated and will be the subject of future studies.
Previous studies have reported that most of the resistant strains carried combinations of several mechanisms for resistance to aminoglycosides (3, 8). In our case, 97.3% (36/37) of the aac(3)-IIg-positive isolates expressed more than four enzymes (Table 3). This can explain the multiple-aminoglycoside-resistance patterns of these strains and suggests that the coexistence of several AMEs may contribute to the broadening of aminoglycoside resistance spectra. In addition, PFGE and dendrogram analysis revealed genetic diversity (including 23 genotypes, ranging from PFGE types 1 to 23) among the 37 aac(3)-IIg-positive isolates, and a small clonal outbreak was observed, although in some instances the resistance phenotypes or genotypes of the strains showing similar PFGE patterns were not exactly the same. Possible explanations for the discrepancy may be point mutations, the differential expression of specific resistance genes, or the presence or absence of a certain plasmid(s) harboring different resistance genes in these isolates.
Further, PCR and sequence analyses revealed that aac(3)-IIg was cotransferred with other types of aminoglycoside resistance genes in all transconjugants of the E. cloacae strains (Table 3). This may indicate a plasmid-mediated intimate association between aac(3)-IIg and other antibiotic resistance genes. Of note, the co-occurrence and cotransfer of multiple resistance genes on the same plasmid may result in the appearance and dissemination of multidrug-resistant (MDR) or even pan-drug-resistant (PDR) strains, especially when AMEs are combined together with other classes of antibiotic-resistant determinants (32, 33). A typical example of this is the extended-spectrum β-lactamase (ESBL) and carbapenemase genes, which are often collocated with aminoglycoside resistance genes on mobile genetic elements (20, 34, 35). Significantly, acquisition of MDR by Enterobacteriaceae members, including E. cloacae complex isolates, is emerging as a global, diversifying threat (9, 10, 12). Additional molecular work, like whole-genome sequencing and comparative genomics, transcriptomics, and/or metabolomics, will be an effective way to better understand the emergence and spread of variable resistance phenotypes.
Finally, our data suggest that the aac(3)-IIg gene is part of a class 1 integron with a gene cassette array of aac(6′)-IIc–IS1247–aac(3)-IIg–arr7, with IS26 playing an important role in its mobilization. The aac(6′)-IIc gene encodes an aminoglycoside 6′-acetyltransferase that confers resistance to gentamicin and tobramycin (3). Furthermore, aac(6′)-IIc has been reported to be located within a class 1 integron in two plasmids (pEC-IMPQ and pEC-IMP) from clinical E. cloacae isolates (36). The insertion element IS1247 encodes an open reading frame (464 amino acids) that shows a high degree of sequence identity to a putative transposase (37). The arr7 gene encodes an ADP-ribosyltransferase, conferring rifampin resistance, and was found to be associated with a class 1 integron in Pseudomonas aeruginosa (38). Moreover, we observed that the IS1247–aac(3)-IIg–arr7 array formed a transposition unit with 4-bp DRs at both ends. It is possible that IS1247 may also play an important role in the mobilization of aac(3)-IIg. Taken together, these features indicate the potential horizontal transmission of these genes and are consistent with the view that AMEs are often associated with integrons or mobile genetic elements to confer aminoglycoside resistance as well as to efficiently disseminate among bacteria (1, 3). However, the origin of aac(3)-IIg is still unclear. An organized and more large-scale surveillance effort is required to better understand this issue and to limit the transmission of aminoglycoside resistance genes like aac(3)-IIg in clinical and environmental settings.
MATERIALS AND METHODS
Bacterial isolates and identification.
In this study, a total of 170 nonduplicate E. cloacae complex strains were isolated from various clinical specimens from patients admitted to the First Affiliated Hospital of Wenzhou Medical University in Wenzhou, Zhejiang, China, during 2005 to 2007. All isolates were initially identified using a Vitek-60 microorganism autoanalysis system (bioMérieux Corporation, Craponne, France). Further species identification was performed by the combination of hsp60 and rpoB genotyping as described previously (21, 22). The species assignment of the 37 aac(3)-IIg-carrying E. cloacae complex isolates is shown in Table S1 in the supplemental material.
Genome sequencing, assembly, annotation, and bioinformatic analysis.
For pooled sequencing, each of the purified 170 isolates was freshly cultured in Luria-Bertani (LB) broth at 37°C for 16 h. These bacterial cultures were then pooled and the genomic DNA was extracted using an AxyPrep bacterial genomic DNA miniprep kit (Axygen Scientific, Union City, CA, USA). Pooled sequencing was performed by the Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China), to conduct 150-bp paired-end sequencing using a HiSeq X Ten platform (Illumina Inc., San Diego, CA, USA). In addition, the whole-genome DNA of the individual E. cloacae complex isolates (isolates Y233, Y323, and Y2152) was extracted as described above and was sequenced using Illumina HiSeq 2500 and Pacific Bioscience (PacBio) systems by the Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China).
Genome assembly of the pooled sequencing data was performed using the MegaHit program (10). The full-length genomes of the Y233, Y323, and Y2152 isolates were assembled from PacBio sequencing reads of ∼10 to 20 kb in length using Canu software (39). Error correction of tentative complete circular sequences was performed using the Pilon (version 1.18) program with short reads derived from HiSeq 2500 sequencing. Potential open reading frames (ORFs) of pooled sequence data were predicted using the Prodigal program with the default parameters. Antibiotic resistance genes were identified using both the Comprehensive Antibiotic Resistance Database (CARD) and ResFinder database. The relative abundance (sequencing depth) of a certain gene was calculated using the BBMap short read aligner (http://sourceforge.net/projects/bbmap/). ORF prediction and annotations for the genomes of Y233, Y323, and Y2152 were determined using the RAST pipeline (40). In silico multilocus sequence typing (MLST) of the three sequenced isolates was performed with the MLST (version 1.8) online server utilizing seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB) (41, 42). Multiple-sequence alignments were performed using the Clustal Omega program (43). The MEME program was used for discovering conserved protein sequence motifs (44). Phylogenetic trees were constructed by the maximum likelihood method using the MEGA (version X) program with the default parameters (45), and the resulting trees were visualized using the Interactive Tree of Life (iTOL) (46). Gene organization diagrams were drawn with the Inkscape program (https://inkscape.org). The sequence retrieval, statistical analysis, and other bioinformatics tools used in this study were written using the Python (https://www.python.org/) and Biopython (47) languages.
Detection of aminoglycoside resistance genes.
Genomic DNA was extracted from each of the 170 clinical E. cloacae complex isolates as described above. The presence of 13 aminoglycoside resistance genes, including aac(3)-IIg, aac(3)-IId, aac(6′)-Ie–aph(2′')-Ia, aac(6′)-IIc, aadA2, aadA5, aadB, aadA1, aphA-1, strA, strB, armA, and rmtB, was determined using PCR. The specific primers used are listed in Table S2. Positive amplification products were confirmed by sequencing, and the resulting sequence of each gene was analyzed and compared with the sequences in the NCBI nucleotide sequence database using the BLAST program (https://www.ncbi.nlm.nih.gov/BLAST).
Cloning experiments.
To clone the aminoglycoside resistance genes, genomic DNA was extracted as described above. The ORF of each resistance gene together with the predicted promoter region (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) was PCR amplified using the primers listed in Table S2. The specific PCR fragment was isolated and inserted into the vector pMD19T. Plasmids were introduced into Escherichia coli DH5α by the calcium chloride method, and the cells were plated on selective LB agar plates supplemented with ampicillin (100 μg/ml). The recombinants harboring the target gene were validated by restriction enzyme digestion and further confirmed by PCR and sequencing.
Antimicrobial susceptibility testing.
MICs were determined using the agar dilution method with Mueller-Hinton agar. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines (48). Since CLSI lacks breakpoints for streptomycin, neomycin, and spectinomycin, the breakpoint values of streptomycin (susceptible, ≤32 μg/ml; resistant, ≥64 μg/ml), neomycin (susceptible, ≤8 μg/ml; resistant, >16 μg/ml), and spectinomycin (128 μg/ml) were used according to criteria proposed by the U.S. Food and Drug Administration (FDA), the Comite de L'Antibiogramme de la Société Française de Microbiologie (http://www.sfm-microbiologie.org/) (49), and Chuanchuen and Padungtod (50), respectively. E. coli ATCC 25922 was used as the quality control strain.
Conjugation experiments.
Conjugation experiments to determine whether aminoglycoside resistance determinants were located on conjugative plasmids were performed as described previously with slight modifications (51, 52). Briefly, candidate donor strains were mated with the rifampin-resistant E. coli C600 recipient strain on sterile nitrocellulose filters. The transconjugants were selected on LB agar plates containing 1,024-μg/ml rifampin and 4-μg/ml gentamicin. Conjugation plates were incubated at both 25°C and 37°C. After confirmation, the MICs of several representative antibiotics for positive transconjugants were assessed. The existence of resistance genes on the transferred plasmid was also detected by PCR and sequencing. The sizes of the large aac(3)-IIg-positive plasmids were further estimated by S1 nuclease pulsed-field gel electrophoresis (PFGE) techniques (53), and the presence of the aac(3)-IIg gene was subsequently confirmed via Southern blot analysis.
PFGE typing.
The clonal relatedness of aac(3)-IIg-positive E. cloacae complex isolates was evaluated by PFGE. In brief, DNA samples were digested with XbaI at 37°C for 2.5 h. Restriction fragments were separated in a 1% SeaKem Gold agarose gel for 18 h at a constant voltage of 6 V/cm with a pulse time gradient of from 2.16 to 54.17 s, using a CHEF-Mapper system (Bio-Rad, Hercules, CA, USA). Chromosomal DNA of Salmonella enterica serovar Braenderup H9812 digested with XbaI was used as a molecular size marker. The banding profiles were analyzed using the Bio-Rad Quantity One program, and cluster analysis was performed using the unweighted pair-group method with arithmetic average (UPGMA). PFGE pulsotypes were interpreted according to previously established guidelines (54), with a similarity of <88% upon dendrogram analysis being considered representative of different PFGE types.
Southern blot analysis.
To confirm the presence of the aac(3)-IIg gene, DNA fragments from the PFGE gel were transferred onto a nylon membrane by Southern blotting. Hybridization analysis was performed with a digoxigenin-labeled aac(3)-IIg gene fragment labeled with a DIG High Prime DNA labeling kit and Detection starter kit II (Roche, Germany), according to the manufacturer's instructions. The aac(3)-IIg-specific probe was obtained by PCR amplification with the primer pairs listed in Table S2.
Data availability.
The sequences of Y233 (CP049046.1), Y323 (CP049188.1), Y2152 (CP049192.1), pIHI2-233 (CP049047.1), p233-142 (CP049048.1), p233-2 (CP049049.1), pIHI2-323 (CP049189.1), pY323-2 (CP049190.1), pY323-3 (CP049191.1), pIHI2-2152 (CP049193.1), pDC2152-6 (CP049194.1), pDC2152-2 (CP049195.1), and aac(3)-IIg (MT090547) have been deposited in GenBank, and the GenBank accession numbers are given in parentheses.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge all study participants and individuals who contributed to this study.
This work was supported by grants from the Natural Science Foundation of Zhejiang Province (grants LY19C060002 and LQ17H190001), the National Natural Science Foundation of China (grants 31500109 and 81973382), and the Science and Technology Project of Inner Mongolia Autonomous Region, China (grant 201802125).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Serio AW, Keepers T, Andrews L, Krause KM. 16 November 2018, posting date. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal Plus 2018 doi: 10.1128/ecosalplus.ESP-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garneau-Tsodikova S, Labby KJ. 2016. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7:11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. doi: 10.1128/MMBR.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause KM, Serio AW, Kane TR, Connolly LE. 2016. Aminoglycosides: an overview. Cold Spring Harb Perspect Med 6:a027029. doi: 10.1101/cshperspect.a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho PL, Wong RC, Lo SW, Chow KH, Wong SS, Que TL. 2010. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J Med Microbiol 59:702–707. doi: 10.1099/jmm.0.015032-0. [DOI] [PubMed] [Google Scholar]
- 7.Rather PN, Mierzwa R, Hare RS, Miller GH, Shaw KJ. 1992. Cloning and DNA sequence analysis of an aac(3)-Vb gene from Serratia marcescens. Antimicrob Agents Chemother 36:2222–2227. doi: 10.1128/aac.36.10.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/cmr.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezzatesta ML, Gona F, Stefani S. 2012. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 10.Davin-Regli A, Pagès J-M. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer A, Hoffmann H. 2012. Prevalences of the Enterobacter cloacae complex and its phylogenetic derivatives in the nosocomial environment. Eur J Clin Microbiol Infect Dis 31:2951–2955. doi: 10.1007/s10096-012-1646-2. [DOI] [PubMed] [Google Scholar]
- 12.Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. 2019. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol 10:44. doi: 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestourie N, Garnier F, Barraud O, Bedu A, Ploy MC, Mounier M. 2014. Outbreak of AmpC beta-lactamase-hyper-producing Enterobacter cloacae in a neonatal intensive care unit in a French teaching hospital. Am J Infect Control 42:456–458. doi: 10.1016/j.ajic.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Dai W, Sun S, Zhang X, Zhang L. 2012. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PLoS One 7:e47636. doi: 10.1371/journal.pone.0047636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SY, Park YJ, Yu JK, Kim YS, Han K. 2009. Prevalence and characteristics of aac(6')-Ib-cr in AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens: a multicenter study from Korea. Diagn Microbiol Infect Dis 63:314–318. doi: 10.1016/j.diagmicrobio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, Park YJ, Yu JK, Kim YS. 2011. Aminoglycoside susceptibility profiles of Enterobacter cloacae isolates harboring the aac(6')-Ib gene. Korean J Lab Med 31:279–281. doi: 10.3343/kjlm.2011.31.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galimand M, Courvalin P, Lambert T. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother 47:2565–2571. doi: 10.1128/aac.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi Y, Yokoyama K, Yamane K, Wachino J-I, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother 48:491–496. doi: 10.1128/aac.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WY, Wong CF, Chung KM, Jiang JW, Leung FC. 2013. Comparative genome analysis of Enterobacter cloacae. PLoS One 8:e74487. doi: 10.1371/journal.pone.0074487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavda KD, Chen L, Fouts DE, Sutton G, Brinkac L, Jenkins SG, Bonomo RA, Adams MD, Kreiswirth BN. 2016. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio 7:e02093-16. doi: 10.1128/mBio.02093-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/aem.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paauw A, Caspers MP, Schuren FH, Leverstein-van Hall MA, Deletoile A, Montijn RC, Verhoef J, Fluit AC. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS One 3:e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill AJ. 2008. New antibacterial agents for treating infections caused by multi-drug resistant Gram-negative bacteria. Expert Opin Investig Drugs 17:297–302. doi: 10.1517/13543784.17.3.297. [DOI] [PubMed] [Google Scholar]
- 24.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M. 2010. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis 29:1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 26.Wu Q, Zhang Y, Han L, Sun J, Ni Y. 2009. Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob Agents Chemother 53:271–272. doi: 10.1128/AAC.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu YS, Zhou H, Yang Q, Chen YG, Li LJ. 2007. Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother 60:454–455. doi: 10.1093/jac/dkm208. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, Jiang HX. 2007. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother 59:880–885. doi: 10.1093/jac/dkm065. [DOI] [PubMed] [Google Scholar]
- 29.Costello SE, Deshpande LM, Davis AP, Mendes RE, Castanheira M. 2019. Aminoglycoside-modifying enzyme and 16S ribosomal RNA methyltransferase genes among a global collection of Gram-negative isolates. J Glob Antimicrob Resist 16:278–285. doi: 10.1016/j.jgar.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Vliegenthart JS, Ketelaar-van Gaalen PA, van de Klundert JA. 1989. Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob Agents Chemother 33:1153–1159. doi: 10.1128/aac.33.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubois V, Arpin C, Dupart V, Scavelli A, Coulange L, Andre C, Fischer I, Grobost F, Brochet JP, Lagrange I, Dutilh B, Jullin J, Noury P, Larribet G, Quentin C. 2008. Beta-lactam and aminoglycoside resistance rates and mechanisms among Pseudomonas aeruginosa in French general practice (community and private healthcare centres). J Antimicrob Chemother 62:316–323. doi: 10.1093/jac/dkn174. [DOI] [PubMed] [Google Scholar]
- 32.Xia J, Fang LX, Cheng K, Xu GH, Wang XR, Liao XP, Liu YH, Sun J. 2017. Clonal spread of 16S rRNA methyltransferase-producing Klebsiella pneumoniae ST37 with high prevalence of ESBLs from companion animals in China. Front Microbiol 8:529. doi: 10.3389/fmicb.2017.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y, Yang J, Zhang Y, Ye L, Wang L, Guo L. 2011. Prevalence of beta-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents 37:352–355. doi: 10.1016/j.ijantimicag.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Chavda KD, Melano RG, Hong T, Rojtman AD, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2289–2294. doi: 10.1128/AAC.02749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Simmonds A, Annavajhala MK, Wang Z, Macesic N, Hu Y, Giddins MJ, O’Malley A, Toussaint NC, Whittier S, Torres VJ, Uhlemann A-C. 2018. Genomic and geographic context for the evolution of high-risk carbapenem-resistant Enterobacter cloacae complex clones ST171 and ST78. mBio 9:e00542-18. doi: 10.1128/mBio.00542-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. 2009. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother 53:1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ploeg J, Willemsen M, van Hall G, Janssen DB. 1995. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol 177:1348–1356. doi: 10.1128/jb.177.5.1348-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuelsen O, Toleman MA, Sundsfjord A, Rydberg J, Leegaard TM, Walder M, Lia A, Ranheim TE, Rajendra Y, Hermansen NO, Walsh TR, Giske CG. 2010. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother 54:346–352. doi: 10.1128/AAC.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME suite. Nucleic Acids Res 43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 47.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 49.Hu Y, Liu L, Zhang X, Feng Y, Zong Z. 2017. In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant Enterobacteriaceae clinical strains. Front Microbiol 8:2275. doi: 10.3389/fmicb.2017.02275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuanchuen R, Padungtod P. 2009. Antimicrobial resistance genes in Salmonella enterica isolates from poultry and swine in Thailand. J Vet Med Sci 71:1349–1355. doi: 10.1292/jvms.001349. [DOI] [PubMed] [Google Scholar]
- 51.Xu T, Wang J, Ying J, Zhu T, Liu Y, Xu L, Li P, Li P, Ying J, Li K, Yi H, Lu J, Hu Y, Zhou T, Bao Q. 2018. Characterisation of a class 1 integron associated with the formation of quadruple blaGES-5 cassettes from an IncP-1beta group plasmid in Pseudomonas aeruginosa. Int J Antimicrob Agents 52:485–491. doi: 10.1016/j.ijantimicag.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Zhu M, Yang G, Li A, Zong L, Dong Z, Lu J, Zhang K, Cheng C, Chang Q, Wu X, Ying J, Li X, Ding L, Zheng H, Yu J, Ying J, Xu T, Yi H, Li P, Li K, Wu S, Bao Q, Wang J. 2017. Identification and molecular characterization of Escherichia coli blaSHV genes in a Chinese teaching hospital. Gene 600:29–35. doi: 10.1016/j.gene.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 53.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 54.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences of Y233 (CP049046.1), Y323 (CP049188.1), Y2152 (CP049192.1), pIHI2-233 (CP049047.1), p233-142 (CP049048.1), p233-2 (CP049049.1), pIHI2-323 (CP049189.1), pY323-2 (CP049190.1), pY323-3 (CP049191.1), pIHI2-2152 (CP049193.1), pDC2152-6 (CP049194.1), pDC2152-2 (CP049195.1), and aac(3)-IIg (MT090547) have been deposited in GenBank, and the GenBank accession numbers are given in parentheses.